Abstract
Selenoprotein P (SELENOP) is a serum glycoprotein that is required for proper selenium distribution in mammals, particularly in supplying selenium to the brain and testes. As the sole mechanism for providing essential selenium to developing spermatozoa, SELENOP metabolism is central to male fertility in all mammals. In addition, this process is important for proper brain function, especially under conditions of limited dietary selenium. Several specific and nonspecific mechanisms for SELENOP uptake in target tissues have been described, but the utilization of SELENOP as a source of selenium for intracellular selenoprotein production has not been systematically characterized. In this report we examine the process of SELENOP uptake using a robust selenium uptake assay that measures selenium utilization in cells fed 75Se-SELENOP. Using a series of inhibitors and modulators we have identified specific regulators of the process and found that SELENOP must be in an oxidized state for uptake. This assay also demonstrates that the proposed SELENOP receptor APOER2 is not required for selenium delivery by SELENOP to cells in culture.
Keywords: selenocysteine, selenoprotein P, selenium, oxidation
Introduction
Selenium is an essential trace element that plays a critical role in a multitude of biological processes. The only known biological function for selenium in mammals is in its incorporation as the 21st amino acid, selenocysteine (Sec). Mammalian cells are thought to obtain selenium through one of three pathways: 1) uptake of inorganic selenium through anion transport; 2) uptake of selenomethionine through methionine transporters and 3) uptake of the selenium-rich glycoprotein, selenoprotein P (SELENOP; previous designations include SelP and SEPP1), which harbors 10 Sec residues.
SELENOP was first identified as a plasma selenoprotein that was distinct from glutathione peroxidase[1, 2], and it was later determined to contain more than one mole of selenium per mole of protein[3]. Cloning of the cDNA encoding SELENOP revealed that that rat gene contained 10 in-frame UGA codons that could potentially be used as Sec codons[4]. Early hypotheses proposed a selenium transport role for SELENOP, which was confirmed when mice lacking the Selenop gene that encodes SELENOP were found to have selenium deficiency in the brain and testis, especially when fed lower selenium diets[5, 6]. The link between SELENOP and the brain-testis axis is phenotypically manifested as male sterility and severe neurologic dysfunction, the latter occurring only under conditions of limited selenium supply[5, 6]. Recent studies have shown, however, that even under selenium replete conditions, mice that lack SELENOP have persistent neurologic defects[7–9]. Interestingly, SELENOP expression is absolutely required for male fertility as supplemental selenium cannot reverse the defects in sperm motility and morphology, which is due to lack of GPX4 expression[10].
Evidence suggests that SELENOP delivers selenium to cells through receptor mediated endocytosis via Megalin in the kidney[11], APOER2 in the testis and brain[12], or by pinocytosis during maternal to fetal transfer[13]. Further, inhibitor studies have implicated clathrin-dependent delivery to the lysosomes as the initial processing event for SELENOP[14], but the fate of the protein and its selenium cargo in the lysosome has not been determined. It has long been proposed that SELENOP is degraded and that the resulting Sec residues are metabolized by Scly[15]. If processing by SCLY were the primary mechanism by which selenium is released from SELENOP, then animals lacking the gene encoding Scly should harbor a SELENOP null (SELENOP−/−) phenotype. However, although Scly−/− mice have reduced selenoprotein expression, they do not harbor any of the SELENOP−/− phenotypes like male sterility or neurologic defects[16]. This indicates that the Sec lyase is not involved in recovering selenium from internalized SELENOP and strongly implicates an alternative pathway. These questions demand the development of a robust SELENOP utilization assay and systematic investigation of the requirements for efficient SELENOP uptake and processing.
In this study we have identified novel and specific inhibitors of SELENOP uptake and processing. These results build on prior evidence that SELENOP processing takes place in the lysosome. Using the zebrafish version of SELENOP, we also demonstrate mammalian cells are able to utilize SELENOP from a phylogenetically distant organism.
Materials and Methods
Cell Culture
All of the experiments in which endogenous selenoprotein expression was assessed were performed in HeLa cells grown in EMEM (Life Technologies) plus 10% fetal bovine serum (except where indicated). HepG2 cells used for SELENOP production were grown in EMEM with 10% fetal bovine serum.
Production and quantitation of 75Se-SELENOP
HepG2 cells were grown in 100 mm dishes in 10 mL of growth media. After the cells reached appropriate confluence their media was aspirated and they were washed with 1X PBS. Serum free media was added, and after 24 hours of serum starvation, 100 nM 75Se selenite was added to the cells. Two batches of 75Se (University of Missouri) were employed in these studies with specific activities ranging from 175 to 700 Ci/g. The media from the cells was collected at 24 or 72 hours and concentrated 50-fold by ultrafiltration (Amicon). Prior to use in experiments free selenium was removed from the concentrated media by spin column gel filtration (P30, BioRad).
For SELENOP quantitation, a 75Se-SELENOP range was run on an SDS-PAGE gel for quantification. The gel was dried, spotted with known concentrations of 75Se selenite, and exposed to a phosphorimage screen. The signal intensity from the range of 75Se-SELENOP was then analyzed and compared to the intensity of the spotted 75Se selenite using ImageQuant software. Using the spotted range of 75Se Selenite as a standard, the concentration of 75Se Selenite in the protein samples was calculated. Since we typically observed the major radioactive band corresponding to the expected molecular weight of glycosylated full length SELENOP, we assumed for the purposes of quantitation that all of the protein contained the full 10 moles of selenium per mole of protein.
Cell labeling with 75Se
For all cell types, endogenous selenoproteins from the cell types indicated were labeled with either 75Se selenite or the 75Se-SELENOP described above. Cells were plated in appropriate growth media until desired confluence was reached. Cells were left in full serum throughout the labeling process unless otherwise noted. Cells were then incubated with 100 nM [75Se]selenite, or various amounts of 75Se- SELENOP. After 24 hours of incubation with either form of 75Se, cells were washed with 1x PBS and lysed in 2X SDS sample buffer (100 mM Tris-HCl pH 6.8, 4% (w/v) SDS, 0.2% (w/v) bromophenol blue and 20% (v/v) glycerol) at room temp for 10 minutes. When the lysis was complete, samples were collected and boiled at 95 °C for 5 minutes and spun at max speed for 1 minute. The samples were then run on an SDS-Page gel for analysis. For the analysis of inhibitors, either the cells or the 75Se-SELENOP preparation was pre-incubated with inhibitor as indicated in the figure legends. For inhibitors pre-incubated with 75Se-SELENOP, the protein was passed through a desalting column (P30, Bio-Rad) prior to its application to cells.
Western Blotting and immunoprecipitation
Total proteins from conditioned medium derived from 75Se labeled HepG2 cells were resolved by 12% SDS-PAGE. The proteins were transferred onto a nitrocellulose membrane using a semi-dry apparatus (Bio-Rad) at 18V for 30 minutes. When the transfer finished the membrane was blocked in 5% non-fat dried milk made in 1x TBS w/ 0.05%Tween 20 for 1 hour at room temperature. The blot was then incubated overnight at 4 °C with a 1:1000 dilution of a monoclonal SELENOP antibody (37A1;Thermo Scientific). The blot was washed 4 times for 8 minutes/wash with TBST and following the washes incubated with HRP-goat anti mouse secondary antibody (Life Technologies) for 1 hour at room temp. The blot was then washed again following the same procedure and following the final wash it was developed using SuperSignal West Femto Kit (Thermo Scientific). The membrane was sealed in plastic after development and exposed to a phosphorscreen overnight.
For immunoprecipitation, HeLa cells were labeled with 75Se-SELENOP or 75Se Selenite for 24 hours. After labeling, cells were placed on ice and washed three times with ice cold PBS, then lysed for 10 min on ice with IP lysis buffer (10 mM Tris-HCl pH 7.6, 150 mM NaCl, 0.5% NP40, 5% Glycerol and 1 mM PMSF). Lysates were centrifuged at 17,000 × g for 10 min and supernatant incubated overnight at 4 °C with 1:100 Thioredoxin Reductase 1 antibody (GenWay) or 1:50 SELENOP antibody (Epitomics). Bound proteins were pulled down using the Dynabeads Protein G Kit (Life Technologies). After pull-down, beads were boiled for 5 minutes at 95 °C in SDS sample buffer and proteins were resolved on a 12% SDS-PAGE gel. Gels were fixed for 15 minutes at room temp in 10% acetic acid, 25% ethanol then dried and exposed to a phosphorscreen overnight.
SELENOP degradation assay
Concentrated conditioned medium from 75Se-selenite labeled HepG2 cells was buffer exchanged into 1X PBS and incubated with varying concentrations of acetic acid in the presence or absence of 2 mM dithiothreitol (DTT). Samples (20 µl total volume) containing 2 µl of the 75Se-SELENOP preparation were incubated at 37 °C for 30 min after which 7 ul of 4X SDS sample buffer was added. Those samples where the bromophenol blue had turned yellow were neutralized with 1 µl of 50 mM NaOH.
Uptake of zebrafish SELENOP
The zebrafish SELENOP cDNA (a gift from V. Gladyshev, Harvard) was subcloned into pCDNA3.1 with a 3’ FLAG tag. HEK-293 cells were transfected with this plasmid and stable maintenance of the plasmid was selected by culturing cells in 630 µg/ml G418. A pool of clones was maintained in 630 µg/ml G418 and two 10 cm2 dishes of nearly confluent cells were labeled with 75Se-selenite for 24 hours. 2 ml of concentrated conditioned medium was incubated with 100 µl of anti-FLAG Sepharose beads (Sigma) for 2 hours at 4 °C. The radiolabeled SELENOP was eluted from the beads with 3X FLAG peptide (Sigma) for 1 hour at 4 °C. Eluted protein as well as an 75Se-selenite (equal cpms) were used to label naive HEK-293 cells for 24 hours. Cell lysates were made in NP-40 lysis buffer (10mM Tris-Cl, pH 7.4, 1%NP40, protease inhibitors), and the 75Se-selenite labeled endogenous selenoproteins were analyzed by SDS-PAGE followed by phosphorimager analysis.
Results and Discussion
Kinetics of selenium utilization
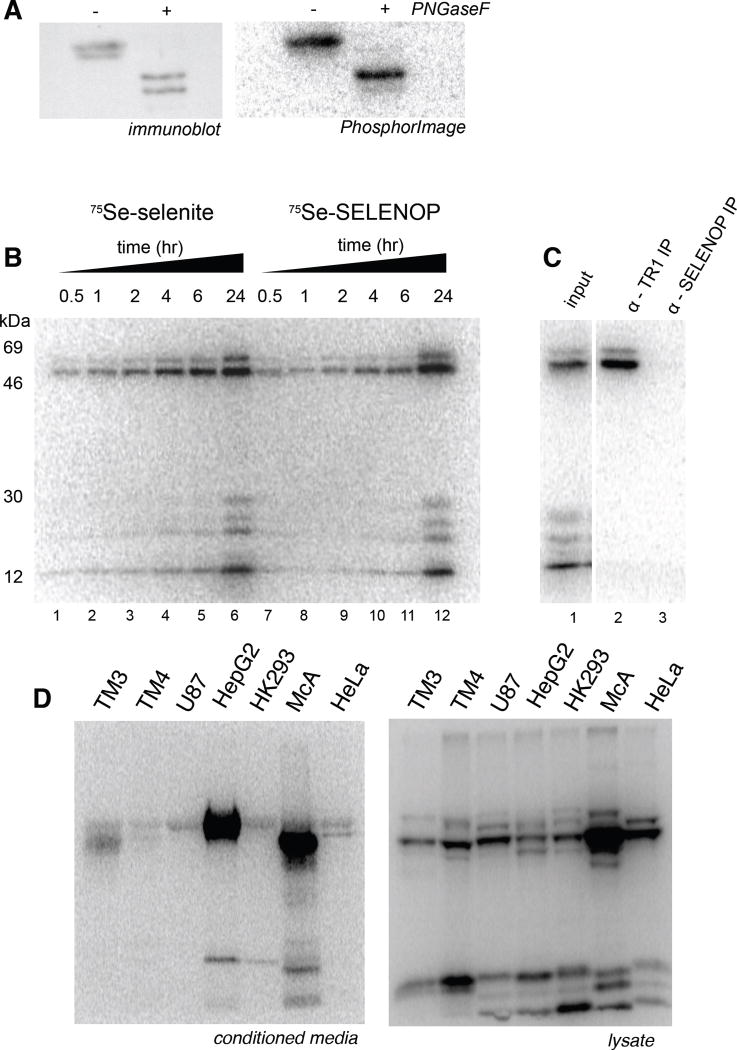
In order to visualize the uptake and utilization of SELENOP by cells in culture, we generated 75Se radio-labeled SELENOP (75Se-SELENOP) by culturing human liver hepatoma cells (HepG2) in the presence of 100 nM 75Se sodium selenite. As a secreted protein, 75Se-SELENOP accumulates in the medium as a glycosylated protein migrating as two species. The major ~63 kDa species corresponds to full-length SELENOP while the slightly faster-migrating species likely corresponds to products resulting from premature translation termination[17]. These products were concentrated approximately 50-fold by ultrafiltration, and the sample was subjected to PNGase F treatment, which fully deglycosylates SELENOP, permitting it to be resolved at its predicted molecular mass(es). Figure 1A shows the analysis of untreated and PNGaseF-treated concentrated conditioned medium by western blot analysis and autoradiography. This is representative of the material used for all downstream assays for SELENOP uptake. Free selenium was removed by gel filtration and the 75Se-SELENOP was then added to cells growing in culture in order to study SELENOP uptake and processing. We have used this method to estimate the amount of each form of selenium that is required to saturate selenoprotein expression in HeLa cells. This corresponds to ~20 nM for sodium selenite and, as expected, about 2 nM for SELENOP, which contains 10 moles of selenium per mole of protein (data not shown). Figure 1B shows a time course in which HeLa cells were incubated with 75Se-SELENOP or 75Se selenite for various amounts of time followed by media replacement. In this experiment we observe that both 75Se-selenite and 75Se-SELENOP are able to support the incorporation of 75Se into endogenous selenoproteins, of which six are easily observed. The two largest proteins correspond to isoforms of thioredoxin reductase as confirmed by immunoprecipitation (Figure 1C). Interestingly, the kinetics of inorganic 75Se utilization are very similar, suggesting a rapid process for selenium assimilation for both selenite and SELENOP uptake. This is further demonstrated by the fact that we are not able to immunoprecipitate 75Se- SELENOP from the labeled cell lysate (Figure 1C). We likely do not observe intracellular SELENOP because as shown below in figure 4B, SELENOP processing is apparently rapid and must be inhibited in order to observe intact intracellular SELENOP. In addition, HeLa cells synthesize significantly less than the liver derived cell lines such as HepG2. Figure 1D shows 75Se-selenite labeling of various cell lines to illustrate the variable amounts of SELENOP secreted and the lack of obvious intracellular SELENOP accumulated in cell lysates. Overall, these results demonstrate that SELENOP can be used as a selenium source for cells in culture, and in terms of generalizability, it is notable that we have used this method to label endogenous selenoproteins in cell types from human (HeLa, primary Sertoli, HepG2, PC-3, LNCaP, SK-N-MC and U87, HEK-293), dog (MDCK and D17) and mouse (N2A, TM3, TM4) (data not shown).
Figure 1. 75Se-SELENOP can be used as a selenium source for intracellular selenoprotein production.
(A) Conditioned medium from HepG2 cells grown in the presence of 75Se-selenite was concentrated ~50-fold and a sample was treated with PNGaseF. The samples were resolved by SDS-PAGE and SELENOP was detected by immunoblot (left) and phosphorimage analysis (right). (B) HeLa cells were incubated with ~2 pmol of 75Se-SELENOP or 100 nM 75Se selenite for the time points indicated then switched to normal growth medium and allowed to grow for a total of 24 hours. Endogenous selenoproteins that utilized the 75Se were resolved by SDS-PAGE and detected by PhosphorImage analysis. (C) HeLa cells were labeled with 75Se-SELENOP as described in (A) and lysates were incubated with either an anti-thioredoxin reductase or anti-SELENOP antibody. Immunoprecipitated 75Se-labeled proteins were resolved by SDS-PAGE and detected by PhosphorImage analysis. (D) 75Se-selenite was used to label the endogenous selenoproteins in the cell types indicated: mouse Leydig cells [TM3], mouse Sertoli cells [TM4], human glioblastoma [U87], human hepatoma [HepG2], rat hepatoma [McArdle 7777], human epithelial [HeLa]. After 24 hours of labeling, 4% of the media (left) and 10% of the lysate (right) was resolved by SDS-PAGE and detected by PhosphorImage analysis.
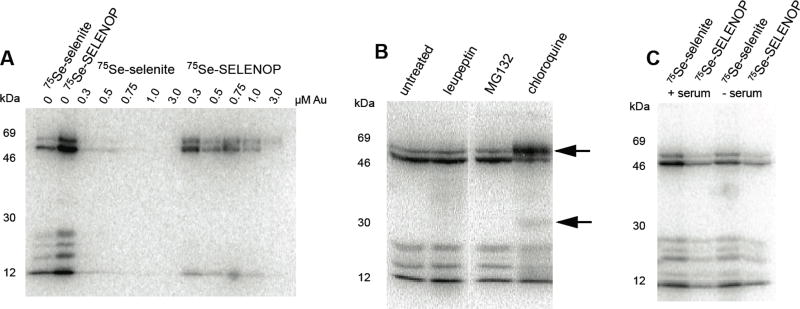
Figure 4. Inhibitors reveal mechanistic details about SELENOP uptake and processing.
(A) HeLa cells were treated with the indicated concentrations of Auranofin for 1 hour prior to the addition of 75Se-selenite or 75Se-SELENOP. After 24 hours, the cells were lysed and 75Se labeled proteins were resolved by SDS-PAGE and detected by PhosphorImage analysis. (B) (C) HeLa cells were labeled with 75Se selenite or 75Se-SELENOP in the presence or absence of 10% fetal bovine serum as indicated. Note that the relative intensity of endogenous selenoproteins labeled by 75Se-selenite or 75Se-SELENOP varied due to the use of two different batches of 75Se that differed in specific activity.
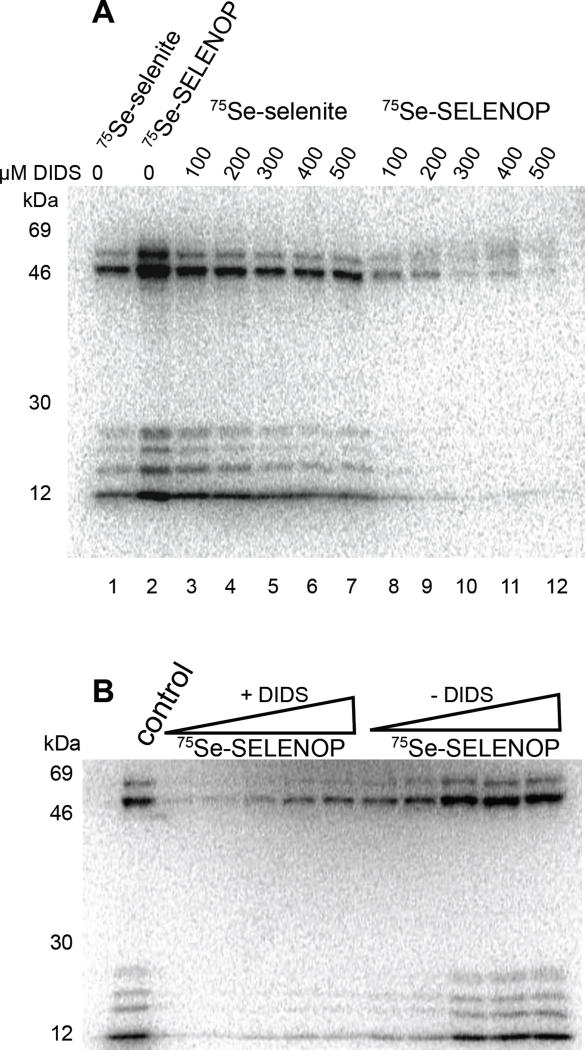
Regulators of selenium uptake and utilization 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid (DIDS)
Prior work has shown that the anion transporter (band 3) inhibitor 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid (DIDS) inhibits selenite uptake[18, 19]. With the idea that we would be able to use DIDS to specifically inhibit 75Se-selenite but not 75Se-SELENOP uptake, we tested a range of DIDS concentrations in our selenoprotein labeling assay. Surprisingly, we found that when DIDS was added to cells, the labeling of endogenous selenoproteins with SELENOP was substantially blocked (Figure 2A, lanes 8-12), but the uptake and utilization of 75Se-selenite was not affected (Figure 2A, lanes 3-7). Although this result was in direct conflict with the prior data, it is likely that the difference in methodology can explain the discrepancy. Prior DIDS inhibition was demonstrated in a 75Se-selenite uptake experiment where low concentrations of selenium were added to cells and uptake was measured by scintillation. Considering that our endpoint was selenoprotein synthesis at 24 hours, it is likely that reduced anion transport rates are not sufficient to significantly alter selenoprotein synthesis. Although the mechanism for DIDS inhibition of SELENOP uptake is unknown, we hypothesize that DIDS, which is known to modify primary amines and thiols, is likely to be directly modifying SELENOP residues and thereby inhibiting recognition and uptake. To test this directly, SELENOP was pre-incubated with DIDS, which was then removed by gel filtration prior to 75Se-SELENOP labeling of cells. Figure 2B shows a range of untreated and DIDS-treated SELENOP applied to HeLa cells. This result shows that DIDS exposure significantly impaired selenoprotein labeling, which suggests that amino acid modification is preventing SELENOP uptake.
Figure 2. DIDS is a specific inhibitor of SELENOP uptake.
(A) HeLa cells were labeled with 75Se selenite or 75Se-SELENOP in the presence of a range of 4,4-Diisothiocyanatostilbene-2,2-Disulfonic Acid (DIDS) as indicated. 75Se labeled proteins were resolved by SDS-PAGE and detected by PhosphorImage analysis. (B) SELENOP was pre-treated with DIDS. Excess DIDS was removed by gel filtration and varying amounts of the DIDS-treated or untreated control protein was used to label endogenous HeLa selenoproteins as above.
Oxidation
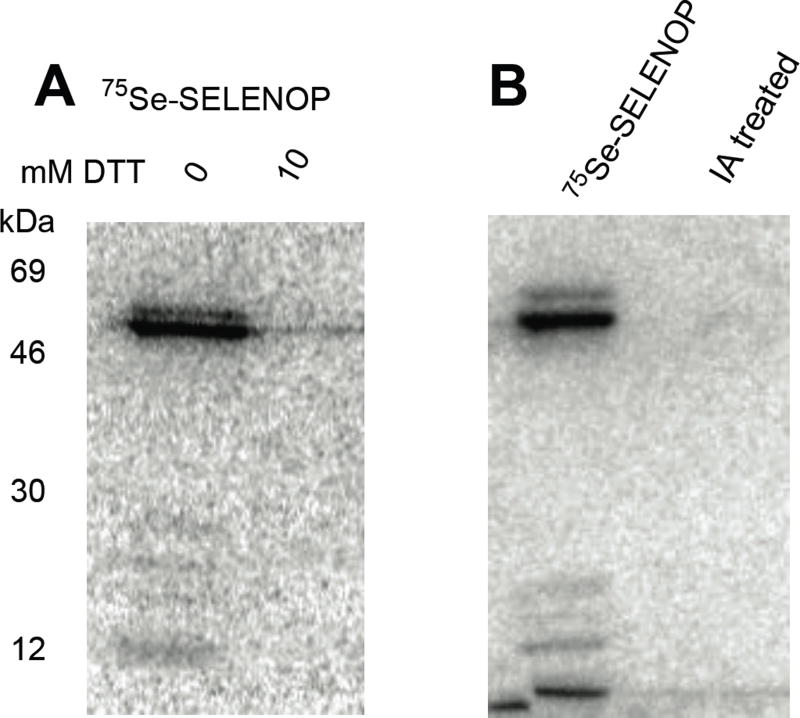
Considering that DIDS might be modifying Cys and Sec residues in SELENOP and that oxidized Cys and Sec residues have previously been mapped in SELENOP[20], we set out to determine if oxidizing conditions are necessary for SELENOP uptake and processing. To that end, we pre-incubated 75Se-labeled SELENOP with 10 mM dithiothreitol (DTT) and then removed excess DTT by buffer exchange. This process did not affect the integrity of SELENOP (see figure 5 below). As shown in Figure 3A, the reduced version of SELENOP was apparently not taken up by the cells as no endogenous selenoprotein labeling is observed and no intact SELENOP can be detected. We further analyzed the role of Cys and Sec residues in regulating SELENOP uptake by modifying the protein with iodoacetamide. As with the use of DTT, iodoacetamide modified SELENOP was not taken up by HeLa cells (Figure 3B). These results clearly suggest that SELENOP must be in an oxidized state for uptake, indicating that a specific conformation is required. With 10 Sec and 19 Cys residues, it is likely that multiple disulfide, diselenide and mixed selenosulfide linkages are required in order to adopt a conformation that is favorable for uptake.
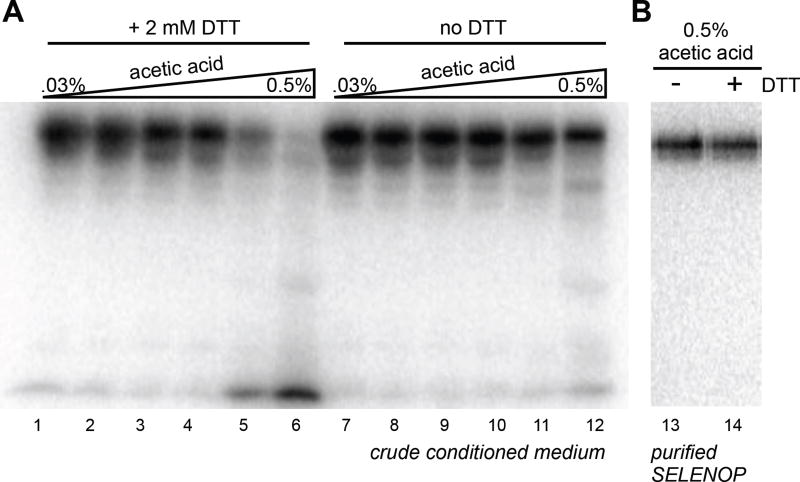
Figure 5. Degradation of SELENOP occurs under acidic and reducing conditions.
(A) HepG2 derived conditioned media containing 75Se-SELENOP was incubated at 37 °C for 30 min in varying concentrations of acetic acid and/or 2 mM DTT as indicated. (B) 75Se-SELENOP was purified by nickel affinity chromatography and incubated at 37 °C for 30 min.
Figure 3. Reduced SELENOP cannot deliver selenium to cells.
(A) 75Se-SELENOP was incubated with 10 mM DTT for 1h at 37 °C. DTT was removed by gel filtration and the protein was then used to label endogenous HeLa selenoproteins as described in Figure 1A. (B) Reduced SELENOP was incubated with 25 mM iodoacetamide (IA). Excess IA was removed by gel filtration and the protein was used to label endogenous HeLa selenoproteins as above.
Selenium chelation
In order to further study the parameters required for SELENOP uptake and processing, we also tested the gold compound auranofin, which is known to chelate selenium[8] and has been found to be a potent inhibitor of thioredoxin reductase[21]. Since this compound is known to interact with both free selenium as well as selenocysteine, we expected significant inhibition for both selenite and 75Se-SELENOP labeling. Interestingly, however, we found that while selenite labeling was completely inhibited at all concentrations tested, 75Se-SELENOP was still able to deliver usable selenium to cells at the lower concentrations of auranofin (~0.3 uM) with the caveat that only the higher molecular weight selenoprotein, TXNRD1, was made (Figure 4A). This is consistent with the compound causing selenium deficiency by chelation, under which conditions TXNRD1 has been shown to be resistant to suppression[22]. This result provides further evidence that TXNRD1 expression is retained at the translational level at the expense of other selenoproteins. Deciphering the underlying mechanism for preferential translation will be key to fully understanding the hierarchy of selenoprotein expression that governs selenium utilization under conditions of stress or selenium deprivation.
Lysosomal inhibitors
Prior work has shown that SELENOP processing is inhibited by the lysosome inhibitor chloroquine[14]. To further the analysis of lysosomal processing, we incubated cells with the lysosomal protease inhibitor, leupeptin, the proteasomal inhibitor MG-132 as well as chloroquine (Figure 4B). While we were able to demonstrate that chloroquine inhibits SELENOP processing and thus endogenous selenoprotein production, neither of the protease inhibitors had an effect. These results reduce the likelihood of action by lysosomal serine, cysteine and threonine proteases in the processing of SELENOP. The use of MG-132 served as a control, showing that the proteasome is also not involved in SELENOP processing. In the case of chloroquine, it is of interest to note that an apparent degradation intermediate does appear (lower arrow), suggesting that proteolytic processing may in fact play a role in the recovery of selenium from SELENOP, but that the majority of the 75Se signal is observed as full- length SELENOP (upper arrow). This is the same result previously obtained, which also demonstrated that the bands that appear to be full length and proteolyzed SELENOP are bona fide as they can be immunoprecipitated with an anti-SELENOP antibody[14].
Fetal bovine serum
In general, the labeling of cells with radioactive selenium is performed in the absence of serum in order to minimize the non-radioactive selenium content during the uptake of 75Se. We set out to determine whether the selenium species present in fetal bovine serum were effective competitors for labeling with either 75Se-SELENOP or 75Se-selenite. Figure 4C shows that the amount of 75Se incorporated into endogenous selenoproteins in HeLa cells is the same regardless of the presence or absence of 10% serum. This data suggests that the selenium species that are present in serum are not effective competitors for selenium uptake either due to low concentrations or poor bioavailability.
In vitro processing
To gain insight into the mechanism by which SELENOP may be processed in cells, we examined the stability of the protein in the context of crude conditioned medium. Since evidence thus far suggests that SELENOP may be degraded in the lysosome, we analyzed the stability in the presence of acidifying and reducing conditions. Figure 5 (lanes1-12) shows that a crude preparation of 75Se-SELENOP is apparently degraded only in the presence of 0.5% acetic acid and 2 mM DTT. This result suggests the presence of an acid protease in the conditioned medium of HepG2 cells. Purification of SELENOP by nickel metal affinity chromatography eliminates its susceptibility to degradation under these conditions (Figure 5, lanes 13-14). These results indicate that SELENOP is very stable in standard conditions, but is susceptible to proteolysis by an acid protease only under acidic and reducing conditions. This may provide clues as to the conditions required to achieve efficient proteolysis of SELENOP, operating under the assumption that proteolysis is a key step in the recovery of selenium from SELENOP.
Zebrafish Selenop can be taken up by human embryonic kidney cells
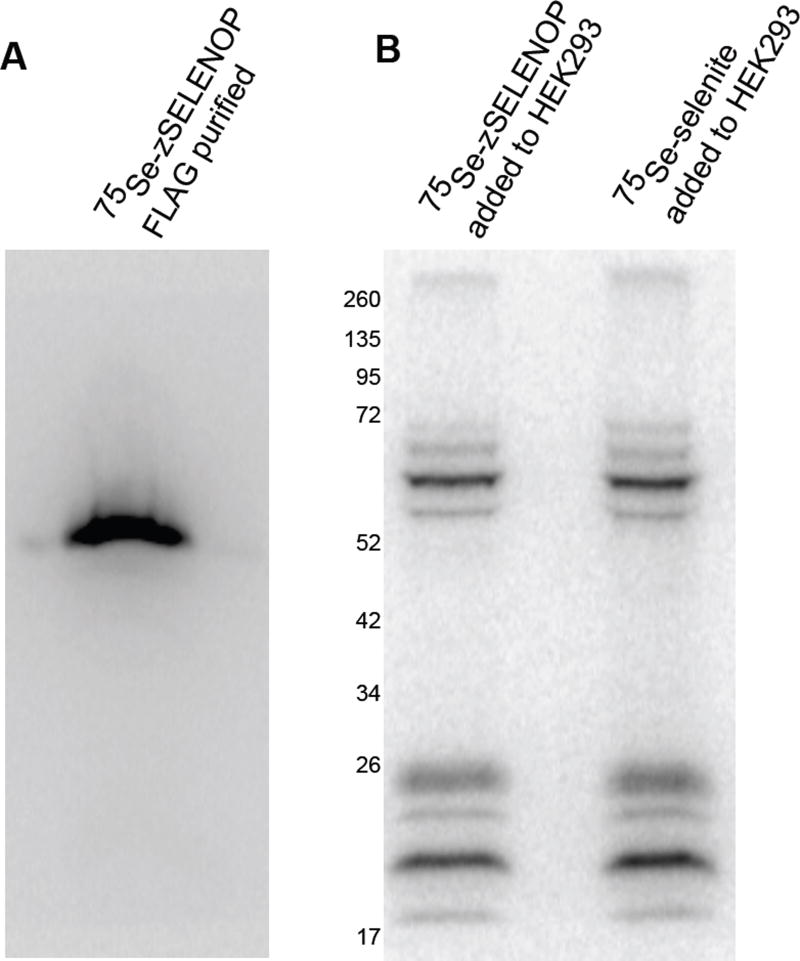
In order to examine the specificity of SELENOP uptake, we took advantage of our ability to produce zebrafish Selenop in human embryonic kidney cells (HEK-293). We stably transfected HEK-293 cells with the cDNA encoding zebrafish SELENOP harboring a C-terminal FLAG tag, labeled these cells with 75Se-selenite and purified the radiolabeled SELENOP from the conditioned medium with anti-FLAG affinity beads, which also removed the free 75Se-selenite. Figure 6A shows the purified zebrafish SELENOP recovered after purification which was free of contaminating selenoproteins and free selenium. This preparation was then added to naive HEK-293 cells and the labeling of endogenous selenoproteins was compared to that obtained with 75Se-selenite. Figure 6B clearly shows that HEK293 cells are able to utilize zebrafish SELENOP in a manner similar to 75Se-selenite. Under these conditions, therefore, the uptake and processing of SELENOP is not species-specific since these human cells are able to utilize fish SELENOP. These data are in contrast to those that previously reported that HEK-293 cells were unable to bind and take up SELENOP without over-expression of exogenous APOER2, one of the proposed SELENOP receptors[23]. It is important to note, however, that the assays utilized were fundamentally different. In the case of Kurokawa et al, they looked exclusively at binding to cells expressing APOER2 after a 3-hour incubation. This is in contrast to our study, which exclusively analyzed the transfer of selenium from SELENOP to the synthesis of endogenous selenoproteins over a 24 hour period. Thus it may be that APOER2 enhances the binding and subsequent uptake for HEK-293 cells, but our results clearly demonstrate that HEK-293 cells can utilize SELENOP in the absence of exogenously expressed APOER2. In addition, an alternative receptor mediated pathway may be taking place in these human kidney derived cell lines since in kidney it has been proposed that a different receptor, Megalin, is responsible for SELENOP uptake in that tissue[11].
Figure 6. Fish SELENOP can also be taken up and processed.
Conditioned medium derived from HEK-293 cells stably expressing C-terminally FLAG tagged zebrafish SELENOP were labeled with equal 75Se-selenite and the labeled protein purified by anti-FLAG immunoaffinity purification. The resulting purified protein was analyzed by SDS-PAGE followed by PhosphorImage analysis. (B) The purified SELENOP shown in (A) was applied to naive HEK-293 cells and lysates were analyzed by SDS-PAGE after 24 hours. The labeling of HEK 293 cells with an equal of 75Se-selenite is shown as a control as indicated.
Conclusions
Here we have described some of the characteristics of selenoprotein P and the conditions that are required for uptake and processing in mammalian cells. These studies lay the groundwork for future work deciphering the biochemical mechanism by which selenium is recovered from SELENOP for use as a precursor to cellular selenoproteins, a process that is essential for male fertility and proper neurologic function in mammals.
Acknowledgments
Thanks to Mark Pinkerton for critical reading of this manuscript. This work was supported by the National Institutes of Health (NIH R01-GM077073 and R21HD083616) and the Found Animals Foundation, Los Angeles, CA (D0910-W11).
References
- 1.Burk RF, Gregory PE. Some characteristics of 75Se-P, a selenoprotein found in rat liver and plasma, and comparison of it with selenoglutathione peroxidase. Arch Biochem Biophys. 1982;213(1):73–80. doi: 10.1016/0003-9861(82)90441-6. [DOI] [PubMed] [Google Scholar]
- 2.Motsenbocker MA, Tappel AL. A selenocysteine-containing selenium-transport protein in rat plasma. Biochim Biophys Acta. 1982;719(1):147–153. doi: 10.1016/0304-4165(82)90318-x. [DOI] [PubMed] [Google Scholar]
- 3.Motchnik PA, Tappel AL. Multiple selenocysteine content of selenoprotein P in rats. J Inorg Biochem. 1990;40(3):265–269. doi: 10.1016/0162-0134(90)80060-b. [DOI] [PubMed] [Google Scholar]
- 4.Hill KE, Lloyd RS, Yang JG, Read R, Burk RF. The cDNA for rat selenoprotein P contains 10 TGA codons in the open reading frame. J Biol Chem. 1991;266(16):10050–10053. [PubMed] [Google Scholar]
- 5.Hill KE, Zhou J, McMahan WJ, Motley AK, Atkins JF, Gesteland RF, Burk RF. Deletion of selenoprotein P alters distribution of selenium in the mouse. J Biol Chem. 2003;278(16):13640–13646. doi: 10.1074/jbc.M300755200. [DOI] [PubMed] [Google Scholar]
- 6.Schomburg L, Schweizer U, Holtmann B, Flohé L, Sendtner M, Köhrle J. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem J. 2003;370(Pt 2):397–402. doi: 10.1042/BJ20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pitts MW, Raman AV, Hashimoto AC, Todorovic C, Nichols RA, Berry MJ. Deletion of selenoprotein P results in impaired function of parvalbumin interneurons and alterations in fear learning and sensorimotor gating. Neuroscience. 2012:20858–68. doi: 10.1016/j.neuroscience.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson-Rosario S, Cowart D, Myers A, Tarrien R, Levine RL, Scott RA, Self WT. Auranofin disrupts selenium metabolism in Clostridium difficile by forming a stable Au-Se adduct. J Biol Inorg Chem. 2009;14(4):507–519. doi: 10.1007/s00775-009-0466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caito SW, Milatovic D, Hill KE, Aschner M, Burk RF, Valentine WM. Progression of neurodegeneration and morphologic changes in the brains of juvenile mice with selenoprotein P deleted. Brain Res. 2011:13981–12. doi: 10.1016/j.brainres.2011.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ursini F, Heim S, Kiess M, Maiorino M, Roveri A, Wissing J, Flohé L. Dual function of the selenoprotein PHGPx during sperm maturation. Science. 1999;285(5432):1393–1396. doi: 10.1126/science.285.5432.1393. [DOI] [PubMed] [Google Scholar]
- 11.Olson GE, Winfrey VP, Hill KE, Burk RF. Megalin mediates selenoprotein P uptake by kidney proximal tubule epithelial cells. J Biol Chem. 2008;283(11):6854–6860. doi: 10.1074/jbc.M709945200. [DOI] [PubMed] [Google Scholar]
- 12.Olson GE, Winfrey VP, Nagdas SK, Hill KE, Burk RF. Apolipoprotein E receptor-2 (ApoER2) mediates selenium uptake from selenoprotein P by the mouse testis. J Biol Chem. 2007;282(16):12290–12297. doi: 10.1074/jbc.M611403200. [DOI] [PubMed] [Google Scholar]
- 13.Burk RF, Olson GE, Hill KE, Winfrey VP, Motley AK, Kurokawa S. Maternal-fetal transfer of selenium in the mouse. FASEB J. 2013 doi: 10.1096/fj.13-231852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurokawa S, Hill KE, McDonald WH, Burk RF. Long Isoform Mouse Selenoprotein P (Sepp1) Supplies Rat Myoblast L8 Cells with Selenium via Endocytosis Mediated by Heparin Binding Properties and Apolipoprotein E Receptor-2 (ApoER2) J Biol Chem. 2012;287(34):28717–28726. doi: 10.1074/jbc.M112.383521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burk RF, Hill KE, Read R, Bellew T. Response of rat selenoprotein P to selenium administration and fate of its selenium. Am J Physiol. 1991;261(1 Pt 1):E26–E30. doi: 10.1152/ajpendo.1991.261.1.E26. [DOI] [PubMed] [Google Scholar]
- 16.Raman AV, Pitts MW, Seyedali A, Hashimoto AC, Seale LA, Bellinger FP, Berry MJ. Absence of selenoprotein P but not selenocysteine lyase results in severe neurological dysfunction. Genes Brain Behav. 2012;11(5):601–613. doi: 10.1111/j.1601-183X.2012.00794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Méplan C, Nicol F, Burtle BT, Crosley LK, Arthur JR, Mathers JC, Hesketh JE. Relative abundance of selenoprotein P isoforms in human plasma depends on genotype, se intake, and cancer status. Antioxid Redox Signal. 2009;11(11):2631–2640. doi: 10.1089/ARS.2009.2533. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki KT, Shiobara Y, Itoh M, Ohmichi M. Selective uptake of selenite by red blood cells. Analyst. 1998;123(1):63–67. doi: 10.1039/a706230c. [DOI] [PubMed] [Google Scholar]
- 19.Ganyc D, Self WT. High affinity selenium uptake in a keratinocyte model. FEBS Lett. 2008;582(2):299–304. doi: 10.1016/j.febslet.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma S, Hill KE, Burk RF, Caprioli RM. Mass spectrometric determination of selenenylsulfide linkages in rat selenoprotein P. J Mass Spectrom. 2005;40(3):400–404. doi: 10.1002/jms.801. [DOI] [PubMed] [Google Scholar]
- 21.Gromer S, Arscott LD, Williams CH, Schirmer RH, Becker K. Human placenta thioredoxin reductase. Isolation of the selenoenzyme, steady state kinetics, and inhibition by therapeutic gold compounds. J Biol Chem. 1998;273(32):20096–20101. doi: 10.1074/jbc.273.32.20096. [DOI] [PubMed] [Google Scholar]
- 22.Crane MS, Howie AF, Arthur JR, Nicol F, Crosley LK, Beckett GJ. Modulation of thioredoxin reductase-2 expression in EAhy926 cells: implications for endothelial selenoprotein hierarchy. Biochim Biophys Acta. 2009;1790(10):1191–1197. doi: 10.1016/j.bbagen.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 23.Kurokawa S, Bellinger FP, Hill KE, Burk RF, Berry MJ. Isoform-specific Binding of Selenoprotein P to the β-Propeller Domain of Apolipoprotein E Receptor 2 mediates Selenium Supply. J Biol Chem. 2014;289(13):9195–9207. doi: 10.1074/jbc.M114.549014. [DOI] [PMC free article] [PubMed] [Google Scholar]