Abstract
Hydroxyapatite (HA) is the principal inorganic component of bones and teeth and has been widely used as a bone repair material because of its good biocompatibility and bioactivity. Understanding the interactions between proteins and HA is crucial for designing biomaterials for bone regeneration. In this study, we evaluated the effects of atomic-level nano-structured HA (110) surfaces on the adsorption of bone morphogenetic protein-7 (BMP-7) and its derived peptide (KQLNALSVLYFDD) using molecular dynamics and density functional theory methods. The results indicated that the atomic-level morphology of HA significantly affected the interaction strength between proteins and HA substrates. The interactions of BMP-7 and its derived peptide with nano-concave and nano-pillar HA surfaces were stronger than those with flat or nano-groove HA surfaces. The results also revealed that if the groove size of nano-structured HA surfaces matched that of residues in the protein or peptide, these residues were likely to spread into the grooves of the nano-groove, nano-concave, and nano-pillar HA, further strengthening the interactions. These results are helpful in better understanding the adsorption behaviors of proteins onto nano-structured HA surfaces, and provide theoretical guidance for designing novel bioceramic materials for bone regeneration and tissue engineering.
Introduction
Hydroxyapatite [HA, Ca10(PO4)6(OH)2] is the major inorganic component of bones and teeth. As a bone implant material, HA has received increasing attention because of its exceptional interface or surface properties and excellent biocompatibility1. It is well-known that, in vivo, the HA surface is rapidly covered by proteins and other biomolecules, leading to the regulation of precipitated calcium phosphate crystal growth or alterations to crystal morphology2. Therefore, it is essential to study the surface properties of HA and interaction behaviors between HA and various proteins. Recently, numerous experimental and theoretical studies have been conducted to study the interaction mechanisms between HA and proteins, such as the bone morphogenetic protein-7 (BMP-7)3, BMP-24, amelogenin5, osteopontin6, fibronectin7, bovine serum albumin (BSA)8, and sialoprotein9, among others. These studies suggest that the surface topology and composition, the pH values of protein solutions and mechanical properties of the biomaterials affect the interactions to some extent10,11. To achieve excellent biocompatibility and osteoinductivity, micro-nano structured HA surfaces were established.
Micro-structured HA generally resulted in cell morphology change and consequently altered cell function changes12–15. Henry et al.16 demonstrated that mouse pre-osteoblast cells (MC3T3-E1) were better arranged along micro-grooves with a groove width of 5 μm for the poly(ε-caprolactone) triacrylate/ HA substrates. Wang et al.17 also showed that the groove width of micro-patterned HA has a prominent effect on the orientated growth of osteoblast cells. Lu et al.18 found that HA micro-grooves with a width of 8 μm significantly affected contact guidance of osteoblasts and myoblasts, whereas those of 24 μm affected myoblasts only. Iwamoto et al.19 obtained a micrometer scale flat HA and nanometer scale convexo-concave HA and found that MC3T3-E1 and mouse fibroblast cells (L929) adhered to the flat HA surface but not to the convexo-concave HA surface.
Nanoscale HA also plays a significant role in regulating cell functions and adsorbing proteins. Wu et al.20 reported that the nano-HA porous structure is appropriate for blood vessels and fibrous connective tissue to enter, providing mesenchymal cells for implementing osteoinductivity of rhBMP-2 in the material. Jouve et al.21 indicated that the porous structure of nano-HA and “nanometer effect” of the nanomaterial significantly enhance HA surface area, provide good support for revascularization of the material, and formation of the bone matrix. Kasaj et al.22 demonstrated that nano-HA enhanced adhesion and proliferation in human periodontal ligament cells. Webster23 found that vitronectin adsorption has more obvious enhancement on nano-HA materials than that on micro-HA materials. Zhu et al.24 reported that the nano-HA/rhBMP-2 composite is more likely to stimulate new bone formation and had more excellent repairing ability for bone defect compared to the nano-HA.
BMP-7 is a member of the transforming growth factor-beta superfamily and comprises 112 residues. BMP-7 plays a crucial role in the induction and regulation of cartilage and bone formation and has an important influence on skeletal development and growth25. This protein has been widely examined in previous studies. Luo et al.26 reported that a lack of BMP-7 can result in murine skeletal defects. Schwarting et al.27 investigated the effects of BMP-7 on tendon bone repair and indicated that BMP-7 is extremely important for tendinous fibroblast transformation and differentiation at the tendon-bone interface. Cheng et al.28 suggested that BMP-7 promotes osteogenesis in mature osteoblasts. Tasnim et al.29 found that BMP-7 or BMP-2 improves the functional performance of primary human renal proximal tubule cells, particularly in the case of BMP-7. Zhou et al.3 used molecular dynamics (MD) and steered molecular dynamics (SMD) methods to investigate the adsorption mechanism of BMP-7 on HA (001) surfaces, and found that –COO– and -NH2 play vital roles in the adsorption of BMP-7 onto the HA surfaces by forming Ca-O connections and hydrogen bonds (H-bonds). Dong et al.30 investigated the dynamic behaviors of BMP-7 on HA (001) and HA (010) surfaces and found that both the type and number of differences of the adsorbed residues and interaction strengths are relevant to the specific texture of the HA surfaces.
BMP-7 not only has strong osteoinductivity, but also affects the growth and physiological function of the reproductive system. However, the structure of BMP-7 is complicated, it degrades easily in the body, and requires considerable time to be produced on a large scale. Thus, BMP-7-derived peptides have attracted attention because of their low-cost and excellent bio-functions31. Studies have shown that these peptides from the BMP-7 or BMP-7 receptors can be used to treat bone-related diseases. Kim et al.32 found that a novel bone forming peptide-1 (GQGFSYPYKAVFSTQ) from the BMP-7 has greater activity in osteogenic differentiation than that of whole BMP-7. Furthermore, the authors suggested that the BMP-7 peptide can also serve as a valuable adjuvant in medical therapies for bone-related diseases. Tao et al.33 prepared a new type of functional, self-assembling peptide nanofiber-hydrogel scaffold with the sequence RADKPS (ARG-ALA-ASP-LYS-PRO-SER); they clearly showed that RADKPS promoted cell proliferation and activity of human degenerative nucleus pulposus cells. Another peptide, KQLNALSVLYFDD (LYS-GLN-LEU-ASN-ALA-LEU-SER-VAL-LEU-TYR-PHE-ASP-ASP), is synthesized from the BMP-7 receptor I- and receptor II-binding active domains31 and, in addition, has excellent osteogenic activity and induces mineralization of bone marrow stromal stem cells. The structures of BMP-7-derived peptides are simple and easy to integrate into biomaterials while maintaining their biological activities during synthesis. Furthermore, these peptide molecules are small and the active sites can be easily exposed.
Although numerous studies have been devoted to the interactions between proteins and biological nanomaterials HA, it is nevertheless difficult to quantitatively comprehend the detailed interaction mechanisms involving nanomaterials and proteins by conventional experimental methodologies. Computer simulation is a favorable method to investigate such interaction mechanisms at the atomic and molecular levels. Therefore, here, we have utilized MD and density functional theory (DFT) simulation methods to investigate interactions between nanometer-scale HA and proteins/peptides, and focused on the effect of nano-morphology on protein/peptide adsorption. The HA (110) crystallographic surface was used to study the interaction with BMP-7 because it not only has high adsorption strength for molecules34, but also is one of the dominant surfaces observed in numerous experimental and theoretical studies35–37. The adsorption behaviors of BMP-7-derived peptide (KQLNALSVLYFDD) on these nano-structured HA (110) surfaces were also evaluated because of its excellent osteogenic activity. Our results are expected to advance understanding of the bioactivity mechanism of calcium phosphate biomaterials and offer theoretical guidance for HA applications in materials design and tissue engineering.
Simulation Methods
Building Models
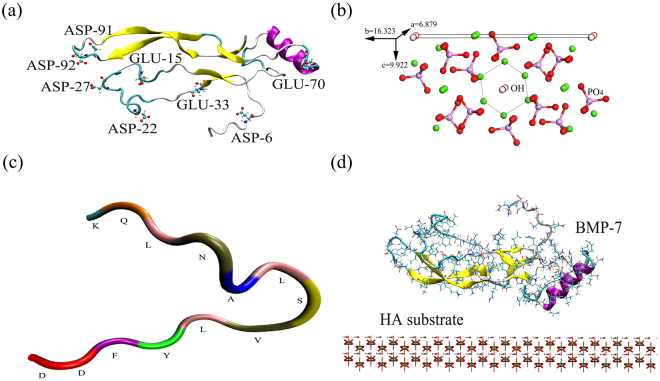
The primary conformation of BMP-7 was obtained from the RCSB protein data bank (http://www.rcsb.org/pdb/, ID: 1M4U); the Newcartoon structure of BMP-7 is illustrated in Fig. 1a. BMP-7-derived peptide (KQLNALSVLYFDD) containing 13 amino acids was built according to the literature (Fig. 1c)31. BMP-7 and its derived peptide molecules were immersed in TIP3P water boxes and charges balanced with sodium and chloride ions. In order to obtain optimized geometries of BMP-7 and its derived peptide, a 20 ps energy minimization and a 1500 ps MD relaxation in the water box were performed with a time step of 0.5 fs in the NPT ensemble using NAMD package38. A constant temperature of 310.6 K and constant pressure of 101.3 kPa were used in the simulations, employing the modified Nose-Hoover method, in which Langevin dynamics was used to control fluctuations in the barostat.
Figure 1.
Newcartoon structures of (a) BMP-7 and (c) BMP-7-derived peptide (KQLNALSVLYFDD), (b) three-dimensional structure of HA (110) cell and (d) BMP-7 interacting with the flat HA surface before MD simulation.
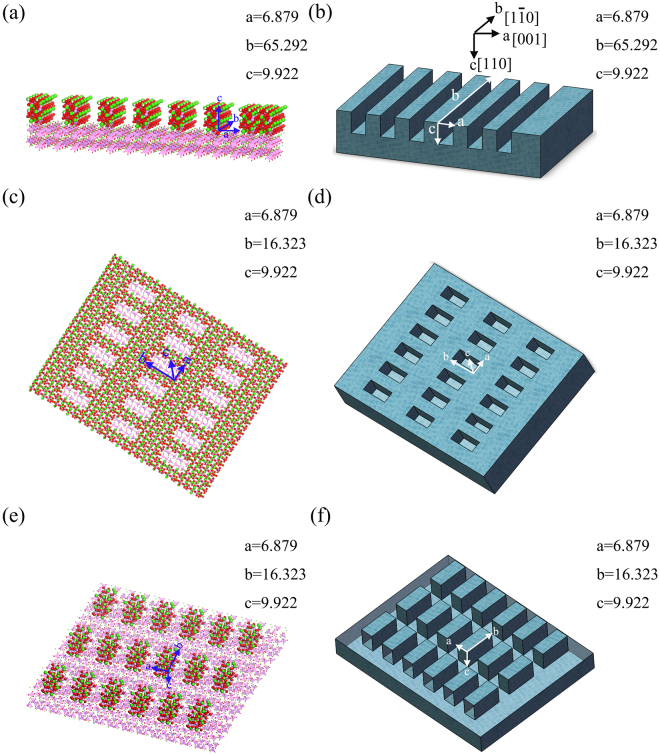
The initial model of HA (P63/m) was acquired from the American Mineralogist Crystal Structure database39. The unit cell parameters of the HA crystal are a = b = 9.424 Å and c = 6.879 Å. The processes used to build nano-structured HA (110) surfaces were as follows. First, the flat HA (110) cell was built with dimensions of a = 6.879 Å, b = 16.323 Å, and c = 9.922 Å (Fig. 1b), and then a flat HA (110) surface was constructed with a size of 96.288 × 65.292× 19.844 Å3. Finally, nine types of nano-structured HA (110) surfaces (three nano-grooves, three nano-concaves, and three nano-pillars) were prepared based on the flat HA (110) surface (Fig. 2). The nano-pillar HA (110) surfaces were similar to the nano-concave HA (110) surfaces except that the concaves became the pillars in the HA surfaces. The sizes and numbers of nano-grooves, nano-concaves, and nano-pillars of HA (110) are illustrated in Table 1. Note that only the HA (110) surface was present in this study, and hence the HA (110) surface was simplified as the HA surface. HA (110) surfaces with different terminations were also constructed, as shown in Supplementary Information. BMP-7 was laid on the flat HA surface (Fig. 1d), which illustrated the interaction between BMP-7 and the flat HA surface before MD simulation. The initial orientation of BMP-7 was chosen according to results from the literature3. The sizes of the interaction systems are shown in Table 1.
Figure 2.
Structures of (a) and (b) groove1a-HA surfaces, (c) and (d) concave1a1b-HA surfaces, (e) and (f) pillar1a1b-HA surfaces. Note that the units of a, b, and c are Å.
Table 1.
Sizes of atomic-level nano-structured HA (110) and its interaction with BMP-7.
| Models | Number of groove/concave/pillar | Volume of groove/concave/pillar (Å3) | increment of surface areas (Å2) |
|---|---|---|---|
| Groove1a-HA | 6 | 6.879 × 65.292 × 9.922 | 6954.88 |
| Groove2a-HA | 4 | 13.758 × 65.292 × 9.922 | 4090.55 |
| Groove3a-HA | 3 | 20.637 × 65.292 × 9.922 | 2658.40 |
| Concave1a1b-HA | 18 | 6.879 × 16.323 × 9.922 | 8287.57 |
| Concave2a1b-HA | 12 | 13.758 × 16.323 × 9.922 | 7163.13 |
| Concave2a2b-HA | 8 | 13.758 × 32.646 × 9.922 | 7363.08 |
| Pillar1a1b-HA | 18 | 6.879 × 16.323 × 9.922 | 8287.57 |
| Pillar2a1b-HA | 12 | 13.758 × 16.323 × 9.922 | 7163.13 |
| Pillar2a2b-HA | 8 | 13.758 × 32.646 × 9.922 | 7363.08 |
| Flat-HA-BMP-7 | 96.288 × 70.706 × 71.674 | ||
| Groove1a-HA-BMP-7 | 96.951 × 98.742 × 80.817 | ||
| Groove2a-HA-BMP-7 | 96.951 × 98.742 × 80.817 | ||
| Groove3a-HA-BMP-7 | 96.951 × 98.742 × 80.817 | ||
| Concave1a1b-HA-BMP-7 | 96.306 × 98.742 × 80.914 | ||
| Concave2a1b-HA-BMP-7 | 96.306 × 98.742 × 80.659 | ||
| Concave2a2b-HA-BMP-7 | 96.306 × 98.742 × 80.914 | ||
| Pillar1a1b-HA-BMP-7 | 96.306 × 98.742 × 80.914 | ||
| Pillar2a1b-HA-BMP-7 | 96.306 × 98.742 × 80.914 | ||
| Pillar2a2b-HA-BMP-7 | 96.306 × 98.742 × 80.914 |
Methods
All simulations were conducted with NAMD using CHARMM27 force field40 with the HA parameters added. Furthermore, three different nano-topographies of HA in our models used the same force field with the flat HA, because these nano-structured HA removed the geometric multiple of HA double cell atoms (Fig. 1b) and did not result in HA atoms/ions with different atomistic environments. Periodic boundary conditions were applied to all models. Particle-mesh Ewald summation was utilized to calculate the long-range electrostatic interactions with a cutoff distance of 14 Å to separate the direct and reciprocal space. Van der Waals interactions were cut off at 12 Å. The force field parameters for HA were fitted based on the results of Bhowmik, Hauptmann and Zhou3,41,42. Additionally, in order to maintain the stability of the systems, the combinational arrangements of protein and HA were firstly underwent 3000 ps MD simulations with a time step of 1 fs in the NPT ensemble. Then, 27000 ps MD simulations were conducted with a time step of 2 fs in the NVT ensemble. For simulation of the adsorption behavior of protein or peptide, tens of thousands of picoseconds simulations are considered suitable43–45.
The molecular graphics package VMD was employed to analyze the static and dynamic structure information after MD simulation. Snapshots of interaction systems, interaction energies, and interaction sites, among others, were acquired from VMD46.
The interaction energies were calculated to evaluate the interaction strength between proteins and the flat, nano-groove, nano-concave, and nano-pillar HA surfaces. The interaction energy () was calculated according to the equation (1)
| 1 |
where ,, and represent the total interaction energy of the HA and protein, the energy of HA, and the energy of protein, respectively. Note that the more negative interaction energy indicates a greater interaction strength between the protein and HA47. The average interaction energies of the simulation systems over the final 100 ps are displayed in Table 2.
Table 2.
Electrostatic and interaction energies between BMP-7 and HA surfaces.
| Systems | ELEC Kcal·mol−1 | Interaction energy Kcal·mol−1 | Main absorbed residues after the MD simulation |
|---|---|---|---|
| Flat-HA-BMP-7 | −1883.96 | −1873.59 | ASP27 GLU70 |
| Groove1a-HA-BMP-7 | −2053.36 | −2029.57 | ASP27 ASP91 GLU70 |
| Groove2a-HA-BMP-7 | −2210.45 | −2166.63 | ASP27 ASP91 GLU70 |
| Groove3a-HA-BMP-7 | −1580.31 | −1560.77 | ASP91 GLU33 VAL60 |
| Concave1a1b-HA-BMP-7 | −3171.04 | −3177.8 | ASP27 ASP92 LIE30 GLU70 |
| Concave2a1b-HA-BMP-7 | −4720.71 | −4660.49 | ASP6 ASP27 ASP92 GLU1 GLU33 |
| Concave2a2b-HA-BMP-7 | −3770.16 | −3738.27 | ASP6 ASP27 GLU33 |
| Pillar1a1b-HA-BMP-7 | −2965.53 | −2924.22 | ASP6 ASP22 ASP27 GLU33 GLU70 |
| Pillar2a1b-HA-BMP-7 | −4157.33 | −4098.97 | ASP6 ASP22 GLU15 GLU33 GLU70 |
| Pillar2a2b-HA-BMP-7 | −3511.39 | −3442.33 | ASP6 ASP27 GLU15 GLU70 SER50 TYR51 |
| Flat-HA-BMP-7-derived-peptide | −236.46 | −241.04 | GLN2 ASN4 |
| Groove2a-HA-BMP-7-derived-peptide | −411.07 | −408.40 | LYS1 GLN2 ASN4 |
| Concave2a1b-HA-BMP-7-derived-peptide | −1213.18 | −1200.36 | LYS1 ASN4 ASP12 |
| Pillar2a1b-HA-BMP-7-derived-peptide | −918.35 | −904.65 | ASP12 ASP13 GLN2 LYS1 |
| Flat-HA-BMP-7-no water | −7407.30 | −7349.66 | Many residues |
| Flat-HA-BMP-7-derived-peptide-no-water | −2861.26 | −2825.55 | Many residues |
From the results of MD simulations, DFT based on the quantum mechanics of the Flat-HA-BMP-7/Flat-HA-BMP-7-derived peptide models was used to investigate atomic charges between the main adsorption residues of proteins and Ca atoms of the flat HA surfaces using the Dmol3 program in Materials Studio (Accelrys, San Diego, CA, USA)48. During simulation, the DNP double numerical basis set was used, which is comparable to the 6–31 G** basis set. A DFT semi-core pseudo potential with double-numerical basis set plus d functions was utilized49. The exchange-correlation energy was determined using the Perdew–Burke–Ernzerhof generalized gradient approximation50. Integrations over reciprocal space were based on the Monkhorst-Pack scheme with a 2 × 2 × 1 k-point mesh. Fermi smearing was 0.005 Ha (1 Ha = 27.211 eV). All atoms in our systems were relaxed during simulation. A vacuum thickness of 25 Å was chosen along the c-direction to avoid interactions among the adjacent cells. Convergence in self-consistent field tolerance, energy tolerance, a maximum force tolerance, and a maximum displacement tolerance were optimized at 1.0 × 10–6 Ha/atom, 1.0 × 10–5 Ha/atom, 0.002 Ha∕ Å, and 0.005 Å, respectively.
Results and Discussions
Adsorption behaviors between BMP-7 and atomic-level nano-structured HA surfaces
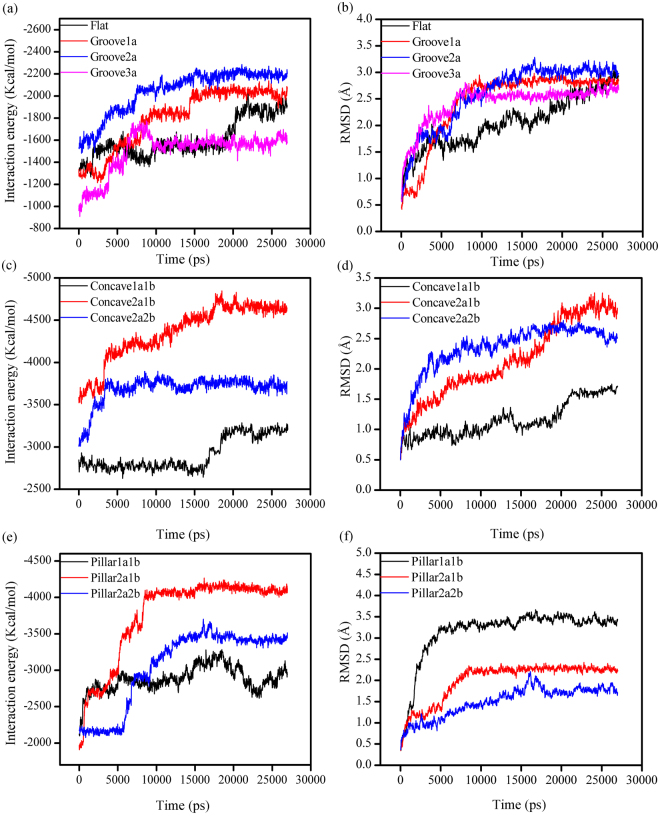
The interaction energy and the root mean square deviation (RMSD) are usually adopted to evaluate whether the system has achieved stability or not45,51. The graph of interaction energy and RMSD versus simulation time is shown in Fig. 3. The results indicated that protein adsorption on various HA surfaces fluctuated mildly in the end of the simulation and had eventually achieved equilibrium.
Figure 3.
The interaction energy and RMSD between the BMP-7 and HA surfaces over the last 27000 ps simulation time: flat and groove HA surfaces, (a) and (b); concave HA surfaces, (c) and (d); pillar HA surfaces, (e) and (f).
The adsorptions of BMP-7 on all nano-structured HA surfaces were thermodynamically favorable because the interaction energies were all negative, as shown in Table 2 and Fig. 4. The interaction energy was -1873.59 Kcal·mol-1 between BMP-7 and the flat HA surface. When BMP-7 interacted with nano-groove HA surfaces, the interaction energies indicated that adsorptions initially became strong and then became weak as the grooves of the HA surfaces gradually widened. Moreover, the interaction energy on the groove2a HA surface was 17.3% more negative than that on the flat HA surface. When BMP-7 was adsorbed on the nano-concave and nano-pillar HA surfaces, the same results were observed. However, the interaction energies suggested that there were stronger interactions between BMP-7 and nano-concave and nano-pillar HA surfaces than those between BMP-7 and flat or nano-groove HA surfaces. In particular, the interaction energies of BMP-7 on the concave2a1b and Pillar2a1b HA surfaces were nearly twice that of BMP-7 on the flat HA surface. These results demonstrated that the atomic-level nano-structured HA surfaces noticeably affected BMP-7 adsorption. Furthermore, nano-concave and nano-pillar HA surfaces greatly enhanced BMP-7 adsorption.
Figure 4.
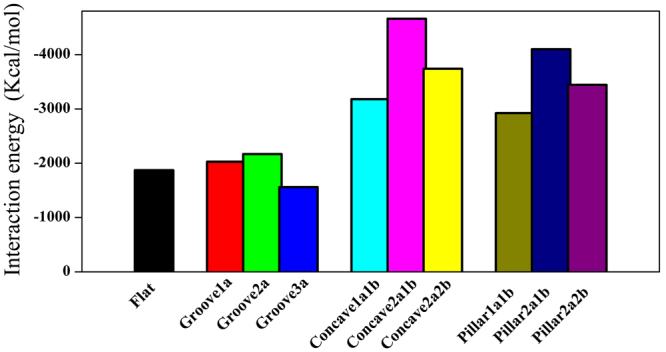
Average interaction energies between BMP-7 and flat, nano-groove, nano-concave and nano-pillar HA surfaces in the last 100 ps simulation time.
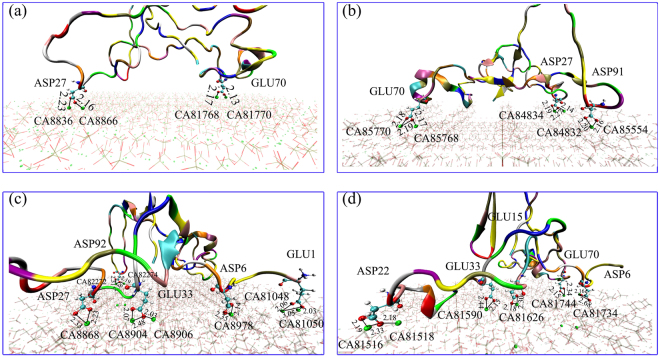
The interactions between BMP-7 and HA mainly depended on electrostatic interaction (E elec) (Table 2). Based on the trajectory animation, BMP-7 adsorptions on different atomic-level HA morphology surfaces involved similar adsorption residues (Fig. 5). The primary interaction residues were ASP and GLU when BMP-7 interacted with the various HA surfaces (Fig. 1a). Both aspartic acid and glutamic acid residues are acidic amino acids and have one carboxyl group. Therefore, strong electrostatic interactions play an important role involving Ca atoms of HA and O atoms of carboxyl groups. Azzopardi et al.52 also indicated that the adsorption of the acidic protein osteopontin to Ca-rich HA (100) surfaces is regulated by electrostatic interactions. In general, the strength of such interactions is closely related to the interaction distance. Moreover, shorter interaction distances or greater numbers of interaction sites will result in larger interaction energies. The distances within 3 Å of Ca-O and H-bonds between BMP-7 and nano-structured HA surfaces are presented in Table 3 and Supplementary Table S1. The quantity and length of these distances indicated that their interactions were stronger on nano-groove, nano-concave, and nano-pillar HA surfaces (except groove 3a) than that on the flat HA surface. We also observed the strongest interaction to be between BMP-7 and the concave2a1b HA surface because of the shorter distances and greatest number of interaction sites within 3 Å (Fig. 5c). In addition, there were five O-Ca-O “bidentate” interactions between the O atoms of GLU1, GLU33, ASP6, ASP27, and ASP92 and Ca atoms of concave2a1b HA surfaces. When the number of interaction distances within 3 Å of BMP-7 on the groove3a HA surfaces reached its lowest value, the interaction energy also exhibited its lowest value (Supplementary Table S1). These results are comparable to those previous investigations of interactions between biomolecules and biomaterials. Wang et al.53 found that the adsorption of Gly on graphene oxide (GO) is stronger than that on graphene (G) because the H-bond distance between the Gly and GO is smaller than that from Gly to the center of G.
Figure 5.
Interaction distances of BMP-7 adsorbing on the (a) flat, (b) groove2a, (c) concave2a1b, and (d) pillar2a1b HA surfaces after 30000 ps MD simulation. Water molecules are omitted for clarity.
Table 3.
Distances of Ca-O interaction or H-bonds between BMP-7 and flat, nano-groove, nano-concave, and nano-pillar HA surfaces. The unit of distance is Å.
| Models | Distances (Å) | Models | Distances (Å) |
|---|---|---|---|
| Flat-HA-BMP-7 | ASP27:OD2-CA8866:CAL = 2.18 | Groove2a-HA-BMP-7 | GLU70:OE2-CA85770 = 2.18 |
| GLU70:OE1-CA85770 = 2.19 | |||
| ASP27:OD1-CA8836:CAL = 2.23 | GLU70:OE1-CA85768 = 2.17 | ||
| GLU70:OE2-CA81768:CAL = 2.17 | ASP27:OD2-CA84834 = 2.19 | ||
| GLU70:OE1-CA81770:CAL = 2.13 | |||
| ASP27:OD1-CA84834 = 2.27 | |||
| ASP27:OD1-CA84832 = 2.14 | |||
| ASP91:OD2-CA85554 = 2.20 | |||
| ASP91:OD1-CA85554 = 2.16 | |||
| Concave2a1b-HA-BMP-7 | GLU1:OE1-CA81050 = 2.03 | Pillar2a1b-HA-BMP-7 | GLU15:OE1-CA81626 = 2.30 |
| GLU1:OE1-CA81048 = 3.05 | GLU15:OE2-CA81626 = 2.18 | ||
| GLU1:OE2-CA81048 = 2.06 | GLU33:OE1-CA81590 = 2.44 | ||
| GLU33:OE1-CA8906 = 2.03 | GLU33:OE2-CA81590 = 2.15 | ||
| GLU33:OE1-CA8904 = 2.48 | GLU70:OE2-CA81744 = 2.34 | ||
| GLU33:OE2-CA8904 = 2.07 | GLU70:OE1-CA81744 = 2.15 | ||
| ASP6:OD1-CA8978 = 2.14 | ASP6:OD1-CA81734 = 2.16 | ||
| ASP6:OD2-CA8978 = 2.35 | ASP6:OD2-CA81734 = 2.68 | ||
| ASP27:OD2-CA8868 = 2.33 | ASP22:OD2-CA81518 = 2.18 | ||
| ASP27:OD1-CA8868 = 2.23 | ASP22:OD1-CA81516 = 2.19 | ||
| ASP92:OD1-CA82274 = 2.19 | ASP22:OD2-CA81516 = 2.33 | ||
| ASP92:OD1-CA82272 = 2.36 | |||
| ASP92:OD2-CA82272 = 2.31 |
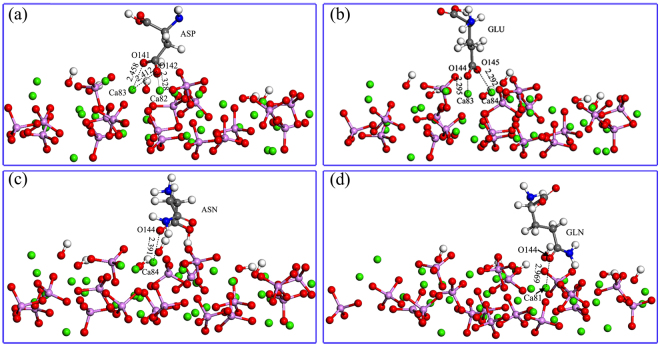
The DFT method is particularly appropriate for investigating physical properties based on electron density by measuring changes of atomic charges54. Electrostatic interactions between BMP-7 and all HA surfaces were dominant in this study. Thus, the atomic charges of the main adsorbed residues on flat HA surfaces changed before and after adsorption. The charge distributions of O atoms of –COO– in ASP27 and GLU70 and Ca atoms of HA were evaluated. The atomic charges of O atoms increased and atomic charges of Ca atoms decreased (Table 4), which indicated that the charge transfer mainly occurred from Ca atoms in HA to O atoms of –COO−. Furthermore, the distances between the O atoms of –COO− and Ca atoms were 2.458, 2.412, 2.328, 2.295, and 2.292 Å, which are typical of O-Ca-O “bidentate” or Ca-O “monodentate” electrostatic interactions (Fig. 6a and b). The DFT results are in accordance with the above MD conclusions.
Table 4.
Change in atomic charge of BMP-7 and its derived peptide on flat HA surfaces.
| Flat-HA-BMP-7 | Flat-HA-BMP-7-derived peptide | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| atoms | Before adsorption | After adsorption | △e | atoms | Before adsorption | After adsorption | △e | ||
| ASP27 | Ca82 | 1.524 | 1.538 | 0.014 | ASN4 | Ca84 | 1.524 | 1.564 | 0.040 |
| Ca83 | 1.558 | 1.561 | 0.003 | O144 | −0.463 | −0.605 | −0.142 | ||
| O141 | −0.362 | −0.614 | −0.252 | O145 | −0.375 | −0.388 | −0.013 | ||
| O142 | −0.340 | −0.757 | −0.417 | ||||||
| GLU70 | Ca83 | 1.541 | 1.566 | 0.025 | GLN2 | Ca84 | 1.539 | 1.542 | 0.003 |
| Ca84 | 1.558 | 1.580 | 0.022 | O144 | −0.463 | −0.514 | −0.051 | ||
| O144 | −0.356 | −0.629 | −0.273 | ||||||
| O145 | −0.356 | −0.664 | −0.417 | ||||||
Figure 6.
(a) and (b) ASP and GLU of BMP-7 and (c) and (d) ASN and GLN of BMP-7-derived peptide interacting with flat HA surfaces.
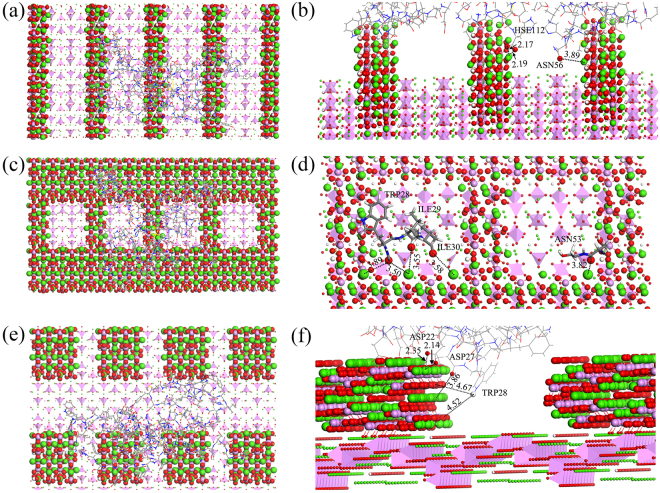
Variation in adsorption behavior of BMP-7 may be related to the HA surface morphology and to additional surfaces (HA (001) or HA ()). The implementation of atomic-level morphology may be achieved by increasing the surface area. In this study, the changes in the surface areas of the nano-groove, nano-concave, and nano-pillar HA surfaces are shown in Table 1. Although all surface areas increased compared to that of the flat HA surface, the change in surface area was disproportionate to the change in the interaction energy, suggesting that an increased surface area was not the only factor affecting interaction energies. The extent of the size matches, such as between groove and protein residues, also plays a crucial role in the interactions. For the groove2a-HA surface, in addition to ASP27, ASP91 and GLU70 absorbed Ca atoms of the HA surface within 3 Å (Fig. 5b), the O atoms of ASN56 and HSE112 residues fell into the grooves and interacted with the Ca atoms of HA (001) surface (Fig. 7b). This may be because ASN56 and HSE112 were approximately 6.816 and 7.600 Å in size, respectively, whereas the width of goove2a was 13.758 Å. Thus, ASN56 and HSE112 can spread into the groove. For the concave2a1b-HA surface, the TRP28, ILE29, ILE30, and ASN53 residues fell into the concaves and interacted with the Ca atoms of the HA (001) surface (Fig. 7d). For the Pillar2a1b-HA surface, ASP22, ASP27, and TRP28 residues could firmly bind the HA (001) surfaces (Fig. 7f). The sizes of TRP28, ILE29, and ASP22 were approximately 8.565, 6.285, and 5.351 Å, respectively. These sizes allowed them to spread into the nanopore and interact with the HA (001) or (10) surface.
Figure 7.
Residues of BMP-7 adsorbing on the (a) and (b) groove2a, (c) and (d) concave2a1b, (e) and (f) pillar2a1b HA (110) side surfaces after 30000 ps MD simulation, side and top views, respectively. Water molecules are omitted for clarity.
There were clearly not only more interaction sites between BMP-7 and atomic-level nano-structured HA surfaces than those between BMP-7 and the flat HA surface, but also the sizes of several residues matched the lengths and widths of the nanopores in the nano-groove, nano-concave and nano-pillar HA surfaces. Therefore, the atomic-level HA morphology significantly affected BMP-7 adsorption. These results may be confirmed by experimental studies. For example, Coata et al.55 and Mao et al.56 have shown that the surface morphology of HA has obvious effects on the adhesion, proliferation, and differentiation of cells.
Adsorption behaviors between BMP-7-derived peptide and atomic-level nano-structured HA surfaces
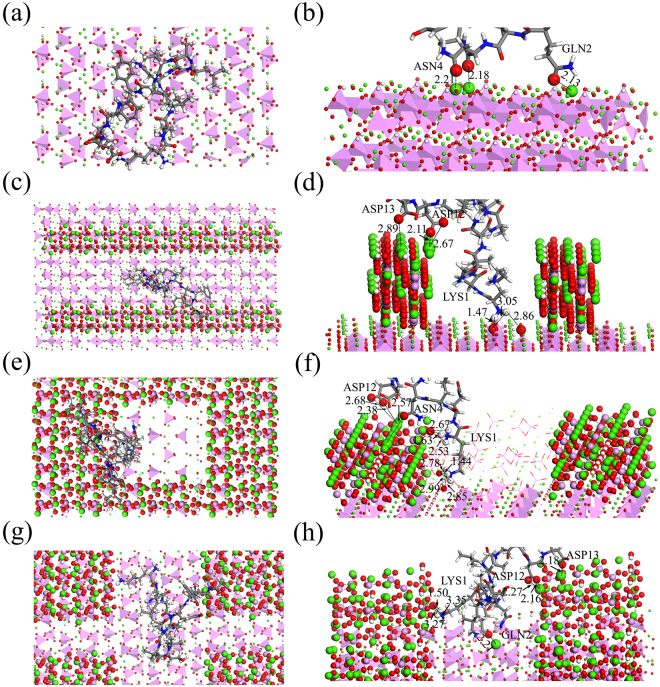
The interaction behaviors between BMP-7-derived peptide (KQLNALSVLYFDD) and the flat, nano-groove2a, nano-concave2a1b, and nano-pillar2a1b HA surfaces were studied. Interactions energies between the peptide and various HA surfaces are shown in Table 2. The interaction energy was -241.04 Kcal/mol when BMP-7-derived peptide interacted with the flat HA surface, and two residues (ASN4 and GLN2) bound the HA surfaces tightly. Interaction distances between O atoms of ASN4 and GLN2 and Ca atoms of HA were 2.21, 2.18, and 2.13 Å, respectively. The interaction increased when BMP-7-derived peptide was adsorbed onto the nano-groove2a HA surface (-408.40 Kcal/mol), and three residues (LYS1, ASP12, and ASP13) were firmly absorbed onto the HA surface. The energy further decreased when the derived peptide was absorbed onto the nano-concave2a1b and nano-pillar2a1b HA surfaces (−1200.36 Kcal/mol and -904.65 Kcal/mol). These results showed that there were stronger adsorptions of BMP-7-derived peptide on the nano-groove, nano-concave, and nano-pillar HA surfaces than that on the flat HA surface. The strongest adsorption was observed for BMP-7-derived peptide on the nano-concave2a1b HA surface.
Furthermore, we found that the KQLNALSVLYFDD peptide expanded slowly on the nano-groove, nano-concave, and nano-pillar HA surfaces with an increase in adsorption time, and there were more carboxyl or amino groups interacting with the Ca atoms, phosphate and hydroxyl groups of HA surfaces, as shown in Fig. 8. Electrostatic interactions remained dominant (Table 2). The charge transfer of the –COO−and ketone group of ASN4 and GLN2 in the derived peptide and Ca atoms of HA were also investigated by the DFT method, and similar simulation results with BMP-7 adsorption on the flat HA surface were obtained (Fig. 6c and d, and Table 4). Figure 6c and d showed that there were Ca-O electrostatic interactions between the O atoms of ASN4 and GLN2 and Ca atoms in HA. The data presented in Table 4 clearly indicate that O atoms of ASN4 and GLN2 acquired electrons and Ca atoms lost electrons, forming Ca-O electrostatic interactions. Therefore, electrostatic interactions between the derived peptide and HA are dominant, which is consistent with reported interactions between other peptides and HA surfaces. For example, Almora-barrios et al.57 employed DFT to investigate the interaction of HA and PRO- hydroxyproline (HYP)-GLY, HYP-PRO-GLY, PRO-LYS-GLY, and PRO-hydroxylysine (HYL)-GLY, and discovered that Ca–O electrostatic interactions prevail between O atoms of the carboxyl and Ca atoms of HA.
Figure 8.
Residues of BMP-7-derived peptide adsorbing on the (a) and (b) flat, (c) and (d) groove2a, (e) and (f) concave2a1b, (g) and (h) pillar2a1b HA surfaces after 30000 ps MD simulation, side and top views, respectively. Water molecules are omitted for clarity.
Alteration trends of interaction energies between the BMP-7-derived peptide and nano-structured HA were similar to those between BMP-7 and nano-structured HA. Thus, nano-groove, nano-concave and nano-pillar HA surfaces largely enhanced BMP-7-derived peptide adsorption. Additionally, we found that the active sites of BMP-7-derived peptide (a small molecule) may easily spread out and stretch inside the trenches to absorb firmly on the side surfaces of nano-groove, nano-concave, and nano-pillar HA, such as the HA (001) surface or HA (10) surface, as illustrated in Fig. 8. In the groove2a-HA model, the LYS1 residue fell into the bottom of the groove; we observed interactions between the amino group of LYS1 and O atoms of the hydroxyl group of the HA (110) surface (Fig. 8d). In the concave2a1b-HA model, the LYS1 residue slipped into the groove and possibly pulled the ASP12 and ASN4 close to the nano-concave2a1b HA surfaces. There were Ca-O interactions between the O atoms of ASP12 and ASN4 and Ca atoms of the HA (110) surface. Additionally, the LYS1 residue was surrounded by the HA (001) and (10) surfaces; thus, the H atoms of LYS1 interacted with the O atoms of -PO4 and -OH on the HA (001) surface (Fig. 8f). In the pillar2a1b-HA models, both the LYS1 and GLN2 residues contacted the HA (001) and (110) surfaces upon entering the groove of the nano-pillar HA surface, and the H atoms of LYS1 or O atoms of GLN2 interacted with the O atoms of -PO4 in HA (001) or Ca atoms of the HA (110) surface, respectively (Fig. 8h).
The sizes of LYS1 and GLN2 are approximately 9.40 and 6.67 Å, respectively, which completely match the width of the grooves (13.758 Å) in goove2a, concave2a1b, and pillar2a1b HA models. These phenomena also demonstrated that the atomic-level surface morphology of HA is pivotal for peptide adsorption. Thus, a compatible groove size of surface morphology would be beneficial for promoting specific peptide adsorption. These results are consistent with those of Zhang et al.58, who found that the interaction was strengthened when the RGD peptide fell into the grooves between the step edges of TiO2.
Water Effects
The effects of water on BMP-7 and its derived peptide adsorptions on flat HA surfaces were also considered by MD and DFT studies. The MD conclusions suggested that the water layer inhibited BMP-7 and its derived peptide absorbing on flat HA surfaces quickly and steadily through intermolecular H-bonds between the H2O and HA or between the H2O and protein (Supplementary Fig. S5). The DFT results suggested that “–COO–…H2O…OPO3” water-bridged H-bonds between the anionic Asp molecule, H2O molecule and PO4 in HA and H-bonds between the -NH2 and H2O molecule were formed, moreover, the water molecules hindered the main “Ca-O” interaction between the ASP and Ca atoms in HA (Supplementary Fig. S6d and f). These results are in accordance with previous literature59,60. Wang et al.45 also reported that water molecules on HA (110) surfaces could prevent BSA protein from migrating close to the surface. Furthermore, there are water-bridged H-bonds between amino acid residues, H2O and HA substrate. The MD and DFT conclusions confirmed that water environments seriously affected the adsorption of amino acids on HA (110) surfaces. Details are presented in the Supplementary Information.
Consideration of other influencing factors
It should be noted that, except for the micro-nano structured surface morphology, other influencing factors should also be considered during MD simulations, including the effects of pH, ionic strength, and surface chemistry, which renders the simulation and analysis of protein structures more complicated10. Researchers suggest that the change of pH, ionic strength, surface chemistry and other factors will eventually result in changes to surface charge intensity and distribution, and further affect the interaction between proteins and HA. For example, when the change of pH or ionic strength causes the Zeta-potential value to decrease, most proteins are prone to adsorbing on HA surfaces due to the decrease in electrostatic repulsions between the HA and the negatively charged protein61. Yin et al.62 found that pH increases resulted in a decreasing adsorption of BSA onto HA surfaces, and NaCl concentration increases caused the adsorption to increase. These effects are attributed to the difference in the negative Zeta-potential of HA being increased due to the pH increase, whereas increases in ionic strength resulted in a decreased negative Zeta-potential of HA. Furthermore, the HA surface environment can be altered by dopants, further affecting interactions with protein. Chen et al.63 investigated the adsorption and desorption behaviors of leucine-rich amelogenin protein (LRAP) on a series of SiHA (100) surfaces. They discovered that the Si doping into the HA (100) surface increases the surface negative charge due to induction of SiO4 4-, which leads to the -NH3 +, -NH2, and -OH moving towards the surface, and the negatively charged -COO− being repelled from the surface. Therefore, the interaction of –COO− and Ca2+ becomes gradually weaker due to the effects of charge rejection and steric hindrance of large-sized SiO4 4- molecules.
Much work involving these factors has been reported, whereas a consensus on which are critical factors has not been reached so far. The interaction behaviors between materials and organic molecules are often the result of the joint actions of different factors. In this study, we mainly considered the effects of different nanoscale surface morphologies of HA substrates on the adsorption of proteins, which is rarely reported. Our results revealed that surface nano-morphology is also an important factor that affects the interaction between HA and protein, in addition to previously-reported factors. In addition, we implemented an abstract model that represents those nano-morphology materials used in previous experimental studies. Although the conclusions drawn from computer simulations cannot fully reflect experimental results, they can still provide meaningful explanations for experimental phenomena within a limited simulation scale and offer possible theoretical guidance for further study on the design of biological nanomaterials.
Conclusions
To explore the effects of different surface morphologies of HA materials on the adsorption of biomolecules, we elucidated the interaction effects of BMP-7 and BMP-7-derived peptide on flat, nano-groove, nano-concave and nano-pillar HA (110) surfaces using MD and DFT methods. Our results indicated that the atomic-level morphology of HA significantly affects the interaction strength between protein/peptide and HA substrates. The interactions of BMP-7 and its derived peptide with the nano-concave and nano-pillar HA surfaces were stronger than those with flat and nano-groove HA surfaces. At the same time, the results also revealed that if the groove size of the nano-structured HA surfaces matches that of residues in the protein or peptide, the residues are likely to spread into the grooves in nano-groove, nano-concave, and nano-pillar HA, further strengthening the interactions. Our results improve the understanding of the adsorption behaviors of proteins on nano-structured HA surfaces. In addition, our results provide valuable theoretical guidance for designing new types of bioceramic materials useful for bone regeneration and tissue engineering applications.
Electronic supplementary material
Acknowledgements
This study was financially supported by the 863 Program (2015AA034202), the National Key Research and Development Program of China (2016YFB0700800), Key Technology Research and Development Program of the Bureau of Science and Technology of Sichuan Province (2014GZ0174), Special Program for Applied Research on Super Computation of the NSFC-Guangdong Joint Fund (the second phase) under Grant (U1501501), Education Department Program of Sichuan Province (16ZB0313), Teaching Reform Program of Mianyang Teachers’ College (Mnu-JY1512), Natural Science Project of Mianyang Teachers’ College (MYSY2017JC09), and Innovation and entrepreneurship training program of university students in Sichuan province (201610639077).
Author Contributions
Q.W. carried out all of the simulation calculations and contributed to data analysis, results interpretation, and manuscript writing. M.H.W. designed the project, analyzed the data and wrote the manuscript. X.L. and K.F.W. contributed to the initial conception of the project and editing the manuscript. L.M.F. provided the calculation time on the MilkyWay II supercomputer and contributed to data interpretation. F.Z.R. supervised the data and analyzed the results. G.M.L. reviewed the final article.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-15219-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Suchanek W, Yoshimura M. Processing and properties of hydroxyapatite-based biomaterials for use as hard tissue replacement implants. J. Mater. Res. 1998;13:94–117. doi: 10.1557/JMR.1998.0015. [DOI] [Google Scholar]
- 2.Kandori K, Horigami N, Kobayashi H, Yasukawa A, Ishikawa T. Adsorption of lysozyme onto various synthetic hydroxyapatites. J. Colloid Interf. Sci. 1997;191:498–502. doi: 10.1006/jcis.1997.4943. [DOI] [Google Scholar]
- 3.Zhou HL, Wu T, Dong X, Wang Q, Shen J. Adsorption mechanism of BMP-7 on hydroxyapatite (001)surfaces. Biochem. Bioph. Res. Co. 2007;361:91–96. doi: 10.1016/j.bbrc.2007.06.169. [DOI] [PubMed] [Google Scholar]
- 4.Tamagawa H, Tenkumo T, Sugaya T, Kawanami M. Effect of nano-hydroxyapatite on bone morphogenetic protein-2-induced hard tissue formation and dentin resorption on a dentin surface. Appl. Surf. Sci. 2012;262:140–145. doi: 10.1016/j.apsusc.2012.03.103. [DOI] [Google Scholar]
- 5.Tarasevich BJ, Lea S, Bernt W, Engelhard M, Shaw WJ. Adsorption of amelogenin onto self-assembled and fluoroapatite surfaces. J. Phys. Chem. B. 2009;113:1833. doi: 10.1021/jp804548x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai ZB, Wang M, Yan C, Oloyede A. Molecular dynamics simulation of mechanical behavior of osteopontin-hydroxyapatite interfaces. J. Mech. Behav. Biomed. 2014;36:12–20. doi: 10.1016/j.jmbbm.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Liao C, Xie Y, Zhou J. Computer simulations of fibronectin adsorption on hydroxyapatite surfaces. RSC Adv. 2014;4:15759–15769. doi: 10.1039/c3ra47381c. [DOI] [Google Scholar]
- 8.Swain SK, Sarkar D. Study of BSA protein adsorption/release on hydroxyapatite nanoparticles. Appl. Surf. Sci. 2013;286:99–103. doi: 10.1016/j.apsusc.2013.09.027. [DOI] [Google Scholar]
- 9.Yang Y, Cui Q, Sahai N. How does bone sialoprotein promote the nucleation of hydroxyapatite? A molecular dynamics study using model peptides of different conformations. Langmuir. 2010;26:9848–9859. doi: 10.1021/la100192z. [DOI] [PubMed] [Google Scholar]
- 10.Wang Q, et al. Computer simulation of biomolecule–biomaterial interactions at surfaces and interfaces. Biomed. Mater. 2015;10:032001. doi: 10.1088/1748-6041/10/3/032001. [DOI] [PubMed] [Google Scholar]
- 11.Emami FS, Puddu V, Berry RJ, Varshney VS, Patwardhan V. Prediction of Specific Biomolecule Adsorption on Silica Surfaces as a Function of pH and Particle Size. Chem. Mater. 2016;26:5725–5734. doi: 10.1021/cm5026987. [DOI] [Google Scholar]
- 12.Rovensky YA, Slavnaja IL, Vasiliev JM. Behavior of fibro blast-like cells on grooved surfaces. Exp. Cell Res. 1971;65:193–201. doi: 10.1016/S0014-4827(71)80066-6. [DOI] [PubMed] [Google Scholar]
- 13.Chaubey A, Ross KJ, Leadbetter RM, Burg KJL. Surface patterning: tool to modulate stem cell differentiation in an adipose system. J. Biomed. Mater. Res. B. 2008;84B:70–78. doi: 10.1002/jbm.b.30846. [DOI] [PubMed] [Google Scholar]
- 14.Yim EKF, Pang SW, Leong KW. Synthetic nanostructures inducing differentiation of human mesenchymal stem cells into neuronal lineage. Exp. Cell Res. 2007;313:1820–1829. doi: 10.1016/j.yexcr.2007.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y, Wang X, Chen L, Ding JD. Preparation of hydroxyapatite micropatterns for the study of cell-biomaterial interactions. J. Mate. Chem. B. 2014;2:2220–2227. doi: 10.1039/c4tb00146j. [DOI] [PubMed] [Google Scholar]
- 16.Henry MG, et al. Roles of hydroxyapatite allocation and microgroove dimension in promoting preosteoblastic cell functions on photocured polymer nanocomposites through nuclear distribution and alignment. Langmuir. 2015;31:2851–2860. doi: 10.1021/la504994e. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z, Xiao ZW, Fan SH, Zhang XD. Fabrication of micro-grooved patterns on hydroxyapatite ceramics and observation of earlier response of osteoblasts to the patterns. J. Inorg. Mater. 2013;28:51–57. doi: 10.3724/SP.J.1077.2013.12093. [DOI] [Google Scholar]
- 18.Lu X, Leng Y. Comparison of the osteoblast and myoblast behavior on hydroxyapatite microgrooves. J. Biomed. Mater. Res. Part B. 2009;90B:438–445. doi: 10.1002/jbm.b.31304. [DOI] [PubMed] [Google Scholar]
- 19.Iwamoto T, Hieda Y, Kogai Y. Effect of hydroxyapatite surface morphology on cell adhesion. Mat. Sci. Eng. C-Mater. 2016;69:1263–1267. doi: 10.1016/j.msec.2016.07.056. [DOI] [PubMed] [Google Scholar]
- 20.Wu F, et al. Self-setting bioactive calcium-magnesium phosphate cement with high strength and degradability for bone regeneration. Acta Biomater. 2008;4:1873–1884. doi: 10.1016/j.actbio.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 21.Jouve JL, Mottet V, Cottalorda J, Frayssinet P, Bollini G. Reimplantation of growth plate chondrocyte cultures in central growth plate defects: Part I. Characterization of cultures. Journal of Pediatric Orthopedics Part B. 1998;7:167–173. doi: 10.1097/01202412-199804000-00016. [DOI] [PubMed] [Google Scholar]
- 22.Kasaj A, Willershausen B, Junker R, Stratul SI, Schmidt M. Human periodontal ligament fibroblasts stimulated by nanocrystalline hydroxyapatite paste or enamel matrix derivative. An in vitro assessment of PDL attachment, migration, and proliferation. Clin. Oral Invest. 2012;16:745–754. doi: 10.1007/s00784-011-0570-7. [DOI] [PubMed] [Google Scholar]
- 23.Webster T. Specific proteins mediate enhanced osteoblast nanophase ceramics. J. Biomed. Mat. Res. 2000;51:475–483. doi: 10.1002/1097-4636(20000905)51:3<475::AID-JBM23>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 24.Zhu W, et al. Experimental Study of Nano-Hydroxyapatite/Recombinant Human Bone Morphogenetic Protein-2 Composite Artificial Bone. Artif. Cell. Blood Sub. 2010;38:150–156. doi: 10.3109/10731191003712756. [DOI] [PubMed] [Google Scholar]
- 25.Hogan BL. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Gene. Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 26.Luo G, et al. BMP-7 is an inducer of nephrogenesis, and is also required for eye development and skeletal patterning. Gene. Dev. 1995;9:2808–2820. doi: 10.1101/gad.9.22.2808. [DOI] [PubMed] [Google Scholar]
- 27.Schwarting T, et al. Bone morphogenetic protein 7 (BMP-7) influences tendon-bone integration in vitro. Plos One. 2015;10:1–17. doi: 10.1371/journal.pone.0116833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng H, et al. Osteogenic activity of the fourteen types of human bone morphogenetic proteins (BMPs) J. Bone. Joint Surg. Am. 2004;22:79–80. doi: 10.2106/00004623-200308000-00017. [DOI] [PubMed] [Google Scholar]
- 29.Tasnim F, et al. Effects of bone morphogenetic proteins on primary human renal cells and the generation of bone morphogenetic protein-7-expressing cells for application in bioartificial kidneys. Langmuir. 2012;19:4063–4069. doi: 10.1089/ten.TEA.2011.0149. [DOI] [PubMed] [Google Scholar]
- 30.Dong XL, Zhou HL, Wu T, Wang Q. Behavior regulation of adsorbed proteins via hydroxyapatite surface texture control. J. Phys. Chem. B. 2008;112:4751–4759. doi: 10.1021/jp0768672. [DOI] [PubMed] [Google Scholar]
- 31.Feng B, Hu D, Zhang Y. Accelerated Bone Regeneration by Chitosan/Nanometer Hydroxyapatite/Collagen Composite Incorporating BMP-7 Mimetic Peptide. J. Hard Tissue Biol. 2012;21:481–487. doi: 10.2485/jhtb.21.481. [DOI] [Google Scholar]
- 32.Kim HK, et al. Osteogenesis induced by a bone forming peptide from the prodomain region of BMP-7. Biomaterials. 2012;33:7057–7063. doi: 10.1016/j.biomaterials.2012.06.036. [DOI] [PubMed] [Google Scholar]
- 33.Tao H, et al. BMP7-Based Functionalized Self-Assembling Peptides for Nucleus PulposusTissue Engineering. Acs Appl. Mater. Inter. 2015;7:17076–17087. doi: 10.1021/acsami.5b03605. [DOI] [PubMed] [Google Scholar]
- 34.Zhang HP, et al. Molecular dynamics simulations on the interaction between polymers and hydroxyapatite with and without coupling agents. Acta Biomater. 2009;5:1169–1181. doi: 10.1016/j.actbio.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Wei Q, Pan F, Yang M, Wei S. Molecular dynamics simulations for the examination of mechanical properties of hydroxyapatite/ poly α-n-butyl cyanoacrylate under additive manufacturing. Biomed. Mater. Eng. 2013;23:S857–S865. doi: 10.3233/BME-130874. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, Mkhonto D, Cui Q, Sahai N. Theoretical study of bone sialoprotein in bone biomineralization. Cells Tissues Organs. 2011;194:182–187. doi: 10.1159/000324648. [DOI] [PubMed] [Google Scholar]
- 37.Xin RL, Leng Y, Wang N. In situ TEM examinations of octacalcium phosphate to hydro xyapatite transformation. J. Cryst. Growth. 2006;289:339–344. doi: 10.1016/j.jcrysgro.2005.11.010. [DOI] [Google Scholar]
- 38.Phillips JC, et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson RM, Elliott JC, Dowker SEP. Rietveld refinement of the crystallographic structure of human dental enamel apatites. Am. Mineral. 1999;84:1406–1414. doi: 10.2138/am-1999-0919. [DOI] [Google Scholar]
- 40.Nelson MT, et al. NAMD: a parallel, object-oriented molecular dynamics program. Int. J. High Perform C. 1996;10:251–268. [Google Scholar]
- 41.Bhowmik R, Katti KS, Katti D. Molecular dynamics simulation of hydroxyapatite-polyacrylic acid interfaces. Polymer. 2007;48:664–674. doi: 10.1016/j.polymer.2006.11.015. [DOI] [Google Scholar]
- 42.Hauptmann S, Dufner H, Brickmann J, Kast SM, Berry RS. Potential energy function for apatites. Phys. Chem. Chem. Phys. 2003;5:635–639. doi: 10.1039/b208209h. [DOI] [Google Scholar]
- 43.Oliveira AF, Gemming S, Seifert G. Conformational analysis of aqueous BMP-2 using atomistic molecular dynamics simulations. J. Phys. Chem. B. 2011;115:1122–1130. doi: 10.1021/jp106092d. [DOI] [PubMed] [Google Scholar]
- 44.Chen X, Wang Q, Shen JW, Pan HH, Wu T. Adsorption of Leucine-Rich Amelogenin Protein on Hydroxyapatite (001) Surface through −COO- Claws. J. Phys. Chem. C. 2007;111:1284–1290. doi: 10.1021/jp0646630. [DOI] [Google Scholar]
- 45.Wang KF, Wang MH, Wang QG, Lu X, Zhang XD. Computer simulation of proteins adsorption on hydroxyapatite surfaces with calcium phosphate ions. J. Eur. Ceram. Soc. 2017;37:2509–2520. doi: 10.1016/j.jeurceramsoc.2017.02.013. [DOI] [Google Scholar]
- 46.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 47.Filgueiras MRT, Mkhonto D, De Leeuw NH. Computer simulations of the adsorption of citric acid at hydroxyapatite surfaces. J. Cryst. Growth. 2006;294:60–68. doi: 10.1016/j.jcrysgro.2006.05.077. [DOI] [Google Scholar]
- 48.Delley B. DMol3 DFT studies: from molecules and molecular environments to surfaces and solids. Comp. Mater. Sci. 2000;17:122–126. doi: 10.1016/S0927-0256(00)00008-2. [DOI] [Google Scholar]
- 49.Delley B. Hardness conserving semilocal pseudopotentials. Phys. Rev. B. 2002;66:155125. doi: 10.1103/PhysRevB.66.155125. [DOI] [Google Scholar]
- 50.Perdew JP, Burke K, Ernzerhof M. Generalized gradient approximation made simple. Phys. Rev. Lett. 1996;77:3865–3868. doi: 10.1103/PhysRevLett.77.3865. [DOI] [PubMed] [Google Scholar]
- 51.Shen JW, Wu T, Wang Q, Pan HH. Molecular simulation of protein adsorption and desorption on hydroxyapatite surfaces. Biomaterials. 2008;29:513–532. doi: 10.1016/j.biomaterials.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 52.Azzopardi PV, et al. Roles of Electrostatics and Conformation in Protein-Crystal Interactions. Plos One. 2010;5:e9330. doi: 10.1371/journal.pone.0009330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang MH, et al. Density functional theory study of interactions between glycine and TiO2/graphene nanocomposites. Chem. Phys. Lett. 2014;599:86–91. doi: 10.1016/j.cplett.2014.03.024. [DOI] [Google Scholar]
- 54.Suganthi S, Kannappan V, Sathyanarayanamoorthi V, Karunathan R. Quantum mechanical investigation of vibrational and electronic spectra of some 5-substituted isoquinolines. Indian J. Pure Ap. Phy. 2016;54:15–34. [Google Scholar]
- 55.Costa DO, et al. The differential regulation of osteoblast and osteoclast activity by surface topography of hydroxyapatite coatings. Biomaterials. 2013;34:7215–7126. doi: 10.1016/j.biomaterials.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 56.Mao LX, et al. Effect of micro-nano-hybrid structured hydroxyapatite bioceramics on osteogenic and cementogenic differentiation of human periodontal ligament stem cell via Wnt signaling pathway. Int. J. Nanomed. 2015;10:7031–7044. doi: 10.2147/IJN.S90343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Almora-barrios N, De Leeuw NH. A density functional theory study of the interaction of collagen peptides with hydroxyapatite surfaces. Langmuir. 2010;26:14535–14542. doi: 10.1021/la101151e. [DOI] [PubMed] [Google Scholar]
- 58.Zhang HP, et al. Effects of aqueous environment and surface defects on Arg-Gly-Asp peptide adsorption on titanium oxide surfaces investigated by molecular dynamics simulation. J. Biomed. Mater. Res. A. 2011;96A:466–476. doi: 10.1002/jbm.a.33003. [DOI] [PubMed] [Google Scholar]
- 59.Dong X, Wang Q, Wu T, Pan HH. Understanding adsorption-desorption dynamics of BMP-2 on hydroxyapatite (001) surface. Biophys. J. 2007;93:750–759. doi: 10.1529/biophysj.106.103168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pham TT, et al. Properties of water confined in hydroxyapatite nanopores as derived from molecular dynamics simulations. Theor. Chem. Acc. 2015;134:1–14. doi: 10.1007/s00214-015-1653-3. [DOI] [Google Scholar]
- 61.Tercinier L, Ye A, Singh A, Anema SG, Singh H. Effects of Ionic Strength, pH and Milk Serum Composition on Adsorption of Milk Proteins on to Hydroxyapatite Particles. Food Biophys. 2014;9:341–348. doi: 10.1007/s11483-014-9360-5. [DOI] [Google Scholar]
- 62.Yin G, Liu Z, Zhan J, Ding F, Yuan N. Impacts of the surface charge property on protein adsorption on hydroxyapatite. Chem. Eng. J. 2002;87:181–186. doi: 10.1016/S1385-8947(01)00248-0. [DOI] [Google Scholar]
- 63.Chen X, Wu T, Wang Q, Shen JW. Shield effect of silicate on adsorption of proteins onto silicon-doped hydroxyapatite (100) surface. Biomaterials. 2008;29:2423–2432. doi: 10.1016/j.biomaterials.2008.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.