Abstract
Meiotic recombination is essential to sexual reproduction and the generation of genetic diversity. Variation in recombination rates is presently of particular interest due to efforts being made to increase the rate of genetic gain in agricultural crops by breaking up large linkage disequilibrium blocks containing both beneficial and detrimental alleles. Here, a high-density genetic linkage map of Setaria was constructed using tunable genotyping by sequencing (tGBS) analysis of a population of recombinant inbred lines (RILs). Several regions of the Setaria genome exhibited significant levels of segregation distortion (SD), and recombination crossovers (COs) were also detected. The regions with high SD generally tended to have fewer COs, particularly for pericentromeric chromosomal areas. Recombination crossovers detected in Setaria were unevenly distributed across the genome and occurred more often in intergenic regions. Quantitative trait loci (QTLs) contributing towards the recombination frequency (Type I) and occurrence of COs in designated loci (Type II) were identified, and Type II QTLs garnered higher statistical power. The result of this study suggest that QTLs analysis of Type II traits using RILs might provide an opportunity to further understand meiotic recombination using high throughput genome sequencing and genotyping technologies.
Introduction
Recombination (or chromosome crossover) plays a vital role in DNA damage repair, chromosome segregation, and the creation of novel haplotypes1, all of which contribute towards genetic diversity, allele assortment under natural selection2, and eukaryotic genome evolution3. Chromosomal double-strand breaks (DSBs) that occur during meiosis can be classified as crossovers (COs) or non-crossovers (NCOs), and lead to genomic exchange and non-exchange with synthesis-based strand annealing, respectively4. The rate and distribution of COs across the whole genome largely determines allele combinations and haplotype structure in the progeny of a segregating cross, and exhibits notable intraspecific5 and interspecific6 variation.
The pyramiding of favorable alleles in a single line is a primary objective in the selection breeding of several crop species, but is time consuming and restricted by the frequency of genetic recombination events. In maize, about 30–50 recombination events were expected in offspring derived from two parental hybrids7,8, and chromosomal COs were concentrated in the 5′ end of transcript9. Genetic factors including Rad5110, Ku7011, Rad5012, PHS113 etc., have been found to contribute towards recombination events in plant and mammalian cells. Many quantitative trait loci (QTLs) contributing towards recombination frequency in plants have been identified using recombinant inbred lines (RILs) in recent years14,15. However, the genetic basis of the recombination events is presently still not clearly understood owing to the limited number of cloned genes and low statistical significance of detected QTLs8.
Foxtail millet (Setaria italica L.) is a globally cultivated cereal crop that was originally domesticated in China 11,500 years ago16,17. Recently, foxtail millet and its wild ancestor green foxtail (Setaria viridis L.) have been developed as novel models (named Setaria) for deciphering biological processes including C4 photosynthesis18,19, stress tolerance20, plant architecture21 and panicle development22. The benefits of using Setaria as a model species include its small genome, reproductive convenience, abundant genetic variation, and the availability of multiple reference genomes23–26. Numerous linkage maps have been constructed for foxtail millet using RFLP27,28, InDel24, SSR29–31 and SNP23,32 markers, and genomic features including segregation distortion (SD) and chromosome translocation have been identified in individual crosses. However, the low density of molecular markers used for linkage analysis in Setaria remains an obstacle for further genetic analysis of this emerging model.
In this trial, a RIL population of foxtail millet was genotyped using tunable genotyping by sequencing (tGBS) and a high-density linkage map was constructed. Genomic features including SDs and recombination event distributions were characterized and QTLs contributing towards variation in overall recombination frequency (Type I) as well as the frequency of COs at specific genomic loci (Type II) were systemically analyzed. The results of this study may provide an opportunity to identify the genomic regions involved in modulating the frequency of recombination events in plants.
Results
Construction of genetic linkage groups
A total of 4,377 SNP markers (Fig. 1A) were used for constructing the linkage map employed here, of which 1,943 displayed significant segregation distortion (<0.05). A total of 1,304 recombination events were detected, of which 495 recombination loci were involved in segregation distortion. Twelve linkage groups with a total genetic distance of 1,069 cM were finally constructed, with an average spacing of 0.24 cM and maximum spacing of 12.1 cM between adjacent markers (Fig. 1B). Comparisons between individual linkage groups and their corresponding chromosomes revealed that nearly all SNP (99.9%) markers were ordered according to the reference genome sequence of “Yugu1” with high coverage (Fig. 1C), except for two loci mapped to chromosomes 2 and 9.
Figure 1.
Characteristics and validation of the Setaria genetic map. (A) Venn diagram indicating the number of markers with significant SD (blue circle); number of marker pairs related to recombination loci (yellow circle); and total markers used for map construction (red circle). (B) Linkage groups constructed using SNP markers. (C) Comparisons between genetic linkage groups and the genome of “Yugu1”. The markers with consistent genetic and physical positions are indicated in black, while the markers with inconsistent genetic and physical positions are shown in red.
Segregation distortion identified in the RIL population
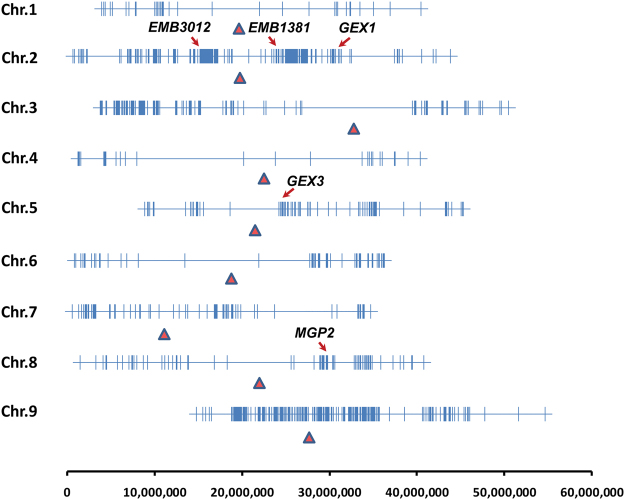
Given that the distorted segregation of SNP markers was observed in this trial (Fig. 1A), the distributions of markers exhibiting SD across all nine chromosomes were analyzed (Fig. 2). More SNP markers exhibiting significantly distorted segregation were mapped to chromosomes 2 and 9, and several genes including EMB3012, EMB1381, GEX1, GEX3, and MGP2 that control embryo development, pollen development, and pollen germination were co-localized with groups of markers with distorted segregation. These observations suggested that seed developmental defectiveness might contribute towards gamete SD in the RIL population.
Figure 2.
Distribution of SNPs exhibiting significant SD across the foxtail millet genome. Vertical lines mark the position of the SNP markers; the solid red triangles mark the centromeres; and the positions of specific genes of interest that overlap with regions of SD are marked by red arrows.
Detection of whole-genome recombination events
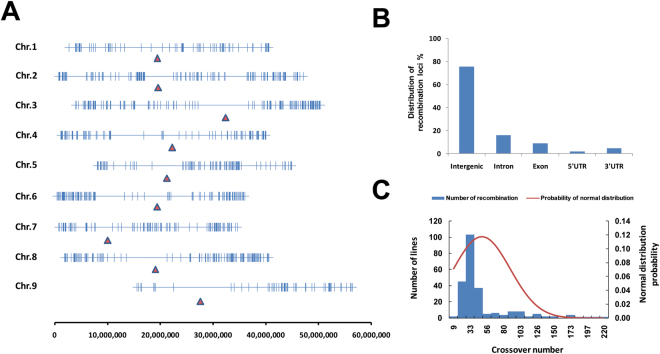
A total of 1,304 recombination events were detected in this trial (Fig. 1A). Chromosome crossover occurred most frequently in the gene rich euchromatic arms of each chromosome, while no recombination events were identified in the pericentromeric regions of each chromosome (Fig. 3A). Further CO events were detected in the intergenic regions, followed by the intron, exon, 3′UTR, and 5′UTR genomic areas (Fig. 3B).
Figure 3.
Recombination events detected in RILs. (A) Distribution of COs across foxtail millet genome. Positions of the COs are represented by vertical lines, while solid red triangles show the position of the centromeres. (B) Location of the COs relative to the annotated gene models in foxtail millet. (C) Distribution of the number of COs per RIL.
The chromosomal recombination events observed per RIL in this study revealed that the crossover number detected in each segregating line was abnormally distributed (Fig. 3C), with an average of 49 recombination events observed per RIL. In more than half of the RILs, the observed number of crossovers ranged from 20 to 44. However, across the entire population, this number varied substantially from 9 to 220.
Identification of Quantitative trait loci (QTLs) controlling overall recombination frequency (Type I) and recombination frequency at specific genomic loci (Type II)
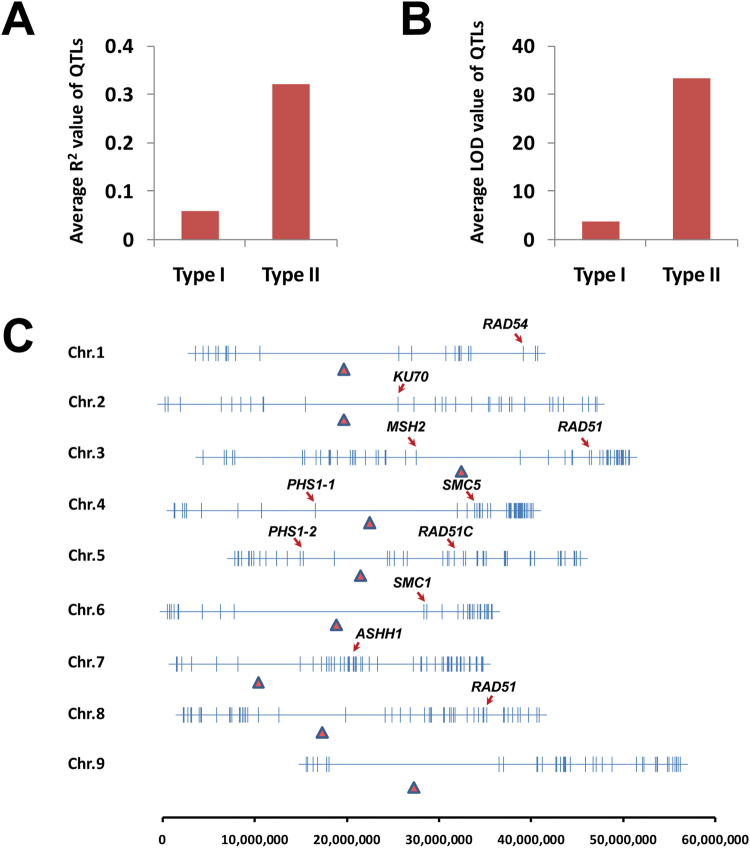
The 914 crossover loci identified in each recombined line were scored for QTL mapping analysis. A total of 617 QTLs (Table S1), including 3 cis- and 614 trans- loci, were identified to control the recombination (Type II) occurring in 341 crossover loci. Notably, in 180 of the genomic recombination loci, the crossovers in each locus were controlled by a QTL, with an average R2 value of 0.32 (ranging from 0.02 to 0.87; Fig. 4A). In terms of the recombination frequency of each line (Type I), three QTLs were identified on chromosome 8, with an average R2 value of 0.06 (Table 1; Fig. 4A and B). QTLs detected in this trial were distributed across all nine chromosomes. Several genes including SMC5, SMC1, RAD51, KU70, and ASHH1 that are known to contribute towards recombination, chromosome segregation, and DNA repair were co-localized with QTLs determined to be contributors towards recombination occurrences in target genomic loci (Fig. 4C).
Figure 4.
QTLs contributing towards recombination events in foxtail millet. (A) Average R2 value and (B) LOD value comparisons between Type I and Type II trait QTLs. (C) Distribution of QTLs detected for Type II traits across the foxtail millet genome. Peak positions of QTLs are marked by vertical lines; solid red triangles indicate the centromere positions; and the positions of particular genes of interest that overlap with QTL are marked by red arrowheads.
Table 1.
QTLs detected that contribute towards the total number of recombination events (Type I) in the RILs.
| QTL name | Chr. | Left marker (bp) | Right marker (bp) | LOD | A | R2 |
|---|---|---|---|---|---|---|
| qrf1 | 8 | 7,085,610 | 7,222,415 | 3.6 | 9.42 | 0.06 |
| qrf2 | 8 | 9,120,263 | 11,153,920 | 4.08 | 9.67 | 0.06 |
| qrf3 | 8 | 12,436,515 | 23,848,966 | 3.93 | 9.42 | 0.06 |
Discussion
In this trial, a high density genetic linkage map consisting of 4,377 SNP markers was constructed using tGBS analysis of a RIL population derived from a cross between “Yugu1”, an elite Setaria line, and “Hongmiaodabaigu”, a Setaria landrace. The density of genetic markers was significantly higher than previous reports of linkage maps constructed in Setaria23,24,27–32, suggesting that tGBS is effective for increasing marker density in linkage maps of Setaria, and is consistent with the findings in maize8,33 and other cereal crops. These types of linkage maps could provide an essential tool for dissecting genomic features in Setaria, an emerging model for deciphering the genetic basis of C4 photosynthesis18, plant and panicle architecture34 and water/nutrition usage19,20 in cereal crop species. The linkage map constructed in this trial includes markers with approximately 44% segregation distortion (Table S2), which were eliminated in skeleton maps constructed in a previous report32. This might explain why the map used in this study is denser than those reported previously.
The distorted segregation of molecular markers is common in segregating populations, and can be attributed to gametic and zygotic selection influenced by physiological or genetic factors35. In Setaria, the segregation distortion of markers has also been observed in many segregating populations23,24,29–32. In this trial, segregation distortion was observed in 44% of the SNP markers used for linkage analysis, which is consistent with results derived from maize studies using both CN-NAM and US-NAM populations characterized by GBS approaches8. The conclusions of this study suggest that segregation distortion does not affect molecular marker order determination in Setaria, which agrees with previous investigations23. Furthermore, the same conclusions have also been reported in other gramineous species, including maize36, as well as in maximum-likelihood models construction studies37. The distorted markers identified in this trial were unevenly distributed across all nine chromosomes of foxtail millet, while limited regions overlapped with known genes related to gametic and zygotic selection. These observations suggest that further research is essential to dissect the mechanisms and processes of genomic SD in Setaria. Interestingly, the markers with distorted segregation clustered in genomic regions where crossover events are rarely observed (Figs 3 and 4A), especially in the pericentromeric area, indicating that SD might affect the occurrence of recombination in foxtail millet; a finding also observed in maize8. Another possible explanation for this is that large linkage disequilibrium (LD) blocks with no recombination are more likely to harbor groups of deleterious alleles than areas of high recombination, where it is easy to select against deleterious alleles during long-term cultivation and breeding processes in Setaria. However, this conjecture requires further evaluation.
In previous work, recombination events were quantified by re-sequencing the recombined gametes sourced from meiosis, such as in humans38–40, yeast41,42, Arabidopsis43,44 and maize9. Chromosomal crossover events can be scored by counting the breakpoints between the stretches of marker alleles in segregating lines derived from the same F1 hybrid8,14,15. In this study, the second approach was applied in the dissection of the recombining features of the foxtail millet genome. We discovered that the majority of crossover events occurred in the intergenic regions, which is similar to the pattern observed in humans39,40,45. Analysis of the number of crossovers also suggested that there were 20–44 recombination events for most of the RILs in foxtail millet, which is in line with the 30–50 crossover events detected in maize lines derived from two parents via continuous selfing7. Our results suggest that genotyping a wider range of Setaria RIL populations in the future could be used to further explore the map loci responsible for the within-species variation in recombination frequency, as well as to investigate the mechanisms responsible for recombination hotspots in this species. Moreover, the genotyping of individual Setaria microgametophytes (i.e., pollen) from F1 individuals may elucidate the areas of segregation distortion that arise from pre-fertilization selection, and those that arise from selection during or after fertilization.
Recombinant inbred lines can be used to identify QTLs that contribute towards recombination events8,14. This is because over the course of single-seed descent, the genetic differences that impact recombination frequency segregate and become fixed in RILs. This differs from F2 individuals where all recombination events observed during meiosis in the F1 parent contain the same consistent set of recombination factors14. In this trial, the recombination frequency of each RIL was used as a trait (Type I) for QTLs analysis. Three QTLs were detected with relatively low statistical power, and similar results have also been observed in maize8,14,15. This might be explained by the rapid reduction in scorable recombination events during successive self-pollinating in RILs5, or parental differences in recombination frequency not being obviously reflected in the number of recombination events occurring in the RILs. Additionally, QTLs that control the occurrence of crossover events at specific loci (Type II) across the population were also tested in this trial. Surprisingly, a few QTLs that contribute to crossover events in many genome loci were identified with high statistical power, and some of the QTL hotspots may overlap with known genes related to meiosis in higher plants (Fig. 4C). This result suggests that the differences in recombination frequency between parental lines might be reflected in the occurrences of crossover events in a few diverse genomic regions in Setaria.
According to studies conducted in Arabidopsis, variations in the distribution of crossover events are controlled by both cis and trans genetic factors1. In this trial, most of the identified Type II QTLs were trans-acting factors (Fig. S1), which is consistent with reported results in maize8. It is possible that trans-acting proteins might play more exclusive roles in controlling recombination events in panicoid grasses. Given the high statistical power of Type II QTLs that control the occurrence of designated genomic loci, it can be inferred that the cloning or pyramiding of Type II QTLs controlling the recombination of target loci would aid precision molecular designing of crop cultivars in the future by increasing the recombination rates in regions recalcitrant to recombination, enabling an increase in the rate of genetic gain by breaking up positive and deleterious alleles that are currently tightly linked.
Materials and Methods
Recombinant inbred lines (RILs) population construction
The foxtail millet cultivar “Yugu1” and the landrace “Hongmiaodabaigu” (collected from Shanxi province of China) were used as parental lines. Hybridization was conducted according to the protocol described previously27. Cultivar “Yugu1” was pollinated using pollen grains collected from “Hongmiaodabaigu” and F1 hybrids were screened in the field based on morphological markers from the male parent “Hongmiaodabaigu”, and later verified using SSRs. A single validated F1 individual was bagged to produce an F2 population consisting of 288 plants. Each F2 plant was self-pollinated, and progenies of each F2 plant were advanced independently by single seed descent to generate a single RIL. The samples used in this study were from the lines advanced to the F8 generation.
Genotyping of RILs
Fresh leaf tissue was collected from each RIL in the field during the foxtail millet growth period in Beijing in 2014. High quality DNA was extracted using the CTAB method46, and the samples were genotyped using tGBS47. Version 2.2 of the genome sequence of “Yugu1” as downloaded from Phytozome 1223 was used as a reference for the alignment of reads and SNP discovery.
Construction of genetic linkage maps
A total of 4,680 Low Missing Data (LMD; missing data rate <30%) filtered among the 236 RILs (individuals that were heterozygous for >5% of SNP sites were removed) were used for constructing the genetic maps. The grouping, ordering, and genetic distance between the linked markers were inferred using the R/qtl software package48, using the following parameters: rf = 0.25, lod = 16, and minimum SNPs of each LinkageGroup (LG) = 50. All SNPs genotyped as heterozygous were treated as missing values in subsequent analyses.
Segregation distortion analysis
The proportion of “Yugu1” or “Hongmiaodabaigu” alleles at each locus were tested for SD against the 1:1 segregation ratio expected of a RIL population using a chi-square test. SNP markers exhibiting SD (p < 0.05) were scored as SD markers.
Detection of recombination events
Recombination events were scored by counting the breakpoints between stretches of marker alleles from one parent to another in the RIL mapping data according to the marker positions ordered by linkage analysis. The total number of recombination events summarized in each RIL was used as a quantitative trait. The location of COs was considered to be the mid-point between the physical positions of the two flanking SNP markers. Genome annotations of all COs were retrieved from Phytozome 12 (https://phytozome.jgi.doe.gov/jbrowse/index.html?data=genomes%2FSitalica/).
QTLs identification
The genetic map, consisting of 4,377 SNP markers, was used for QTLs analysis using composite interval mapping (CIM) and the standard model (Model 6) as implemented by Windows QTL Cartographer V. 2.549, with a walk speed of 1.0 cM and window size of 10.0 cM. An LOD score of 3.0 was used as the significance threshold. The number of recombination events observed in each RIL was used as the phenotype for mapping Type I QTL. For type II QTL, QTLs controlling recombination at each CO locus were mapped separately. RILs with a CO at a given location were scored as “2” and RILs without a crossover at the location were scored as “1”. Only COs that occurred in more than two different lines were selected for Type II QTL mapping analysis.
Data availability
All QTL data are available in the manuscript.
Electronic supplementary material
Acknowledgements
We thank Dr. James C. Schnable from the Department of Agronomy & Horticulture, University of Nebraska Lincoln, USA, for discussion and modification of the manuscript. This work was supported by the National Natural Science Foundation of China (31522040 and 31301328), Fundamental Research Funds of CAAS (Grant to Guanqing Jia, 1610092016116, Y2017JC15), China Agricultural Research System (CARS07-13.5-A02), and the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences.
Author Contributions
X.D. and G.J. designed the experiments. G.J., H.W., S.T., S.L., Q.W., and Z.Q. performed the experiments. H.Z. and X.D. provided materials. G.J. and H.W. analyzed the data. G.J. wrote the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Guanqing Jia and Haigang Wang contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-15576-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Li J, Hsia A, Schnable PS. Recent advances in plant recombination. Current Opinion in Plant Biology. 2007;10:131–135. doi: 10.1016/j.pbi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Gaut BS, Wright SI, Rizzon C, Dworak J, Anderson LK. Recombination: an underappreciated factor in the evolution of plant genomes. Nature Review Genetics. 2007;8:77–84. doi: 10.1038/nrg1970. [DOI] [PubMed] [Google Scholar]
- 3.Coop G, Przeworski M. An evolutionary view of human recombination. Nature Review Genetics. 2007;8:23–34. doi: 10.1038/nrg1947. [DOI] [PubMed] [Google Scholar]
- 4.Phadnis N, Hyppa RW, Smith GR. New and old ways to control meiotic recombination. Trends of Genetics. 2011;27:411–421. doi: 10.1016/j.tig.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer E, et al. Intraspecific variation of recombination rate in maize. Genome Biology. 2013;14:R103. doi: 10.1186/gb-2013-14-9-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smukowski CS, Noor MA. Recombination rate variation in closely related species. Heredity. 2011;107:496–508. doi: 10.1038/hdy.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai J, et al. Genome-wide patterns of genetic variation among elite maize inbred lines. Nature Genetics. 2010;42:1027–1030. doi: 10.1038/ng.684. [DOI] [PubMed] [Google Scholar]
- 8.Li C, et al. Analysis of recombination QTLs, segregation distortion, and epistasis for fitness in maize multiple populations using ultra-high-density markers. Theoretical and Applied Genetics. 2016;129:1775–1784. doi: 10.1007/s00122-016-2739-6. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Li L, Yan J. Dissecting meiotic recombination based on tetrad analysis by single-microspore sequencing in maize. Nature Communications. 2015;6:6648. doi: 10.1038/ncomms7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen C, Kurimasa A, Brennemann MA, Chen DJ, Nickoloff JA. DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3758–3763. doi: 10.1073/pnas.052545899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopfner KP, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 13.Pawlowski WP, et al. Coordination of meiotic recombination, pairing, and synapsis by PHS1. Science. 2004;303:89–92. doi: 10.1126/science.1091110. [DOI] [PubMed] [Google Scholar]
- 14.Esch E, Szymaniak JM, Yates H, Pawlowski WP, Buckler ES. Using crossover breakpoints in recombinant inbred lines to identify quantitative trait loci controlling the global recombination frequency. Genetics. 2007;177:1851–1858. doi: 10.1534/genetics.107.080622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, et al. Genetic analysis of QTL affecting recombination frequency in whole genome of maize and rice. Sci Agric Sin. 2009;42:2262–2270. [Google Scholar]
- 16.Lu H, et al. Earliest domestication of common millet (Panicum miliaceum) in east Asia extended to 10,000 years ago. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7367–7372. doi: 10.1073/pnas.0900158106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, et al. Early millet use in northern china. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3726–3730. doi: 10.1073/pnas.1115430109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brutnell TP, et al. Setaria viridis: a model for C4 photosynthesis. Plant Cell. 2010;22:2537–2544. doi: 10.1105/tpc.110.075309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diao X, Schnable J, Bennetzen JL, Li J. Initiation of Setaria as a model plant. Frontiers of Agricultural Science and Engineering. 2014;1:16–20. doi: 10.15302/J-FASE-2014011. [DOI] [Google Scholar]
- 20.Muthamilarasan M, Prasad M. Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses. Theoretical and Applied Genetics. 2015;128:1–14. doi: 10.1007/s00122-014-2399-3. [DOI] [PubMed] [Google Scholar]
- 21.Doust AN, Devos KM, Gadberry M, Gale MD, Kellogg EA. Genetic control of branching in the foxtail millet. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9045–9050. doi: 10.1073/pnas.0402892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doust AN, Devos KM, Gadberry M, Gale MD, Kellogg EA. The genetic basis for inflorescence variation between foxtail and green millet (Poaceae) Genetics. 2005;169:1659–1672. doi: 10.1534/genetics.104.035543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennetzen JL, et al. Reference genome sequence of the model plant Setaria. Nature Biotechnology. 2012;30:555–561. doi: 10.1038/nbt.2196. [DOI] [PubMed] [Google Scholar]
- 24.Zhang G, et al. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nature Biotechnology. 2012;30:549–554. doi: 10.1038/nbt.2195. [DOI] [PubMed] [Google Scholar]
- 25.Jia G, et al. A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica) Nature Genetics. 2013;45:957–961. doi: 10.1038/ng.2673. [DOI] [PubMed] [Google Scholar]
- 26.Tsai KJ, et al. Assembling the Setaria italica L. Beauv. genome into nine chromosomes and insights into regions affecting growth and drought tolerance. Scientific Reports. 2016;6:35076. doi: 10.1038/srep35076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Devos KM, Liu C, Wang R, Gale MD. Construction of RFLP-based maps of foxtail millet. Setaria italica. Theoretical and Applied Genetics. 1998;96:31–36. doi: 10.1007/s001220050705. [DOI] [Google Scholar]
- 28.Devos KM, Wang ZM, Beales J, Sasaki Y, Gale MD. Comparative genetic maps of foxtail millet (Setaria italica) and rice (Oryza sativa) Theoretical and Applied Genetics. 1998;96:63–68. doi: 10.1007/s001220050709. [DOI] [Google Scholar]
- 29.Jia X, et al. Development and genetic mapping of SSR markers in foxtail millet [Setaria italica (L.) P. Beauv.] Theoretical and Applied Genetics. 2009;118:821–829. doi: 10.1007/s00122-008-0942-9. [DOI] [PubMed] [Google Scholar]
- 30.Qie L, et al. Mapping of quantitative trait locus (QTLs) that contribute to germination and early seedling drought tolerance in the interspecific cross Setaria italica×Setaria viridis. PLoS One. 2014;9:e101868. doi: 10.1371/journal.pone.0101868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang X, et al. A high density genetic map and QTL for agronomic and yield traits in Foxtail millet [Setaria italica (L.) P. Beauv] BMC Genomics. 2016;17:336. doi: 10.1186/s12864-016-2628-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang K, et al. Identification of QTLs for 14 agronomically important traits in Setaria italica based on SNPs generated from high-throughput sequencing. G3 (Bethesda, Md.). 2017;7:1587–1594. doi: 10.1534/g3.117.041517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elshire RJ, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One. 2011;6:e19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doust AN, Kellogg EA, Devos KM, Bennetzen JL. Foxtail millet: A sequence-driven grass model system. Plant Physiol. 2009;149:137–141. doi: 10.1104/pp.108.129627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu JJ, Qu LJ. Meiotic and mitotic cell cycle mutants involved in gametophyte development in Arabidopsis. Molecular Plant. 2008;1:564–574. doi: 10.1093/mp/ssn033. [DOI] [PubMed] [Google Scholar]
- 36.Lu H, Romero-Severson J, Bernardo R. Chromosomal regions associated with segregation distortion in maize. Theoretical and Applied Genetics. 2002;105:622–628. doi: 10.1007/s00122-002-0970-9. [DOI] [PubMed] [Google Scholar]
- 37.Lorieux M, Perrier X, Goffinet B, Lanaud C, González LD. Maximum likelihood models for mapping genetic markers showing segregation distortion. 2. F2 populations. Theoretical and Applied Genetics. 1995;90:81–89. doi: 10.1007/BF00220999. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150:402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu S, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole genome sequencing. Science. 2012;338:1627–1630. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou Y, et al. Genome analyses of single human oocytes. Cell. 2013;155:1492–1506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 41.Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ludlow CL, et al. High-throughput tetrad analysis. Nature Methods. 2013;10:671–675. doi: 10.1038/nmeth.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wijnker E, et al. The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. eLife. 2013;2:e01426. doi: 10.7554/eLife.01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu P, et al. Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by resequencing Landsberg erecta and all four products of a single meiosis. Genome Research. 2012;22:508–518. doi: 10.1101/gr.127522.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–1398. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- 46.Doyle, J. J. DNA protocol for plants CTAB total DNA isolation in Molecular techniques in taxonomy (ed. Hewitt, G. M.) 283–293 (Springer, 1991).
- 47.Ott, A. et al. Tunable genotyping-by-sequencing (tGBS®) enables reliable genotyping of heterozygous loci. bioRxiv. 100461; 10.1101/100461 (2017). [DOI] [PMC free article] [PubMed]
- 48.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 49.Wang, S., Basten, C. J. & Zeng, Z. B. Windows QTL cartographer 2.5. Department of Statistics, Raleigh, NC (2012).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All QTL data are available in the manuscript.