Abstract
Background:
Although aberrant expression of cytokines and small molecules (analytes) is well documented in acute myeloid leukaemia (AML), their co-expression patterns are not yet identified. In addition, plasma baselines for some analytes that are biomarkers for other cancers have not been previously reported in AML.
Methods:
We used multiplex array technology to simultaneously detect and quantify 32 plasma analyte (22 reported analytes and 10 novel analytes) levels in 38 patients.
Results:
In our study, 16 analytes are found to be significantly deregulated (13 higher, 3 lower, Mann–Whitney U-test, P-value <0.005), where 5 of them have never been reported before in AML. We predicted a seven-analyte-containing multiplex panel for diagnosis of AML and, among them, MIF could be a possible therapeutic target. In addition, we observed that circulating analytes show five co-expression signatures.
Conclusions:
Circulating analyte expression in AML significantly differs from normal, and follow distinct expression patterns.
Keywords: cytokines, chemokines, growth factors, plasma baseline, expression patterns, acute myeloid leukaemia
Acute myeloid leukaemia (AML) is a heterogeneous disease with complex interplay of genetic and/or cytogenetic alterations in haematopoietic progenitors, together with aberrant cytokines and small molecule (analyte) expression in the tumour microenvironment, that contribute to AML pathogenesis. Circulating analytes are a heterogeneous group of soluble small polypeptides or glycoproteins that mediate crosstalk between normal and neoplastic cells. Their roles in the pathophysiology at different stages of carcinogenesis are increasingly recognised (Hanahan and Weinberg, 2000). Although deregulated expression of circulating analytes in AML is well documented (Van Etten, 2007; Seruga et al, 2008; Tsimberidou et al, 2008; Kornblau et al, 2010; Brenner et al, 2017), considerable baseline variability of inter- and intra-assay plasma analytes levels are reported in AML (Kornblau et al, 2010; Kupsa et al, 2014). In addition, plasma baselines for some analytes that are biomarkers for other cancers have not been previously reported in AML. A comprehensive profiling of plasma analytes might give us a greater insight of baseline variability and co-expression signature, potentially leading to the identification of novel diagnostic biomarkers and therapeutic targets in AML.
In this study, we aim to validate the baseline of multiple analytes and report baseline of novel analytes in AML. We also aim to examine the differential expression and categorise co-expression patterns of analytes, followed by exploratory analysis and validation using The Cancer Genome Atlas (TCGA) data (Cancer Genome Atlas Research Network, 2013).
Materials and methods
Peripheral blood samples were collected and fresh plasma was separated from 38 individuals: 19 AML cases and 19 healthy controls (Table 1 and Supplementary Table S1). Cytogenetic studies were performed as a routine diagnostic test for all AML cases. The diagnostic leftover samples were used for further study. Based on Malaysian guideline on the use of human biological specimen for research, no patient consent was required for this study. The study was approved by the Medical Ethics Committee (NMRR-16-1384-31900 S1 R0). Two sets of analytes were selected for this study (Figure 1A): (1) 22 reported analytes (baselines have been previously reported in AML) and (2) 10 novel analytes (baselines have not been previously reported in AML). Reported analytes are AFP, CA 15-3, Leptin, IL-6, sFasL, CEA, CA 125, IL-8, HGF, sFas, TNFα, Prolactin, SCF, OPN, FGF2, bHCG, TGFα, VEGF, Galectin-3, MPO, IGFBP3, and Ferritin. Novel analytes are tPSA, CA 19-9, MIF, TRAIL, CYFRA 21-1, HE4, Cathepsin D, FAPa, MIA, and SHBG. Using multiplex array technique, all 32 analytes (Supplementary Table S2) were simultaneously detected and quantified from 38 plasma samples (details of the methods are shown in Supplementary File).
Table 1. Demographics of 19 AML cases and 19 healthy controls.
| Category | Number |
|---|---|
| No. of AML cases | 19 |
| Sex (F/M) | 7/12 |
| Median age (years) | 38±15.24 |
| Race (Malay/Chinese) | 13/6 |
| Cytogenetics | |
| T(8;21) | 3 |
| T(15;17) | 1 |
| Inversion 16 | 2 |
| Trisomy 11 | 1 |
| Trisomy 21 | 1 |
| Normal | 11 |
| No. of healthy controls | 19 |
| Sex (F/M) | 8/11 |
| Median age (years) | 47±10.81 |
| Race (Malay/Chinese) | 3/16 |
Abbreviations: AML=acute myeloid leukaemia; F=female; M=male.
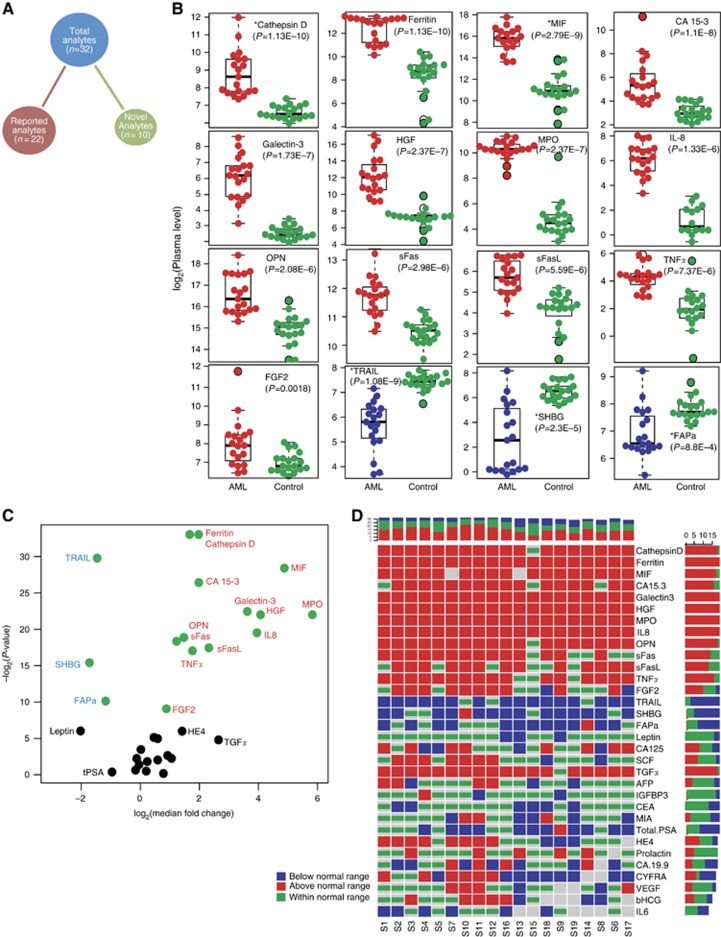
Figure 1.
Deregulated cytokines and small molecules in AML patients. (A) Distribution of the analytes. The total analytes (n=32) are divided into two sets: reported analytes (baselines reported previously in AML) and novel analytes (baselines have not been reported previously in AML). (B) Beeswarm plots of the 16 significantly deregulated (P<0.005, Mann–Whitney U-test) analytes in AML cases (red or blue colour) compared with healthy controls (green colour). Plots for the upregulated (red colour) and downregulated (blue colour) analytes are ordered based on the calculated P-value. Novel analytes are denoted by an asterisk (*). The y-axis shows plasma level in log2 scale. Nonsignificant analytes are shown in Supplementary Figure S1. (C) Volcano plot showing the relationship between median fold change (x-axis) and P-value of the Mann–Whitney U-test (y-axis). Positive values on the x-axis show upregulation, and negative values show downregulation. Green circles denote significantly deregulated (P<0.005) analyets. (D) Patient-specific aberrant expression of analytes. Columns denote the 19 studied AML patients and the 32 interrogated analytes are represented by rows. Horizontal and vertical sidebars summarise the distributions per analyte and patient, respectively. Red colour denotes upregulation, blue downregulation, and green no significant deregulation (within normal/healthy control range). Grey boxes denote missing values.
Results
The plasma expression level of the selected 32 analytes for both healthy controls and AML cases was curated from available peer-reviewed literature (Supplementary Tables S3 and S4). Comparing our study with previously published data, 17 out of 22 reported analytes show similar expression range. The expression range of three analytes (FGF2, MPO, and sFas) are observed higher, whereas two analytes (SCF and sFasL) are observed lower in our study (Supplementary Table S4). All the novel analytes were within the detectable range in AML plasma. In total, 16 analytes are found to be significantly deregulated in AML compared with healthy control (Mann–Whitney U-test, P-value <0.005, Figure 1B) and 5 of them (Cathepsin D, MIF, TRAIL, SHBG, and FAPa) are novel analytes. In all, 13 analytes are upregulated and 3 are downregulated. The remaining 16 analytes are found to be nonsignificant (Supplementary Figure S1). Although some of the nonsignificant analytes (like TGFα, HE4, Leptin, and Total PSA) pointed large median fold changes, they are not significant probably because of inconsistent expression. Fold change and corresponding P-values are shown as volcano plot in Figure 1C. We next investigated the consistency of the expression levels across the AML cohort (Figure 1D). The first 9 analytes in Figure 1D are above the normal expression range for >80% of AML patients. Based on consistent expression, fold change, and P-value, seven analytes (Cathepsin D, Ferritin, MIF, Galectin-3, HGF, MPO, and IL8) are predicted to be a multiplex panel for AML diagnosis.
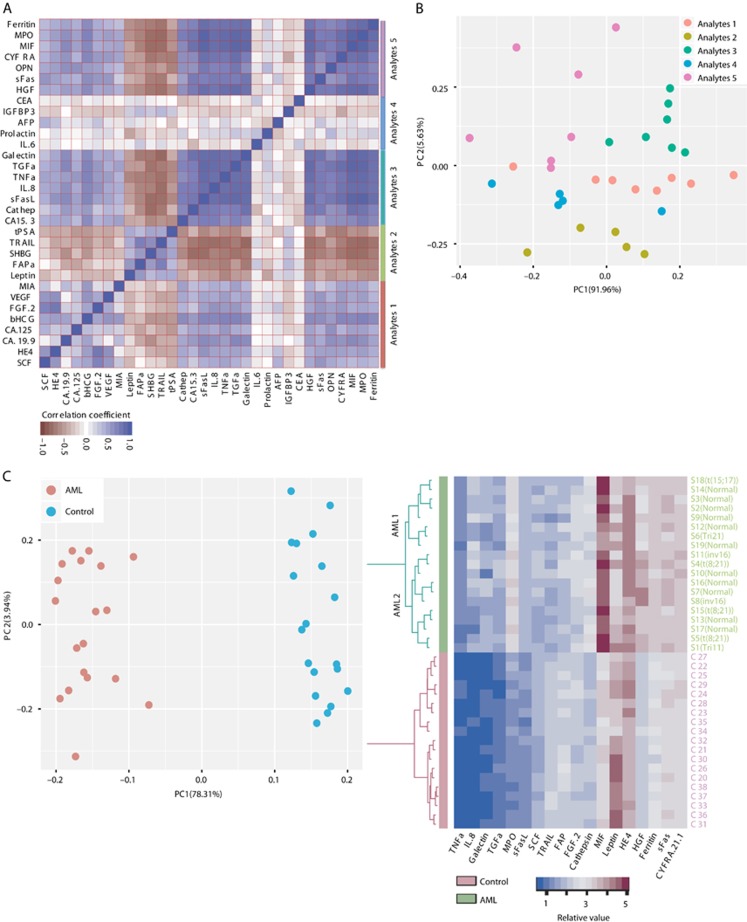
To determine co-expression patterns, we calculated pairwise Pearson’s correlation coefficients between analytes in AML and observed five distinct groups (analytes 1–5, Figure 2A). Although analytes in the second group are uniquely co-expressed, the underlying biology behind the observed co-expression groups is yet to be explored. We use principal component analysis (PCA) for 32 analytes and track the co-expressed groups and 5 groups are plotted separately (Figure 2B). Then, we check the distribution of each group’s aggregated median across the samples and analyte 3 could significantly differentiate AML from control (Supplementary Figure S2A). PCA is also applied to 38 samples (Figure 2C); AML and control are distinctive and two groups of AML are visible by PC2. Using ‘Bimodality Index’ (Wang et al, 2009), 18 analytes are identified as informative (Supplementary Table S5, BI >1.4) for sample clustering and 2 distinct groups (AML1 and AML2) are also visible in AML patients by unsupervised hierarchical clustering dendrogram (Figure 2D). These data support the previous reports (Bruserud et al, 2007; Kornblau et al, 2009, 2010; Brenner et al, 2017) that demonstrated the circulating analytes expression-based AML patient clustering. Both MPO and HGF are differentially expressed between AML1 and AML2 (Supplementary Figure S2B and C). Interestingly, 5 out of 6 patients with favourable karyotypes are clustered in AML2. The CA-125 and TGFα are deregulated (up and down, respectively) in the favourable group compared with other karyotypes (nonsignificant, Supplementary Figure S2D and E). The TGFα has also exhibited similar pattern in TCGA (Cancer Genome Atlas Research Network, 2013) (P<0.005, Supplementary Figure S2F) that demonstrates prognostic potential.
Figure 2.
Cytokine and small molecule expression-based clustering. (A) Heatmap of pairwise Pearson’s correlation coefficients between analytes. Five distinct groups of analytes (Analytes 1–5) are identified. Analyte 2 is uniquely correlated to each other only, whereas Analyte 4 shows opposite trend. (B) Principal component analysis (PCA) plot of the first 2 PCs of the 32 analytes. The five groups and their colours are as defined in (A). (C) The PCA plot of the first 2 PCs of the 38 samples (AML=19, control=19). Components were calculated from the expression profiles of all 32 analytes. The AML and control groups are clearly separated. Two broad groups of AML could be inferred based on the PC2 coordinates. (D) Heatmap displaying the expression levels of 18 analytes that show bimodal distribution patterns (Bimodality Index >1.4). Unsupervised hierarchical clustering performed on patients (rows) is displayed as a dendrogram and suggests two subgroups of AML (AML1 and AML2). Five of the six AML patients with favourable karyotypes belong to the AML2 group.
The MIF is a novel analyte, known as a proinflammatory cytokine, and is found 30-fold upregulated in AML plasma compared with control (Figure 1B). Other cancer types, including breast, prostate, and gastric cancer, also reported elevated MIF level that prevents apoptosis and promotes tumour cells survival by directly activating the phosphoinositide-3-kinase (PI3K)/Akt pathway (Lue et al, 2007). The same pathway is also constitutively activated in 50% of AML blasts (Park et al, 2010). Another novel analyte is TRAIL, a potent inducer of apoptosis (Henry and Martin, 2017), found 2.75-fold downregulated in AML plasma. A negative correlation is noticed between MIF and TRAIL in AML in both our study and TCGA (Supplementary Figure S3A and B, respectively), but not in controls (Supplementary Figure S3C and D). We speculate that MIF is promoting survival of AML blasts using PI3K/Akt pathway through the anti-apoptotic mechanism.
Next, we sought to explore the analyte corresponding genes in TCGA AML data (Cancer Genome Atlas Research Network, 2013). We extracted promoter methylation status of the targeted 32 analyte corresponding genes from 194 AML cases and 30 healthy controls and identified 3 clusters of analytes based on promoter CpG site methylation (Supplementary Figure S4A). Interestingly, the promoters of FASLG and LGALS3 genes are highly methylated in the TCGA data set, and these analytes are abnormally highly expressed in our cohort, suggesting that perhaps feedback loops in the cells repress these genes when overexpressed. Both MPO and HGF show distinct promoter methylation patterns between cases and controls where MPO is a known prognostic factor for AML (Matsuo et al, 2003). We checked mRNA expression using mRNAseq and found three clusters but noticed some inconsistencies (Supplementary Figure S4B) compared with methylation clusters. In addition, we noticed significant differential expression for MPO, HGF, and LGALS3 (log2 fold change 7.26, 5.60, and 3.50, respectively) compared with healthy controls in TCGA mRNAseq data.
Discussion
The altered baseline of circulating analytes and their receptors has been found in numerous studies of patients with various types of cancer, including AML, both at primary and metastases stages compared with healthy people (Seruga et al, 2008). Analytes secreted in the bone marrow microenvironment form a complex functional network and play important roles in modulating cell survival, proliferation, differentiation, and the immune response (Welner et al, 2015). A comprehensive profiling of secreted proteins is important to understand their function during tumour development and progression and to identify common and actionable extrinsic pathways independent of mutation status in AML that might lead to developing targeted therapies with clinical efficacy (Carey et al, 2017).
In this study, we detected and quantified 32 different analytes in plasma using highly sensitive Bioplex multiplex technology and documented comparative plasma baseline variability between different studies in AML. This is the first published report of baseline for one-third of these analytes in AML.
We curated baseline from available peer-reviewed literature for both AML cases and healthy controls. We noticed that the baseline was more consistent with studies that used the Bioplex method that has been reported as having better sensitivity (Kornblau et al, 2010) as compared with the studies that used other methodologies. We observed that baseline expression ranges are higher for three analytes (FGF2, MPO, and sFas) and lower for two analytes (SCF and sFasL) compared with previously published reports.
In our study, 16 analytes are found to be significantly deregulated (13 higher and 3 lower), whereas other 16 analytes are found to be nonsignificant (Mann–Whitney U-test, P-value <0.005). We used higher cutoff for P-value to avoid false positive error. We also performed power analysis and found that almost 80% of the analytes in the significant group have the power of 1 (almost 100% probability of getting a significant P-value despite the sample size).
We reported for the first time plasma baselines for 10 novel analytes in AML, where 5 of them (Cathepsin D, MIF, TRAIL, SHBG, and FAPa) were statistically significant with high median fold changes.
We predicted a seven-analyte-containing multiplex panel (Cathepsin D, Ferritin, MIF, Galectin-3, HGF, MPO, and IL8) for diagnosis of AML, where they are almost always upregulated, regardless of the heterogeneity of the disease. Further validation is required for this panel before clinical use.
Interestingly, we found a negative correlation between MIF and TRAIL in AML that was also supported by TCGA data. We speculated that MIF is promoting survival of leukaemic blasts using PI3K/Akt pathway through attenuation of apoptosis. Based on existing literature (O'Reilly et al, 2016), we think MIF could be a possible therapeutic target in AML.
In addition, we observed that circulating analytes show co-expression and form five distinct groups in PCA. These co-expressed groups were differentially expressed across the AML cohort. We also clustered AML patients based on 18 informative analyte expression profile and noticed distinct groups. Previously, we reported that systematic aggregation of methylation and mutation profile improve AML subclassification (Islam et al, 2017). Our current observation lead us to speculate that circulating analyte expression signatures have the potential to be used for subclassification of AML, complementing cytogenetic, genetic, and epigenetic information.
One of the major limitation of this study is the small sample size, although we showed that the sample size is not affecting some of our major and notable findings like significant and consistent deregulation. In addition, some studies reported a notable baseline difference between plasma, serum, and bone marrow aspirate (Stroncek et al, 2005).
In conclusion, our current study demonstrates that circulating analyte expression in AML significantly differs from normal. We validated the baseline of multiple reported-analyte and reported novel-analyte levels. We also identified the differential analyte expression and categorise the co-expression patterns in AML. In future study, different sources of samples (serum and marrow aspirate), a larger sample size, and more elaborate analyte panels could be used to extend our current findings.
Acknowledgments
We thank the Director General of Health, Malaysia, for the permission to publish this study. This study was supported by the University of Malaya HIR Research Grant: HIR H20001-E000025 and Ministry of Health Grant: NMRR-16-1384-31900.
Author contributions
MI designed the study, conducted the experiments, analysed the data, and wrote the manuscript. YA analysed epigenomics data. EHM, EE, NRK, SHZ, YMY helped for sample collection. ZM and ZZ supervised the project. All the authors reviewed the manuscript.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Brenner AK, Tvedt THA, Nepstad I, Rye KP, Hagen KM, Reikvam H, Bruserud Ø (2017) Patients with acute myeloid leukemia can be subclassified based on the constitutive cytokine release of the leukemic cells; the possible clinical relevance and the importance of cellular iron metabolism. Expert Opin Ther Targets 21(4): 357–369. [DOI] [PubMed] [Google Scholar]
- Bruserud Ø, Ryningen A, Olsnes AM, Stordrange L, Øyan AM, Kalland KH, Gjertsen BT (2007) Subclassification of patients with acute myelogenous leukemia based on chemokine responsiveness and constitutive chemokine release by their leukemic cells. Haematologica 92(3): 332–341. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2013) Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368(22): 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey A, Edwards DK, Eide CA, Newell L, Traer E, Medeiros BC, Pollyea DA, Deininger MW, Collins RH, Tyner JW (2017) Identification of interleukin-1 by functional screening as a key mediator of cellular expansion and disease progression in acute myeloid Leukemia. Cell Rep 18(13): 3204–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100(1): 57–70. [DOI] [PubMed] [Google Scholar]
- Henry CM, Martin SJ (2017) Caspase-8 acts in a non-enzymatic role as a scaffold for assembly of a pro-inflammatory ‘FADDosome’ complex upon TRAIL stimulation. Mol Cell 65(4): 715–729.e5. [DOI] [PubMed] [Google Scholar]
- Islam M, Mohamed Z, Assenov Y (2017) Differential analysis of genetic, epigenetic, and cytogenetic abnormalities in AML. Int J Genomics 2017: 2913648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblau SM, McCue D, Singh N, Chen W, Estrov Z, Coombes KR (2010) Recurrent expression signatures of cytokines and chemokines are present and are independently prognostic in acute myelogenous leukemia and myelodysplasia. Blood 116(20): 4251–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblau SM, Tibes R, Qiu YH, Chen W, Kantarjian HM, Andreeff M, Coombes KR, Mills GB (2009) Functional proteomic profiling of AML predicts response and survival. Blood 113(1): 154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupsa T, Vasatova M, Karesova I, Zak P, Horacek J (2014) Baseline serum levels of multiple cytokines and adhesion molecules in patients with acute myeloid leukemia: results of a pivotal trial. Exp Oncol 36(4): 252–257. [PubMed] [Google Scholar]
- Lue H, Thiele M, Franz J, Dahl E, Speckgens S, Leng L, Fingerle-Rowson G, Bucala R, Lüscher B, Bernhagen J (2007) Macrophage migration inhibitory factor (MIF) promotes cell survival by activation of the Akt pathway and role for CSN5/JAB1 in the control of autocrine MIF activity. Oncogene 26(35): 5046–5059. [DOI] [PubMed] [Google Scholar]
- Matsuo T, Kuriyama K, Miyazaki Y, Yoshida S, Tomonaga M, Emi N, Kobayashi T, Miyawaki S, Matsushima T, Shinagawa K (2003) The percentage of myeloperoxidase-positive blast cells is a strong independent prognostic factor in acute myeloid leukemia, even in the patients with normal karyotype. Leukemia 17(8): 1538–1543. [DOI] [PubMed] [Google Scholar]
- O'Reilly C, Doroudian M, Mawhinney L, Donnelly SC (2016) Targeting MIF in cancer: therapeutic strategies, current developments, and future opportunities. Med Res Rev 36(3): 440–460. [DOI] [PubMed] [Google Scholar]
- Park S, Chapuis N, Tamburini J, Bardet V, Cornillet-Lefebvre P, Willems L, Green A, Mayeux P, Lacombe C, Bouscary D (2010) Role of the PI3K/AKT and mTOR signaling pathways in acute myeloid leukemia. Haematologica 95(5): 819–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seruga B, Zhang H, Bernstein LJ, Tannock IF (2008) Cytokines and their relationship to the symptoms and outcome of cancer. Nat Rev Cancer 8(11): 887–899. [DOI] [PubMed] [Google Scholar]
- Stroncek D, Slezak S, Khuu H, Basil C, Tisdale J, Leitman SF, Marincola FM, Panelli MC (2005) Proteomic signature of myeloproliferation and neutrophilia: analysis of serum and plasma from healthy subjects given granulocyte colony-stimulating factor. Exp Hematol 33(10): 1109–1117. [DOI] [PubMed] [Google Scholar]
- Tsimberidou AM, Estey E, Wen S, Pierce S, Kantarjian H, Albitar M, Kurzrock R (2008) The prognostic significance of cytokine levels in newly diagnosed acute myeloid leukemia and high–risk myelodysplastic syndromes. Cancer 113(7): 1605–1613. [DOI] [PubMed] [Google Scholar]
- Van Etten RA (2007) Aberrant cytokine signaling in leukemia. Oncogene 26(47): 6738–6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wen S, Symmans WF, Pusztai L, Coombes KR (2009) The bimodality index: a criterion for discovering and ranking bimodal signatures from cancer gene expression profiling data. Cancer Inform 7: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welner RS, Amabile G, Bararia D, Czibere A, Yang H, Zhang H, Pontes LLDF, Ye M, Levantini E, Di Ruscio A (2015) Treatment of chronic myelogenous leukemia by blocking cytokine alterations found in normal stem and progenitor cells. Cancer Cell 27(5): 671–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.