Abstract
Background:
Resistance to chemotherapy is a major obstacle in the treatment of human hepatocellular carcinoma (HCC). Despite playing an important role in chemoprevention, nuclear factor erythroid 2-related factor 2 (NRF2) also contributes to chemo- and radio-resistance. The current study focusses on camptothecin as a novel NRF2 inhibitor to sensitise HCC to chemotherapy.
Methods:
The expression and transcriptional activity of NRF2 in human HCC biopsies and camptothecin-treated culture cells were determined using immunostaining, western blot, reverse-transcription quantitative real-time PCR (RT–qPCR) and luciferase reporter assay. The effect of camptothecin on chemosensitivity of cancer cells was assessed in vitro and in xenografts.
Results:
The expression and transcriptional activity of NRF2 were substantially elevated in HCC biopsies compared with corresponding adjacent tissues, and positively correlated with serum α-fetoprotein, a clinical indicator of pathological progression. In searching chemicals targeting NRF2 for chemotherapy, we discovered that camptothecin is a potent NRF2 inhibitor. Camptothecin markedly suppressed NRF2 expression and transcriptional activity in different types of cancer cells including HepG2, SMMC-7721 and A549. As a result, camptothecin sensitised these cells to chemotherapeutic drugs in vitro and in xenografts.
Conclusions:
Camptothecin is a novel NRF2 inhibitor that may be repurposed in combination with other chemotherapeutics to enhance their efficacy in treating high NRF2-expressing cancers.
Keywords: camptothecin, NRF2, hepatocellular carcinoma, ARE, chemosensitivity
Hepatocellular carcinoma (HCC) is a common human cancer and one of the leading causes of cancer mortality worldwide (El-Serag, 2012; Attwa and El-Etreby, 2015). Most cases of HCC are secondary to viral hepatitis infection or cirrhosis resulting from chronic alcoholism. The extremely poor prognosis of HCC is mainly attributed to the lack of effective therapeutic options (Ferenci et al, 2010). Although surgical resection and liver transplantation can be effective for early detected HCC, they only benefit ∼25% of patients because of scarce matching liver donors and the fact that HCC is often diagnosed after metastasis occurs (Llovet et al, 2003). Only a small subset of HCC patients benefit from radiotherapy, interventional radiotherapy, and targeted therapy, whereas patients with metastasis have a 5-year survival rate of <5% (Bertino et al, 2014). Because most HCC patients are diagnosed at advanced stages, chemotherapeutic treatment is the most commonly used strategy of intervention, and realistically one of the few options available. However, chemoresistance can be a major challenge in advanced-stage HCC. The exact nature of chemoresistance in HCC is not completely understood. Presently, efforts are being directed to discovering new therapeutic targets within HCC cells, and toward developing new chemotherapeutic drugs that can circumvent chemoresistance.
Many cancer chemotherapeutic agents cause oxidative stress as part of their mode of action. Therefore, cancer cells may develop chemoresistance through augmenting their antioxidant capacity. Nuclear factor-E2-related factor 2 (NRF2) is a CNC-bZIP transcription factor that is well established as a master regulator of cytoprotection against oxidative stress. It is negatively regulated by the redox sensor protein, Kelch-like ECH associated protein (KEAP1) (Zhang et al, 2004). In parallel with repression by KEAP1, NRF2 is also negatively controlled via the β-transducin repeat-containing protein (β-TrCP)/glycogen synthase kinase-3β (GSK3β) nexus, and thus NRF2 can be partially suppressed by AKT inhibitors (Rada et al, 2011; Chowdhry et al, 2013; Tebay et al, 2015). The NRF2 recognises the antioxidant response elements (AREs) located in the promoters of many antioxidant and metabolising enzymes (Maher and Yamamoto, 2010; Pi et al, 2010). It is conceivable that once cancer cells are subject to drug-induced oxidative stress, NRF2 can be activated to diminish the effects of chemotherapy by means of inducing a battery of antioxidant and metabolising enzymes and transporters. Indeed, recent studies showed that persistently high level and activity of NRF2–ARE can promote cancer formation and contribute to chemoresistance (Lau et al, 2008; Hayes and McMahon, 2009; Kensler and Wakabayashi, 2010; Ren et al, 2011). In human HCC, elevated expression of NRF2 is positively correlated with metastasis (Wang et al, 2016). Studies have revealed that many NRF2 target genes, including drug-metabolising enzymes and transporters, play crucial roles in determining drug response and resistance (Ren et al, 2011). The NRF2 can lower intracellular drug accumulation by inducing multidrug resistance proteins (MRPs) (Maher et al, 2007; Chen et al, 2012). Multiple members of MRPs have been found to be elevated in KEAP1-mutated cancer cells (Mahaffey et al, 2009; Sasaki et al, 2012). Because of multidrug resistance, the efficacy of chemotherapy of HCC can be <30% (Van Thiel et al, 2002). Given the significant role of NRF2 in mediating chemoresistance, it has become a potential target of drug development for HCC and other cancers with the goal of identifying specific NRF2 inhibitors to enhance their sensitivity to chemotherapeutics.
In our previous studies using an ARE luciferase reporter stably expressed in human cell lines including HepG2 and HaCaT cells (Zhao et al, 2011), we discovered several effective NRF2 inhibitors, such as isoniazid (INH) (Chen et al, 2013) and ethionamide (ETH) (Peng et al, 2016). Although INH and ETH both exhibit clear suppressive effects on the NRF2–ARE pathway, the high concentrations required for these compounds to achieve effective NRF2 inhibition in vitro raise doubts regarding their clinical usefulness in this regard. In the present study, we identified and characterised camptothecin (CPT), an antitumour drug for gastrointestinal and head and neck cancers, as a novel chemical inhibitor of NRF2–ARE activity. We found that CPT can enhance the sensitivity of different types of cancer cells, hepatocyte carcinoma cell HepG2 in particular, to a variety of chemotherapeutic agents.
Materials and methods
Human tissue selection, tissue handling and pathological analysis
Sixteen hepatectomised specimens including the HCC tissues and adjacent tissues (AT) to carcinoma were collected from 13 male and 3 female patients at The First Affiliated Hospital of China Medical University (Shenyang, Liaoning, China). The age of patients ranged from 43 to 69 years with a mean of 53.6 years. None of the 16 patients had a history of prior radiation or chemotherapy. Each tissue was divided into two pieces: one was formalin fixed and paraffin embedded for immunohistochemistry (IHC) and the other was stored at −80 °C for subsequent total mRNA and protein extraction. Patients were informed about and consented with the research in accordance with the Declaration of Helsinki. The study was approved by the Institutional Board of China Medical University (Shenyang, Liaoning, China). All methods were performed in accordance with the relevant guidelines and regulations.
Immunohistochemical staining and analysis
Human HCC and AT samples were fixed in formalin (10%, pH 7.4) and embedded in paraffin wax. Serial sections of 4 μm thickness were cut and mounted on charged glass slides, and then stained with haematoxylin and eosin (H&E) or used for IHC staining as described previously (Pi et al, 2009). Rabbit polyclonal antibody against NRF2 (sc-13032, Santa Cruz Biotechnology, Santa Cruz, CA, USA) was used at dilutions of 1 : 500. After counterstaining slides with H&E or IHC, cover slips were placed over the slides using Crystal Mount (Electron Microscopy Sciences, Hatfield, PA, USA). Quantitative analysis of NRF2 staining was carried out using the Image-Pro Plus v6.0 image analysis software (Media Cybernetics, Bethesda, MD, USA). Integrated optical density (IOD) value of NRF2 immunostaining was measured in five randomly acquired areas of one visual field in HCC and AT. Mean of IOD was used for further analyses.
Cell culture and reagents
The HepG2 (hepatocellular carcinoma) cells and A549 (lung carcinoma) cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). The SMMC-7721 (hepatocellular carcinoma) cells were purchased from KeyGEN BioTECH Co. Ltd (Nanjing, China). The HepG2 cells, A549 cells and SMMC-7721 cells were grown in EMEM medium (ATCC), DMEM medium (Life Technologies, Carlsbad, CA, USA) and RPMI1640 (Life Technologies), respectively. All the media were supplemented with 10% foetal bovine serum (FBS, Biological Industries, Cromwell, CT, USA), 100 U ml−1 penicillin and 100 μg ml−1 streptomycin. Exponentially growing cultures were maintained in a humidified atmosphere of 5% CO2 at 37 °C. Phosphate-buffered saline (PBS, pH 7.4) and supplements for cell culture were purchased from Life Technologies. Arsenic trioxide (As2O3), fluorouracil (5-FU) and epirubicin (EPI) were purchased from Beijing Shuanglu Pharmaceutical Co. (Beijing, China), Tianjin Jinyao Amino Acid Co. (Tianjin, China) and Pfizer Co. (Wuxi, China), respectively. Sodium arsenite (NaAsO2, purity ⩾90%), cisplatin (purity ⩾99%) and cycloheximide (CHX, purity ⩾94%) were purchased from Sigma-Aldrich (St Louis, MO, USA). The CPT (purity ⩾99%) was purchased from Cayman Chemical (Ann Arbor, MI, USA) and was dissolved in DMSO (Sigma-Aldrich).
Lentivirus-based shRNA transduction
The Mission shRNA lentiviral particles were purchased from Sigma-Aldrich. Lentiviral transduction of HepG2 cell with particles for shRNAs targeting NFE2L2 (NRF2 gene, SHVRS-NM_006164) or scrambled nontarget negative control (sh-Scr/SCR, SHC002V) to achieve NFE2L2 knockdown (NFE2L2-KD) or Scramble cells was performed as described previously (Woods et al, 2009). The selection media for HepG2 cells contained 3.0 μg ml−1 of puromycin (Life Technologies). Stable cell lines were continuously grown in the media containing the same concentration of puromycin.
Cell viability assay
Sensitivity of HepG2, A549 and SMMC-7721 cells to chemotherapeutic drug cytotoxicity was assessed by MTS as described previously (Pi et al, 2005; Peng et al, 2016). CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay Kits were purchased from Promega (Madison, WI, USA). Cells were exposed to various concentrations of As2O3, 5-Fu, EPI or cisplatin with or without CPT at a determined concentration for 24 or 48 h. Measurements were expressed as percentage change from untreated control (Vehicle) of appropriate cells. The lethal concentration 50 (LC50) values were determined from analysis of the log-linear phase of the curves.
The cell growth and proliferation of HepG2 cells were determined using the RTCA system (ACEA Biosciences, San Diego, CA, USA). Cell culture media (50 μl) were placed in each well of the E-plate 16 (ACEA Biosciences, San Diego, CA, USA). The E-plate 16 was then connected to the RTCA system to obtain background impedance readings. A total of 20 000 cells in 100 μl were incubated in the RTCA system overnight. Cells were treated with 10 μM As2O3, 200 μM 5-FU or 4 μM EPI with or without 0.1 μM CPT for 24 h on the RTCA SP Station located in an incubator at 37 °C with 5% CO2. Cell index values for cell activities were measured by continuous impedance recording every 5 min (Ren et al, 2011).
Reverse-transcription quantitative real-time PCR
Total RNA was isolated and extracted from clinical samples with Trizol reagent (Life Technologies) using TissueLyser II (Retsch, Newtown, PA, USA). Total RNA in HepG2 cells was extracted with Trizol reagent directly. The RT–qPCR was performed as described previously (Yang et al, 2012). The primers (sequences are listed in Supplementary Table S1) were designed by using Primer Express 4 (Life Technologies) and synthesised by Bioneer, Inc. (Alameda, CA, USA). Total RNA was reversely transcribed with the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). A GoTaq qPCR master mix (Promega) was used for qPCR. Real-time fluorescence detection was performed by using a QuantStudio 6 Flex Real-time PCR System (Life Technologies). The GAPDH expression was used for loading control and normalisation.
Western blot analysis
Collection of sample lysates and western blotting were performed as detailed previously (Xue et al, 2013). Antibodies for NRF2 (sc-13032; 1 : 500), NRF1 (sc-13031X, 1 : 1000) and β-actin (A1978; 1 : 3000) were purchased from Santa Cruz Biotechnology. β-Actin was used as loading control. The molecular weight (MW) of each protein shown on the immunoblot was estimated based on the MagicMark XP Western Protein Standard (Life Technologies) on 10% Tris-Glycine Gel. Quantification of the results normalised to β-actin was conducted using Image J (NIH, Bethesda, MD, USA).
ARE luciferase reporter assay
The ARE luciferase reporter assay was established previously (Zhao et al, 2012). Cignal Lenti ARE reporter, which expresses a luciferase gene driven by multiple ARE (5′-TCACAGTGACTCAGCAAAATT-3′) repeats, was obtained from SA Biosciences (Frederick, MD, USA). Cells were grown to ∼90% confluence and subcultured in medium containing 3.0 μg ml−1 of puromycin. The luciferase activity was measured using the Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol. The luciferase activity was normalised to cell viability that was determined by the MTS assay.
Relative NRF2 score calculation and associations with clinical pathological parameters
Relative NRF2 score calculated based on the cumulative expression of classic NRF2 target genes was used as our NRF2 activity index (Liu et al, 2010; DeNicola et al, 2015). In this study, the NRF2 score was calculated based on seven classic NRF2 target genes: GCLC, GCLM, NQO1, HMOX-1, Aldoketo Reductase family 1 member C1 (AKR1C1), C2 (AKR1C2) and C3 (AKR1C3) (Tebay et al, 2015; MacLeod et al, 2016). The expression of each gene was normalised by GAPDH and the results for these seven genes were added together for HCC and AT, respectively, to obtain total relative NRF2 scores. The following clinical data were collected to explore the potential correlation with relative NRF2 score in HCC: serum α-fetoprotein (AFP) levels, longest diameters measured with computed tomography (CT, Discovery CT750 HD, GE Health-Care Biosciences, Pittsburgh, PA, USA) or magnetic resonance imaging (MRI, MAGNETOM Skyra 3.0T, Siemens Healthineers, Erlangen, Germany) in mm, Child–Pugh classification (A/B/C), Barcelona Clinic Liver Cancer (BCLC) classification (0, A, B, C, D), tumour pathological differentiated degrees and plasma biochemical indexes, including alanine transaminase, aspartate transaminase, albumin, prothrombin time, prothrombin activity, prothrombin time activity of pre- and post-operation.
Subcutaneous tumour xenografts
BALB/Cnu/nu mice (4–6 weeks, male) were purchased from Beijing Huafukang Bioscience Co. Inc. (Beijing, China). Mice were inoculated subcutaneously to the right upper back region with SMMC-7721 cells (5 × 106 cells per mouse) that were suspended in 0.1 ml of serum-free RPMI-1640. The tumour xenografts sizes were measured and calculated by a standard formulate (V=length × width2 × π/6). Once the tumours reached 1000 mm3, mice were randomly allocated into four groups and treated i.p. with Vehicle, CPT (3 mg kg−1 bodyweight), EPI (1 mg kg−1 bodyweight) or in combination twice a week for a total of three times. All mice were maintained under standard animal housing conditions with a 12 h dark cycle and allowed access ad libitum to sterilised water and chow diet. All protocols for animal were approved by the Animal Ethics Committee of China Medical University.
Statistical analysis
All statistical analyses were performed using Graphpad Prism 5 software (San Diego, CA, USA). Statistical significance was defined as P<0.05. Data are expressed as mean±s.d. For comparisons between human HCC and AT results, paired t-tests were performed. For comparisons among multiple groups, one-way ANOVA followed by Tukey’s multiple comparison tests or two-way ANOVA with the Bonferroni post hoc testing were performed. Spearman’s analysis was used to derive possible correlations between relative NRF2 score and parameters of clinical pathology.
Results
The levels of NRF2 and ARE-dependent genes are elevated in human HCC tissues and correlate with clinical parameters
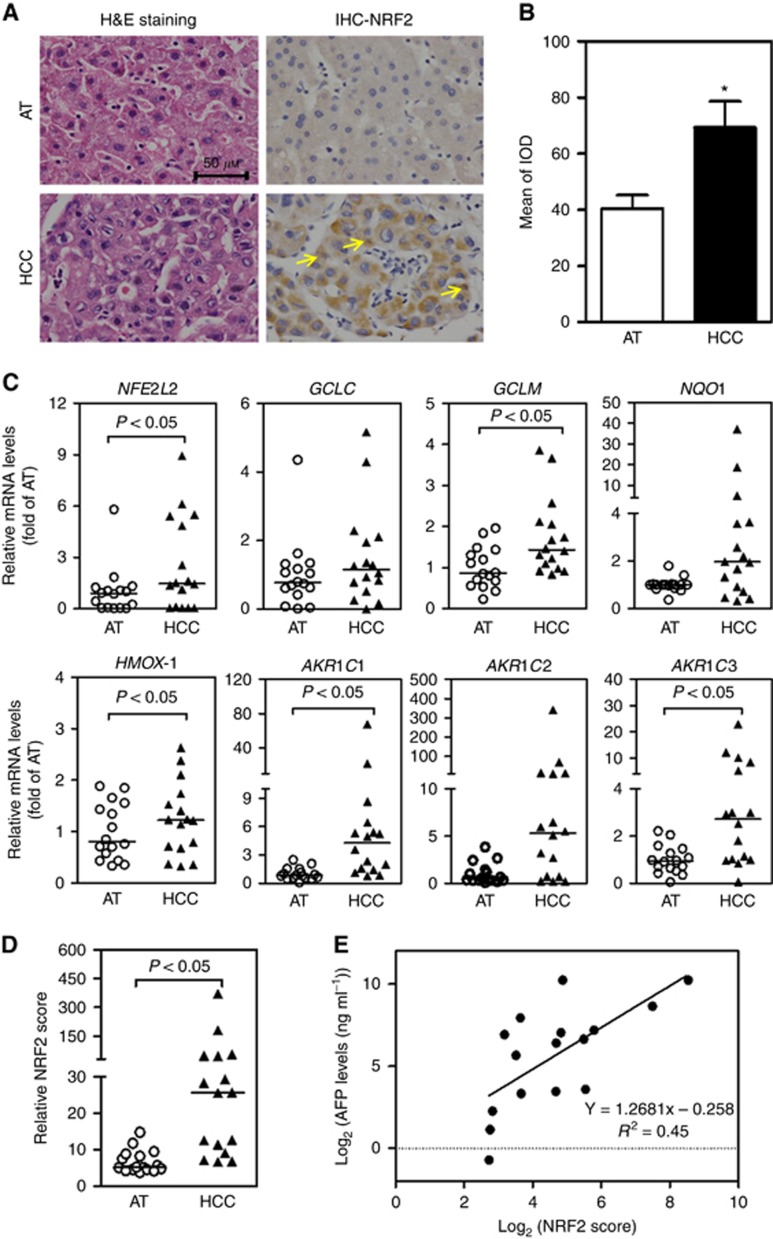
Histological examination was used to distinguish the type of HCC. Among the 16 cases studied, 10 were moderately differentiated, 4 poorly differentiated and 2 between moderately and poorly differentiated. The clinical data from all 16 patients with HCC are shown in Supplementary Table S2. Serial tissue sections of HCC and corresponding AT were analysed for NRF2 expression by IHC. The expressions of NRF2 were clearly higher in HCC than those in AT with nuclear accumulation (Figure 1A and B and Supplementary Figure S1). The mRNA levels of NFE2L2 and ARE-dependent genes (GCLC, GCLM, NQO1, HMOX-1, AKR1C1, AKR1C2 and AKR1C3) were also higher in HCC than those in AT (Figure 1C).
Figure 1.
The NRF2 activity and ARE-dependent gene expression are elevated in a subgroup of human HCC tissues and correlate with serum AFP. AT=adjacent tissue; HCC=hepatocellular carcinoma; n=16. *P<0.05 vs AT. (A) Representative images of H&E (left) and IHC (right) staining with a polyclonal NRF2 antibody in HCC and AT. Images were taken by a microscope with a Nikon 80i camera (40 × magnification). Arrows indicate nuclear NRF2 staining. (B) Quantification of NRF2 IHC staining. IOD=integrated optical density. (C) Elevated mRNA levels of NFE2L2 and ARE-dependent genes in HCC. (D) Relative NRF2 score was calculated by summing up the relative mRNA expression of NQO1, GCLC, GCLM, HMOX-1, AKR1C1, AKR1C2 and AKR1C3. (E) The NRF2 score in HCC is positively correlated with serum levels of AFP. AFP=α-fetoprotein. Linear correlation analysis: R2=0.45. A full colour version of this figure is available at the British Journal of Cancer journal online.
We then calculated relative NRF2 score by adding the relative mRNA expression levels of seven well accepted NRF2 targeted genes: GCLC, GCLM, NQO1, HMOX-1, AKR1C1, AKR1C2 and AKR1C3 (DeNicola et al, 2015; Tebay et al, 2015; MacLeod et al, 2016). Each patient has two relative NRF2 scores: one for HCC and the other for AT. In agreement with the expression of ARE-dependent genes, relative NRF2 scores in HCC were higher than those in AT (Figure 1D). Meanwhile, the correlations of relative NRF2 scores and clinical HCC pathological parameters were assessed. Using Spearman’s correlation analysis, we found that among these pathology markers, relative NRF2 score was positively correlated with serum AFP in HCC (r=0.71, P<0.05), indicating that relative NRF2 score could predict tumour severity (Figure 1E). Other measured clinical parameters, including BCLC classification (Supplementary Figure S2A), differentiation status (Supplementary Figure S2B), longest diameter and plasma biochemical indexes, showed no significant correlations with relative NRF2 scores.
Silence of NFE2L2 sensitises HepG2 cells to chemotherapeutic drug-induced cytotoxicity
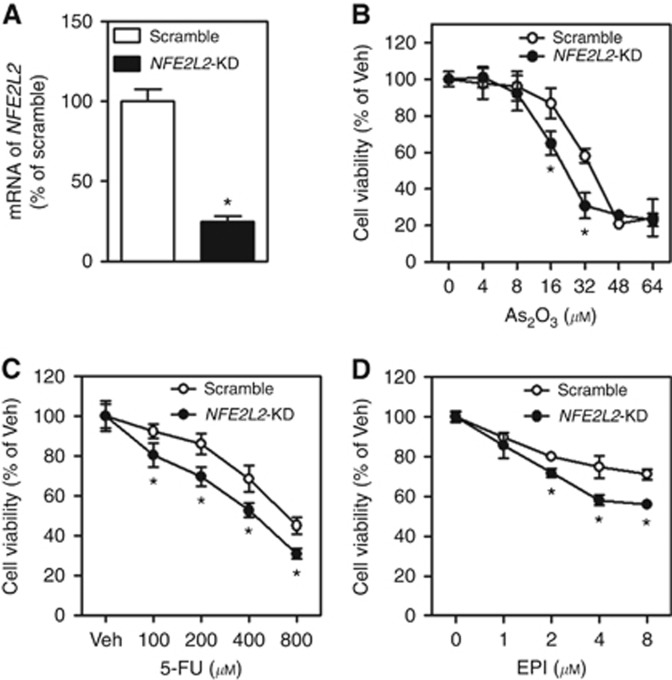
To explore the potential role of NRF2 in chemosensitivity, we developed a HepG2 cell line with stable knockdown of NFE2L2 (NFE2L2-KD) using a lentiviral shRNA against NFE2L2. As shown in Figure 2A, mRNA expression of NFE2L2 was effectively silenced in NFE2L2-KD cells. In agreement with the critical role of NRF2–ARE in chemoresistance, silencing of NFE2L2 increased the susceptibility of HepG2 cells to the acute cytotoxicity of multiple chemotherapeutic drugs, including As2O3, 5-FU and EPI (Figure 2B–D). This finding clearly demonstrates that reducing NRF2 expression can in principle sensitise HCC cells to chemotherapeutic agents.
Figure 2.
Effect of stable knockdown of NFE2L2 on chemotherapeutic drug-induced cytotoxicity in HepG2 cells. EPI=epirubicin; 5-FU=fluorouracil; NFE2L2-KD=stable knockdown of NFE2L2; Scramble=scrambled non-target negative control; Veh=Vehicle. (A) mRNA expression of NFE2L2. n=3. *P<0.05 vs Scramble. Cell viability was assessed by MTS assay following 24 h of treatment with indicated concentrations of As2O3 (B), 5-FU (C) and EPI (D); n=6. *P<0.05 vs Scramble with the same treatment.
Identification of CPT as a novel inhibitor of the NRF2–ARE signalling pathway
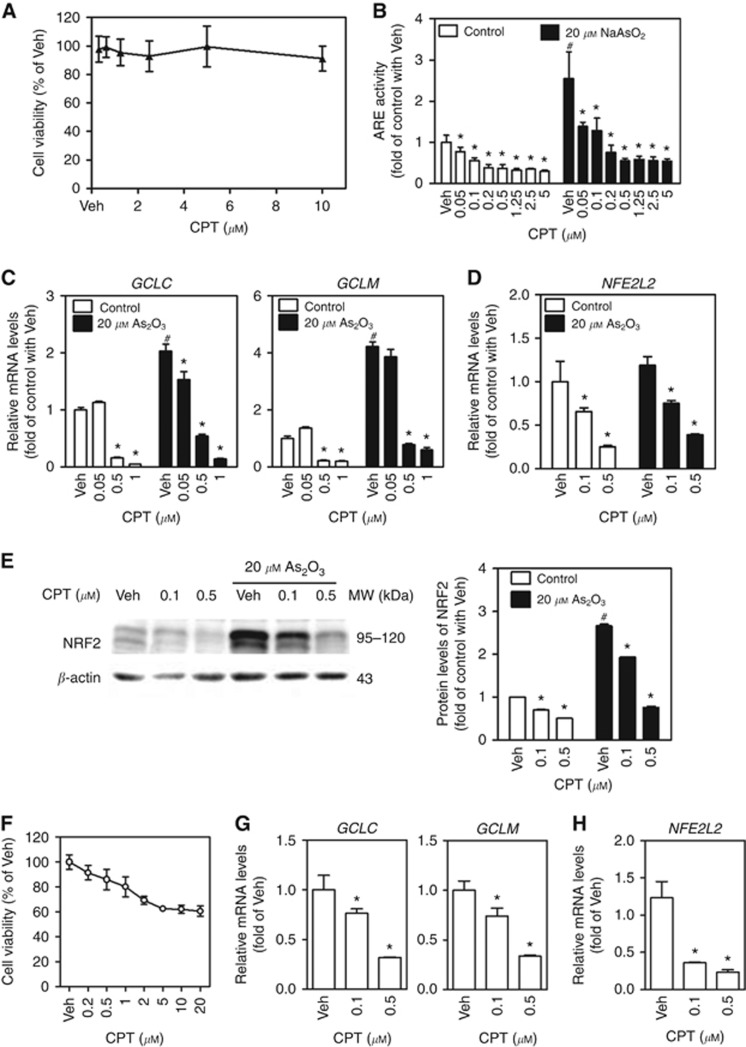
Our previous studies have shown that INH and ETH, two widely used anti-tubercular drugs, displayed substantial inhibitory properties against ARE activities in diverse mouse and human cells (Chen et al, 2013; Peng et al, 2016). To discover novel clinically relevant inhibitors of NRF2, a subset of clinical drugs was tested and several potent inhibitors were identified. Here we found noncytotoxic concentrations of CPT (Figure 3A), a potent antitumour agent for gastrointestinal and head and neck cancers, suppressed the activity of ARE luciferase in a concentration-dependent manner in HepG2 cells under basal (no stressor) and NaAsO2-challenged (20 μM for 6 h) conditions (Figure 3B). We further confirmed the suppression of ARE activity by noncytotoxic levels of CPT in HepG2 cells by showing attenuated mRNA expression of multiple ARE-dependent genes, including GCLC, GCLM and NFE2L2 (Figure 3C and D). In addition, a western blot analysis showed that CPT suppressed NRF2 protein levels under basal and As2O3-treated (20 μM for 4 h) conditions in HepG2 cells (Figure 3E). These results revealed that CPT inhibited NRF2 at both transcript and protein levels. To verify that the inhibition of CPT on NRF2 expression and activity is not HepG2 cell specific, the effects of CPT were determined in another HCC cell line SMMC-7721 cells. Noncytotoxic concentrations of CPT (Figure 3F) concentration-dependently reduced the mRNA expression of NFE2L2 (Figure 3H) and ARE-dependent genes GCLC and GCLM (Figure 3G).
Figure 3.
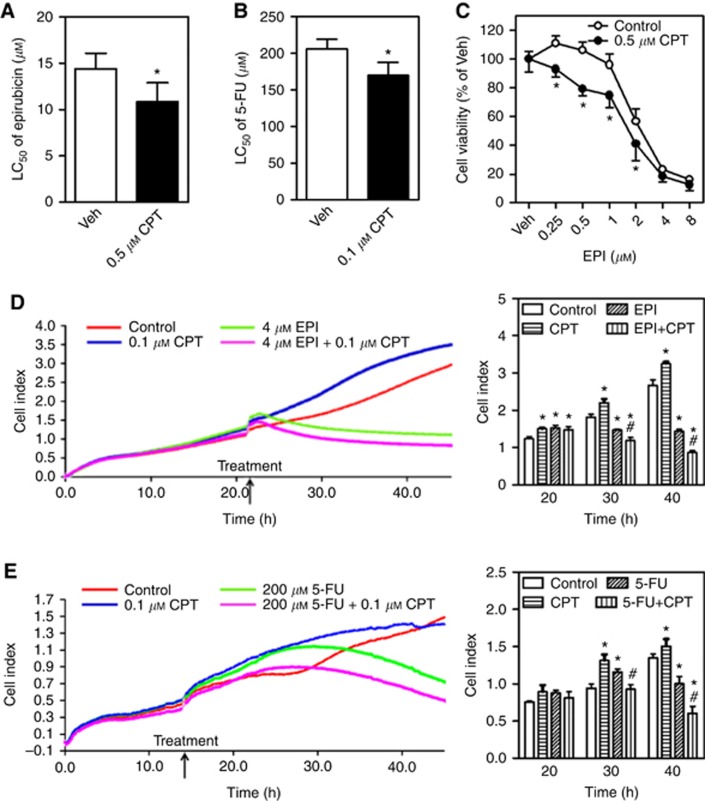
Camptothecin suppresses NRF2–ARE-dependent transcriptional activity in HepG2 and SMMC-7721 cells. CPT=camptothecin; Veh=Vehicle (medium). *P<0.05 vs Veh with the same arsenite concentration; #P<0.05 vs Control (Cont) with Veh. (A) Cytotoxicity of CPT determined by MTS assay in HepG2 cells. Cells were treated with CPT at indicated concentrations for 24 h; n=8. (B) The ARE luciferase activity under control and NaAsO2-treated conditions in HepG2 cells. Cells were exposed to various concentrations of CPT in the presence or absence of NaAsO2 for 6 h; n=6. (C) The mRNA expression of ARE-dependent genes under control or As2O3-treated conditions in HepG2 cells. Cells were treated with CPT at indicated concentrations in the presence or absence of As2O3 for 6 h; n=3. (D) The mRNA levels of NFE2L2 under control or As2O3-treated conditions in HepG2 cells; n=3. (E) Protein levels of NRF2 determined by western blot in HepG2 cells. Cells were treated with CPT at indicated concentrations in the presence or absence of As2O3 for 4 h. Whole-cell lysates were used for western blot. Left panels: western blots. Right panels: quantification of western blots of NRF2 normalised by β-actin. Experiments were repeated three times. (F) Cytotoxicity of CPT determined by MTS assay in SMMC-7721 cells. Cells were treated with CPT at indicated concentrations for 24 h; n=8. (G) The mRNA expression of ARE-dependent genes in SMMC-7721 cells. Cells were treated with CPT at indicated concentrations for 6 h; n=3. *P<0.05 vs Veh. (H) The mRNA levels of NFE2L2 in SMMC-7721 cells. Cells were treated with CPT at indicated concentrations for 6 h; n=3. *P<0.05 vs Veh.
To explore the mechanisms underlying the inhibition of CPT on the protein expression of NRF2, a cycloheximide chase assay was performed. As shown in Supplementary Figure S3, a CHX chase assay in HepG2 cells showed that CPT had no significant effect on the protein degradation of NRF2 induced by arsenite pretreatment (Supplementary Figure S3). To check the specificity of inhibition of CPT on NRF2, we assessed protein and mRNA levels of NRF1 in HepG2 cells. Both NRF1 and NRF2 belong to the CNC-bZIP transcription factor family and regulate ARE-driven genes and display similar degradation processes (Zhang et al, 2006; Zhao et al, 2011; Tsujita et al, 2015; Kim et al, 2016). In contrast to the strong inhibitory effect of CPT on NRF2, even higher concentrations of CPT had no significant effect on NRF1 protein and mRNA levels in HepG2 cells under basal (no stressor) and As2O3-challenged (20 μM) conditions (Supplementary Figure S4A and B), indicating that CPT is not a global protein synthesis inhibitor in HepG2 cells.
To exclude the possibility that the suppression of CPT on NRF2–ARE is cell type specific, we examined the effect of CPT on NRF2 expression and activity in human lung carcinoma A549 cell that harbours mutations in KEAP1. As shown in Supplementary Figure S5, the inhibition of CPT on NRF2 protein levels and expression of ARE-dependent genes was confirmed in A549 cells (Supplementary Figure S5). This finding indicates that the inhibitory effect of CPT on NRF2 is not limited in HepG2 cells and the inhibition appears to be KEAP1 independent.
CPT shows chemosensitising effects in a variety of cancer cells and a xenograft HCC model in mice
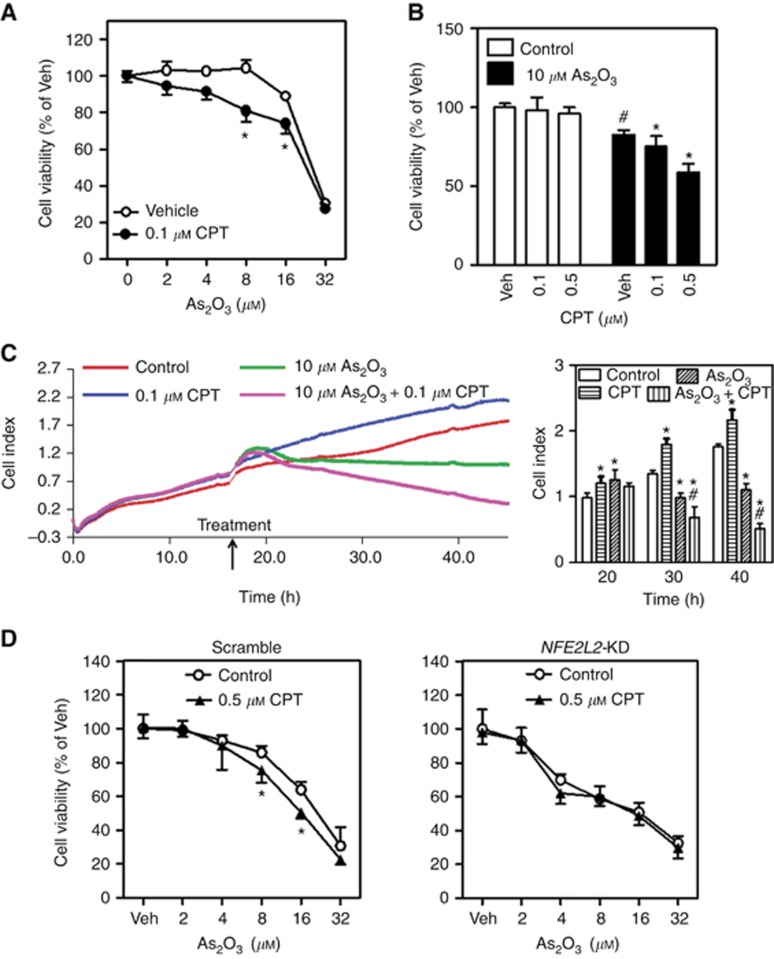
To test whether CPT might be used as a chemosensitising agent in cancer treatment, we determined the effects of CPT on various chemotherapeutic drug-induced cytotoxicity in multiple cancer cell models. As shown in Figures 4 and Figures 5, CPT at noncytotoxic concentrations sensitised HepG2 cells (Figure 4 and Figure 5A, B, D and E) and SMMC-7721 cells (Figure 5C) to chemotherapeutic drugs, including As2O3 (Figure 4A, C and D), EPI (Figure 5A and D) and 5-FU (Figure 5B and E), induced inhibition of cell viability (Figures 4A, B, D and 5A–C) and/or proliferation (Figures 4C and 5D, 5E). Importantly, the sensitisation of HepG2 cells to As2O3-induced cytotoxicity by CPT was NRF2 dependent as no further increased sensitivity to As2O3 was observed in NFE2L2-KD cells treated with CPT (Figure 4D). To further exclude the possibility that the effects of CPT on chemotherapeutic drug-induced cytotoxicity are cell type specific, the efficacy of CPT was also investigated in other type of cancer cells. As shown in Supplementary Figure S6, 0.5 μM CPT significantly sensitised A549 cells to cisplatin-induced cytotoxicity.
Figure 4.
Camptothecin (CPT) sensitises HepG2 cells to As2O3-induced cytotoxicity. Veh=Vehicle (medium). (A) The HepG2 cells were exposed to various concentrations of As2O3 in the presence or absence of 0.1 μM CPT for 24 h and subsequently the cell viability was measured by MTS assay; n=6. *P<0.05 vs Control with the same As2O3 treatment. (B) CPT enhances the cytotoxicity of As2O3 in a concentration-dependent manner. The HepG2 cells were treated with 10 μM As2O3 in the presence of different concentrations of CPT; n=6. *P<0.05 vs Control with the same As2O3 treatment; #P<0.05 vs Control with Veh. (C) The HepG2 cells were treated with 10 μM As2O3 in the presence or absence of 0.1 μM CPT for 24 on a xCELLigence real-time cell analysis system to detect cell growth and proliferation. The proliferation curves (left panel) and cumulative data (right panel) of cells at different times; n=3. *P<0.05 vs Control at the same time. #P<0.05 vs As2O3 at the same time. (D) Cytotoxicity of As2O3 in the Scramble and NFE2L2-KD cells. Cells were treated with various concentrations of As2O3 in the presence or absence of 0.5 μM CPT in Scramble or NFE2L2-KD cells for 24 h; n=6. *P<0.05 vs Control with the same As2O3 treatment. A full colour version of this figure is available at the British Journal of Cancer journal online.
Figure 5.
Camptothecin (CPT) sensitises HCC cells to epirubicin- or 5-FU-induced cytotoxicity. Veh=Vehicle (medium). (A) The HepG2 cells were exposed to various concentrations of epirubicin (EPI) in the presence or absence of 0.5 μM CPT for 24 h and subsequently the cell viability was measured by MTS assay; n=6. *P<0.05 vs Control with the same epirubicin treatment. The LC50 of epirubicin in MTS assay. (B) The HepG2 cells were exposed to 5-FU at different concentrations in the presence or absence of 0.1 μM CPT for 48 h and subsequently the cell viability was measured by MTS assay; n=6. *P<0.05 vs Control with the same 5-FU treatment. The LC50 of 5-FU in MTS assay. (C) The SMMC-7721 cells were exposed to various concentrations of EPI in the presence or absence of 0.5 μM CPT for 24 h and subsequently the cell viability was measured by MTS assay; n=6. *P<0.05 vs Control with the same EPI treatment. (D and E) Cell growth and proliferation were detected by a xCELLigence real-time cell analysis system. The HepG2 cells were treated with 4 μM EPI (D) or 200 μM 5-FU (E) in the presence or absence of 0.1 μM CPT. The proliferation curves (left panels) and cumulative data (right panels) of cells at different times; n=3. *P<0.05 vs Control at the same time. #P<0.05 vs EPI (D) or 5-FU (E) at the same time. A full colour version of this figure is available at the British Journal of Cancer journal online.
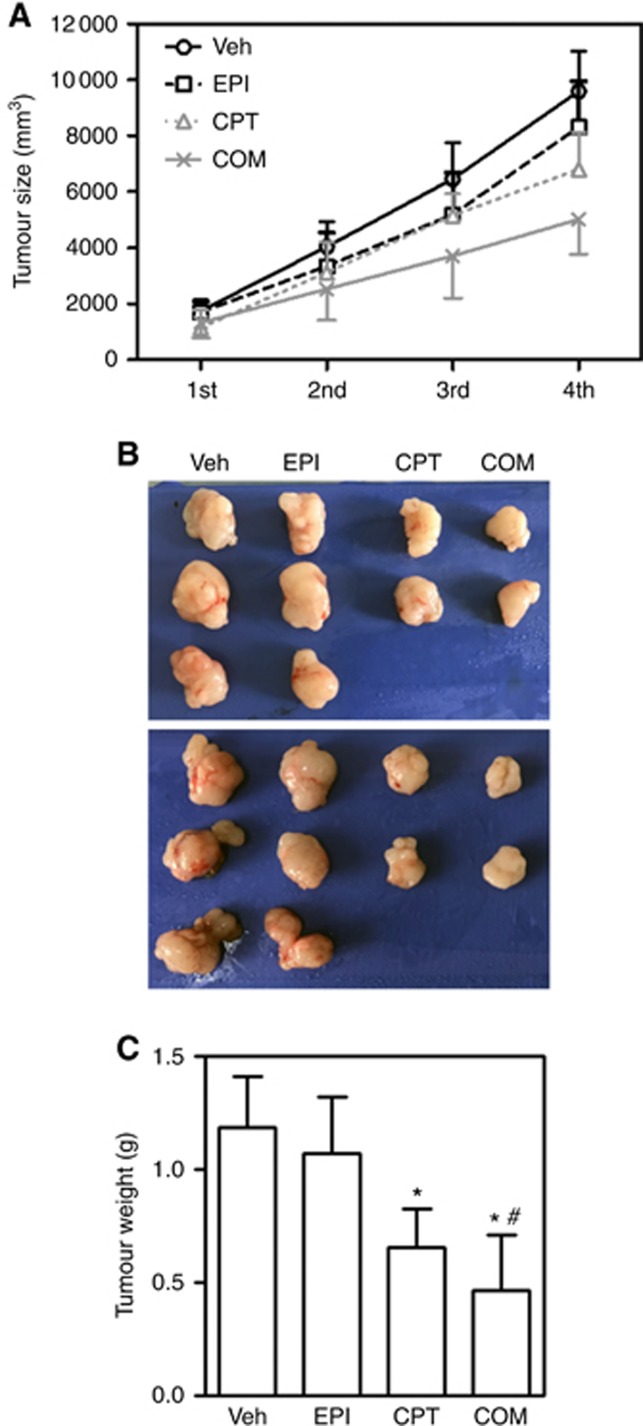
To further confirm the chemosensitising effect of CPT in HCC treatment, we used a HCC xenograft model developed by SMMC-7721 cell inoculation in nude mice to test CPT in vivo. Compared with EPT alone, the combination of CPT and EPI substantially reduced the tumour growth (Figure 6A) and tumour mass (Figure 6B), indicating that CPT may improve the efficacy of EPI.
Figure 6.
Camptothecin (CPT) sensitised HCC xenografts to EPI treatment. Nude mice were injected with SMMC-7721 cells. CPT=camptothecin; EPI=epirubicin. Mice were treated with CPT, EPI or in combination (COM) twice a week for a total of three times. At the end of the experiments, tumours were excised and weighed at the end of the experiment. (A) Tumour growth curves for SMMC-7721 xenografts. (B) Representative images of excised tumours. (C) Excised xenograft tumours weights at the end of the experiment; n=8–10; *P<0.05 vs Veh control. #P<0.05 vs EPI. A full colour version of this figure is available at the British Journal of Cancer journal online.
Discussion
The present study showed that NRF2 was highly expressed at both transcript and protein levels in a subgroup of human HCC. In accordance with previous studies, we demonstrated that suppressing NRF2 may increase the sensitivity of different types of solid tumour cells with high basal NRF2 expression to chemotherapeutic chemicals. We identified CPT as a novel inhibitor of NRF2–ARE activation that, at μM concentrations, can sensitise HepG2, SMMC-7721 and A549 cells to clinically important chemotherapeutic drugs. Compared with other known NRF2–ARE inhibitors such as INH and ETH, CPT has the advantages of being effective at lower concentrations and is already a conventional antitumour drug. Thus, CPT can be potentially used with other anticancer drugs in combination chemotherapy to treat HCC expressing high levels of NRF2.
Acquired or inherent chemoresistance is a major obstacle in therapeutic oncology. Multiple mechanisms, including increased metabolism and efflux of chemotherapeutic drugs, enhanced DNA repair and decreased cancer cell apoptosis, have been identified (Borowski et al, 2005; Deeley et al, 2006; Gao et al, 2013; Tebay et al, 2015), and NRF2 may be involved in most of the mechanisms (Hu et al, 2013). However, this dark side of NRF2 needs to be viewed in light of the fact that NRF2 plays an apparent dual but conflicting role in cancer. The NRF2-knockout mice are much more susceptible to carcinogens than genetically normal mice (Khor et al, 2008) and activation of NRF2 by chemopreventive compounds can prevent oxidative DNA damage from carcinogens (Jeong et al, 2006; Bishayee et al, 2010; Itoh et al, 2010). In contrast, accumulating data strongly indicate that constitutive activation of NRF2 also protects cancer cells from damage induced by chemotherapeutic drugs (Moon and Giaccia, 2015; Zhu et al, 2016). The NRF2 can promote tumour cell growth by modulating the signal transduction pathway mediated through EGFR-MEK1/2-ERK (Yamadori et al, 2012). The NRF2 is also able to directly activate the transcription of anti-apoptotic proteins Bcl-2 and Bcl-XL, increasing cell survival by desensitising many apoptosis-inducing drugs (Niture and Jaiswal, 2012, 2013). Furthermore, high expression of various NRF2–ARE genes, including antioxidant and detoxification enzymes, such as HMOX-1, NQO1 and MRP, has been implicated in chemotherapeutic resistance (Maher et al, 2007; Rushworth et al, 2010; Ganan-Gomez et al, 2013). It is also possible that some anticancer drugs can directly activate NRF2-mediated adaptive response leading to chemoresistance (Wang et al, 2014). Thus, NRF2–ARE is a reasonable target to tackle inherent and acquired chemoresistance in cancer treatments.
Our study showed that a subgroup of human HCC tissue samples had high levels of both NRF2 transcript and protein leading to augmented expression of many NRF2-dependent antioxidant and detoxification genes, including GCLC, GCLM, NQO1, HMOX-1, AKR1C1, AKR1C2 and AKR1C3. Several mechanisms have been reported concerning persistent activation of NRF2 in HCC. Recent studies using whole-exome sequencing have revealed mutations of NFE2L2 (NRF2) (6.4%) or KEAP1 (8%) genes in human HCC (Guichard et al, 2012; Cleary et al, 2013; Schulze et al, 2015). These somatic mutations alter the interaction between KEAP1 and NRF2 and result in persistent activation of NRF2 (Sporn and Liby, 2012; Ganan-Gomez et al, 2013). Moreover, the mutations tend to occur in pathologically advanced HCC, indicating that NRF2 hyperactivation may contribute to HCC progression (Nault et al, 2014; Schulze et al, 2015). In addition to KEAP1 and NFE2L2 mutation, p62 accumulation is associated with augmented activation of NRF2 in human HCC (Inami et al, 2011; Ichimura et al, 2013). The p62 binds with KEAP1 and inhibits KEAP1-driven ubiquitination of NRF2, resulting in stabilisation of NRF2 (Jiang et al, 2015). Dysregulation of the p62–KEAP1–NRF2 axis (e.g., increased aggregation of phosphorylated p62 and KEAP1) has been frequently observed in HCC (Ichimura et al, 2013). Moreover, a recent study indicated that p62-caused persistent activation of NRF2 is involved in sorafenib and cisplatin resistance in human HCC (Saito et al, 2016). Apart from gene mutations and p62 accumulation, epigenetic modifications of NRF2 and KEAP1 may lead to NRF2 upregulation as well (Guo et al, 2015). In addition, oncogenes such as Kirsten rat sarcoma viral oncogene (kRas), B-Raf proto-oncogene (bRaf), myelocytomatosis viral oncogene (Myc) and various microRNAs can also affect the expression of NRF2 in HCC (Imbeaud et al, 2010; Tew, 2011; Giordano and Columbano, 2013). The high expression of NRF2 in HCC likely plays a critical role in the development of chemoresistance in these tumours by mediating the expression of ARE-dependent genes. It would be of interest to compare these results with livers from nontumour-bearing donors, but these were not available to our group at the time of this study. Although genetically identical, AT likely receives inflammatory mediators from the nearby HCC that could partially activate the NRF2 pathways (Bishayee et al, 2013), and hence these data should be seen in light of this potential issue.
In the present study there was a strong, positive correlation between relative NRF2 scores in human HCC and serum AFP levels. However, there is no consensus on how to most accurately reflect the activity of NRF2. The genes regulated by NRF2 may be distinct under different metabolic conditions or in different kinds of cells. In particular, large differences were detected among clinical samples in the expression of various NRF2–ARE downstream genes. However, as a group, the expression of NRF2–ARE downstream genes are relatively stable. Therefore, relative NRF2 score calculated based on the cumulative expression of classic NRF2 target genes has been applied as a NRF2 activity index. DeNicola et al (2015) proposed the definition of NRF2 score and calculated the NRF2 score based on 20 NRF2 target genes. Before the NRF2 score was defined, we utilised eight NRF2 target genes in the NCI-60 tumour cell panel to calculate NRF2 activity (Liu et al, 2010). In the calculation process, the use of specific NRF2 target genes were different, but the concept used to quantify NRF2 activity is the same. The AFP is a fairly sensitive and specific biomarker for detecting HCC and is also considered an indicator of HCC prognosis (Dwyer et al, 2014). A serum surrogate indicator of liver NRF2 could be quite valuable in the therapeutic decision-making process. If a circulating biomarker (e.g., AFP for HCC) suggests high levels of NRF2 activity in cancer cells, then inhibiting NRF2 can be recommended as part of the treatment regimen. If the NRF2 score can be obtained through biopsy, it could be used together with serum indicators to predict the effectiveness of a NRF2 inhibition approach.
The CPT is a natural alkaloid isolated from the Chinese medicinal plant Camptotheca acuminate, and is regarded as an effective antitumour agent for a wide spectrum of cancers, such as colon and gastric cancers (Wall and Wani, 1996). It inhibits cancer growth by inhibiting DNA replication through binding to DNA topoisomerase I using its acyl position of the lactone ring during the S stage of mitosis (Tang et al, 2014). The clinical applications of CPT have been limited by its poor solubility (3.122 μM in water) and side effects, such as diarrhoea, haemorrhagic cystitis and unpredictable myelosuppression (Tang et al, 2014). The present study demonstrated that CPT is an effective NRF2 inhibitor at much lower concentrations than is needed as a direct antitumour agent. The inhibited NRF2 expression in HepG2, SMMC-7721 and A549 cells in a concentration-dependent manner with a minimum effective concentration of 0.1 μM. In contrast, the half-maximal inhibitory concentration (IC50) of INH on ARE luciferase activity is above 10 mM. The minimum concentration of ETH for suppressing ARE activity and sensitising leukaemia cells to As2O3-induced cytotoxicity is ∼0.1 mM (Chen et al, 2013; Peng et al, 2016). Thus, CPT is a relatively strong inhibitor of NRF2–ARE activity. Nonetheless, there are scattered reports of activation of NRF2-mediated adaptive response by CPT in human and rodent cells. For example, CPT has been shown to upregulate NRF2-mediated HMOX-1 expression in human prostate cancer DU145 cells (Jayasooriya et al, 2015) and activate NRF2 pathway in rat pheochromocytoma PC12 cells (Zhang et al, 2015). Therefore, more accurate and detailed preclinical studies need to be done to elucidate dose- and cell type-dependent effects of various NRF2 inhibitors (Zhu et al, 2016).
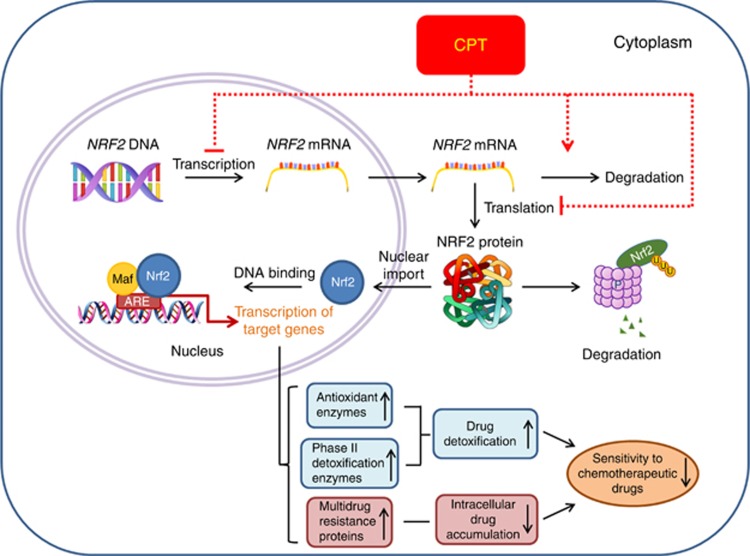
Regarding CPT-induced reduction of NRF2 protein expression, our data of CHX chase assay do not support that CPT has direct effect in promoting NRF2 protein degradation. Thus, we propose that the inhibitory effect of CPT on NRF2 expression may be through suppressing its transcription, translation and/or promoting mRNA degradation (Figure 7). For CPT to decrease NFE2L2 mRNA levels, one possible mechanism is that NRF2 autoregulates its own expression through an ARE-like element located on its promoter (Kwak et al, 2002). Reduced NRF2 protein levels may unavoidably lead to diminished NRF2 nuclear accumulation and thus compromised binding to ARE-like elements on the NFE2L2 promoter, and subsequently results in less NFE2L2 transcription. The inhibitory effect of CPT on NRF2 could be boosted by this positive loop (Figure 7). Of note, it needs further investigation to clarify how CPT suppresses the expression of NRF2 in cancer cells.
Figure 7.
Schematic description of the mechanisms by which CPT sensitises HCC cells to chemotherapeutic drugs. The NRF2 is highly expressed in a subgroup of HCC cells. The activated NRF2 upregulates the expression of ARE-dependent antioxidant, phase II enzymes and MRPs. This activation essentially protects cancer cells from the effects of cancer chemotherapeutics. The CPT likely downregulates the transcription and protein expression of NRF2 that, in turn, suppresses ARE-dependent gene expression, and thus sensitises HCC cells to chemotherapeutics. A full colour version of this figure is available at the British Journal of Cancer journal online.
In summary, this study demonstrated that CPT has a strong inhibitory effect on the NRF2–ARE pathway in vitro. By lowering NRF2 activity, CPT enhances the cytotoxicity of chemotherapeutic drugs in HCC cells. Human HCCs exhibit a positive correlation between the levels of NRF2–ARE activity and serum AFP, suggesting that the latter could potentially be used as a circulating biomarker to help determine whether usage of NRF2 inhibitors such as CPT can be useful on top of conventional chemotherapeutic drugs for HCC. Clearly, further study on repurposing CPT as a chemotherapeutic adjuvant for chemoresistant cancers requires more detailed testing in vivo and in humans.
Acknowledgments
This research was supported in part by National Natural Science Foundation of China 81502841 (to HW) and 81573106 (to JP), the Startup Funding of China Medical University (to JP), Liaoning Pandeng Scholar (to JP) and Program for Liaoning Innovative Research Team in University (LT2015028). The content is solely the responsibility of the authors. All authors have agreed to its content. We thank Dr Rong Zhang for providing arsenic trioxide, Dr Yanan Ma for his help in statistical analysis and Dr Michael P Waalkes and Henry H Kou for their editorial supports.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Attwa MH, El-Etreby SA (2015) Guide for diagnosis and treatment of hepatocellular carcinoma. World J Hepatol 7(12): 1632–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertino G, Demma S, Ardiri A, Proiti M, Gruttadauria S, Toro A, Malaguarnera G, Bertino N, Malaguarnera M, Malaguarnera M, Di Carlo I (2014) Hepatocellular carcinoma: novel molecular targets in carcinogenesis for future therapies. Biomed Res Int 2014: 203693. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bishayee A, Barnes KF, Bhatia D, Darvesh AS, Carroll RT (2010) Resveratrol suppresses oxidative stress and inflammatory response in diethylnitrosamine-initiated rat hepatocarcinogenesis. Cancer Prev Res 3(6): 753–763. [DOI] [PubMed] [Google Scholar]
- Bishayee A, Thoppil RJ, Darvesh AS, Ohanyan V, Meszaros JG, Bhatia D (2013) Pomegranate phytoconstituents blunt the inflammatory cascade in a chemically induced rodent model of hepatocellular carcinogenesis. J Nutr Biochem 24(1): 178–187. [DOI] [PubMed] [Google Scholar]
- Borowski E, Bontemps-Gracz MM, Piwkowska A (2005) Strategies for overcoming ABC-transporters-mediated multidrug resistance (MDR) of tumor cells. Acta Biochim Pol 52(3): 609–627. [PubMed] [Google Scholar]
- Chen Q, Li W, Wan Y, Xia X, Wu Q, Chen Y, Lai Z, Yu C, Li W (2012) Amplified in breast cancer 1 enhances human cholangiocarcinoma growth and chemoresistance by simultaneous activation of Akt and Nrf2 pathways. Hepatology 55(6): 1820–1829. [DOI] [PubMed] [Google Scholar]
- Chen Y, Xue P, Hou Y, Zhang H, Zheng H, Zhou T, Qu W, Teng W, Zhang Q, Andersen ME, Pi J (2013) Isoniazid suppresses antioxidant response element activities and impairs adipogenesis in mouse and human preadipocytes. Toxicol Appl Pharmacol 273(3): 435–441. [DOI] [PubMed] [Google Scholar]
- Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, Hayes JD (2013) Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 32(32): 3765–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary SP, Jeck WR, Zhao X, Chen K, Selitsky SR, Savich GL, Tan TX, Wu MC, Getz G, Lawrence MS, Parker JS, Li J, Powers S, Kim H, Fischer S, Guindi M, Ghanekar A, Chiang DY (2013) Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology 58(5): 1693–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeley RG, Westlake C, Cole SP (2006) Transmembrane transport of endo- and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiol Rev 86(3): 849–899. [DOI] [PubMed] [Google Scholar]
- DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, Wistuba II, Minna JD, DeBerardinis RJ, Cantley LC (2015) NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet 47(12): 1475–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer JP, Hosking P, Lubel J (2014) Multiple liver lesions in a patient with positive hepatitis C serology and elevated AFP: is it HCC? Gastroenterology 147(3): E12–E13. [DOI] [PubMed] [Google Scholar]
- El-Serag HB (2012) Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 142(6): 1264–1273 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenci P, Fried M, Labrecque D, Bruix J, Sherman M, Omata M, Heathcote J, Piratsivuth T, Kew M, Otegbayo JA, Zheng SS, Sarin S, Hamid S, Modawi SB, Fleig W, Fedail S, Thomson A, Khan A, Malfertheiner P, Lau G, Carillo FJ, Krabshuis J, Le Mair A World Gastroenterology Organisation Guidelines and Publications Committee (2010) World Gastroenterology Organisation Guideline. Hepatocellular carcinoma (HCC): a global perspective. J Gastrointestin Liver Dis 19(3): 311–317. [PubMed] [Google Scholar]
- Ganan-Gomez I, Wei Y, Yang H, Boyano-Adanez MC, Garcia-Manero G (2013) Oncogenic functions of the transcription factor Nrf2. Free Radic Biol Med 65: 750–764. [DOI] [PubMed] [Google Scholar]
- Gao AM, Ke ZP, Shi F, Sun GC, Chen H (2013) Chrysin enhances sensitivity of BEL-7402/ADM cells to doxorubicin by suppressing PI3K/Akt/Nrf2 and ERK/Nrf2 pathway. Chem -Biol Interact 206(1): 100–108. [DOI] [PubMed] [Google Scholar]
- Giordano S, Columbano A (2013) MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology 57(2): 840–847. [DOI] [PubMed] [Google Scholar]
- Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, Clement B, Balabaud C, Chevet E, Laurent A, Couchy G, Letouze E, Calvo F, Zucman-Rossi J (2012) Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 44(6): 694–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Yu S, Zhang C, Kong AN (2015) Epigenetic regulation of Keap1-Nrf2 signaling. Free Radic Biol Med 88(Pt B): 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, McMahon M (2009) NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci 34(4): 176–188. [DOI] [PubMed] [Google Scholar]
- Hu XF, Yao J, Gao SG, Wang XS, Peng XQ, Yang YT, Feng XS (2013) Nrf2 overexpression predicts prognosis and 5-FU resistance in gastric cancer. Asian Pac J Cancer Prev 14(9): 5231–5235. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, Hoshii T, Hirao A, Takagi K, Mizushima T, Motohashi H, Lee MS, Yoshimori T, Tanaka K, Yamamoto M, Komatsu M (2013) Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell 51(5): 618–631. [DOI] [PubMed] [Google Scholar]
- Imbeaud S, Ladeiro Y, Zucman-Rossi J (2010) Identification of novel oncogenes and tumor suppressors in hepatocellular carcinoma. Semin Liver Dis 30(1): 75–86. [DOI] [PubMed] [Google Scholar]
- Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, Watanabe S, Ando J, Iwadate M, Yamamoto M, Lee MS, Tanaka K, Komatsu M (2011) Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol 193(2): 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Mimura J, Yamamoto M (2010) Discovery of the negative regulator of Nrf2, Keap1: a historical overview. Antioxid Redox Signal 13(11): 1665–1678. [DOI] [PubMed] [Google Scholar]
- Jayasooriya RG, Park SR, Choi YH, Hyun JW, Chang WY, Kim GY (2015) Camptothecin suppresses expression of matrix metalloproteinase-9 and vascular endothelial growth factor in DU145 cells through PI3K/Akt-mediated inhibition of NF-kappaB activity and Nrf2-dependent induction of HO-1 expression. Environ Toxicol Pharmacol 39(3): 1189–1198. [DOI] [PubMed] [Google Scholar]
- Jeong WS, Jun M, Kong AN (2006) Nrf2: a potential molecular target for cancer chemoprevention by natural compounds. Antioxid Redox Signal 8(1-2): 99–106. [DOI] [PubMed] [Google Scholar]
- Jiang T, Harder B, Rojo de la Vega M, Wong PK, Chapman E, Zhang DD (2015) p62 links autophagy and Nrf2 signaling. Free Radic Biol Med 88(Pt B): 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N (2010) Nrf2: friend or foe for chemoprevention? Carcinogenesis 31(1): 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor TO, Huang MT, Prawan A, Liu Y, Hao X, Yu S, Cheung WK, Chan JY, Reddy BS, Yang CS, Kong AN (2008) Increased susceptibility of Nrf2 knockout mice to colitis-associated colorectal cancer. Cancer Prev Res 1(3): 187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HM, Han JW, Chan JY (2016) Nuclear Factor Erythroid-2 Like 1 (NFE2L1): Structure, function and regulation. Gene 584(1): 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak MK, Itoh K, Yamamoto M, Kensler TW (2002) Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol Cell Biol 22(9): 2883–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD (2008) Dual roles of Nrf2 in cancer. Pharmacol Res 58(5-6): 262–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zhang H, Smeester L, Zou F, Kesic M, Jaspers I, Pi J, Fry RC (2010) The NRF2-mediated oxidative stress response pathway is associated with tumor cell resistance to arsenic trioxide across the NCI-60 panel. BMC Med Genomics 3: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet JM, Burroughs A, Bruix J (2003) Hepatocellular carcinoma. Lancet 362(9399): 1907–1917. [DOI] [PubMed] [Google Scholar]
- MacLeod AK, Acosta-Jimenez L, Coates PJ, McMahon M, Carey FA, Honda T, Henderson CJ, Wolf CR (2016) Aldo-keto reductases are biomarkers of NRF2 activity and are co-ordinately overexpressed in non-small cell lung cancer. Br J Cancer 115(12): 1530–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaffey CM, Zhang H, Rinna A, Holland W, Mack PC, Forman HJ (2009) Multidrug-resistant protein-3 gene regulation by the transcription factor Nrf2 in human bronchial epithelial and non-small-cell lung carcinoma. Free Radic Biol Med 46(12): 1650–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher J, Yamamoto M (2010) The rise of antioxidant signaling-The evolution and hormetic actions of Nrf2. Toxicol Appl Pharm 244(1): 4–15. [DOI] [PubMed] [Google Scholar]
- Maher JM, Dieter MZ, Aleksunes LM, Slitt AL, Guo G, Tanaka Y, Scheffer GL, Chan JY, Manautou JE, Chen Y, Dalton TP, Yamamoto M, Klaassen CD (2007) Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology 46(5): 1597–1610. [DOI] [PubMed] [Google Scholar]
- Moon EJ, Giaccia A (2015) Dual roles of NRF2 in tumor prevention and progression: possible implications in cancer treatment. Free Radic Biol Med 79: 292–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nault JC, Calderaro J, Di Tommaso L, Balabaud C, Zafrani ES, Bioulac-Sage P, Roncalli M, Zucman-Rossi J (2014) Telomerase reverse transcriptase promoter mutation is an early somatic genetic alteration in the transformation of premalignant nodules in hepatocellular carcinoma on cirrhosis. Hepatology 60(6): 1983–1992. [DOI] [PubMed] [Google Scholar]
- Niture SK, Jaiswal AK (2012) Nrf2 protein up-regulates antiapoptotic protein Bcl-2 and prevents cellular apoptosis. J Biol Chem 287(13): 9873–9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niture SK, Jaiswal AK (2013) Nrf2-induced antiapoptotic Bcl-xL protein enhances cell survival and drug resistance. Free Radic Biol Med 57: 119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Wang HH, Xue P, Hou YY, Dong J, Zhou T, Qu WD, Peng SQ, Li J, Carmichael PL, Nelson B, Clewell R, Zhang Q, Andersen ME, Pi JB (2016) Suppression of NRF2-ARE activity sensitizes chemotherapeutic agent-induced cytotoxicity in human acute monocytic leukemia cells. Toxicol Appl Pharm 292: 1–7. [DOI] [PubMed] [Google Scholar]
- Pi J, He Y, Bortner C, Huang J, Liu J, Zhou T, Qu W, North SL, Kasprzak KS, Diwan BA, Chignell CF, Waalkes MP (2005) Low level, long-term inorganic arsenite exposure causes generalized resistance to apoptosis in cultured human keratinocytes: potential role in skin co-carcinogenesis. Int J Cancer 116(1): 20–26. [DOI] [PubMed] [Google Scholar]
- Pi J, Zhang Q, Fu J, Woods CG, Hou Y, Corkey BE, Collins S, Andersen ME (2010) ROS signaling, oxidative stress and Nrf2 in pancreatic beta-cell function. Toxicol Appl Pharmacol 244(1): 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi JB, Bai YS, Daniel KW, Liu DX, Lyght O, Edelstein D, Brownlee M, Corkey BE, Collins S (2009) Persistent oxidative stress due to absence of uncoupling protein 2 associated with impaired pancreatic beta-cell function. Endocrinology 150(7): 3040–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, Cuadrado A (2011) SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol 31(6): 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren DM, Villeneuve NF, Jiang T, Wu TD, Lau A, Toppin HA, Zhang DD (2011) Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc Natl Acad Sci USA 108(4): 1433–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushworth SA, Bowles KM, Raninga P, MacEwan DJ (2010) NF-kappaB-inhibited acute myeloid leukemia cells are rescued from apoptosis by heme oxygenase-1 induction. Cancer Res 70(7): 2973–2983. [DOI] [PubMed] [Google Scholar]
- Saito T, Ichimura Y, Taguchi K, Suzuki T, Mizushima T, Takagi K, Hirose Y, Nagahashi M, Iso T, Fukutomi T, Ohishi M, Endo K, Uemura T, Nishito Y, Okuda S, Obata M, Kouno T, Imamura R, Tada Y, Obata R, Yasuda D, Takahashi K, Fujimura T, Pi J, Lee MS, Ueno T, Ohe T, Mashino T, Wakai T, Kojima H, Okabe T, Nagano T, Motohashi H, Waguri S, Soga T, Yamamoto M, Tanaka K, Komatsu M (2016) p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat Commun 7: 12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Shitara M, Yokota K, Hikosaka Y, Moriyama S, Yano M, Fujii Y (2012) MRP3 gene expression correlates with NRF2 mutations in lung squamous cell carcinomas. Mol Med Rep 6(4): 705–708. [DOI] [PubMed] [Google Scholar]
- Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, Calatayud AL, Pinyol R, Pelletier L, Balabaud C, Laurent A, Blanc JF, Mazzaferro V, Calvo F, Villanueva A, Nault JC, Bioulac-Sage P, Stratton MR, Llovet JM, Zucman-Rossi J (2015) Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 47(5): 505–U106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporn MB, Liby KT (2012) NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer 12(8): 564–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang XJ, Han M, Yang B, Shen YQ, He ZG, Xu DH, Gao JQ (2014) Nanocarrier improves the bioavailability, stability and antitumor activity of camptothecin. Int J Pharmaceut 477(1-2): 536–545. [DOI] [PubMed] [Google Scholar]
- Tebay LE, Robertson H, Durant ST, Vitale SR, Penning TM, Dinkova-Kostova AT, Hayes JD (2015) Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic Biol Med 88(Pt B): 108–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tew KD (2011) NrF2/Keap1 as gatekeepers of redox homeostasis – do they prevent or cause cancer? Pigment Cell Melanoma Res 24(6): 1078–1079. [DOI] [PubMed] [Google Scholar]
- Tsujita T, Baird L, Furusawa Y, Katsuoka F, Hou Y, Gotoh S, Kawaguchi S, Yamamoto M (2015) Discovery of an NRF1-specific inducer from a large-scale chemical library using a direct NRF1-protein monitoring system. Genes Cells 20(7): 563–577. [DOI] [PubMed] [Google Scholar]
- Van Thiel DH, Brems J, Holt D, Hamdani R, Yong S (2002) Chemosensitivity of primary hepatic neoplasms: a potential new approach to the treatment of hepatoma. Hepatogastroenterology 49(45): 730–734. [PubMed] [Google Scholar]
- Wall ME, Wani MC (1996) Camptothecin and taxol: from discovery to clinic. J Ethnopharmacol 51(1-3): 239–253, ; discussion 253-4. [DOI] [PubMed] [Google Scholar]
- Wang H, Liu X, Long M, Huang Y, Zhang L, Zhang R, Zheng Y, Liao X, Wang Y, Liao Q, Li W, Tang Z, Tong Q, Wang X, Fang F, Rojo de la Vega M, Ouyang Q, Zhang DD, Yu S, Zheng H (2016) NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Sci Transl Med 8(334): 334ra51. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Li Y, Luo L, Wang H, Chi Z, Xin A, Li X, Wu J, Tang X (2014) Oxaliplatin activates the Keap1/Nrf2 antioxidant system conferring protection against the cytotoxicity of anticancer drugs. Free Radic Biol Med 70: 68–77. [DOI] [PubMed] [Google Scholar]
- Woods CG, Fu JQ, Xue P, Hou YY, Pluta LJ, Yang LL, Zhang Q, Thomas RS, Andersen ME, Pi JB (2009) Dose-dependent transitions in Nrf2-mediated adaptive response and related stress responses to hypochlorous acid in mouse macrophages. Toxicol Appl Pharm 238(1): 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue P, Hou Y, Chen Y, Yang B, Fu J, Zheng H, Yarborough K, Woods CG, Liu D, Yamamoto M, Zhang Q, Andersen ME, Pi J (2013) Adipose deficiency of Nrf2 in ob/ob mice results in severe metabolic syndrome. Diabetes 62(3): 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamadori T, Ishii Y, Homma S, Morishima Y, Kurishima K, Itoh K, Yamamoto M, Minami Y, Noguchi M, Hizawa N (2012) Molecular mechanisms for the regulation of Nrf2-mediated cell proliferation in non-small-cell lung cancers. Oncogene 31(45): 4768–4777. [DOI] [PubMed] [Google Scholar]
- Yang B, Fu J, Zheng H, Xue P, Yarborough K, Woods CG, Hou Y, Zhang Q, Andersen ME, Pi J (2012) Deficiency in the nuclear factor E2-related factor 2 renders pancreatic beta-cells vulnerable to arsenic-induced cell damage. Toxicol Appl Pharmacol 264(3): 315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Chen S, Bao J, Zhang Y, Huang B, Jia X, Chen M, Wan JB, Su H, Wang Y, He C (2015) Low doses of camptothecin induced hormetic and neuroprotective effects in PC12 cells. Dose Response 13(2): 1559325815592606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M (2004) Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol 24(24): 10941–10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Crouch DH, Yamamoto M, Hayes JD (2006) Negative regulation of the Nrf1 transcription factor by its N-terminal domain is independent of Keap1: Nrf1, but not Nrf2, is targeted to the endoplasmic reticulum. Biochem J 399(3): 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Hou Y, Zhang Q, Woods CG, Xue P, Fu J, Yarborough K, Guan D, Andersen ME, Pi J (2012) Cross-regulations among NRFs and KEAP1 and effects of their silencing on arsenic-induced antioxidant response and cytotoxicity in human keratinocytes. Environ Health Perspect 120(4): 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Hou YY, Xue P, Woods CG, Fu JQ, Feng B, Guan DW, Sun GF, Chan JY, Waalkes MP, Andersen ME, Pi JB (2011) Long isoforms of NRF1 contribute to arsenic-induced antioxidant response in human keratinocytes. Environ Health Perspect 119(1): 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Wang H, Chen F, Fu J, Xu Y, Hou Y, Kou HH, Zhai C, Nelson MB, Zhang Q, Andersen ME, Pi J (2016) An overview of chemical inhibitors of the Nrf2-ARE signaling pathway and their potential applications in cancer therapy. Free Radic Biol Med 99: 544–556. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.