Abstract
Genome-wide association studies (GWAS) can serve as strong evidence in correlating biological pathways with human diseases. Although ischemic stroke has been found to be associated with many biological pathways, the genetic mechanism of ischemic stroke is still unclear. Here, we performed GWAS for a major subtype of stroke—small-vessel occlusion (SVO)—to identify potential genetic factors contributing to ischemic stroke. GWAS were conducted on 342 individuals with SVO stroke and 1,731 controls from a Han Chinese population residing in Taiwan. The study was replicated in an independent Han Chinese population comprising an additional 188 SVO stroke cases and 1,265 controls. Three SNPs (rs2594966, rs2594973, rs4684776) clustered at 3p25.3 in ATG7 (encoding Autophagy Related 7), with P values between 2.52 × 10−6 and 3.59 × 10−6, were identified. Imputation analysis also supported the association between ATG7 and SVO stroke. To our knowledge, this is the first GWAS to link stroke and autophagy. ATG7, which has been implicated in autophagy, could provide novel insights into the genetic basis of ischemic stroke.
Introduction
Stroke is known to be the second leading cause of death and a major cause of disability worldwide1. Although traditional vascular risk factors such as hypertension, diabetes, atrial fibrillation, and cigarette smoking are common in stroke, stroke incidence2 and subtype distribution3 are different among ethnicities. It is possible that a non-traditional risk factor such as genetic predisposition might be important. Data from twin and family history studies have suggested a role for genetic factors in stroke risk4,5. A previous study of vascular disease reported that family history is an independent risk factor for SVO, especially in cases presenting before the age of 65, suggesting the involvement of underlying genetic components in the development of SVO6. A GWAS conducted using a Japanese cohort with ischemic stroke identified a genetic variant in PRKCH 7; however, a meta-analysis of GWAS data from a Caucasian population found no association between ischemic stroke and PRKCH genetic variants8.
Compared with recent advances in high throughput genotyping for other subtypes of ischemic stroke9–11, gene discovery for SVO has progressed slowly because of etiologic heterogeneity and variations among different ethnic backgrounds. By exploiting these phenotypically more homogeneous classifications, a GWAS conducted using a single population may identify a genetic association specific to SVO. Thus, in the present study, we aimed to identify genetic correlations for SVO using a GWAS within a Han Chinese population.
Methods
Ethical statement
All methods were performed in accordance with the relevant guidelines and regulations. The study was approved by the Institutional Review Board and the Ethics Committee of the Institutional Review Board of Chang Gung Healthcare System and Academia Sinica, Taiwan. Written informed consents were obtained from the subjects or their family members in accordance with institutional requirements and Declaration of Helsinki principles.
Study subjects and phenotype definitions
Individuals with SVO stroke (n = 530) (comprising 342 SVO stroke cases from the GWAS and 188 SVO stroke cases from the replication study) were recruited from the three branch hospitals of Chang Gung Healthcare System, Linkou, Chiayi and Kaohsiung, in collaboration with the Translational Resource Center (TRC) for Genomic Medicine of Taiwan. These three branch hospitals cover a population of six million in Taiwan with a total of 3,300 annual ischemic stroke patients. SVO was defined by the presence of subcortical, hypodense lesions with a diameter of <15 mm with accompanying clinical lacunar syndrome. The medical information and blood samples of all cases were centralized in Linkou CGMH, and the SVO stroke subtype was classified according to modified TOAST criteria12 by single physician, TH Lee, to prevent from interobserver discrepancy. Besides the criteria of clinical presentations and lacunas in brain images, only cases with diameter stenosis <30% in extracranial carotid artery confirmed by carotid ultrasound and/or in intracranial carotid artery by angiography (magnetic resonance, computed tomography or digital subtraction) were included for analysis. The control subjects (1,731 in the discovery study and 1,265 in the replication study) from the GWAS were randomly selected from the Taiwan Han Chinese Cell and Genome Bank in Taiwan. These controls were presumably disease-free as reported previously13.
Genotyping and quality control
Genomic DNA was extracted from blood using a Puregene DNA Isolation Kit (Gentra Systems). Each individual was genotyped using the Axiom Genome-Wide CHB (with a total of 642,832 SNPs) according to manufacturer’s protocols by the National Center for Genome Medicine (NCGM) at Academia Sinica. All sample call rates were >98.69%, and the mean individual sample call rate was 99.5 ± 0.26%. First-degree relatives (parent-offspring and full sibling pairs) in SVO stroke cases and in control samples were identified by kinship analysis and were excluded from further analysis. Genotyping quality control for each SNP was further determined by the total call rate (successful call rate) and MAF in SVO stroke cases and controls. SNPs were excluded from further analysis if only one allele appeared in SVO stroke cases and controls, if the total call rate was <0.95, or if the total MAF was <0.05 and the total call rate was <0.99.
Statistical analysis
The statistical method used for GWAS analysis has been well-established in our previous study9,14. Detection of possible population stratification that could influence association analysis was carried out using EIGENSTRAT 2.0. We estimated the variance inflation factor for genomic controls. Genome-wide association analysis and GC correction were carried out to compare allele and genotype frequencies between cases and controls using the Cochran-Armitage trend test. A quantile-quantile (Q-Q) plot was used to determine P value distribution (Fig. 1). The adjustment for principle components suggested that inflation was not due to population stratification.
Figure 1.
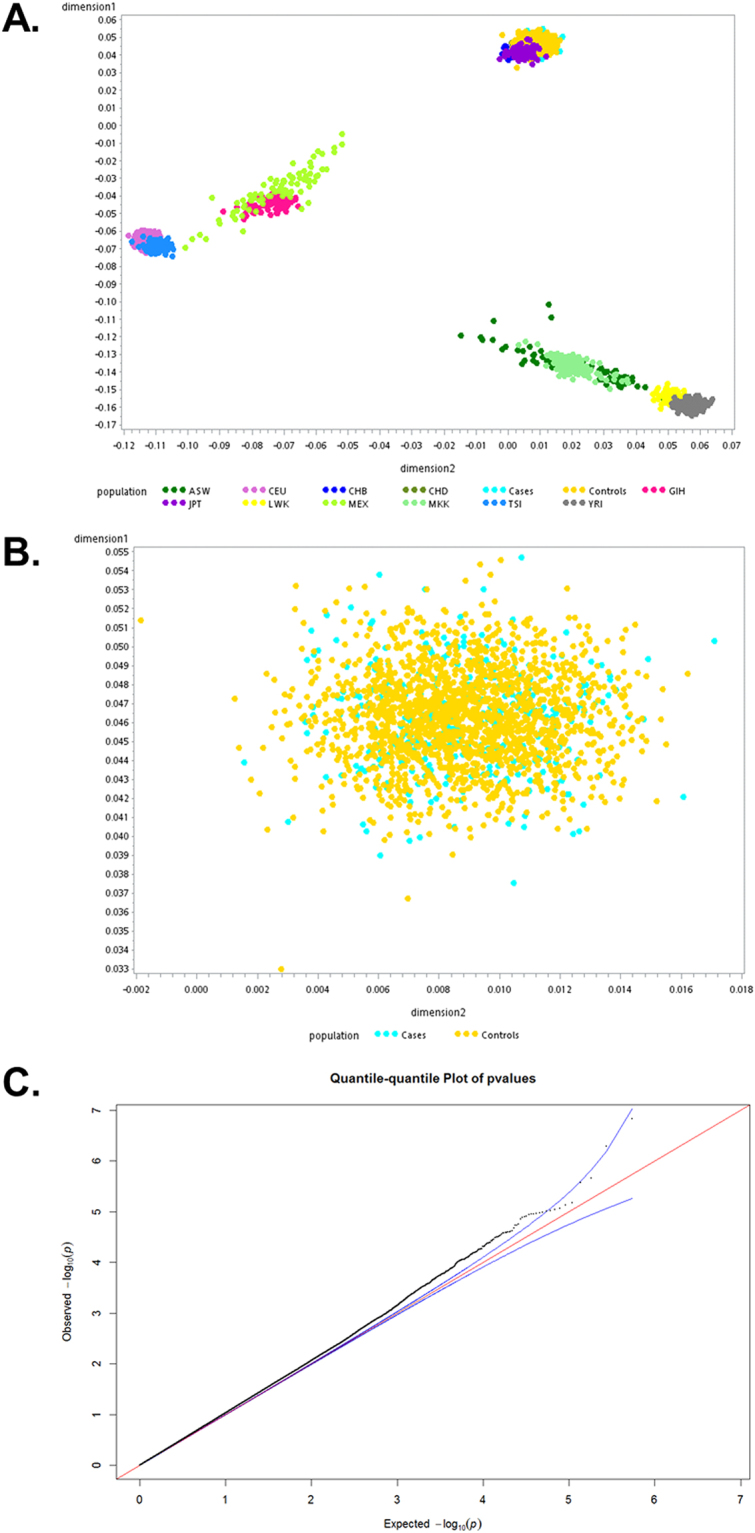
Multidimensional scaling analysis. (A) Results of the multidimensional scaling analysis of the GWAS samples with HapMap populations. (B) Results of the multidimensional scaling analysis of the GWAS samples with the GWAS samples only. (C) Q-Q plot of the P values in a Cochran-Armitage trend test. Lambda value is 1.09.
GWAS validation and replication
The top three SNPs (P < 1 × 10−4) from the genome-wide association analysis of the 342 SVO stroke cases and controls were further validated using MALDI-TOF mass spectrometry (MassARRAY, Sequenom) (Supplementary Table 1). In addition, we also validated other 30 SNPs (P < 1 × 10−2) in the discovery stage (Supplementary Table 1). SNP genotypes with over 98% success rate and over 98% concordance between the two platforms were then genotyped. An additional 188 SVO stroke cases were used for the replication study.
Imputation
For enhancement of the coverage, untyped SNPs were imputed by IMPUTE2 using 1000 Genomes reference panel15–17. In the pre-phasing step, we set up the haplotypes inferences via SHAPEIT method for optimizing the imputation procedure18. For elimination of edge effects, we expanded 500 kb buffer region on each side of imputation region. We determined the uncertainty of imputed genotypes based on likelihood scoring in SNPTEST v2 and frequentist association test of the additive model. We further validated the top imputed SNP by direct genotyping.
Data availability
The datasets generated during and/or analysed during the current study are available in the International Stroke Genetics Consortium repository, http://www.cerebrovascularportal.org/. The name of the dataset is “SVO-Han-population Taiwan-NCGM”.
Results
Study populations
Characteristics of the SVO stroke groups are shown in Table 1. The mean age of the SVO group in the GWAS (discovery) group and replication group was 57.9 years old and 56.0 years old, respectively. The ratio of males to females in the GWAS group and replication groups was 67.3% and 67.6%, respectively. For other stroke risk factors, there was no significant difference between the GWAS group and the replication group.
Table 1.
Baseline Demographic Summary of Patients
| n | Discovery (n = 342) | n | Replication (n = 188) | |||
|---|---|---|---|---|---|---|
| Median Age (IQR) | 342 | 57.9 | (48.3 − 67.0) | 183 | 56 | (44.0 − 67.0) |
| Sex-Male (%) | 342 | 67.30% | 188 | 67.60% | ||
| Hypertension (%) | 340 | 77.60% | 187 | 78.10% | ||
| Diabetes mellitus (%) | 340 | 29.10% | 187 | 32.60% | ||
| Alcohol (%) | 329 | 22.20% | 173 | 15.00% | ||
| Family history of stroke (%) | 327 | 41.90% | 166 | 42.80% | ||
| Median HDL-C (IQR) | 331 | 44 | (36.0 − 53.0) | 175 | 42 | (35.0 − 49.0) |
| Median LDL-C (IQR) | 328 | 110 | (93.0 − 135.0) | 174 | 115 | (91.0 − 137.0) |
| Median VLDL (IQR) | 133 | 26 | (19.0 − 42.0) | 91 | 30 | (20.0 − 37.3) |
| Median Triglyceride (IQR) | 341 | 131 | (99.0 − 188.0) | 188 | 145 | (105.0 − 185.5) |
| Median Cholesterol (IQR) | 341 | 185 | (164.0 − 212.0) | 188 | 187.5 | (162.8 − 216.3) |
| Median Uric acid (IQR) | 329 | 5.9 | (4.9 − 7.1) | 177 | 6.6 | (5.1 − 7.6) |
Assessment of population stratification
We performed a case-control GWAS to identify loci associated with increased risk of small-vessel ischemic stroke in the Han Chinese population using an Affymetrix Axiom CHB array containing 642,832 SNP probes. We initially enrolled 342 SVO stroke and 1,731 controls from a Han Chinese population residing in Taiwan. After kinship analysis and strict quality control filtering, we analyzed 552,090 SNPs (representing 87.9% of array SNPs) for the samples from the GWAS group. Multidimensional scaling analysis (Fig. 1) and results of permutation tests for identity-by-state revealed no differences between the SVO and control groups, providing no evidence for strong population stratification. Quantile–quantile (Q–Q) plots were used to examine P value distributions (Fig. 1), and the lambda value was 1.09. In total, we found three top validated SNPs associated with SVO (P < 1 × 10−4) (Supplementary Table 1). The IBS sharing method implemented in PLINK showed no cryptic family relationships among SVO stroke cases and controls.
GWAS and cross-platform validation
The analysis was first performed with samples from 342 individuals with SVO stroke and 1,731 controls (Fig. 2 and Table 2). Qualified Affymetrix calling (>99%) of clustering in both SVO stroke cases and controls, located in (or within) 50 kb of known genes, was selected for cross-platform validation using a Sequenom MassARRAY or direct sequencing.
Figure 2.
Results of genome-wide association analysis (−log10 P) shown in chromosomal order for 552,090 SNPs tested for association in initial samples from 342 patients with SVO stroke and 1,731 controls. X-axis represents each of the SNPs used in the primary scan. Y-axis represents the −log10 P value of the trend test. Signals in the ATG7 loci are indicated.
Table 2.
SNPs with P values < 1 × 10−5 in the Joint Analysis.
| Chr. | SNP | Position | Gene | Allele format | Risk allele | Stage | control/case | RAF controls | RAF cases | Trend P | OR | 95%CI | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | rs2594966 | 11325276 | ATG7 | GA | GWAS | 1731/342 | 0.6155 | 0.6959 | 7.00E-05 | 1.43 | 1.198 | 1.706 | |
| GA | A | Replication | 1265/188 | 0.6105 | 0.6765 | 1.31E-02 | 1.334 | 1.059 | 1.681 | ||||
| GA | Combined | 2996/530 | 0.6133 | 0.689 | 2.52E-06 | 1.397 | 1.214 | 1.607 | |||||
| 3 | rs2594973 | 11395821 | ATG7 | CG | GWAS | 1731/342 | 0.5948 | 0.6795 | 3.24E-05 | 1.445 | 1.212 | 1.722 | |
| CG | G | Replication | 1265/188 | 0.5918 | 0.6489 | 3.38E-02 | 1.275 | 1.017 | 1.598 | ||||
| CG | Combined | 2996/530 | 0.5935 | 0.6686 | 3.81E-06 | 1.381 | 1.203 | 1.587 | |||||
| 3 | rs4684776 | 11443223 | ATG7 | CT | GWAS | 1731/342 | 0.6096 | 0.6877 | 1.08E-04 | 1.41 | 1.183 | 1.681 | |
| CT | T | Replication | 1265/188 | 0.6067 | 0.6729 | 1.19E-02 | 1.334 | 1.06 | 1.678 | ||||
| CT | Combined | 2996/530 | 0.6084 | 0.6824 | 3.59E-06 | 1.383 | 1.203 | 1.59 | |||||
| 3 | rs4686799 | 186451236 | KNG1 | TC | GWAS | 1731/342 | 0.6598 | 0.7108 | 9.95E-03 | 1.268 | 1.057 | 1.521 | |
| TC | C | Replication | 1265/188 | 0.6454 | 0.7486 | 1.06E-04 | 1.637 | 1.276 | 2.099 | ||||
| TC | Combined | 2996/530 | 0.6537 | 0.7244 | 8.91E-06 | 1.392 | 1.202 | 1.612 | |||||
| 4 | rs78868369 | 126077143 | ANKRD50, FAT4 | TC | GWAS | 1731/342 | 0.08449 | 0.1268 | 4.81E-04 | 1.574 | 1.219 | 2.033 | |
| TC | T | Replication | 1265/188 | 0.08796 | 0.133 | 4.55E-03 | 1.59 | 1.146 | 2.208 | ||||
| TC | Combined | 2996/530 | 0.08595 | 0.129 | 7.61E-06 | 1.575 | 1.288 | 1.927 | |||||
| 6 | rs536348 | 168098215 | LOC441178, C6orf123 | TC | GWAS | 1731/342 | 0.6407 | 0.7132 | 2.52E-04 | 1.395 | 1.165 | 1.67 | |
| TC | C | Replication | 1265/188 | 0.6395 | 0.7234 | 1.26E-03 | 1.474 | 1.16 | 1.874 | ||||
| TC | Combined | 2996/530 | 0.6402 | 0.7169 | 1.12E-06 | 1.423 | 1.232 | 1.643 | |||||
| 8 | rs17201317 | 104096256 | ATP6V1C1, BAALCOS | CT | GWAS | 1731/342 | 0.1096 | 0.1485 | 3.45E-03 | 1.417 | 1.118 | 1.796 | |
| CT | C | Replication | 1265/188 | 0.1093 | 0.1738 | 2.99E-04 | 1.715 | 1.276 | 2.304 | ||||
| CT | Combined | 2996/530 | 0.1095 | 0.1575 | 6.67E-06 | 1.521 | 1.265 | 1.829 | |||||
| 14 | rs11846182 | 80872021 | DIO2-AS1 | CT | GWAS | 1731/342 | 0.602 | 0.6754 | 2.58E-04 | 1.376 | 1.156 | 1.638 | |
| CT | T | Replication | 1265/188 | 0.6135 | 0.6818 | 9.98E-03 | 1.35 | 1.07 | 1.702 | ||||
| CT | Combined | 2996/530 | 0.6069 | 0.6777 | 9.48E-06 | 1.362 | 1.185 | 1.565 | |||||
Chr., chromosome; gene, genes containing the SNP or the closest gene up to 50 kb upstream or downstream of the SNP; RAF controls, risk allele frequency in controls; RAF cases, risk allele frequency in SVO stroke; OR, odds ratio; 95% CI, 95% confidence interval.
Replication of top variants for SVO
In the replication stage, 33 SNPs (Supplementary Table 1) were replicated in an independent cohort of 188 patients with SVO stroke and 1,265 controls (Supplementary Table 2). In a combined analysis of the GWAS and replication cohorts, P values for 8 of the identified SNPs were lower than 10−5 (Table 2).
We found that the SNPs rs2594966 (P = 2.52 × 10−6), rs2594973 (P = 2.52 × 10−6), and rs4684776 (P = 2.52 × 10−6) located at 3p25.3 in ATG7 (encoding Autophagy Related 7). Other 5 SNPs were located at 3q27.3 (rs4686799, P = 8.9 × 10−6) in KNG1 (encoding Kininogen-1), at 4q28.1 (rs78868369, P = 7.6 × 10−6) in ANKRD50 (encoding Ankyrin repeat domain-containing protein 50), at 6q27 (rs536348, P = 1.1 × 10−6) in LOC441178, at 8q22.3 (rs17201317, P = 6.7 × 10−6) in ATP6V1C1 (encoding ATPase H + transporting V1 subunit C1), and at 14q31.1 (rs11846182, P = 9.5 × 10−6) in DIO2-AS1 (encoding DIO2 antisense RNA 1), respectively. These were all replicated in the independent Han Chinese population (Table 1).
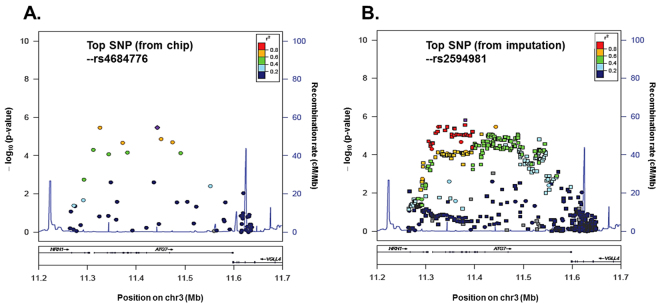
In addition, to enhance the SNP coverage, whole gene region of ATG7 was identified using discovery GWAS dataset (Fig. 3). The imputation demonstrated a strong association within one LD with identified the top SNPs including rs2594966, rs2594973, and rs4684776. The top imputed SNP, rs2594981, was further validated by direct genotyping in discovery GWAS. These data also supported the association between ATG7 and SVO stroke.
Figure 3.
Association plots for the ATG7 locus. Regional association plot for the ATG7 locus on chromosome 3 (A) with gene annotations superimposed. Each SNP is plotted with respect to its chromosomal location (x-axis) and its −log10 P values (left y-axis) for the trend test from the primary GWAS scan at that region of the chromosome. After imputation (B), squares represent imputed SNPs, and circles represent genotyped SNPs. Colors denote the strength of the linkage disequilibrium of the SNPs to ATG7.
Discussion
In this study, we identified novel genetic variants associated with SVO stroke susceptibility. This represents the first report of a GWAS for SVO stroke conducted on a Han Chinese population. Based on two independent Han Chinese groups and without significant difference in other risk factors, several novel loci for SVO stroke were identified and replicated. These findings suggest that SVO is a heritable trait and provide new insights into the genetic basis of SVO stroke.
Although there have been studies showing that autophagy may be involved in stroke19,20, the genetic association between autophagy-related genes and SVO stroke was never reported. In the present study, we identified five SVO stroke-associated SNPs—rs2594966, rs2594973, rs4684776, rs34843621, and rs12637318—in the same linkage disequilibrium (LD) of chromosome region 3p25.3. The SNPs are located in the ATG7 gene (Supplementary Fig. 1), which encodes an ubiquitin-activating E1-like enzyme critical for autophagy21. This newly identified genetic link may reveal novel molecular insights into the pathogenesis of SVO stroke. A recent study revealed that deletion of ATG7 was strongly protective against neuronal damage in the brain22. A megakaryocyte- and platelet-specific deletion of ATG7 caused modest defects in platelet aggregation and granule cargo packaging in a mouse model23, suggesting that ATG7 may be a contributing factor for thrombosis. Moreover, ATG7-dependent autophagy has been related to hepatic lipid metabolism24, which has in turn been found to be associated with stroke25,26. It is therefore possible that enhancement of SVO stroke by alteration of the lipid profile may be mediated by ATG7-dependent autophagy. The potential role of ATG7 in occlusion and SVO stroke pathogenesis will require further investigation. Another SVO-associated SNP, rs4686799, was identified within an intron of the KNG1 gene. KNG1 was identified from a GWAS for plasma factor XI levels as a genetic determinant of activated partial thromboplastin time27. Elevated plasma levels of FXI have been correlated with venous thrombosis and ischemic stroke; therefore, KNG1 could be involved in the pathogenesis of SVO stroke via regulation of plasma FXI levels.
A meta-analysis showed that SNP 1425 G/A in PRKCH was associated with ischemic stroke, particularly lacunar infarction, in Chinese and Japanese populations28 and ALDH2 29,30 and FOXF2 29,30 with small vessel disease and white matter hyperintensity in Caucasians, respectively. The disparity in risk genes between Asian and Caucasian populations could be due to the inconsistency in diagnostic and stroke subtyping criteria among centers and nations. The current study was conducted in a single healthcare system using similar diagnostic tools, and the SVO subtype was classified by single doctor.
A major limitation of the current study could be the sample size. However, because SVO was reported to be common in patients with intracranial artery stenosis31, the current study used strict recruitment criteria to exclude cases with extracranial and/or intracranial artery stenosis; hence, the sample size was reduced. An additional independent larger group, such as the International Stroke Genetics Consortium, CHARGE, or METASTROKE, would strengthen our findings with detailed stroke subtyping. Further genome-wide association tests assessing whether the potential susceptibility loci have genome-wide significance in different populations will elucidate the genetic contribution in SVO stroke pathogenesis.
In this study, we provide the first genome-wide evidence showing in two independent cohorts, thirty-three SNPs located in novel genetic loci that were found to be associated with SVO stroke in a Han Chinese population. The novel risk loci for SVO stroke contained genes, especially for ATG7, that have been implicated in autophagy and thrombosis, which may provide insights into future studies to identify the therapeutic targets for SVO stroke.
Electronic supplementary material
Acknowledgements
We thank all affected individuals and their families who devoted their time and effort to participate in this study. We thank doctors in the three branch hospitals of Chang Gung Healthcare System, Taiwan, for their contributions in recruiting stroke patients. We gratefully acknowledge the members of the Translational Resource Center (TRC) (NSC102-2325-B-001-023) of National Research Program for Biopharmaceuticals (NRPB) and the National Center for Genome Medicine (NCGM) (NSC102-2319-B-001-001) of National Core Facility Program for Biotechnology (NCFPB), National Science Council, at Academia Sinica for their support in subject recruitment, genotyping, and statistical analysis. This study was supported by the Academia Sinica Genomic Medicine Multicenter Study, Taiwan (40-05-GMM) and Chang Gung Memorial Hospital (CMRPG35072, 35073, 39082, 39083, CMRPG3B0611, CMPRG3A0352, CMRPG3B0111 and BMRP 274). The funders had no role in study design, data collection or analysis, the decision to publish or preparation of the manuscript.
Author Contributions
T.-H.L., Y.-T C., and J.-Y.W. are the principal investigators who conceived and obtained funding for this project. T.-M.K., C.-H.C. and J.-Y.W. organised and supervised the GWAS and replication genotyping pipeline and devised the overall analysis plan. T.-M.K. wrote the first draft of the manuscript. T.-H.L., T.-M.K., Y.-T.C., and J.-Y.W. contributed to the writing of the manuscript. Replication and validation data were provided by Y.-T.C and L.-S.L., T.-H.L., Y.-J.C., C.-H.C., K.-L.H., T.-Y.C., J.-D.L., K.-C.C., J.-T.Y., M.-S.W., C.-Y.W., T.-C.C., S.-Y.C., M.-T.L., Y.-T C., and J.-Y.W. coordinated and contributed subject and database.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Tsong-Hai Lee and Tai-Ming Ko contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14355-3.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yuan-Tsong Chen, Email: chen0010@ibms.sinica.edu.tw.
Jer-Yuarn Wu, Email: jywu@ibms.sinica.edu.tw.
References
- 1.Feigin VL, et al. Global and regional burden of stroke during 1990-2010: findings from the Global Burden of Disease Study 2010. Lancet. 2014;383:245–54. doi: 10.1016/S0140-6736(13)61953-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feigin VL, Lawes CM, Bennett DA, Barker-Collo SL, Parag V. Worldwide stroke incidence and early case fatality reported in 56 population-based studies: a systematic review. Lancet Neurol. 2009;8:355–69. doi: 10.1016/S1474-4422(09)70025-0. [DOI] [PubMed] [Google Scholar]
- 3.White H, et al. Ischemic stroke subtype incidence among whites, blacks, and Hispanics: the Northern Manhattan Study. Circulation. 2005;111:1327–31. doi: 10.1161/01.CIR.0000157736.19739.D0. [DOI] [PubMed] [Google Scholar]
- 4.Flossmann E, Schulz UG, Rothwell PM. Systematic review of methods and results of studies of the genetic epidemiology of ischemic stroke. Stroke. 2004;35:212–27. doi: 10.1161/01.STR.0000107187.84390.AA. [DOI] [PubMed] [Google Scholar]
- 5.Markus HS. Unravelling the genetics of ischaemic stroke. PLoS Med. 2010;7:e1000225. doi: 10.1371/journal.pmed.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jerrard-Dunne P, Cloud G, Hassan A, Markus HS. Evaluating the genetic component of ischemic stroke subtypes: a family history study. Stroke. 2003;34:1364–9. doi: 10.1161/01.STR.0000069723.17984.FD. [DOI] [PubMed] [Google Scholar]
- 7.Kubo M, et al. A nonsynonymous SNP in PRKCH (protein kinase C eta) increases the risk of cerebral infarction. Nat Genet. 2007;39:212–7. doi: 10.1038/ng1945. [DOI] [PubMed] [Google Scholar]
- 8.Traylor M, et al. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11:951–62. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee TH, et al. Identification of PTCSC3 as a Novel Locus for Large-Vessel Ischemic Stroke: A Genome-Wide Association Study. J Am Heart Assoc. 2016;4:e003003. doi: 10.1161/JAHA.115.003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holliday EG, et al. Common variants at 6p21.1 are associated with large artery atherosclerotic stroke. Nat Genet. 2012;44:1147–51. doi: 10.1038/ng.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.International Stroke Genetics, C. et al. Genome-wide association study identifies a variant in HDAC9 associated with large vessel ischemic stroke. Nat Genet44, 328-33 (2012). [DOI] [PMC free article] [PubMed]
- 12.Adams HP, Jr., et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24:35–41. doi: 10.1161/01.STR.24.1.35. [DOI] [PubMed] [Google Scholar]
- 13.Pan WH, et al. Han Chinese cell and genome bank in Taiwan: purpose, design and ethical considerations. Hum Hered. 2006;61:27–30. doi: 10.1159/000091834. [DOI] [PubMed] [Google Scholar]
- 14.Lee YC, et al. Two new susceptibility loci for Kawasaki disease identified through genome-wide association analysis. Nat Genet. 2012;44:522–5. doi: 10.1038/ng.2227. [DOI] [PubMed] [Google Scholar]
- 15.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–70. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 18.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–9. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian F, et al. In vivo imaging of autophagy in a mouse stroke model. Autophagy. 2010;6:1107–14. doi: 10.4161/auto.6.8.13427. [DOI] [PubMed] [Google Scholar]
- 20.Puyal J, Clarke PG. Targeting autophagy to prevent neonatal stroke damage. Autophagy. 2009;5:1060–1. doi: 10.4161/auto.5.7.9728. [DOI] [PubMed] [Google Scholar]
- 21.Hong SB, et al. Insights into noncanonical E1 enzyme activation from the structure of autophagic E1 Atg7 with Atg8. Nat Struct Mol Biol. 2011;18:1323–30. doi: 10.1038/nsmb.2165. [DOI] [PubMed] [Google Scholar]
- 22.Xie C, et al. Neuroprotection by selective neuronal deletion of Atg7 in neonatal brain injury. Autophagy. 2016;12:410–23. doi: 10.1080/15548627.2015.1132134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouseph MM, et al. Autophagy is induced upon platelet activation and is essential for hemostasis and thrombosis. Blood. 2015;126:1224–33. doi: 10.1182/blood-2014-09-598722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao, Y. et al. MAPK1/3 regulate hepatic lipid metabolism via ATG7-dependent autophagy. Autophagy, 1-2 (2016). [DOI] [PMC free article] [PubMed]
- 25.Adibhatla RM, Dempsy R, Hatcher JF. Integration of cytokine biology and lipid metabolism in stroke. Front Biosci. 2008;13:1250–70. doi: 10.2741/2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, et al. Proprotein convertase subtilisin/kexin type 9 (PCSK9) in lipid metabolism, atherosclerosis and ischemic stroke. Int J Neurosci. 2016;126:675–80. doi: 10.3109/00207454.2015.1057636. [DOI] [PubMed] [Google Scholar]
- 27.Sabater-Lleal M, et al. A genome-wide association study identifies KNG1 as a genetic determinant of plasma factor XI Level and activated partial thromboplastin time. Arterioscler Thromb Vasc Biol. 2012;32:2008–16. doi: 10.1161/ATVBAHA.112.248492. [DOI] [PubMed] [Google Scholar]
- 28.Li J, Luo M, Xu X, Sheng W. Association between 1425G/A SNP in PRKCH and ischemic stroke among Chinese and Japanese populations: a meta-analysis including 3686 cases and 4589 controls. Neurosci Lett. 2012;506:55–8. doi: 10.1016/j.neulet.2011.10.047. [DOI] [PubMed] [Google Scholar]
- 29.NINDS Stroke Genetics Network (SiGN) & International Stroke Genetics Consortium (ISGC) Loci associated with ischaemic stroke and its subtypes (SiGN): a genome-wide association study. Lancet Neurol. 2016;15:174–184. doi: 10.1016/S1474-4422(15)00338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neurology Working Group of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium, Stroke Genetics Network (SiGN) & International Stroke Genetics Consortium (ISGC) Identification of additional risk loci for stroke and small vessel disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2016;15:695–707. doi: 10.1016/S1474-4422(16)00102-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon HM, et al. Frequency, Risk Factors, and Outcome of Coexistent Small Vessel Disease and Intracranial Arterial Stenosis: Results From the Stenting and Aggressive Medical Management for Preventing Recurrent Stroke in Intracranial Stenosis (SAMMPRIS) Trial. JAMA Neurol. 2016;73:36–42. doi: 10.1001/jamaneurol.2015.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available in the International Stroke Genetics Consortium repository, http://www.cerebrovascularportal.org/. The name of the dataset is “SVO-Han-population Taiwan-NCGM”.