Abstract
Borrelia burgdorferi (Bb) is the causative agent of Lyme disease in the US, a disease that can result in carditis, and chronic and debilitating arthritis and/or neurologic symptoms if left untreated. Bb survives in the midgut of the Ixodes scapularis tick, or within tissues of immunocompetent hosts. In the early stages of infection, the bacteria are present in the bloodstream where they must resist clearance by the innate immune system of the host. We have found a novel role for OspC from B. burgdorferi and B. garinii in interactions with the complement component C4b and bloodstream survival in vivo. Our data show that OspC inhibits the classical and lectin complement pathways and competes with complement protein C2 for C4b binding. Resistance to complement is important for maintenance of the lifecycle of Bb, enabling survival of the pathogen within the host as well as in the midgut of a feeding tick when ospC expression is induced.
Keywords: Borrelia, adhesins, infection, vasculature, immunity, mouse, OspC, complement, C4b
INTRODUCTION
Borrelia burgdorferi, the causative agent of Lyme disease in the U.S., is transmitted via tick bite and is known to disseminate into host tissues where it can survive for the lifetime of the host. If left untreated, the human infection can become chronic leading to a number of different symptoms that vary in part with the infecting strain and species of Lyme disease Borrelia. Human infection with B. garinii strains, which occur with greater frequency than B. burgdorferi in parts of Europe and Asia, more commonly present with neurologic symptoms (Centers for Disease Control and Prevention, 2015), while infection with B. burgdorferi commonly presents with monoarticular arthritis (Baranton et al., 1992; Strle, Ružić-Sabljić, Cimperman, Lotrič-Furlan, & Maraspin, 2006; van Dam et al., 1993).
Lyme disease Borrelia spp. exist in a tick-vertebrate life cycle in which the bacteria are exposed to immune defenses of both the host and the tick. The spirochetes are able to evade clearance by these systems and can survive and cause a disseminated infection in the mammalian host, in part by modulation of their surface proteome. For example, when entering the tick midgut, the bacteria upregulate the production of the outer surface protein A (OspA) which is promptly downregulated when the tick takes a blood meal from the next host (Schwan, Piesman, Golde, Dolan, & Rosa, 1995). Exposure to blood products inside the tick upregulates the production of OspC, presumably to prepare the bacteria for entry into the host tissues via tick bite (Srivastava & de Silva, 2008).
Disseminated infection by Lyme disease Borrelia involves bacteremia and spread to distant tissues, and these bacteria have evolved mechanisms to evade clearance by the complement cascade present in the host blood and tissues (Alitalo et al., 2002; Brangulis, Petrovskis, Kazaks, Akopjana, & Tars, 2014; Caesar, Johnson, Kraiczy, & Lea, 2013; Hallström et al., 2013; Hammerschmidt et al., 2012; Hammerschmidt et al., 2014; Kenedy, Vuppala, Siegel, Kraiczy, & Akins, 2009; Kraiczy et al., 2004). The complement cascade involves a series of proteolytic cleavage events of inactive precursors into active enzymes in the host serum and tissues that result in the formation a lytic pore complex in the pathogen membrane called the membrane attack complex (Figure S1). The complement cascade can be activated by the classical, lectin, or alternative pathways. The classical and lectin pathways are triggered by recognition of the pathogen surface by antibodies or mannose-binding lectin, respectively, whereas the alternative pathway is activated by spontaneous deposition of the complement component, C3, onto the surface of the bacteria. The host possesses many mechanisms to regulate the activation of the complement cascade to minimize harm to host tissues. Proteins such as Factor H and Factor H-like protein 1 inhibit the alternative complement cascade, while proteins such as C4 binding protein (C4BP), and C1 inhibitor (C1-INH) function to inhibit the classical and lectin pathways (Ratnoff, Pensky, Ogston, & Naff, 1969), reviewed in (Meri & Jarva, 1998); see Figure S1). The host also produces proteins, such as vitronectin, CD59, and clusterin, that inhibit the end stages of the complement cascade common to all branches.
B. burgdorferi is able to recruit many of these host factors to its surface creating a cloak of protection from the activities of the complement cascade. Production of CspA (Hallström et al., 2013; Hammerschmidt et al., 2014; Kenedy et al., 2009) or CspZ (Hartmann et al., 2006; Siegel et al., 2008), have been shown to recruit Factor H and Factor H-like 1 to the spirochete surface, in vitro, and ultimately inhibit the alternative complement cascade. Recently, it was shown that the fibronectin and glycosaminoglycan binding protein, BBK32, is able to bind to the complement component C1 cleavage product, C1r, and inhibit its activation (Garcia, Zhi, Wager, Hook, & Skare, 2016). This binding activity on the surface of B. burgdorferi was found to inhibit in vitro complement activation through the classical pathway, in vitro (Garcia et al., 2016). Interestingly, two studies of mice deficient in complement component C3 suggested that complement-mediated defense diminishes the bacterial burden in a variety of tissues, particularly at 2 weeks after infection (Lawrenz et al., 2003). Nevertheless, other studies describe a relatively similar course of infection in mice deficient in complement components C5 compared to wild type mice, suggesting that B. burgdorferi may encode complement inhibition mechanisms that limit the effectiveness of complement defenses at multiple steps in vivo (Bockenstedt, Barthold, Deponte, Marcantonio, & Kantor, 1993).
OspC is a 22kDa, dimeric lipoprotein on the surface of Lyme disease Borrelia. Production of OspC, induced in the midgut of a feeding tick, has been shown to be critical for the early stages of mammalian infection (Grimm et al., 2004; Tilly et al., 2006). The high immunogenicity and virulence attributes of OspC have made the protein a focus of vaccination studies for several years with a moderate amount of success (Gilmore, Kappel, Dolan, Burkot, & Johnson, 1996; Preac-Mursic et al., 1992; Probert & LeFebvre, 1994; Wilske et al., 1993). OspC displays high variability of the central region of the OspC protein among not only different species of Lyme disease Borrelia but also between different strains within the same species (T. Lin, Oliver, & Gao, 2002), limiting the cross-protective activity of vaccines based on OspC (Probert, Crawford, Cadiz, & LeFebvre, 1997). Based on this variability, OspC from different strains and species of Lyme disease Borrelia have been separated into 22 different classes (named A-U) with alleles having less than 2% variability in nucleotide sequence falling into the same class, and different classes of alleles having greater than 8% sequence variability between them (Earnhart, Buckles, Dumler, & Marconi, 2005; Seinost et al., 1999). Strains that produce class A, B, C, D, I, K, or N have been associated with disseminated infection in humans or mice (Earnhart et al., 2005; Lagal, Postic, Ruzic-Sabljic, & Baranton, 2003; Seinost et al., 1999; Wang et al., 2001; Wang et al., 2002). Due to the differences in invasiveness and disease seen with Borrelia carrying the various OspC classes, and its requirement for B. burgdorferi survival early in infection, it is possible that OspC may be an effector for evasion of innate defenses in the host.
By observing the survival capabilities in vivo of ospC deletion and complemented derivatives of B. burgdorferi in mouse models of Lyme borreliosis, we identified a novel role for OspC in bloodstream survival. By utilizing ospC mutant B. burgdorferi strains producing different classes of OspC protein from two well studied strains of B. burgdorferi, strain B31-A3 (class A) and strain N40 clone D10/E9 (class M), as well as from B. garinii strain PBr (class B), we showed a similarity in afforded bloodstream survival of the bacteria. Through a series of in vitro assays we identified a potential mechanism for the action of the OspC protein from B. burgdorferi but not B. garinii in affording Borrelia resistance to the classical and/or mannose-binding lectin branches of the complement cascade, promoting bloodstream survival of the bacteria.
RESULTS
An OspC-deficient mutant B. burgdorferi exhibits reduced survival in the blood at 30 minutes post inoculation
We previously showed that 60 min. after intravenous (i.v.) inoculation into C3H/HeN mice, the OspC-deficient B. burgdorferi strain B31-A3ΔospC was undetectable in the blood compared to WT infectious B. burgdorferi strain B31-A3 (Caine & Coburn, 2015). To characterize the kinetics of clearance of the ospC mutant from the bloodstream of mice, C3H/HeN mice were i.v. inoculated with infectious B. burgdorferi strain B31-A3 or an isogenic ospC mutant (Table S1) and at serial time points post inoculation (p.i), blood was collected and bacterial genomes were quantified by qPCR. At 5 min p.i. approximately 5.3 × 103/ 100ng DNA of WT and approximately 7.1 × 103 / 100ng DNA of ospC mutant bacteria were detected in the bloodstream of infected mice (Figure 1 and Table S2). At 10 minutes p.i., 3/8 mice infected with the ospC mutant had cleared spirochetes from their blood compared to 1/6 mice infected with WT bacteria (Figure 1 and Table S2). By 20 minutes p.i. 6/8 mice had cleared the ospC mutant bacteria compared to 2/6 mice infected with WT bacteria (Figure 1 and Table S2). By 30 minutes all but one mouse had cleared the ospC mutant bacteria compared to 1/6 of the mice infected with WT bacteria (Figure 1 and Table S2). This result is seen again at 60 minutes p.i. where, in these experiments, 2/6 WT-inoculated mice had cleared bacteria from their blood compared to 6/8 of mice inoculated with the ospC mutant (Figure 1 and Table S2). For each timepoint, the number of bacteria detected was not statistically significantly different between WT and ospC mutant inoculated mice. However, the number of mice that had cleared WT or ospC mutant bacteria at 30 min. p.i. was significantly different by Fisher’s exact test (Table S2). From these data we concluded that the ospC mutant bacteria begin to be cleared from the bloodstream of a mouse within the first 20–30 minutes p.i. compared to WT bacteria which are able to survive throughout the duration of the experiment.
Figure 1. B. burgdorferi ospC deletion mutant is cleared from the bloodstream early after inoculation.
C3H/HeN mice were retro-orbitally inoculated with 1×108 infectious B. burgdorferi strain B31-A3+vector (B31-A3) (n=6) or B31-A3ΔospC+vector (ΔospC) (n=8). At the indicated times post inoculation, blood was collected from the saphenous vein and bacterial burdens were quantified by qPCR. OspC mutant bacteria were quantified using primers to the kanamycin resistance cassette and WT bacteria were quantified using primers to the recA gene. Each time point represents blood collected from the same cohort of mice. The percent of the cohort of mice with detectible levels of bacteria in the blood at each time point are plotted over the timecourse. The number of mice with detectible WT or ospC mutant bacteria burdens was significantly different at 30 minutes p.i. by Fisher’s exact test, *= p-value <0.05.
OspC from B. burgdorferi and B. garinii promotes bloodstream survival in vivo
Different species of Lyme disease Borrelia show varying sensitivity to human serum proteins (van Dam et al., 1997). B. burgdorferi is known to be resistant to killing by human serum in vitro, while the B. garinii strains tested are more sensitive (Kurtenbach et al., 2002; van Dam et al., 1993; van Dam et al., 1997). To determine if OspC from B. garinii has similar roles in bloodstream survival as OspC from B. burgdorferi, C3H/HeN mice were i.v. inoculated with B31-A3ΔospC harboring the control vector or plasmids constitutively expressing ospC alleles from B. burgdorferi strain B31-A3 or N40 D10/E9, or from B. garinii strain PBr, all under control of a constitutive flagellar promoter (Table S1, Figure 2A). Surface localization of the B31-A3 and N40 D10/E9 OspC proteins in the ospC mutant background was confirmed by incubation of intact bacteria with proteinase K or buffer. (Figure 2B). Due to limited cross-reactivity of the antibody generated against B. burgdorferi OspC with OspC from other species of Borrelia, we performed Triton X-114 membrane extraction and silver stain on each of the bacterial strains. Production of OspC from B. burgdorferi strain B31-A3 and B. garinii was observed in the membrane fraction (Figure 2C).
Figure 2. OspC from B. burgdorferi and B. garinii is able to restore bloodstream survival to a B. burgdorferi ospC mutant 1 hour post inoculation.
(A) C3H/HeN mice were retro-orbitally inoculated with infectious B. burgdorferi strain B31-A3 (B31-A3/Vector)(n=15), B31-A3ΔospC+vector (Vector)(n=9), or B31-A3ΔospC constitutively expressing ospC alleles from B. burgdorferi strain B31-A3 (+B31-A3)(n=12), N40 clone D10/E9 (+N40-D10/E9)(n=15), or B. garinii strain PBr (+PBr)(n=10). Production of OspC from B31-A3 and N40 restored bloodstream survival to similar levels at 1 hr. post inoculation, while OspC from B. garinii strain PBr restored bloodstream survival to a reduced level. Statistical significance determined by Mann-Whitney unpaired t-test. *=Statistically significantly different than ospC mutant, p-value <0.05. #= Statistically significant difference between +B31-A3 and +PBr, p-value <0.05. (B) Surface exposure of the exogenously expressed ospC alleles was confirmed by incubating B. burgdorferi expressing each allele, from the flaB promoter, with proteinase K (+) or buffer (−). Confirmation of bacterial cell integrity is shown by the presence of intact flagellin with and without proteinase K. Sensitivity of the membrane protein, P66, is seen in the presence of proteinase K. (C) Silver stain of membrane extracts from B. burgdorferi OspC derivatives separated by 15% SDS-PAGE. Arrow indicates OspC, (M) indicates membrane fraction, (P) indicates non-membrane fraction.
Borrelia genomes were quantified in the blood of mice 1 hour post inoculation (h.p.i.), and showed that production of OspC from B. burgdorferi strain B31-A3 or N40 D10/E9 by B31-A3ΔospC restored bacterial burdens to WT levels (Figure 2A). Production of OspC from B. garinii strain PBr also significantly increased bacterial burdens, but to a lesser extent than OspC from B. burgdorferi strain B31-A3, suggesting that B. burgdorferi OspC provides greater bloodstream survival activity than B. garinii OspC (Figure 2A).
Differences in bloodstream survival afforded by OspC from B. burgdorferi and B. garinii are recapitulated at 3 but not 7 d.p.i
OspC is required for the establishment of mammalian infection (Tilly et al., 2006). Consistent with published results (Tilly et al., 2006), upon quantitation of bacterial loads by qPCR (see Materials and Methods) we found that bacterial burdens in the blood of mice intradermally inoculated with B31-A3ΔospC were significantly lower than WT at both 3 and 7 d.p.i. (Figure 3A). To test whether the B. burgdorferi and B. garinii ospC alleles, which promoted bloodstream survival at 1 hr. post i.v. inoculation, also promote bacteremia at these later time points, we generated strains in B31-A3ΔospC that express ospC alleles from B. burgdorferi strain B31-A3, N40 D10/E9, or B. garinii strain PBr, from the native ospC promoter (Table S1). Flow cytometry was performed on the strains used in these studies to confirm the surface production of OspC proteins produced under the control of the native ospC promoter (Figure 3B,C).
Figure 3. Differences in bloodstream survival afforded by OspC from B. burgdorferi and B. garinii are recapitulated at 3 but not 7 d.p.i.
(A) C3H/HeN mice were intradermally inoculated with 1×104 B. burgdorferi strain B31-A3 (B31-A3/Vector), ospC deficient mutant B31-A3ΔospC (Vector), or B31-A3ΔospC producing OspC from B. burgdorferi strain B31-A3 (B31), N40 clone D10/E9 (N40-D10/E9), or B. garinii strain PBr (PBr) all under control of the native ospC promoter. The bacterial load in the blood collected at 3 and 7 d.p.i. was determined by qPCR. Statistical significance was determined by one-way ANOVA. *=p-value of < 0.05 compared to the normalized spirochete number of B31A3ΔospC/Vector, n=8. (B) Surface exposure of OspC on intact B. burgdorferi B31-A3ΔospC producing OspC variants was determined by flow cytometry. (C) OspC (top) or Flagellin (bottom) on the surface of untreated (solid bars) or methanol-permeabilized bacteria (open bars) was determined by flow cytometry. Values are normalized to the levels of OspC or Flagellin in permeabilized B. burgdorferi strain B31-A3/Vector. Each bar represents the mean ± SEM of four independent experiments. *=p-value <0.05 by student t-test compared to WT bacteria treated in the same way (top panel), or compared to the untreated control sample of the same bacteria strain (bottom panel).
We found that the bloodstream survival phenotypes promoted by the OspC proteins observed at 1 h.p.i. were roughly recapitulated at the later time points tested. All three OspC variants restored blood burdens to WT levels by 7 d.p.i., indicating a role for OspC in bloodstream survival (Figure 3A). However, at 3 d.p.i. only the B. burgdorferi OspC variants from strains B31-A3 and N40 D10/E9 promoted bacteremia (Figure 3A, upper panel). OspC from B. garinii strain PBr, which conferred increased bloodstream survival in the 1 hr infection compared to B31-A3ΔospC, did not promote bacteremia at 3 d.p.i. (Figure 3A, upper panel). Interestingly, blood burdens of strains producing OspC from PBr were significantly lower than burdens of strains producing OspC from B31-A3 (Figure 2A), suggesting differences in the mechanism of bloodstream resistance afforded by B. garinii OspC and B. burgdorferi OspC.
OspC interacts with complement component C4b in vitro
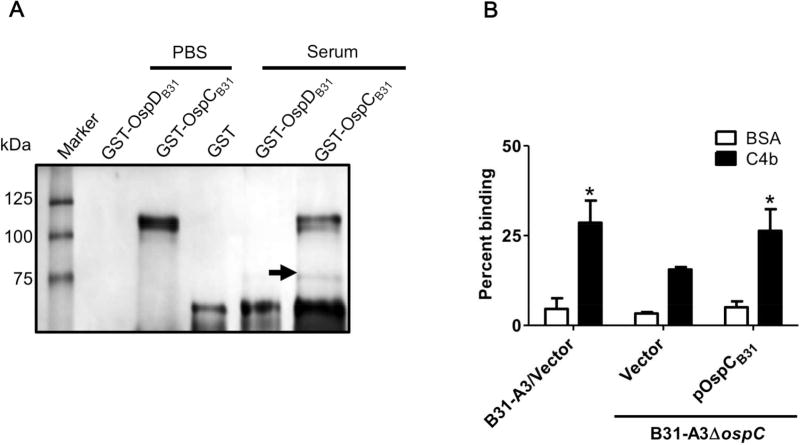
B. burgdorferi produces proteins on its surface that have the capacity to bind to complement regulatory factors such as Factor H and inhibit activation of the alternative complement cascade (Figure S1) (Kraiczy et al., 2006; Kraiczy, Skerka, Brade, & Zipfel, 2001; Kraiczy, Skerka, Kirschfink, Brade, & Zipfel, 2001). To determine which, if any, proteins in human serum interact with OspC to potentially provide B. burgdorferi protection from clearance in the bloodstream, a series of pulldown assays was performed. Human serum was incubated with resin saturated with GST-OspCB31, a purified protein consisting of GST fused to OspC from B. burgdorferi strain B31-A3. As negative controls, resin was saturated with purified GST or GST-OspDB31. Unbound serum proteins were washed from the resins and total bound proteins were eluted and analyzed by reducing SDS-PAGE and silver stain (Figure 4A). A band of approximately 75 kDa present in the elution from the OspC-baited resin and absent from the negative control eluates (Figure 4A) was excised and analyzed by mass spectrometry (Table S3). One of the most abundant proteins detected in the band (per spectral counts) was the β-chain of complement component C4b (which is approximately 75 kDa) identified with 32% sequence coverage (Table S3, Figure S2). Peptides of the β-chain of C4b are shared with the full length pro-protein, C4, as well as several smaller cleavage products generated during C4 processing.
Figure 4. OspCB31 interacts with C4b.
(A) Human serum or PBS buffer was flowed through resin containing immobilized GST, recombinant GST-tagged OspCB31 or OspDB31 (GST-OspDB31, derived from strain B31-A3 ospD) as a negative control. The band indicated by the arrow was excised and shown, by mass spectrometry, to contain the C4b β-chain (approximately 75 kDa). (B) The binding of radiolabeled B. burgdorferi strain B31-A3 containing empty vector (B31-A3/Vector), B31-A3ΔospC containing empty vector (Vector), or B31-A3ΔospC producing OspC from strain B31-A3 (B31) to purified human C4b or BSA. The data are graphed as percent of inoculum, and each bar represents the mean ± standard deviation of four independent experiments. Statistical significance was determined by student’s t-test *=p-value of < 0.05 compared to the ospC deletion strain incubated with C4b.
To confirm a direct interaction between OspCB31 and C4b, an ELISA was performed by incubating increasing concentrations of recombinant GST-tagged OspCB31, OspDB31, and GST (negative control) in C4b coated wells (Figure S3). The KD of the OspCB31-C4b interaction was approximately 414 nM (Table 1, Figure S3), an affinity consistent with a potentially biologically relevant interaction given that C4, the pro-protein from which C4b is produced during proteolytic processing, is present in blood at roughly 500 nM. No binding was detected between OspDB31 and C4b (Table 1, Figure S3). The interaction between OspCB31 and C4b detected in these assays can mediate interactions between intact bacteria and C4b, because radiolabeled B. burgdorferi strain B31-A3 and B31-A3ΔospC producing OspC from strain B31-A3 bound significantly better than B31-A3ΔospC to C4b coated wells compared to BSA (control) (Figure 4B).
Table 1.
Approximate Binding Affinities of Recombinant Proteins to Purified Human C4b.
| OspC variant | KD (µM) ± SD |
|---|---|
| B. burgdorferi | |
| OspCB31 | 0.476±0.071 |
| OspDB31 | n.b. |
| OspCN40-D10/E9 | 0.233±0.042 |
| B. garinii | |
| OspCPBr | n.b. |
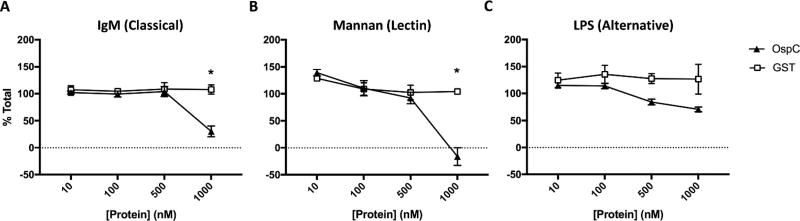
To determine if OspC is able to directly inhibit the complement cascade allowing WT B. burgdorferi to survive in the bloodstream of mice, we performed an ELISA based complement activation assay. Plates were coated with initiators of each pathway of the complement cascade; IgM (classical pathway), mannan (lectin pathway), LPS (alternative pathway), or buffer (negative control). Coated plates were incubated with a mixture of pooled active human serum and various concentrations of purified GST-OspCB31, GST, or buffer and complement deposition was measured in each well by ELISA (Figure 5). Wells coated with IgM or mannan and incubated with 1000 nM GST-OspC have significantly lower levels of deposited complement compared to wells incubated in GST (Figure 5A,B). This affect is not seen in LPS coated wells, where the addition of GST-OspC does not inhibit complement deposition compared to GST at any concentration tested (Figure 5C).
Figure 5. OspC inhibits the classical and lectin complement pathways.
Plates were coated with IgM (A), mannan (B), or LPS (C) and incubated with pooled human serum combined with various concentrations of GST-OspC or GST. GST-OspC significantly inhibits the deposition of complement initiated by the classical and lectin pathways compared to GST. Complement deposition in each well was determined by ELISA for complement component C3b. Data are presented as mean percent of total (complement deposition in coated wells incubated in serum only) ± standard deviation. *=Statistically significant difference between GST-OspC and GST at a particular concentration, p<0.05.
C2 competes with C4b for binding to OspC
Complement factor C2 is a native binding partner of C4b, and this interaction is required for initiation of the subsequent steps of the complement cascade (Figure S1). A plausible hypothesis is that binding of C4b to OspC or C2 are mutually exclusive, so that OspC interrupts the complement cascade, promoting bloodstream survival of B. burgdorferi. To address this, competition experiments were performed in a solid phase assay. Wells coated with purified human C4b were incubated with 1 µM recombinant OspC from B. burgdorferi strain B31-A3 and increasing concentrations of purified human C2 or BSA as a negative control. The percent of recombinant OspC added to the reaction that was bound to each well was determined by ELISA and compared to binding in the presence of the non-competitor BSA control (Figure 6A). C2 was capable of significantly decreasing binding of OspC to C4b with as low as 75 ng/ mL, whereas BSA had no effect (Figure 6A).
Figure 6. The binding of OspC to C4b is inhibited by exogenous C2.
(A) 1 µM GSTOspC from B. burgdorferi strain B31-A3 in the presence of competitor human C2 or BSA (negative control), was added to wells coated with 1 µg of purified human C4b. OspC bound to each well was measured by ELISA. Binding is expressed as the percent of OspC bound to each well, normalized to OspC bound in the absence of competitor (BSA). Data represent the average of four replicates. Statistical significance determined by student’s t-test *=p<0.01 compared to BSA control at the given competitor concentration. (B) Radiolabeled B. burgdorferi strain B31-A3 (B31-A3/Vector), B31-A3ΔospC (Vector), or B31-A3ΔospC producing OspC from B. burgdorferi strain B31-A3 (pOspCB31) was incubated with 1 µg of purified human C2 or BSA (negative control) prior to addition to purified human C4b-coated wells. Radioactive counts in each well were quantified and normalized to counts in the inoculum to calculate the percent bound bacteria. Each bar represents the mean ± standard deviation of four independent experiments. Statistical significance determined by student’s t-test, *=p-value <0.05.
To determine if complement C2 could also block binding of C4b to OspC localized on the spirochetal surface, we assayed the ability of radiolabeled B. burgdorferi strain B31-A3, B31-A3ΔospC, or B31-A3ΔospC with OspC production restored (Table S1) to bind to purified C4b in the presence or absence of competitor. As expected, B31-A3ΔospC did not bind to C4b-coated wells efficiently, and the basal level of binding was not altered by the presence of C2 (Figure 6B). In contrast, compared to control incubation with BSA, incubation of bacteria with 10 ng/ µl purified C2 resulted in a significant decrease in binding to C4b by the ospC mutant producing OspC from B. burgdorferi strain B31-A3 as well as by WT bacteria (Figure 6B, Table S1). The competitive inhibition of the C2-C4b complex formation by OspC on the surface of B. burgdorferi was also observed by immunofluorescence microscopy (Figure S4).
OspCB31 and OspCN40, but not OspCPBr, bind C4b with high affinity
OspC from different strains and species of Lyme disease Borrelia have varying abilities to support bloodstream survival in vivo, and thus, we hypothesized that the binding affinities of these various OspC molecules to C4b may differ. To test this hypothesis, ELISAs were performed as described above, incubating increasing concentrations of purified OspC from B. burgdorferi strain N40 clone D10/E9, and B. garinii strain PBr. The amount of bound recombinant OspC was used to estimate the KD of each interaction (Table 1). OspC from B. garinii strain PBr had no detectable binding (Table 1) compared to OspC from B. burgdorferi strain N40-D10/E9 (KD= ~233 nM), an affinity similar to what was measured for B. burgdorferi strain B31-A3 (Table 1, Figure S3).
DISCUSSION
OspC has been shown to be important in the early stages of mouse infection but is detrimental to the bacteria if produced later during mammalian infection, though this early requirement has never been mechanistically defined (Skare, Shaw, Trzeciakowski, & Hyde, 2016; Tilly et al., 2006). Using an in vivo model for the early stages of infection, we can eliminate the selection bottleneck at the site of inoculation in the skin and inject virulent or avirulent derivatives of B. burgdorferi directly into the bloodstream and monitor their ability to survive in the blood up to 1 h.p.i. By i.v. inoculating a mouse strain commonly used in the Borrelia field, C3H/HeN, with B. burgdorferi strain B31-A3 deficient in OspC production, we were able to observe a role for OspC in bloodstream survival of B. burgdorferi (Caine & Coburn, 2015). In C3H/HeN mice we were unable to detect B31-A3ΔospC in the bloodstream in a majority of the mice, 1 h.p.i. (Figures 1 and 2) (Caine & Coburn, 2015). We previously observed a gain of survival in the bloodstream when inoculating with non-infectious B. burgdorferi strain B31A exogenously expressing BBK32 from B. burgdorferi strain B31-A3 (Caine & Coburn, 2015). These results may indicate an importance of OspC (as presented here) and BBK32 in bloodstream survival and support in vitro findings that BBK32 binds complement component C1r (Garcia et al., 2016).
Differences in serum sensitivity in vitro have been documented for different species of Lyme disease Borrelia. B. burgdorferi was found to be resistant to killing by human serum, where B. garinii is more sensitive, in a series of in vitro assays (van Dam et al., 1997). There have been several Borrelia proteins that have been shown to contribute to Borrelia resistance to killing by the alternative complement cascade, including CspA (Hallström et al., 2013; Hammerschmidt et al., 2014; Kenedy et al., 2009) and CspZ (Hartmann et al., 2006; Siegel et al., 2010), most of them studied in vitro. These proteins are likely produced by the bacteria both within the mammal and the tick to provide resistance to serum complement proteins present in the blood meal as well as within the vertebrate host to perpetuate the enzoonotic lifestyle of Borrelia spp. For example, expression of cspA has been shown to be upregulated within the tick, and downregulated in mammalian infection (Kenedy & Akins, 2011). Conversely, CspZ has been shown to be produced in the mouse throughout the duration of infection (Bykowski et al., 2007). To date, only one Borrelia protein, BBK32, has been found to interact with a protein in the classical complement pathway (Garcia et al., 2016). However, it has recently been shown that the presence of OspC on the surface of B. burgdorferi decreases uptake by professional phagocytes in vivo (Carrasco et al., 2015). Opsonization of the pathogen surface can induce activation of the classical complement cascade, and increases phagocytosis, though a correlation between these two processes was not made in the above mentioned experiments (Carrasco et al., 2015). By using our model of short term Borrelia infection of mice, we showed that OspC from B. burgdorferi and B. garinii restored bloodstream survival to B31-A3ΔospC in mice despite the fact that B. garinii was more susceptible than B. burgdorferi to killing by human serum proteins. At 3 days post intradermal infection of mice, the number of B. burgdorferi cells present in the blood producing OspC from B. garinii was significantly lower than strains expressing alleles from B. burgdorferi. This reiterates the data observed at 1 h.p.i. with significantly fewer bacteria producing OspC from B. garinii in the blood compared to OspC from B. burgdorferi strain B31-A3. It is possible that this may be due to different rates of dissemination and entry into the circulation, or due to different in vivo replication rates, between B. burgdorferi strains producing different OspC proteins. It is possible that the phenotypes of bloodstream survival that we are seeing with strains producing OspC from B. garinii are an artifact of surface production in an unnatural micro-environment on the spirochetal surface. Being that B. garinii is a distinct species from B. burgdorferi, the native proteins present on the surface that OspC from B. garinii might interact with may be absent or in different abundance on the surface of the surrogate, B. burgdorferi. Alternatively, the bloodstream survival of our OspC derivatives that we have observed in our mouse model may be different than expected due to the source of the blood. The increased sensitivity of B. garinii to serum proteins compared to B. burgdorferi was determined by incubation with human serum in vitro, and may differ from what is observed in a living mouse (van Dam et al., 1997). No significant differences in replication rates in vitro were observed between the different B. burgdorferi strains used in this work, but the in vivo environment is considerably different from that of laboratory culture. Taken together, the data presented here and those previously published are consistent with the conclusion that the reduced survival of B. burgdorferi producing OspC from PBr at both 1 hour and 3 days post inoculation is due to the reduced binding to C4b.
From the time course assay performed we found the OspC deficient mutant had decreased bloodstream survival at 30 min p.i. with significantly fewer mice with detectible ospC mutant compared to WT bacteria at this timepoint. Our data suggest that the ospC mutant is being cleared by a rapid innate immune mechanism, such as the complement cascade. These data corroborate the previous findings describing an ability for mice to clear the ospC mutant from the tissues despite a lack of functioning T and B cells as well as NK cells (Carrasco et al., 2015; Grimm et al., 2004).
Using several in vitro approaches, we identified an interaction between OspC and complement component C4b. By immunofluorescence microscopy we show that OspC is able to inhibit deposition of the C3 convertase as measured by C2a deposition, suggesting a possible mechanism by which OspC inhibits the classical and lectin pathways of the complement cascade (Figure 5 and S4). Inhibition of the classical and lectin pathways of the complement cascade afforded by OspC on the surface of B. burgdorferi is likely responsible for the increased survival of WT compared to ospC mutant bacteria in the bloodstream of mice. Further studies will need to be performed to determine the details of this mechanism. Using ELISAs we estimated the strength of the interaction between OspC from several different strains and species of Borrelia, and C4b, to be within a biologically relevant range. Interestingly, though production of OspC from B. garinii affords bloodstream survival to the ospC mutant in vivo, we were unable to detect an interaction of recombinant OspCPBr with purified C4b in vitro by ELISA, suggesting that OspC from B. garinii strain PBr may be enabling bloodstream survival of B. burgdorferi by a different mechanism than OspC from B. burgdorferi strains. We have shown that C2, the natural binding partner of C4b, and OspC from B. burgdorferi strain B31-A3 competes for binding to C4b in vitro, and that this competition may be biologically relevant as it can also be observed with live B. burgdorferi producing OspC from B31-A3. Collectively, these data support the model that OspC binds to C4b, blocking the interaction with its native binding partner, C2, which inhibits the activity of the complement cascade allowing B. burgdorferi to survive in the presence of host blood factors.
Future work will investigate the OspC residues that bind C4b and are important in complement inhibition. With this work, we have identified a novel role for OspC in bloodstream survival by inhibiting the classical and lectin pathways of the complement cascade. These data suggest a common role for OspC from different species of Lyme disease Borrelia in bloodstream survival through distinct mechanisms. This ability is important for bacterial maintenance in the tick-mammalian lifecycle, in which the bacteria must survive both within the host blood and tissues as well as within the midgut of a feeding tick. Pharmacologic inhibitors of ligand interactions with OspC may be effective approaches to inducing bacterial clearance through increased serum sensitivity during a mammalian infection.
MATERIALS AND METHODS
Animals
Female C3H/HeN mice were acquired from Charles River Laboratories (Charles River Laboratories International, Inc., Wilmington, MA) and used at 4 wks of age for long term i.d. inoculation studies, or 8 wks of age or older for i.v inoculation experiments. All work with animals was approved by the IACUC at the Medical College of Wisconsin or Tufts University School of Medicine. All efforts were made to minimize animal suffering. Normal human serum was collected from healthy, anonymous donors. The procedures for collecting human serum were approved by the Institutional Reviewer Board (IRB) of Tufts University School of Medicine, Boston, MA (Protocol docket number 11016).
Bacteria culture and strains
All strains used in this study are described in Table S1. B. burgdorferi strains were grown in Barbour-Stoenner-Kelly (BSKII) medium (Barbour, 1984) at 33°C to a density of 1×108 spirochetes/ mL. When antibiotic selection was required, kanamycin was used at 200 µg/ mL and gentamicin was used at 40 µg/ mL. Presence of genomic plasmids and shuttle vectors was determined in each culture by PCR prior to inoculation into mice (Bunikis, Kutschan-Bunikis, Bonde, & Bergström, 2011; Elias et al., 2002). All vectors were sequenced through the promoter-ospC region to ensure that no mutations had occurred.
E. coli strains (Table S1) were grown in Luria-Burtani (LB) broth (Thermo Fisher Scientific, Carlsbad, CA) for cloning experiments, and 2× YT (BD Biosciences, San Jose, CA) for recombinant protein production. When antibiotic selection was required, kanamycin was used at 50 µg/ mL, spectinomycin and ampicillin were used at 100 µg/mL. Protein production was induced in BL21(DE3) strains (Table S1) with 1 mM final isopropyl β-D-1 thiogalactopyranoside (IPTG) (Gold BioTechnology, Inc., St. Louis, MO). All clones were verified to have the correct sequence before use.
Borrelia strain construction
To generate B. burgdorferi strains constitutively expressing ospC alleles, plasmid pTM61spc-MCS was generated by annealing oMCS3 and oMCS4 (Table S4) and ligating the fragment into pGEM ™-T Easy (Promega corp., Madison, WI). The MCS was excised from pGEM ™-T Easy (Promega corp., Madison, WI) using BspHI and EarI, and ligated into pTM61spc (Kumar et al., 2015) at the BspHI site. The ospC deficient B. burgdorferi strain B31-A3ΔospC, generated previously (Tilly et al., 2006), was engineered to express ospC alleles from various strains and species of Lyme disease Borrelia constitutively from the flagellar promoter (flaB), or from the ospC promoter from B. burgdorferi strain B31-A3, in the shuttle vector, pBSV2G (Table S1) (Elias et al., 2003). To generate ospC alleles constitutively expressed under control of the flaB promoter, alleles containing the native ribosome binding sites were amplified from genomic DNA isolated from B. burgdorferi strain B31-A3 and N40 clone D10/E9, and B. garinii strain PBr using the Wizard SV Genomic DNA Purification System (Promega corp., Madison, WI) and the oligonucleotides described in Table S4. PCR fragments containing the ospC alleles were cloned in pGEM ™-T Easy (Promega corp., Madison, WI), excised using SacI and XmnI (New England BioLabs, Inc., Ipswich, MA), and cloned downstream of the flaB promoter in pTM61spc-MCS. The flagellar promoter was amplified from B. burgdorferi strain B31-A3 genomic DNA and ligated into pGEM ™-T Easy (Promega corp., Madison, WI), excised using NheI and SalI (NEB, Ipswich, MA), and subcloned into the NheI and XhoI sites of pTM61spc-MCS. Promoter-ospC allele fragments were excised from pTM61spc-MCS using HindIII and BamHI and ligated to pBSV2G digested with the same enzymes. All vectors were transformed into E. coli SURE strain (Agilent Technologies, Inc., Santa Clara, CA), isolated using the QIAGEN Plasmid Maxi Kit (Qiagen, Valencia, CA), and transformed into B. burgdorferi by electroporation of 50 µg of CpG methylated DNA (Chen et al., 2008) and plating into semi-solid agar as previously described (Samuels, 1995). All ospC alleles were found to have the correct sequence when carried on pBSV2G in B. burgdorferi with the exception of the ospC allele from B. burgdorferi strain N40 clone D10/E9 which was found to encode the single amino acid mutation at N53S (Shih & Chao, 2002). The constitutively expressed WT ospC sequence from N40-D10/E9 was not successfully maintained in B. burgdorferi B31-A3ΔospC despite multiple independent rounds of cloning and transformation.
To generate the plasmids encoding ospC alleles under control of the native promoter, the ospC genes from B. burgdorferi strain B31-A3 and N40 clone D10/E9, and B. garinii strain PBr were PCR amplified with oligonucleotides containing a SalI site at the 5’ end and a BamHI site at the 3’ end (Table S4). Amplified DNA fragments were inserted into the TA cloning vector pCR2.1-TOPO (Invitrogen, Houston, TX) to generate plasmids pCR2.1-ospCB31, pCR2.1- ospCN40-D10/E9 or pCR2.1-ospCPBr. The plasmids were then digested with SalI and BamHI to release the ospC alleles, and cloned into the SalI and BamHI sites of pBBE22. The promoter region of ospC from B. burgdorferi strain B31-A3, beginning 184 bp upstream from the start codon of ospC (Fischer, LeBlanc, & Leong, 2006), was PCR amplified adding HindIII and SalI sites at the 5’and 3’ ends, respectively, using primers pospCfp and pospCrp (Table S4). Promoter fragments were then inserted into the HindIII and SalI sites of pBBE22 to drive the expression of ospCB31, ospCN40-D10/E9 and ospCPBr.
Confirmation of OspC surface production on B. burgdorferi
To confirm the production of OspC classes on the surface of B. burgdorferi, cultures were grown to a density of > 1×108/ mL in BSKII. Cells were pelleted (644 ×g, 30 min, at ambient temperature), supernatants were pipetted off, and pellets were resuspended in 1 mL of sterile 1× PBS. Cells were centrifuged at 5,900 ×g for 8 min at ambient temperature and supernatants were pipetted off. Bacteria were resuspended in 1 mL of sterile 1× PBS and diluted to 1×108/ mL in 1 mL final volume with or without 10 µg/mL proteinase K (Sigma-Aldrich Co., St. Louis, MO). Bacteria were incubated in the presence or absence of proteinase K for 20 min at 33°C. Proteinase activity was stopped by the addition of 0.1M PMSF to a final concentration of 0.2 nM. Cell pellets were analyzed on a 12.5% SDS-PAGE gel and immunoblotted as described below, 2.5×107 cells/ well. Bacterial cell integrity was confirmed by maintenance of the periplasmic protein, flagellin, in cell lysates after proteinase K treatment. Confirmation of proteinase K activity in the assays was confirmed by detecting a size shift of an outer membrane protein of B. burgdorferi, P66, in the presence of proteinase K.
For verification of surface production of OspC from B. garinii, which is not detected by the antibody used for immunoblotting, membranes were extracted from cultures using Triton X-114 and analyzed by silver stain (Brandt, Riley, Radolf, & Norgard, 1990). Briefly, Borrelia cultures grown in BSKII and collected at a density > 1×108/ mL. 1×109 total bacteria were centrifuged at 1145 ×g, 30 min. at 4°C. Bacterial pellets were washed in room temperature 1× PBS+ 0.2% bovine serum albumin (BSA) and centrifuged at 9,300 ×g for 10 min. at ambient temperature. Cells were resuspended in 1 mL 1× PBS+2% Triton X-114 (Sigma-Aldrich, St. Louis, MO) and incubated overnight at 4°C. Lysates were centrifuged at 16,100 ×g, 30 min at 4°C and supernatants were warmed to 37°C. Warmed supernatants were centrifuged 16,100 ×g, 15 min. at ambient temperature to separate membrane from non-membrane fractions. Both fractions were washed in cold 1× PBS, proteins were precipitated with 95% ethanol and analyzed by 15% SDS-PAGE and silver stain (Shevchenko, Wilm, Vorm, & Mann, 1996).
To determine the surface localization of OspC proteins in B. burgdorferi strains derived from strain B31-A3 under control of the native ospC promoter on pBSV2G, 1 × 108 B. burgdorferi cells were washed three times with HBSCDB buffer (25 mM Hepes acid, 150 mM sodium chloride, 1 mM MnCl2, 1 mM MgCl2, 0.25 mM CaCl2, 0.1% dextrose, and 0.2% BSA, final concentrations) and resuspended into 500 µL of the same buffer. Washed spirochetes were incubated in methanol for permeabilization, or HBSC buffer containing DB. Mouse anti-FlaB or a mixture containing equal amounts of mouse antisera raised against OspC from B. burgdorferi strain B31-A3 and N40 clone D10/E9, and B. garinii strain PBr (generated in our lab) was used as a primary antibody, and Alexa488-conjugated goat anti-mouse IgG (Invitrogen, Houston, TX) was used as a secondary antibody. Bacteria were fixed in 0.1% formalin, and antibody exposure of OspC and FlaB were measured by flow cytometry using the Becton Dickinson FACSCalibur flow cytometer equipped with a 15 mW, 488 nm air-cooled argon laser, a standard three-color filter arrangement, and CELLQuest Software (BD Bioscience, Franklin Lakes, NJ). All flow cytometry experiments were performed two days after B. burgdorferi samples were fixed and incubated with antibodies. Spirochetes in the suspension were distinguished by their distinct light scattering properties. The mean fluorescence index (MFI) of each sample was obtained from the FlowJo software (FlowJo, LLC, Ashland, OR) representing the surface production of the indicated proteins. To compare the surface production of OspC and FlaB in different strains, results are shown as relative production with the MFI normalized to that of permeabilized B. burgdorferi strain B31-A3/Vector and represent the mean of twelve independent determinations ± the standard deviation.
SDS-PAGE, silver stain, and immunoblotting
For analysis of total OspC production by B. burgdorferi strains, bacterial cultures were centrifuged at 11,200 × g for 8 min. at ambient temperature and supernatants were removed. Cell pellets were resuspended in 1 mL of 1× PBS and centrifuged at 11,200 × g for 8 min. at ambient temperature. Cell pellets were resuspended in Laemmli buffer +0.1% final β-mercaptoethanol and boiled at 100°C for 10 min. 1×107 total bacteria were loaded into each well of a 15% SDS-PAGE gels and transferred to PVDF membranes. Non-specific binding sites were blocked with 5% milk in 1× TBS for 1 hour at ambient temperature. Membranes were incubated overnight at 4°C in rabbit anti-OspC (#200-401-C11, Rockland Immunochemicals, Inc., Limerick, PA), rabbit anti-P66, and mouse anti-flagellin antibodies, washed 3 times in 1× TBS, and incubated in alkaline phosphatase (AP)-conjugated anti-rabbit or AP-conjugated anti-mouse antibodies (Promega Corp., Madison, WI), washed, and incubated with chromogenic substrates NBT and BCIP (Sigma-Aldrich, St. Louis, MO). For detection by film, membranes were incubated in HRP-conjugated anti-rabbit, or HRP-conjugated anti-mouse (Promega Corp., Madison, WI), washed, and incubated in SuperSignal™ West Pico or Femto Chemiluminescent substrate (ThermoFischer Scientific Inc., Grand Island, NY).
Mouse infections
Short term i.v. inoculation and perfusion experiments were performed as previously described (Caine & Coburn, 2015). Briefly, C3H/HeN mice under anesthesia were retro-orbitally inoculated with 1×108 B. burgdorferi. Bacteria were allowed to circulate for 1 hr., and blood was collected by cardiac puncture. Animals were perfused with sterile saline to wash away any unbound bacteria from the vasculature. Tissues were harvested, rinsed and frozen on dry ice before genomic DNA isolation and quantitation of Borrelia and mouse genomes by qPCR as previously described (Table S3) (Caine & Coburn, 2015; Liveris et al., 2002).
Time course experiments were performed as described above except that at 5, 10, 20, 30, and 60 min. post inoculation (p.i.) blood was collected from the saphenous vein of each animal. Prior to the first time point, mice were anesthetized and hair was removed with a hair clipper, skin was sterilized with 70% ethanol, and a small amount of petroleum jelly was applied to each leg. The saphenous vein was punctured with a 23-gauge needle, and a maximum of 100 µL of blood was collected at each time point using a sterile pipet and combined with citrate anticoagulant (detailed above) to a 10% final volume and stored on dry ice. The skin of each leg was re-sterilized with 70% ethanol and a new layer of petroleum jelly was applied prior to each time point. Blood collection from the saphenous vein alternated between the left and right leg with each time point. Blood samples were stored at −80°C until DNA was isolated. OspC mutant bacteria were quantified by qPCR using primers that amplified a section of the kanamycin resistance cassette with a detection limit of 61 copies, and WT bacteria were quantified using recA primers with a detection limit of 6.1 gene copies as previously described (Caine & Coburn, 2015; Liveris et al., 2002)
In long term experiments, four-week-old female C3H/HeN mice (Charles River, Wilmington, MA) were used. Mice were infected by intradermal injection as previously described (Weening et al., 2008) with 1×104 B. burgdorferi. To test the bloodstream survival promoted by OspC variants, mice were euthanized at 3 or 7 days post inoculation (d.p.i.), blood was collected, and genomic DNA was extracted for quantification of bacterial and mammalian genomes by qPCR (Table S4) (Caine & Coburn, 2015; Liveris et al., 2002; Ristow et al., 2012).
Generation of recombinant GST proteins and antisera
To generate recombinant glutathione-S-transferase (GST)-tagged OspC proteins, the ospC open reading frames lacking the putative signal sequences from B. burgdorferi strains B31-A3 and N40 clone D10/E9, and B. garinii strain PBr were amplified and cloned in pGEX4T2 (GE Healthcare, Piscataway, NJ) as previously described (Benoit, Fischer, Lin, Parveen, & Leong, 2011) (Table S4). Amplified fragments were engineered to encode a BamHI site at the 5’ end and a stop codon followed by a SalI site at the 3’ end. PCR products were sequentially digested with BamHI and SalI and then inserted into the BamHI and SalI sites of pGEX4T2 (GE Healthcare, Piscataway, NJ). The resulting plasmids were transformed into E. coli strain BL21(DE3) (Table S1) and induced for production of GST-tagged OspC variants. Proteins were purified by glutathione chromatography according to the manufacturer’s instructions (BD Biosciences, San Jose, CA). Antisera against OspCB31-A3, OspCN40-D10/E9, and OspCPBr were generated by immunizing five-week-old BALB/C mice with each of the OspC proteins as described previously (Barthold, Hodzic, Tunev, & Feng, 2006).
Glutathione-S-Transferase (GST) fusion protein pull down assays
A total of 10 µg of GST tagged OspCB31 or OspDB31 (negative control), purified from E. coli, was incubated with a 50% slurry of glutathione agarose beads for 1 hr. at 4°C (BD Bioscience, Franklin Lake, NJ) and centrifuged at 13,000 ×g, 1 min. at 4°C and supernatants were discarded. The resin was washed three times with 1× PBS and incubated with normal human serum or 1× PBS (negative control) for 16 hrs. at 4°C with gentle mixing. Samples were centrifuged at 13,000 ×g, 1 min at 4°C and the resin was washed three times in 1× PBS. All proteins were eluted from the resin with 20 mM reduced glutathione (Sigma, St. Louis, MO). After centrifugation at 13,000 ×g for 1 min., eluted proteins were collected and analyzed by reducing SDS-PAGE using an 8% polyacrylamide gel. A protein band present at about 75 kDa in the lane containing GST-OspCB31 incubated in serum that was absent in both the GST-OspDB31 and GST-control pulldown samples was excised. A control slice in the corresponding area of the gel in the GST-OspDB31 lane was also excised. All gel fragments were washed three times in acetonitrile. Proteins present in the excised bands were identified by HPLC/MS/MS by the Beth Israel Deaconess Medical Center mass spectrometry facility (Boston, MA).
Quantitative C4b binding assay
Dissociation constants of OspC-C4b complexes were determined by ELISA as previously described with the noted changes (Y.-P. Lin et al., 2009). For quantitative ELISA, microtiter plate wells were coated with 1 µg C4b purified from human serum (Complement Technology, Tyler, Texas) or BSA (negative control and data not shown), as described previously (Fischer et al., 2006). Various concentrations (2, 1, 0.5, 0.25, 0.125, 0.0625 µM) of GST (negative control) or GST-OspC proteins were added to the coated wells, incubated 1 hr. at ambient temperature and washed 3 times. OspC bound to each well was detected using goat anti-GST (GE Healthcare, Piscataway, NJ) and HRP-conjugated anti-goat IgG (Promega, Madison, WI). Plates were washed three times with PBST (0.05% Tween 20 in 1× PBS buffer), and tetramethyl benzidine (TMB) solution (KPL, Gaithersburg, MD) was added to each well and incubated for 5 min. The reaction was stopped by adding 100 µL of 0.5% hydrosulfuric acid to each well. Plates were read at 405 nm using an ELISA plate reader (SpectraMAX 250, Molecular Devices; Sunnyvale, CA). To determine the dissociation constant (KD), the following equation was used, with results generated using KaleidaGraph software (Version 2.1.3 Synergy Software, Reading, PA).
Inhibition of C4b-OspC interaction by exogenous C2
96-well microtiter plates were coated with 1 µg of C4b purified from human serum, or with BSA (negative control) similar to previously described methods (Fischer et al., 2006). GST-OspC (1 µM) from B. burgdorferi strain B31-A3 was combined with 300, 150, 75, or 37.5 ng/ mL of soluble C2 purified from normal human serum (#A112, Complement Technology, Inc., Tyler, TX) or BSA (negative control) and incubated at room temperature for 1 hr. prior to addition to C4b-coated wells. Binding of GST-OspC was measured by ELISA as described above.
Binding of radiolabeled B. burgdorferi to purified C4b
Binding of B. burgdorferi to purified C4b was determined in a similar way to a previously described protocol (Benoit et al., 2011) with the following modifications. Briefly, spirochetes were radiolabeled with [35S] methionine, and 1 × 108 radiolabeled bacteria were added to microtiter plate wells previously incubated with 1 µg/mL C4b or BSA as a negative control. After 1 hr. at 37°C, unbound bacteria were removed by washing with 1× PBS containing 0.2% BSA. Plates were air-dried and binding was determined by liquid scintillation counting. The percent bacteria bound was determined by dividing the radioactive counts in the experimental wells by the counts in the inoculum. Four experiments were performed in triplicate.
Inhibition of binding of spirochetes to C4b by exogenous C2
Spirochetes were prepared as described above and incubated for 30 min. at room temperature in BSK-H supplemented with 1 µg of purified C2 (#A112, Complement Technology, Inc., Tyler, TX) or BSA (negative control). After incubation, spirochetes were diluted 1:3 in GHS buffer (10mM glucose, 10mM Hepes, and 50mM NaCl at pH 7.0) before incubating with C4b coated wells. The percent of bound bacteria was calculated as described above.
Complement inhibition ELISAs
Complement inhibition assays were performed as previously described (Garcia et al., 2016) with the following modifications. Human IgM (classical pathway initiator) and Saccharomyces cerevisiae mannan (lectin pathway initiator) were acquired from Sigma-Aldrich Co., St. Louis, MO, and Salmonella enteriditis LPS (alternative pathway initiator) was acquired from Hycult Biotech Inc., Plymouth Meeting, PA. Reaction volumes were 50 µL and consisted of pooled complement human serum (Innovative Research, Inc., Novi, MI) at a final concentration of 1% (classical and lectin pathway assays) or 20% (alternative pathway assays), and 10, 100, 500, or 1000 nM of GST-OspC or GST in CP/LP buffer (classical and lectin pathway assays) (0.1% gelatin, 5nM Veronal, 145 mM NaCl, 0.025% NaN3, 0.15 mM CaCl2, 0.5 mM MgCl2 (pH 7.3), Complement Technologies, Inc., Tyler, TX), or AP buffer (alternative pathway assays) (20mM HEPES (pH 7.3), 0.1% (v/v) gelatin, 140 mM NaCl, 5mM MgCl2, 10mM EGTA). Serum/GST-protein mixtures were incubated in wells coated with pathway initiators or the appropriate buffer only (control) for 1 hr at ambient temperature. Complement deposition in the wells was detected using complement protein C3d monoclonal antibody (clone 10C7) at a 1:2000 dilution (ThermoFischer Scientific Inc., Grand Island, NY) incubated at ambient temperature for 1 hr. Wells were incubated in HRP conjugated anti-mouse antibody (Promega, Madison, WI) at a 1:20,000 dilution for 1 hr. at ambient temperature. HRP-labeled antibody was detected using TMB tablets (Sigma-Aldrich Co., St. Louis, MO) dissolved in DMSO and 3% H2O2 and incubating for 5 min. at ambient temperature. Reactions were stopped by the addition of 6% final sulfuric acid and the absorbance was measured at 450 nm using a SoftMax Pro microplate reader (Molecular Devices, Sunnyvale, CA). All assays were performed with a minimum of 3 biological replicates performed in quadruplicate. The average absorbance in uncoated wells incubated in buffer only was subtracted from average absorbances of coated wells incubated in serum +/− GST or GST-OspC for each plate. All average absorbance values of each plate were normalized to the average absorbance of coated wells incubated with serum only before graphing and statistical analysis using GraphPad Prism 7. Statistical significance was determined using the Holm-Sidak multiple comparisons method.
Immunofluorescence (IF) microscopy
Human complement complexes from pooled human serum deposited on the surface of B. burgdorferi were detected by IF microscopy. WT or ospC mutant B. burgdorferi were grown in serum-free BSKII ((Posey & Gherardini, 2000), without the addition of 3-phosphoglyceric acid) and used at densities > 1×108/ mL. Cultures were incubated on fibronectin-coated coverslips (fibronectin, Sigma-Aldrich Co., St. Louis, MO) for 1 hr. at 33°C. Coverslips were washed in PBS+ 0.2% BSA and incubated with 50% pooled active or heat-inactivated human serum in serum-free BSKII for 15 min. at ambient temperature. Coverslips were washed in PBS+0.2% BSA and fixed in 2% final PFA in PBS. Fixed samples were blocked in PBS+10% BSA for 1 hr, incubated with rabbit anti-human C2a (#bs-10428R, Bioss Antibodies, Woburn, MA) and mouse anti-OspCB31 antibody, raised in our lab against purified GST-OspC from B. burgdorferi strain B31-A3, for 1 hr. at ambient temperature. Coverslips were washed three times in PBS +10% BSA for 10 min. each and incubated in Alexa Fluor™ 549-conjugated goat anti-mouse and Alexa Fluor™ 488-conjugated goat anti-rabbit antibodies for 1 hr. at ambient temperature (both from ThermoFisher Scientific, Grand Island, NY). In the last 15 min. of the incubation, 5 µg/ mL DAPI (ThermoFisher Scientific, Grand Island, NY) was added to each coverslip. Coverslips were washed three times in PBS+10% BSA for 10 min. each and mounted using ProLong™ Gold Antifade mountant with DAPI (ThermoFisher Scientific, Grand Island, NY). The mountant was cured in the dark for 12 h or longer before sealing of the coverslips with nail polish. Fluorescence microscopy images were acquired by a Nikon Eclipse Ti-U inverted microscope equipped with a CoolSNAP ES2 CCD camera (Photometrics, Tucson, AZ) and a multifluorescent Sedat Quad ET filter set (multichroic splitter, Chroma Technology Corp., Bellows Falls, VT) using the oil-immersion 60× Plan Apo VC objective lens (N.A. 1.40, Nikon Instruments Inc., Melville, NY). NIS-Elements software (Nikon Instruments Inc., Melville, NY) was used for image acquisition and processing. Scale bars represent 2 µm.
Supplementary Material
Acknowledgments
We thank George Chaconas and Richard Robinson for valuable advice and support, and Patricia Rosa for providing us with B. burgdorferi strain B31-A3, the ospC mutant strain OspCK1 (ΔospC), and pBSV2G, Jorge Benach for α-FlaB CB1 monoclonal antibody, Allen Parmelee and Stephen Kwok in the Flow Cytometry Core in the Department of Pathology at Tufts University School of Medicine for assisting with flow cytometry, and Ashleigh Labbe for providing necessary protocols and training. This work was supported by NIH R01AI093104, R01AI121401, R01AI118799.
Footnotes
The authors declare no competing financial interests.
References
- Alitalo A, Meri T, Lankinen H, Seppälä I, Lahdenne P, Hefty P, Meri S. Complement Inhibitor Factor H Binding to Lyme Disease Spirochetes Is Mediated by Inducible Expression of Multiple Plasmid-Encoded Outer Surface Protein E Paralogs. J Immunol. 2002;169(7):3847–3853. doi: 10.4049/jimmunol.169.7.3847. [DOI] [PubMed] [Google Scholar]
- Baranton G, Postic D, Saint Girons I, Boerlin P, Piffaretti J, Assous M, Grimont PAD. Delineation of Borrelia burgdorferi Sensu Stricto, Borrelia garinii sp. nov., Group VS461 Associated with Lyme Borreliosis. Int J Syst Bacteriol. 1992;42(3):378–383. doi: 10.1099/00207713-42-3-378. [DOI] [PubMed] [Google Scholar]
- Barbour AG. Isolation and Cultivation of Lyme Disease Spirochetes. Yale J Biol Med. 1984;57(4):521–525. [PMC free article] [PubMed] [Google Scholar]
- Barthold SW, Hodzic E, Tunev S, Feng S. Antibody-Mediated Disease Remission in the Mouse Model of Lyme Borreliosis. Infect Immun. 2006;74(8):4817–4825. doi: 10.1128/iai.00469-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit VM, Fischer JR, Lin Y-P, Parveen N, Leong JM. Allelic Variation of the Lyme Disease Spirochete Adhesin DbpA Influences Spirochetal Binding to Decorin, Dermatan Sulfate, and Mammalian Cells. Infect Immun. 2011;79(9):3501–3509. doi: 10.1128/iai.00163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockenstedt LK, Barthold S, Deponte K, Marcantonio N, Kantor FS. Borrelia burgdorferi infection and immunity in mice deficient in the fifth component of complement. Infect Immun. 1993;61(5):2104–2107. doi: 10.1128/iai.61.5.2104-2107.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt ME, Riley BS, Radolf JD, Norgard MV. Immunogenic integral membrane proteins of Borrelia burgdorferi are lipoproteins. Infect Immun. 1990;58(4):983–991. doi: 10.1128/iai.58.4.983-991.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangulis K, Petrovskis I, Kazaks A, Akopjana I, Tars K. Crystal structures of the Erp protein family members ErpP and ErpC from Borrelia burgdorferi reveal the reason for different affinities for complement regulator factor H. Biochim Biophys Acta. 2014;(0) doi: 10.1016/j.bbapap.2014.12.025. doi: http://dx.doi.org/10.1016/j.bbapap.2014.12.025. [DOI] [PubMed]
- Bunikis I, Kutschan-Bunikis S, Bonde M, Bergström S. Multiplex PCR as a Tool For Validating Plasmid Content of Borrelia burgdorferi. J Microbiol Methods. 2011;86(2):243–247. doi: 10.1016/j.mimet.2011.05.004. doi: http://dx.doi.org/10.1016/j.mimet.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Bykowski T, Woodman ME, Cooley AE, Brissette CA, Brade V, Wallich R, Stevenson B. Coordinated Expression of Borrelia burgdorferi Complement Regulator-Acquiring Surface Proteins during the Lyme Disease Spirochete's Mammal-Tick Infection Cycle. Infect Immun. 2007;75(9):4227–4236. doi: 10.1128/iai.00604-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caesar JJE, Johnson S, Kraiczy P, Lea SM. ErpC, A Member of the Complement Regulator-acquiring Family of Surface Proteins from Borrelia burgdorferi, Possesses an Architecture Previously Unseen in This Protein Family. Acta Crystallogr F Struct Biol. 2013;69(6):624–628. doi: 10.1107/S1744309113013249. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caine JA, Coburn J. A Short-Term Borrelia burgdorferi Infection Model Identifies Tissue Tropisms and Bloodstream Survival Conferred by Adhesion Proteins. Infect Immun. 2015;83(8):3184–3194. doi: 10.1128/iai.00349-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco SE, Troxell B, Yang Y, Brandt SL, Li H, Sandusky GE, Yang XF. Outer Surface Protein OspC Is an Antiphagocytic Factor That Protects Borrelia burgdorferi from Phagocytosis by Macrophages. Infect Immun. 2015;83(12):4848–4860. doi: 10.1128/iai.01215-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Lyme Disease- Data and Statistics. 2015 Sep 24; http://www.cdc.gov/lyme/stats/. Retrieved from http://www.cdc.gov/lyme/stats/
- Chen Q, Fischer JR, Benoit VM, Dufour NP, Youderian P, Leong JM. In Vitro CpG Methylation Increases the Transformation Efficiency of Borrelia burgdorferi Strains Harboring the Endogenous Linear Plasmid lp56. J Bacteriol. 2008;190(24):7885–7891. doi: 10.1128/jb.00324-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnhart CG, Buckles EL, Dumler JS, Marconi RT. Demonstration of OspC Type Diversity in Invasive Human Lyme Disease Isolates and Identification of Previously Uncharacterized Epitopes That Define the Specificity of the OspC Murine Antibody Response. Infect Immun. 2005;73(12):7869–7877. doi: 10.1128/iai.73.12.7869-7877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias AF, Bono JL, Kupko Iii JJ, Stewart PE, Krum JG, Rosa PA. New Antibiotic Resistance Cassettes Suitable for Genetic Studies in Borrelia burgdorferi. J Mol Microbiol Biotechnol. 2003;6(1):29–40. doi: 10.1159/000073406. [DOI] [PubMed] [Google Scholar]
- Elias AF, Stewart PE, Grimm D, Caimano MJ, Eggers CH, Tilly K, Rosa P. Clonal Polymorphism of Borrelia burgdorferi Strain B31 MI: Implications for Mutagenesis in an Infectious Strain Background. Infect Immun. 2002;70(4):2139–2150. doi: 10.1128/iai.70.4.2139-2150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer JR, LeBlanc KT, Leong JM. Fibronectin Binding Protein BBK32 of the Lyme Disease Spirochete Promotes Bacterial Attachment to Glycosaminoglycans. Infect Immun. 2006;74(1):435–441. doi: 10.1128/iai.74.1.435-441.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia BL, Zhi H, Wager B, Höök M, Skare JT. Borrelia burgdorferi BBK32 Inhibits the Classical Pathway by Blocking Activation of the C1 Complement Complex. PLoS Pathog. 2016;12(1):e1005404. doi: 10.1371/journal.ppat.1005404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore RD, Kappel KJ, Dolan MC, Burkot TR, Johnson BJ. Outer surface protein C (OspC), but not P39, is a protective immunogen against a tick-transmitted Borrelia burgdorferi challenge: evidence for a conformational protective epitope in OspC. Infect Immun. 1996;64(6):2234–2239. doi: 10.1128/iai.64.6.2234-2239.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Tilly K, Byram R, Stewart PE, Krum JG, Bueschel DM, Rosa PA. Outer-surface Protein C of the Lyme Disease Spirochete: A Protein Induced in Ticks for Infection of Mammals. PNAS. 2004;101(9):3142–3147. doi: 10.1073/pnas.0306845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallström T, Siegel C, Mörgelin M, Kraiczy P, Skerka C, Zipfel PF. CspA from Borrelia burgdorferi Inhibits the Terminal Complement Pathway. mBio. 2013;4(4):e004891–004813. doi: 10.1128/mBio.00481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt C, Hallström T, Skerka C, Wallich R, Stevenson B, Zipfel PF, Kraiczy P. Contribution of the Infection-associated Complement Regulator-acquiring Surface Protein 4 (ErpC) to Complement Resistance of Borrelia burgdorferi. Clin Dev Immunol, 2012. 2012 doi: 10.1155/2012/349657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt C, Koenigs A, Siegel C, Hallström T, Skerka C, Wallich R, Kraiczy P. Versatile Roles of CspA Orthologs in Complement Inactivation of Serum-Resistant Lyme Disease Spirochetes. Infect Immun. 2014;82(1):380–392. doi: 10.1128/iai.01094-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann K, Corvey C, Skerka C, Kirschfink M, Karas M, Brade V, Kraiczy P. Functional characterization of BbCRASP-2, a distinct outer membrane protein of Borrelia burgdorferi that binds host complement regulators factor H and FHL-1. Mol Microbiol. 2006;61(5):1220–1236. doi: 10.1111/j.1365-2958.2006.05318.x. [DOI] [PubMed] [Google Scholar]
- Kenedy MR, Akins DR. The OspE-Related Proteins Inhibit Complement Deposition and Enhance Serum Resistance of Borrelia burgdorferi, the Lyme Disease Spirochete. Infect Immun. 2011;79(4):1451–1457. doi: 10.1128/iai.01274-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenedy MR, Vuppala SR, Siegel C, Kraiczy P, Akins DR. CspA-Mediated Binding of Human Factor H Inhibits Complement Deposition and Confers Serum Resistance in Borrelia burgdorferi. Infect Immun. 2009;77(7):2773–2782. doi: 10.1128/iai.00318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraiczy P, Hartmann K, Hellwage J, Skerka C, Kirschfink M, Brade V, Stevenson B. Immunological Characterization of the Complement Regulator Factor H-binding CRASP and Erp Proteins of Borrelia burgdorferi. Int J Med Microbiol. 2004;293(37):152–157. doi: 10.1016/S1433-1128(04)80029-9. [DOI] [PubMed] [Google Scholar]
- Kraiczy P, Rossmann E, Brade V, Simon MM, Skerka C, Zipfel PF, Wallich R. Binding of Human Complement Regulators FHL-1 and Factor H to CRASP-1 Orthologs of Borrelia burgdorferi. Wien klin Wochenschr. 2006;118(21–22):669–676. doi: 10.1007/s00508-006-0691-1. [DOI] [PubMed] [Google Scholar]
- Kraiczy P, Skerka C, Brade V, Zipfel PF. Further Characterization of Complement Regulator-Acquiring Surface Proteins of Borrelia burgdorferi. Infect Immun. 2001;69(12):7800–7809. doi: 10.1128/iai.69.12.7800-7809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraiczy P, Skerka C, Kirschfink M, Brade V, Zipfel PF. Immune Evasion of Borrelia burgdorferi by Acquisition of Human Complement Regulators FHL-1/Reconectin and Factor H. Eur J Immunol. 2001;31(6):1674–1684. doi: 10.1002/1521-4141(200106)31:6<1674::AID-IMMU1674>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Kumar D, Ristow LC, Shi M, Mukherjee P, Caine JA, Lee W-Y, Chaconas G. Intravital Imaging of Vascular Transmigration by the Lyme Spirochete: Requirement for the Integrin Binding Residues of the B. burgdorferi P66 Protein. PLoS Pathog. 2015;11(12):e1005333. doi: 10.1371/journal.ppat.1005333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtenbach K, Schäfer SM, Sewell H, Peacey M, Hoodless A, Nuttall PA, Randolph SE. Differential Survival of Lyme Borreliosis Spirochetes in Ticks That Feed on Birds. Infect Immun. 2002;70(10):5893–5895. doi: 10.1128/iai.70.10.5893-5895.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagal V, Postic D, Ruzic-Sabljic E, Baranton G. Genetic Diversity among Borrelia Strains Determined by Single-Strand Conformation Polymorphism Analysis of the ospC Gene and Its Association with Invasiveness. J Clin Micro. 2003;41(11):5059–5065. doi: 10.1128/jcm.41.11.5059-5065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrenz MB, Wooten RM, Zachary JF, Drouin S, Weis JJ, Wetsel RA, Norris SJ. Effect of Complement Component C3 Deficiency on Experimental Lyme Borreliosis in Mice. Infect Immun. 2003;71(8):4432–4440. doi: 10.1128/iai.71.8.4432-4440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Oliver J, J H, Gao L. Genetic Diversity of the Outer Surface Protein C Gene of Southern Borrelia Isolates and Its Possible Epidemiological, Clinical, and Pathogenetic Implications. J Clin Micro. 2002;40(7):2572–2583. doi: 10.1128/jcm.40.7.2572-2583.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-P, Lee D-W, McDonough SP, Nicholson LK, Sharma Y, Chang Y-F. Repeated Domains of Leptospira Immunoglobulin-like Proteins Interact with Elastin and Tropoelastin. J Biol Chem. 2009;284(29):19380–19391. doi: 10.1074/jbc.M109.004531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liveris D, Wang G, Girao G, Byrne DW, Nowakowski J, McKenna D, Schwartz I. Quantitative Detection of Borrelia burgdorferi in 2-Millimeter Skin Samples of Erythema Migrans Lesions: Correlation of Results with Clinical and Laboratory Findings. J Clin Micro. 2002;40(4):1249–1253. doi: 10.1128/jcm.40.4.1249-1253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meri S, Jarva H. Complement Regulation. Vox Sanguinis. 1998;74(S2):291–302. doi: 10.1111/j.1423-0410.1998.tb05434.x. [DOI] [PubMed] [Google Scholar]
- Posey JE, Gherardini FC. Lack of a Role for Iron in the Lyme Disease Pathogen. Science. 2000;288(5471):1651–1653. doi: 10.1126/science.288.5471.1651. [DOI] [PubMed] [Google Scholar]
- Preac-Mursic V, Wilske B, Jauris S, Will G, Reinhardt S, Lehnert G, Klockmann U. Active immunization with pC protein of Borrelia burgdorferi protects gerbils against B. burgdorferi infection. Infect Immun. 1992;20(6):342–349. doi: 10.1007/BF01710681. [DOI] [PubMed] [Google Scholar]
- Probert WS, Crawford M, Cadiz RB, LeFebvre RB. Immunization with Outer Surface Protein (Osp) A, but Not OspC, Provides Cross-Protection of Mice Challenged with North American Isolates of Borrelia burgdorferi. J Infect Dis. 1997;175(2):400–405. doi: 10.1093/infdis/175.2.400. [DOI] [PubMed] [Google Scholar]
- Probert WS, LeFebvre RB. Protection of C3H/HeN mice from challenge with Borrelia burgdorferi through active immunization with OspA, OspB, or OspC, but not with OspD or the 83-kilodalton antigen. Infect Immun. 1994;62(5):1920–1926. doi: 10.1128/iai.62.5.1920-1926.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnoff OD, Pensky J, Ogston D, Naff GB. The inhibition of plasmin, plasma kallikrein, plasma permeability factor, and the C1r subcomponent of the first componenet of complement by serum C1 esterase inhibitor. J Exp Med. 1969;129(2):315–331. doi: 10.1084/jem.129.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow LC, Miller HE, Padmore LJ, Chettri R, Salzman N, Caimano MJ, Coburn J. The β3-Integrin Ligand of Borrelia burgdorferi is Critical for Infection of Mice but Not Ticks. Mol Microbiol. 2012;85(6):1105–1118. doi: 10.1111/j.1365-2958.2012.08160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels DS. Electrotransformation of the Spirochete Borrelia burgdorferi. In: Nickoloff JA, editor. Electroporation Protocols for Microorganisms. Totowa, NJ: Humana Press; 1995. pp. 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan TG, Piesman J, Golde WT, Dolan MC, Rosa PA. Induction of an Outer Surface Protein on Borrelia burgdorferi During Tick Feeding. PNAS. 1995;92(7):2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seinost G, Dykhuizen DE, Dattwyler RJ, Golde WT, Dunn JJ, Wang I, Luft BJ. Four Clones of Borrelia burgdorferi Sensu Stricto Cause Invasive Infection in Humans. Infect Immun. 1999;67(7):3518–3524. doi: 10.1128/iai.67.7.3518-3524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass Spectrometric Sequencing of Proteins Silver-stained Polyacrylamide Gels. Anal Chem. 1996;68(5):850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Shih C-M, Chao L-L. Genetic analysis of the outer surface protein C gene of Lyme disease spirochaetes (Borrelia burgdorferi sensu lato) isolated from rodents in Taiwan. J. Med. Micro. 2002;51(4):318–325. doi: 10.1099/0022-1317-51-4-318. doi: [DOI] [PubMed] [Google Scholar]
- Siegel C, Hallström T, Skerka C, Eberhardt H, Uzonyi B, Beckhaus T, Kraiczy P. Complement Factor H-Related Proteins CFHR2 and CFHR5 Represent Novel Ligands for the Infection-Associated CRASP Proteins of Borrelia burgdorferi. PLoS ONE. 2010;5(10):e13519. doi: 10.1371/journal.pone.0013519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel C, Schreiber J, Haupt K, Skerka C, Brade V, Simon MM, Kraiczy P. Deciphering the Ligand-binding Sites in the Borrelia burgdorferi Complement Regulator-acquiring Surface Protein 2 Required for Interactions with the Human Immune Regulators Factor H and Factor H-like Protein 1. J Biol Chem. 2008;283(50):34855–34863. doi: 10.1074/jbc.M805844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skare JT, Shaw DK, Trzeciakowski JP, Hyde JA. In Vivo Imaging Demonstrates That Borrelia burgdorferi ospC Is Uniquely Expressed Temporally and Spatially throughout Experimental Infection. PLoS ONE. 2016;11(9):e0162501. doi: 10.1371/journal.pone.0162501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava SY, de Silva AM. Reciprocal Expression of ospA and ospC in Single Cells of Borrelia burgdorferi. J Bacteriol. 2008;190(10):3429–3433. doi: 10.1128/jb.00085-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strle F, Ružić-Sabljić E, Cimperman J, Lotrič-Furlan S, Maraspin V. Comparison of Findings for Patients with Borrelia garinii and Borrelia afzelii Isolated from Cerebrospinal Fluid. Clin Infect Dis. 2006;43(6):704–710. doi: 10.1086/506936. [DOI] [PubMed] [Google Scholar]
- Tilly K, Krum JG, Bestor A, Jewett MW, Grimm D, Bueschel D, Rosa P. Borrelia burgdorferi OspC Protein Required Exclusively in a Crucial Early Stage of Mammalian Infection. Infect Immun. 2006;74(6):3554–3564. doi: 10.1128/iai.01950-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam AP, Kuiper H, Vos K, Widjojokusumo A, de Jongh BM, Spanjaard L, Dankert J. Different Genospecies of Borrelia burgdorferi Are Associated with Distinct Clinical Manifestations of Lyme Borreliosis. Clin Infect Dis. 1993;17(4):708–717. doi: 10.1093/clinids/17.4.708. [DOI] [PubMed] [Google Scholar]
- van Dam AP, Oei A, Jaspars R, Fijen C, Wilske B, Spanjaard L, Dankert J. Complement-mediated Serum Sensitivity Among Spirochetes That Cause Lyme Disease. Infect Immun. 1997;65(4):1228–1236. doi: 10.1128/iai.65.4.1228-1236.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Ojaimi C, Iyer R, Saksenberg V, McClain SA, Wormser GP, Schwartz I. Impact of genotypic variation of Borrelia burgdorferi sensu stricto on kinetics of dissemination and severity of disease in C3H/HeJ mice. Infect Immun. 2001;69 doi: 10.1128/iai.69.7.4303-4312.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Ojaimi C, Wu H, Saksenberg V, Iyer R, Liveris D, Schwartz I. Disease severity in a murine model of lyme borreliosis is associated with the genotype of the infecting Borrelia burgdorferi sensu stricto strain. J Infect Dis. 2002;186 doi: 10.1086/343043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weening EH, Parveen N, Trzeciakowski JP, Leong JM, Höök M, Skare JT. Borrelia burgdorferi Lacking DbpBA Exhibits an Early Survival Defect during Experimental Infection. Infect Immun. 2008;76(12):5694–5705. doi: 10.1128/iai.00690-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilske B, Preac-Mursic V, Jauris S, Hofmann A, Pradel I, Soutschek E, Wanner G. Immunological and molecular polymorphisms of OspC, an immunodominant major outer surface protein of Borrelia burgdorferi. Infect Immun. 1993;61(5):2182–2191. doi: 10.1128/iai.61.5.2182-2191.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.