Abstract
Autophagy degrades non-functioning or damaged proteins and organelles to maintain cellular homeostasis in a physiological or pathological context. Autophagy can be protective or detrimental, depending on its activation status and other conditions. Therefore, autophagy has a crucial role in a myriad of pathophysiological processes. From the perspective of autophagy-related (ATG) genes, the molecular dissection of autophagy process and the regulation of its level have been largely unraveled. However, the discovery of long non-coding RNAs (lncRNAs) provides a new paradigm of gene regulation in almost all important biological processes, including autophagy. In this review, we highlight recent advances in autophagy-associated lncRNAs and their specific autophagic targets, as well as their relevance to human diseases such as cancer, cardiovascular disease, diabetes and cerebral ischemic stroke.
Facts
Autophagy degrades non-functioning or damaged components of cells to maintain cellular homeostasis and thus has a very significant role in cell development, differentiation and death.
LncRNAs have emerged as non-canonical regulators that take part in a collection of pathophysiological processes, including autophagy, by directly binding to RNA, DNA or protein.
LncRNAs generally modulate autophagy via regulating the expression of ATG genes. They often function as competing endogenous RNAs (ceRNAs) to modulate autophagy-related microRNAs (miRNAs).
Autophagy may, in turn, regulate lncRNA expression, although only one such lncRNA has been found so far.
Open Questions
Can autophagy-related lncRNAs directly regulate ATG genes apart from indirect modulation via miRNAs or epigenetic modification enzymes, and if so, how?
Among the several ATG genes that are simultaneously regulated by one autophagy-related lncRNA, is there a chief gene that determines the occurrence or repression of autophagy?
How does autophagy regulate the levels of lncRNAs? Does it degrade them directly in a selective manner or regulate them indirectly?
LncRNAs are non-coding RNAs longer than 200 nucleotides.1, 2 They have recently been found to have a crucial role in various fundamental physiopathologic processes, such as carcinogenesis, as well as autoimmune, cardiovascular and neurological diseases.3, 4, 5 LncRNAs can be classified according to their gene locations or functions. On one hand, lncRNAs can be named after their gene loci relative to adjacent protein-coding genes, which are antisense lncRNAs, intronic lncRNAs, bidirectional lncRNAs and intergenic lncRNAs (also named lincRNAs).6, 7, 8, 9 On another, lncRNAs can act as decoys, scaffolds, guides and enhancers to regulate DNA, RNA and proteins, on the basis of their function.10, 11, 12, 13, 14 In addition, lncRNAs have recently been documented to serve as competing endogenous RNAs (ceRNAs), which can sequester microRNAs (miRNAs) from targeted mRNAs sharing the same miRNA response elements (MREs), thereby regulating the expression of the targeted mRNAs.15 Owing to the increasing number of functionally characterized lncRNAs, the classification is constantly being updated. In this review, we will summarize the functions of lncRNAs involved in autophagy, a self-digestive process that helps to maintain cellular homeostasis.16, 17
Macroautophagy (hereafter referred to as autophagy) is a process that delivers cytoplasm components, enclosed in double-membrane vesicles, to lysosomes for degradation.16 By doing so, autophagy has a critical role in maintaining cellular homeostasis in response to various environmental stresses, such as nutrient deprivation and hypoxia, as well as chemical and physical damage.16, 17 Therefore, autophagy is crucial in various pathological and physiological processes such as immunity, cancer, cardiovascular diseases and neurodegenerative diseases.18, 19, 20
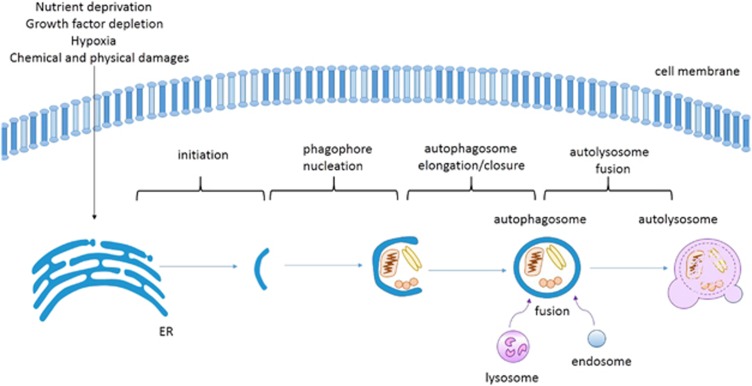
To date, at least 37 autophagy-related (ATG) genes are known to exist in yeast, and many of their orthologs have been identified in mammals.21, 22 We will discuss the crosstalk between autophagy-related lncRNAs and various ATG genes based on the four well-defined steps of autophagy: initiation, phagophore nucleation, autophagosome elongation/closure and autolysosome fusion23 (Figure 1). Certainly, lncRNAs may target several ATG genes at the same time and thus regulate autophagy at different stages. In this case, we will classify them into the category of most relevant stage.
Figure 1.
The simple model of autophagy pathway. Diversities of stimuli for autophagy (such as amino-acid starvation, growth factor depletion, hypoxia, chemical and physical damage) initiate formation of the isolation membrane (or ‘phagophore’) from endoplasmic reticulum (ER, the best recognized origin for autophagosome membrane). Subsequently, the isolation membrane encloses cytoplasmic components as it elongates sequentially and its closure lead to the birth of ‘autophagosome’. Then, autophagosome fuses with endosomes and lysosomes to form autolysome in which the enclosed cytoplasmic materials are degraded by lysosomal
LncRNAs Involved in Regulating Autophagy via ATG Genes
LncRNAs related to autophagy initiation
Adenine monophosphate-activated protein kinase (AMPK) and mammalian target of rapamycin complex 1 (mTORC1) are two major proteins that sense external stimuli for autophagy initiation.24, 25, 26 Upon energy limitation, AMPK is phosphorylated and activated by increased AMP to inhibit mTORC1 and to activate the ATG1/ULK1/2 (yeast/mammal) complex, leading to the initiation of autophagy.24, 25, 26 In addition, mTORC1 can be activated by class I phosphatidylinositol 3-kinase (PI3K)/AKT signaling, and thus the PI3K/AKT/mTOR pathway is negatively linked to autophagy initiation.27, 28 Once autophagy is initiated, ATG17 forms a complex with ATG29 and ATG31, and then interacts with ATG1 and ATG13 to mediate the assembly of pre-autophagosomal structure (PAS), (the site of autophagosome formation) in yeast.29, 30 Similarly, in mammalian cells, ULK1/2 (mammal ortholog of ATG1) forms a stable complex with ATG13, FIP200/RB1CC1 and ATG101 and is subsequently transferred to omegasomes (the cradle for autophagosome biogenesis).31, 32
A study reveals that high glucose reduces the level of lncRNA H19, which subsequently relieves the transcriptional repression of DIRAS3, and ultimately induces autophagy initiation by repressing PI3K/AKT/mTOR pathway.33 As marked activation of autophagy by high glucose has been shown to be detrimental to cardiac function, overexpression of H19 helps to alleviate this tendency.33 In addition, downregulation of H19 can increase Beclin 1 (mammal orthologous of ATG6) and ATG7 expression, which is a good indicator for further investigation of the association between H19 and autophagy.33, 34 Contrarily, another study has found that exogenous overexpression of H19 induces autophagic cell death in cerebral ischemia and reperfusion (I/R) injury.34 Researchers confirmed that H19 induces autophagy by inhibiting DUSP5, a mitogen-activated protein kinase phosphatase. DUSP5 is known to repress phosphorylation of ERK1/2, and activation of the latter has been reported to provoke autophagy.35 The opposite effects of H19 on autophagy in diabetic cardiomyopathy and cerebral I/R injury indicate that conditional gene interference with H19 may be an efficient therapeutic approach to different pathological processes.
A recent study suggests that the lncRNA neighbor of BRCA1 gene 2 (NBR2) can bind to AMPK and promote its activation. Intriguingly, the expression of NBR2 also can be induced by the increasing activation of AMPK under energy stress. Thus, AMPK/NBR2 forms a feed-forward loop to sustain AMPK activation in response to energy stress, which is a protective factor for normal cells to resist tumors.36, 37 Indeed, NBR2 expression is reduced in human cancers, and loss of its function is correlated with poor prognosis for cancer patients.36, 38, 39 This study serves as an interesting example illustrating that lncRNAs can regulate protein activation through direct binding. Future work is needed to demonstrate this functional model of lncRNAs and to address how it takes effect: through changing the conformation of the protein or altering its affinity for other regulatory factors? Another study demonstrates that miR-19a can negatively regulate NBR2 and AMPK, probably by base pairing between miR-19a and NBR2 or miR-19a and PRAA1 (the gene encoding AMPK).40 The upregulation of miR-19a in acute liver failure (ALF) results in the downregulation of NBR2 and AMPK expression, which represses autophagy. In light of the protective role of autophagy in the progression of ALF, activation of the miR-19a-NBR2/AMPK regulatory axis exacerbates ALF.40 This study gave rise to the hypothesis of a ceRNA network in which NBR2 might act as a sponge for miR-19a and prevent it from binding to AMPK mRNA.
As a multitargeted tyrosine kinase inhibitor (TKI), sorafenib has been demonstrated to induce autophagic hepatocellular carcinoma (HCC) cell death, and thus the blockage of autophagy facilitates sorafenib resistance in HCC.41, 42 Several miRNAs, including miR-21, miR-216a, miR-217 and miR-494, have been shown to confer sorafenib resistance in HCC by inhibiting autophagy through targeting phosphatase and tensin homolog (PTEN).42, 43, 44 For the sake of reversing sorafenib resistance, an artificial long non-coding RNA (AlncRNA), Ad5-AlncRNA, was constructed to target these miRNAs simultaneously.45 Overexpression of Ad5-AlncRNA in HCC can sequester these miRNAs from binding to the 3'-UTR of PTEN mRNA. As a result, PTEN is upregulated to repress AKT/mTOR activity and then activates autophagy, sequentially reversing sorafenib resistance.45 Thus, artificial lncRNAs targeting several miRNAs of the same mRNA will be a potent therapeutic strategy for diseases.45, 46
IFN-γ induced by Mycobacterium bovis BCG can repress lncRNA maternally expressed gene 3 (MEG3), which decreases p70-S6K (Thr389) phosphorylation subsequently, a downstream factor of mTOR complex, indicating mTOR inactivation. As a result, autophagy is activated and eradication of intracellular Mycobacterium bovis BCG is enhanced.47 Similar to pathogen infection in macrophages, tumor cells may also trigger autophagy to survive under various stresses through repressing MEG3. Indeed, the MEG3 expression level is significantly reduced in bladder cancer cells, leading to increased autophagy flux and cell proliferation.48 In addition, MEG3 has also been found to promote cisplatin-induced apoptosis by inhibiting autophagy in human glioma cells.49 By contrast, another study shows that overexpression of MEG3 induces autophagy, thereby repressing tumorigenesis and progression of epithelial ovarian carcinoma by regulating ATG3. MEG3 can bind to ATG3 protein and protect ATG3 mRNA from degradation.50 However, more work is needed to elucidate how MEG3 modulates ATG3 protein and mRNA.
Studies have demonstrated that downregulation of the lncRNA HOTARIM1 can block autophagy, and thus inhibit all-trans retinoic acid (ATRA)-induced autophagic degradation of PML-RARA in promyelocytic leukemia (PML) cells.51, 52, 53 HOTAIRM1 may provoke autophagy by preventing miR-20a/106b and miR-125b from decreasing ULK1, E2F1 and DRAM2 expression, thus contributing to autophagy-dependent degradation of PML-RARA.53, 54, 55, 56 This indicates that overexpression of HOTAIRM1 might be a potential therapeutic measure for PML.
The lncRNA PTENP1 shares a similar 3'-UTR with the tumor-suppressor gene PTEN, and protects PTEN from miRNA-mediated silencing.57 Sustained PTENP1 expression in HCC cells upregulates PTEN expression and suppresses the PI3K/AKT signaling cascade, which results in the induction of autophagy, as well as the inhibition of cell proliferation and migration/invasion.58 In addition, PTENP1 can antagonize miR-17 and miR-20a to enhance the expression of ULK1, ATG7 and p62/SQSTM1 (sequestosome 1).58, 59, 60 Furthermore, overexpression of PTENP1 can suppress mTOR phosphorylation and downregulate Bcl-2 expression.58 Nevertheless, it remains to be determined whether molecules in this complicated regulatory network are directly regulated by PTENP1 or indirectly altered secondary to some initial factors. Interestingly, researchers have constructed Sleeping Beauty (SB)-based hybrid baculovirus (BV) vectors for sustained PTENP1 expression.58 This system could be a promising instrument for endogenous overexpression of lncRNAs in specific tissues for therapeutic purposes.
The lncRNA regulator of insulin sensitivity and autophagy (Risa) can affect insulin sensitivity by altering autophagic activity. Indeed, knockdown of Risa increases the phosphorylation of ULK1 (Ser757), which contributes to autophagy activation and attenuates insulin resistance.61 Unfortunately, there are still divergent schools of thought regarding what role autophagy has in insulin resistance or diabetes.62, 63 Thus, whether knockdown of Risa can alleviate insulin resistance by promoting autophagy in patients with diabetes requires further exploration.
The lncRNA AK156230 has been found to repress replicative senescence (RS) in mouse embryonic fibroblasts (MEFs), and one of the downstream pathways involved is autophagy.64 Pathway analysis shows that the mTOR signaling pathway is associated with AK156230 knockdown, and transcriptional levels of ATG genes including ULK2, ATG7 and ATG16L apparently decrease accordingly.64 Vague though the facts are, AK156230 seems to participate in autophagy induction, as its downregulation results in a deficiency of autophagy, which may accelerate aging.65
Another lncRNA, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), has attracted a great deal of interest for its elusive role in autophagy regulation.66, 67, 68, 69, 70, 71 MALAT1 is upregulated and acts as a sponge for miR-26b to upregulate its target ULK2 in cerebral I/R injury.71 In light of the protective effect of autophagy against I/R in brain microvascular endothelial cells, the enhanced activity of the MALAT1/miR-26b/ULK2 regulatory axis appears to be a self-defense mechanism in response to ischemic stroke, the pathological basis of which is I/R injury.71 Another study demonstrates that MALAT1 silencing notably elevates p62 and decreases LAMP2 expression, which is essential for the fusion of autophagosomes and lysosomes.66 Moreover, MALAT1 can act as a 'sponge' for miR-216b to induce autophagy, probably through derepressing Beclin 1 expression, which, in consequence, ameliorates the multidrug resistance of HCC cells.67, 68 Interestingly, a newly published article reports that MALAT1 knockdown can indeed reduce the expression of Beclin 1 in multiple myeloma tissues, although the details of the mechanism are not known.69 In contrast to the studies mentioned above, MALAT1 inhibition has been found to induce autophagy in diffuse large B-cell lymphoma (DLBCL) cells and improve its response to adriamycin treatment.70
As summarized in Figure 2 and Table 1, the lncRNA NBR2 promotes autophagy initiation by directly activating AMPK.36 The lncRNAs Ad5-AlncRNA45 and PTENP158 provoke autophagy initiation by repressing the PI3K/AKT/mTOR pathway, whereas MEG347 and H1933 has the opposite effect. The lncRNAs HOTAIRM1,53 PTENP158 and MALAT171 enhance autophagy initiation by increasing ULK expression. Meanwhile, the lncRNA Risa negatively regulates autophagy initiation by inhibiting ULK1 activation.72 In addition, the lncRNAs H19,33 MEG3,50 AK156230,64 PTENP158 and MALAT171 can also influence other ATG genes and autophagy adaptor proteins involved in later steps of autophagy regulation.
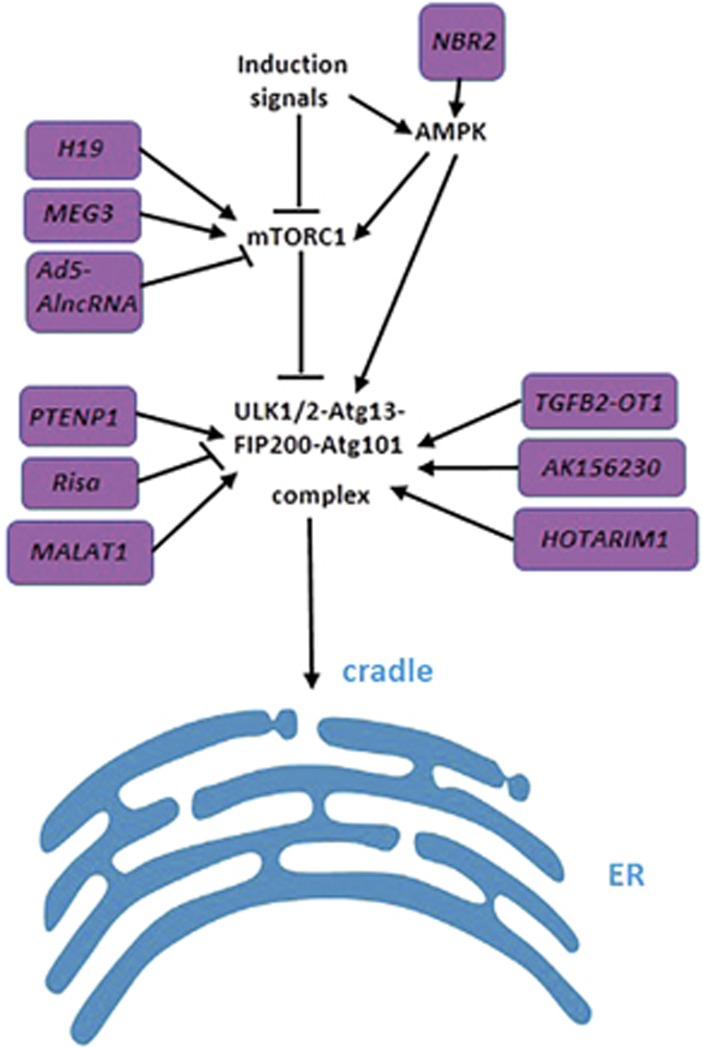
Figure 2.
Brief summarization of the initiation step of mammalian core autophagy machinery and its regulation by lncRNAs. Upon energy limitation, autophagy can be initiated by AMPK and mTORC1. Then, ULK1 complex composed of ULK1, ATG13, FIP200 and ATG101 is activated. lncRNA NBR2 directly modulates AMPK and mTORC1 are positively regulated by MEG3 and H19 and negatively regulated by Ad5-AlncRNA. In addition, lncRNA PTENP1, Risa, MALAT1, TGFB2-OT1, AK156230 and HOTARIM1 collectively regulate ULK1 complex
Table 1. lncRNAs involved in autophagy regulation.
| Autophagy stages | Relevant autophagy-related genes or proteins | lncRNA |
|---|---|---|
| Initiation | AMPK | NBR236, 37 |
| mTORC1 | Ad5-AlncRNA45MEG347H1933 | |
| ATG13 | TGFB2-OT1/FLJ1181289, 90 | |
| ULK1/2 | PTENP158, 59, 60 Risa72 MALAT171 HOTAIRM153, 134, 135 AK15623064 | |
| Vesicle nucleation | Beclin 1 | PCGEM1100H1933, 136GAS594 MALAT169 loc14688083Linc-ROR81 |
| Bcl-2 | PTENP158, 59, 60 | |
| Autophagasome elongation/closure | ATG7 | TGFB2-OT1/FLJ1181289, 90 AK15623064 PTENP158, 59, 60 GAS594APF106HOTAIR109 H1933HULC104 |
| ATG5 | PCGEM1100HNF1A-AS195, 137Chast120GAS594 HULC104 | |
| ATG3 | MEG350 TGFB2-OT1/FLJ1181289, 90 PCGEM1100 GAS594 HOTAIR109 | |
| ATG12 ATG16L | PCGEM1100HNF1A-AS127, 95GAS594HOTAIR108 AK15623064 | |
| p62 | PTENP158, 59, 60 MALAT166 | |
| Fusion | Plekhm1 LAMP2 | Chast120 MALAT166 |
LncRNAs related to phagophore nucleation
Once activated and translocated to PAS/omegasome, the ATG1/ULK1 complex can activate the class III PI3K complex, which mainly comprises Vps34, Vps15, vps30/Beclin 1 (yeast/mammal) and ATG14/Barkor (yeast/mammal), to generate phosphatidylinositol 3-phosphate (PI3P).73, 74, 75 PI3P recruits double FYVE-containing protein 1 (DFCP1) to promote the formation of the omegasome.76 Meanwhile, other essential regulators such as ATG9, ATG18 (WIPI1/2/3/4) and VMP1 are present on the autophagic membrane.30, 77 During the process of class III PI3K complex formation and function, BCL-2 and Rubicon act as two negative regulators.78, 79
Two studies have demonstrated that the inhibition of LincRNA, regulator of reprogramming (Linc-ROR) can elicit autophagy by upregulating Beclin 1 expression, and revise gemcitabine and tamoxifen resistance in breast cancer respectively.80, 81 However, further experiments need to reveal how Linc-ROR regulates Beclin 1 expression and whether knockdown of Linc-ROR could be feasible in clinical practice.
LncRNA loc146880 reveals to be intricately related to autophagy and the biogenesis of lung cancer.82, 83 High expression of reactive oxygen species (ROS) induced by PM2.5 (a class of particulate matters, <2.5μm in diameter) exposure enhances lncRNA loc146880 expression, which results in autophagy activation and subsequently contributes to lung cell migration and invasion. Beclin 1 mRNA is upregulated along with the increase of loc146880 expression, but the underlying mechanism is far from fully explained.83
LncRNA AC023115.3 is upregulated by cisplatin and promotes cisplatin-induced apoptosis by impeding autophagy in glioma. Further mechanistic studies reveal that AC023115.3 can elevate GSK3β expression by sponging miR-26a.84 GSK3β promotes the degradation of Mcl1, a member of BCL-2 family that sequesters Beclin 1 from the class III PI3K complex.85 Further studies are needed to address whether Beclin 1 will activate the class III PI3K complex to a greater extent upon overexpression of AC023115.3 as hypothesized. Regardless, the study reveals that the AC023115.3/miR-26a/GSK3β signaling cascade has a significant role in promoting chemosensitivity in gliomas and serves as an exciting indicator to exploit the association between AC023115.3 and Beclin 1.
Collectively, both Linc-ROR and loc146880 can impact vesicle nucleation by modulating Beclin 1 gene or protein expression81, 83 (Figure 3; Table 1). In addition, the association between AC023115.3 and Beclin 1 still needs further investigation.
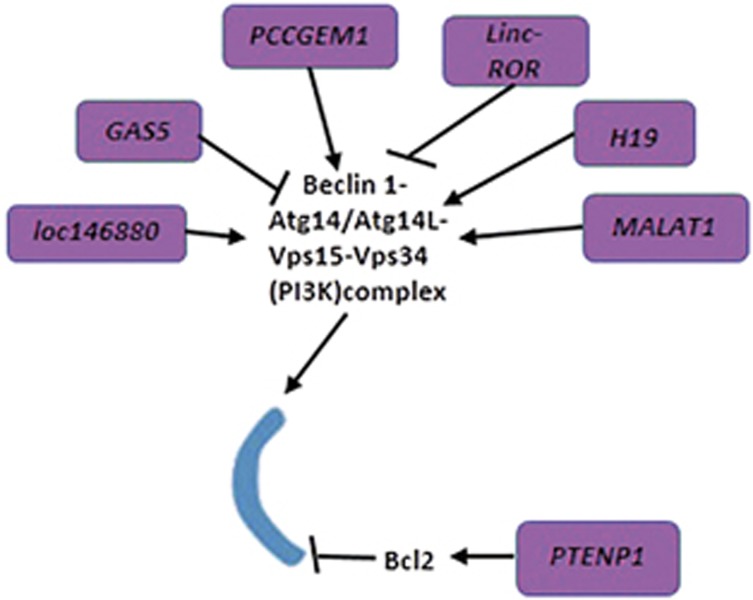
Figure 3.
Brief summarization of the vesicle nucleation step of mammalian core autophagy machinery and its regulation by lncRNAs. Vesicle nucleation is predominantly modulated by class III PI3K complex, which comprises Vps34, Vps15, Beclin 1 and ATG14/ATG14L. During the process of PI3K complex formation and function, BCL-2 and Rubicon act as two negative regulators. lncRNA GAS5, PCGEM1 and Linc-ROR regulate PI3K complex, among which, Beclin 1 itself is positively regulated by lncRNA PCGEM1, MALAT1, H19 and loc146880, whereas negatively regulated by GAS5, Linc-ROR and H19. Moreover, lncRNA PTENP1 can repress autophagy via increasing BCL-2 expression
lncRNAs related to autophagosome elongation/closure
The two unique ubiquitin-like conjugation systems have crucial roles in the elongation and closure of the isolated membrane. Driven by ATG7 (E1-like enzyme) and ATG10 (E2-like enzyme), ATG12 conjugates to ATG5 and then interacts with ATG16 (mammal ortholog is ATG16L) to form the ATG12-ATG5-ATG16 complex.86, 87, 88 Subsequently, the ATG12-ATG5-ATG16 complex, ATG7 and ATG3 (E2-like enzyme) jointly transform ATG8 (LC3 in mammals) from its cytosolic soluble isoform (LC3-I) to its membrane-anchored isoform (LC3-II).23 In addition, adaptor proteins such as ATG19 (an adaptor for the Cvt pathway) and ATG32 in yeast, as well as the neighbor of BRCA1 gene 1 (NBR1), Nix (also called Bnip3L) and p62 in mammals, selectively mediate the degradation of proteins or organelles by recruiting them to autophagosomes via binding to LC3-II.31
A study reveals that the lncRNA TGFB2 overlapping transcript 1 (TGFB2-OT1, also known as FLJ11812) can be upregulated by vascular endothelial cell (VEC) inflammation triggers and function as the sponge for miR-3960, miR-4488 and miR-4459, thereby increasing the expression levels of their targets, such as ATG13, ceramide synthase 1 (CERS1) and La ribonucleoprotein domain family, member 1 (LARP1). In addition, overexpression of TGFB2-OT1 can also increase ATG3, ATG7 and p62 expression, probably through upregulating LARP1 by sponging miR-4459, an RNA-binding protein related to transcript stability and translation of mRNAs.89, 90 Moreover, when TGFB2-OT1 prevents miR-3960-mediated repression of CERS1, production of C18-ceramide is increased to induce mitophagy (a class of selective autophagy targeting dysfunctional mitochondria) by directly interacting with LC3-II-containing autophagolysosomes upon Drp1-dependent mitochondrial fission.89, 91 In summary, ectopic expression of TGFB2-OT1 induced by VEC inflammation triggers activates autophagy via increasing ATG13, ATG3, ATG7 and p62 expression, and a small molecular inhibitor 3BDO significantly decreases TGFB2-OT1 level and inhibits the subsequent autophagic and inflammatory reaction.89 Given that autophagy and inflammation have an intricate correlation, intervening in TGFB2-OT1 could be a possible treatment strategy for infectious and autoimmune diseases.92
The lncRNA growth arrest-specific 5 (GAS5) has been reported to inhibit autophagy and enhance cisplatin sensitivity in NSCLC cells.93 In contrast to its deletion in several species of tumors, GAS5 is upregulated in osteoarthritis (OA), repressing autophagy and stimulating the apoptosis of OA chondrocytes, which is a key determinant responsible for cartilage degradation and thus OA pathogenesis.94 High expression of GAS5 in OA represses autophagy possibly through downregulating Beclin 1, ATG3, ATG5, ATG7, ATG12 and LC3B expression.94 It seems that GAS5-repressed autophagy is beneficial for enhancing drug sensitivity but results in the occurrence of OA. Therefore, different modes of interference with GAS5 may be essential for distinct therapeutic purposes.
The lncRNA HNF1A-AS1 can sequester miR-30b from binding to its target ATG5 and thereby provoke autophagy in HCC.95 In addition, Beclin 1 and ATG12 have also been defined as targets of miR-30b in a previous study, indicating that, aside from ATG5, HNF1A-AS1 might also upregulate Beclin 1 and ATG12 expression to promote vesicle nucleation and autophagosome elongation/closure.95, 96 Considering that autophagy was previously confirmed to promote HCC, it is likely that HNF1A-AS1 facilitates HCC biogenesis by promoting autophagy.97, 98, 99
Exogenous overexpression of the lncRNA, prostate cancer gene expression marker 1 (PCGEM1), can increase the mRNA levels of Beclin 1, ATG3, ATG5 and ATG12, indicating that PCGEM1 may be involved in the induction of autophagy, and promote the proliferation of human synoviocytes.100 PCGEM1 expression is associated positively with mortality rate in African-American patients with prostate cancer, probably because of PCGEM1-induced autophagy.101 However, further analysis is required to confirm whether and how PCGEM1 affects autophagy.
Mounting evidences reveal that LncRNAs harbor much more tissue and developmental stage specificity than coding transcripts.102 In line with this discovery, lncRNA highly upregulated in liver cancer (HULC) has been found to predominantly express in primary HCC and metastatic hepatic carcinoma.103 One study elucidates clearly how the 'HULC/USP22/Sirt1/protective autophagy' pathway attenuates the chemosensitivity of HCC.104 In detail, HULC can upregulate ubiquitin-specific peptidase (USP22) expression level via repressing miR-6825-5p, miR-6845-5p and miR-6886-3p, and ectopic expression of USP22 can stabilize Sirt1 protein, which promotes protective autophagy by deacetylating Atg5 and Atg7.104 Mazy as it seems, this HULC-regulated autophagy pathway makes a promising target to address chemoresistance in HCC. Another work indicates that HULC overexpression increases proliferation and invasion of gastric cancer (GC) cells probably through arousing autophagy activation.105 Clearly, these findings put forward a new topic worthy of studying that how HULC provokes autophagy in GC and whether HULC can be identified as a biomarker both in HCC and GC.
In anoxia/reoxygenation (A/R)-induced autophagy of cardiomyocytes, the lncRNA autophagy promoting factor (APF) is upregulated to protect ATG7 from being downregulated by miR-188-3p, and thereby promotes autophagic death of cardiomyocytes.106 Intriguingly, despite the poor conservation of full-length APF across species, the binding site for miR-188-3p is highly conserved, highlighting the significance of the APF/miR-188-3p/ATG7 regulatory axis in autophagy activation.106
The lncRNA HOTAIR is well characterized for recruiting the epigenetic modification factors, such as polycomb repressive complex 2 (PRC2), to regulate gene expression, thereby promoting tumor cell proliferation and migration.11, 107 Analogously, HOTAIR is upregulated in chondrosarcoma and induces DNA methylation of miR-454-3p promoter regions by recruiting EZH2 and DNA methyltransferase 1 (DNMT1), which markedly silences miR-454-3p expression. ATG12 and STAT3 are targets of miR-454-3p, providing one molecular dissection of HOTAIR deficiency-induced autophagy repression and apoptosis.108 Another study demonstrates that HOTAIR is upregulated in HCC to promote HCC cell proliferation, probably by enhancing ATG3 and ATG7 expression to expedite autophagy flux.109 As HOTAIR can interact with numerous miRNAs, such as miR-34a, miR-331-3P, miR-130a and miR-454-3p, we should recall that HOTAIR regulates autophagy in two ways. First, HOTAIR may serve as a scaffold to recruit epigenetic modification enzymes to inhibit miRNA transcription; second, HOTAIR can act as a sponge to sequester miRNAs from their targets. In both cases, HOTAIR will upregulate specific genes targeted by corresponding miRNAs, possibly including some ATG genes. For instance, miR-130a can repress the transcription of ATG5 and ATG16L, and Atg12 is a target of miR-454-3p.12, 108, 110, 111, 112, 113, 114
Taken together, as shown in Figure 4 and Table 1, the lncRNAs TGFB2-OT1,89, 90 GAS5,94 HNF1A-AS1,95, 96 PCGEM1,100 HULC,104 APF106 and HOTAIR,108, 109 promote autophagosome elongation/closure by elevating the expression of ATG genes involved in the ubiquitin-like conjugation systems. Meanwhile, the lncRNA GAS594 may repress and the lncRNA PCGEM1100 may activate autophagy nucleation by interacting with Beclin 1. The lncRNA TGFB2-OT1 activates autophagy initiation by increasing ATG13 expression.89
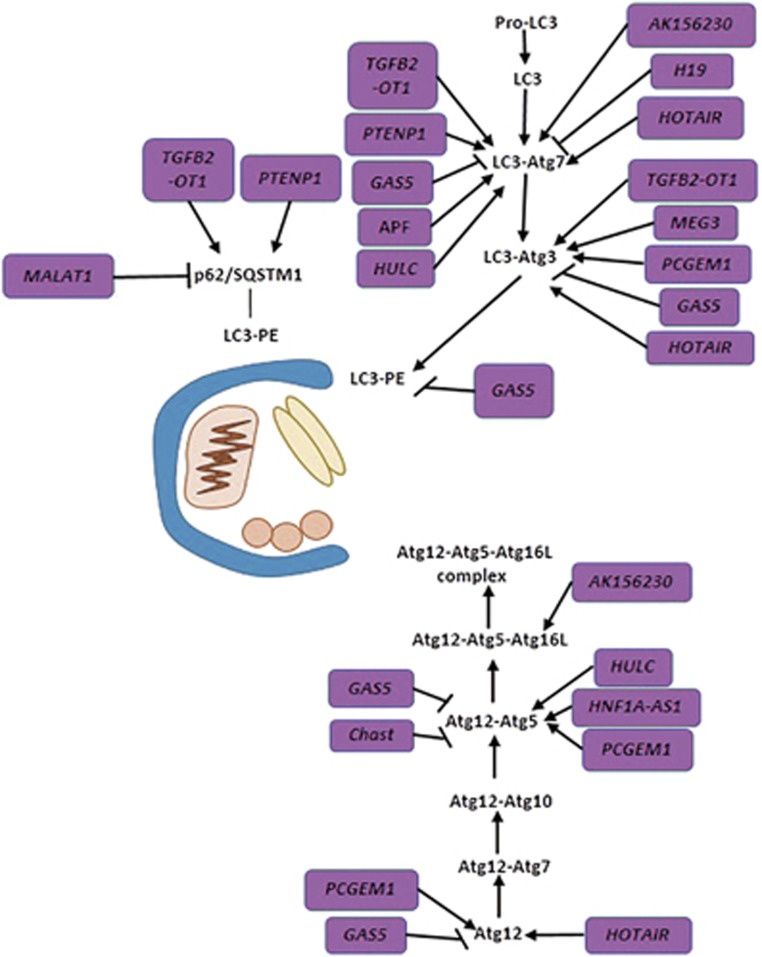
Figure 4.
Brief summarization of the autophagosome elongation/closure step and its regulation by lncRNAs. Autophagosome elongation/closure is controlled by two ubiquitin-like conjugation systems and scaffold proteins, such as p62. ATG12-ATG5-ATG16L complex, ATG7 and ATG3 jointly transform LC3-I to LC3-II. Meanwhile, p62 selectively mediates the degradation of proteins or organelles by recruiting them to autophagosome via its binding to LC3. Numerous lncRNAs are involved in this process, containing TGFB2-OT1, PTENP1, GAS5, APF, AK156230, HOTAIR, HULC and H19 (ATG7), MEG3,TGFB2-OT1, PCGEM1, GAS5 and HOTAIR (ATG3), HNF1A-AS1, PCGEM1, GAS5, HULC and Chast (ATG5), PCGEM1, HOTAIR and GAS (ATG12),AK156230 (ATG16L), PTENP1, TGFB2-OT1 and MALAT1 (p62)
LncRNAs related to autolysosome fusion
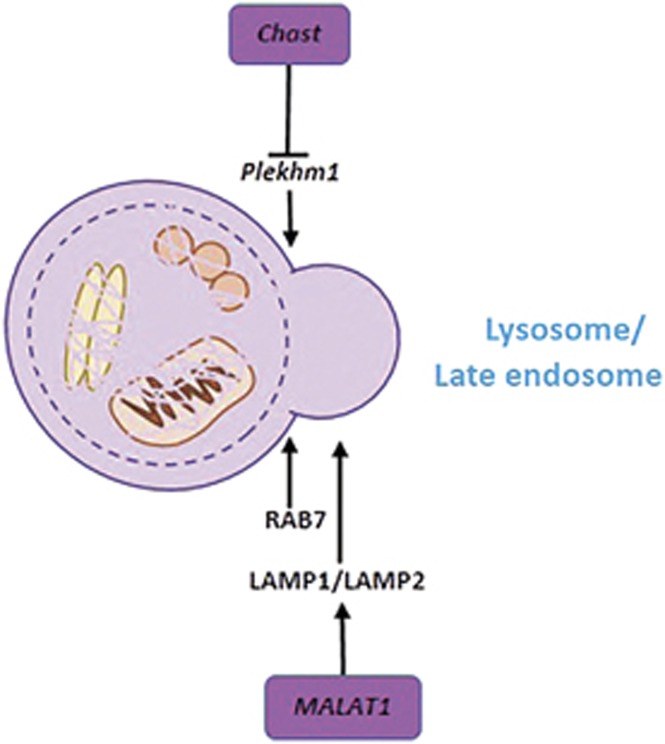
The final step of autophagy flux is the fusion of the autophagosome and the lysosome to form an autolysosome, where the autophagic cargo is degraded.31 The core molecules in this stage include the Rab-SNARE system and the lysosome membrane proteins LAMP1 and LAMP2.115, 116, 117, 118 In addition, adaptor proteins are necessary to link endocytic and autophagic pathways to the lysosome. Pleckstrin homology domain-containing protein family M member 1 (Plekhm1) is one such adaptor protein, which contains an LC3-interacting region and directly interacts with the homotypic fusion and protein sorting complex to mediate the fusion of endosomes and autophagosomes with lysosomes.119
The lncRNA cardiac hypertrophy-associated transcript (Chast) can suppress autophagy via downregulating Plekhm1 and possibly ATG5 expression to induce cardiomyocyte hypertrophy (Figure 5).120 This study sheds light on the mechanism of Chast/Plekhm1 interaction during autolysosome fusion and implies that Chast is a possible target for the prevention of cardiac remodeling.
Figure 5.
Brief summarization of the autolysosome fusion and its regulation by lncRNAs. Fusion is the final step that autophagosome fuses with endosome and lysosome to form autolysosome, where the autophagic cargo is degraded. The lncRNA chast and MALAT1 have been found to regulate the step
Additional lncRNA Regulators of Autophagy
Some lncRNAs, including lncRNA 7SL,121 BANCR,122 PCA3,123 LINC01116124 and CTA,125 also have been found to be related to autophagy, although the evidence of their correlation with ATG genes is lacking. BANCR122 can provoke autophagy flux to facilitate tumor proliferation, whereas 7SL,121 PCA3,123 LINC01116124 and CTA125 have the opposite effect.
LncRNAs Regulated by Autophagy
As an increasing number of lncRNAs have been identified to regulate autophagy, it would be interesting to know whether autophagy could also affect the expression of lncRNAs. Autophagy can degrade several types of RNAs and associated ribonucleoprotein complexes.126 Plasmacytoma variant translocation 1 (PVT1) is the sole lncRNA reported to be regulated by autophagy so far. PVT1 is upregulated in diabetes, and autophagy repression decreases its transcriptional level.127 The elevation of PVT1 mediated by autophagy functions crucially in the development and progression of diabetic nephropathy.128, 129 Clearly, PVT1 is probably not degraded by autophagy, as it is downregulated when autophagy is repressed.127 Thus, extensive further investigations are needed to demonstrate what determinants participate in this process.
Conclusions
Given the limitations of the research that has been conducted to date, we have gained limited knowledge of the underlying mechanisms of regulation between identified lncRNAs and autophagy. The majority of lncRNAs typically function as sponges to sequester autophagy-related miRNAs from their targets.130, 131 Certainly, lncRNAs have more complicated functions in autophagy regulation that are waiting to be elucidated, including but not limited to chromatin and histone remodeling, transcriptional regulation and protein–protein interactions.132 Current studies also indicate that we may need to classify lncRNAs according to their roles in distinct types of autophagy, such as mitophagy, to probe their function more specifically.47, 89 From the growing knowledge based on lncRNAs and autophagy, we have formed the impression that most lncRNAs involved in autophagy have parts in tumorigenesis and that this function can be interpreted through their association with autophagy. In consideration of the intimate linkage between lncRNAs and autophagy, it would be possible to develop lncRNA-based approaches to monitor or interfere with autophagy flux. As lncRNA expression is prevalent along the developmental and spatial axis, and several autophagy-associated lncRNAs described in this review virtually exhibit tissue specificity, such as HULC and PCA3, lncRNAs may serve as biomarkers of specific diseases, and pertinent therapeutic measures may be developed.103, 133 Finally, both lncRNAs and autophagy are involved in a vast range of diseases including cancer; therefore, a joint intervention targeting lncRNAs and autophagy may be a promising therapeutic method.
Acknowledgments
This work was supported by National Natural Science Foundation of China (81301706; 81372178; 81672360).
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M Agostini
The authors declare no conflict of interest.
References
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011; 12: 861–874. [DOI] [PubMed] [Google Scholar]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016; 17: 47–62. [DOI] [PubMed] [Google Scholar]
- Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol 2013; 11: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 2013; 14: 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol 2013; 425: 3723–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012; 81: 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcid EA, Tsukiyama T. Expansion of antisense lncRNA transcriptomes in budding yeast species since the loss of RNAi. Nat Struct Mol Biol 2016; 23: 450–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Li D, Wang G, Zhang C, Zhang M, Zhang W et al. Three intronic lncRNAs with monoallelic expression derived from the MEG8 gene in cattle. Anim Genet 2017; 48: 272–277. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 2009; 106: 11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 2010; 3: ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010; 329: 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007; 129: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464: 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matharu N, Ahituv N. Minor loops in major folds: enhancer-promoter looping, chromatin restructuring, and their association with transcriptional regulation and disease. PLoS Genet 2015; 11: e1005640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov 2013; 3: 1113–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 2000; 290: 1717–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 2007; 8: 931–937. [DOI] [PubMed] [Google Scholar]
- Rebecca VW, Amaravadi RK. Emerging strategies to effectively target autophagy in cancer. Oncogene 2016; 35: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM. Autophagy: in sickness and in health. Trends Cell Biol 2004; 14: 70–77. [DOI] [PubMed] [Google Scholar]
- Shibutani ST, Saitoh T, Nowag H, Munz C, Yoshimori T. Autophagy and autophagy-related proteins in the immune system. Nat Immunol 2015; 16: 1014–1024. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics 2013; 194: 341–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Baskaran S, Schekman R, Hurley JH. The protein-vesicle network of autophagy. Curr Opin Cell Biol 2014; 29: 18–24. [DOI] [PubMed] [Google Scholar]
- Kume S, Koya D. Autophagy: a novel therapeutic target for diabetic nephropathy. Diabetes Metab J 2015; 39: 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop EA, Tee AR. mTOR and autophagy: a dynamic relationship governed by nutrients and energy. Semin Cell Dev Biol 2014; 36: 121–129. [DOI] [PubMed] [Google Scholar]
- Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol 2011; 13: 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011; 13: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell 2007; 129: 1261–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heras-Sandoval D, Perez-Rojas JM, Hernandez-Damian J, Pedraza-Chaverri J. The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration. Cell Signal 2014; 26: 2694–2701. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 2001; 20: 5971–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papinski D, Kraft C. Atg1 kinase organizes autophagosome formation by phosphorylating Atg9. Autophagy 2014; 10: 1338–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011; 27: 107–132. [DOI] [PubMed] [Google Scholar]
- Davies CW, Stjepanovic G, Hurley JH. How the Atg1 complex assembles to initiate autophagy. Autophagy 2015; 11: 185–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo C, Jiang R, Lin X, Shao M. LncRNA H19 inhibits autophagy by epigenetically silencing of DIRAS3 in diabetic cardiomyopathy. Oncotarget 2017; 8: 1429–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cao B, Han D, Sun M, Feng J. Long non-coding RNA H19 induces cerebral ischemia reperfusion injury via activation of autophagy. Aging Dis 2017; 8: 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Fu X, Xie J, Meng Z, Gu Y, Wang X et al. MiR-26a enhances autophagy to protect against ethanol-induced acute liver injury. J Mol Med (Berl) 2015; 93: 1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Xiao ZD, Han L, Zhang J, Lee SW, Wang W et al. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol 2016; 18: 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Xiao ZD, Gan B. An lncRNA switch for AMPK activation. Cell Cycle 2016; 15: 1948–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao ZD, Liu X, Zhuang L, Gan B. NBR2: a former junk gene emerges as a key player in tumor suppression. Mol Cell Oncol 2016; 3: e1187322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov SD, Matouskova E, Forejt J. Expression of BRCA1, NBR1 and NBR2 genes in human breast cancer cells. Folia Biol (Praha) 2001; 47: 120–127. [DOI] [PubMed] [Google Scholar]
- Liu YM, Ma JH, Zeng QL, Lv J, Xie XH, Pan YJ et al. MiR-19a affects hepatocyte autophagy via regulating lncRNA NBR2 and AMPK/PPARalpha in D-GalN/lipopolysaccharide-stimulated hepatocytes. J Cell Biochem 2017. (doi:10.1002/jcb026188). [DOI] [PubMed]
- Tai WT, Shiau CW, Chen HL, Liu CY, Lin CS, Cheng AL et al. Mcl-1-dependent activation of Beclin 1 mediates autophagic cell death induced by sorafenib and SC-59 in hepatocellular carcinoma cells. Cell Death Dis 2013; 4: e485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Dong X, Zhai B, Jiang X, Dong D, Li B et al. MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget 2015; 6: 28867–28881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, Ooi LL, Hui KM. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology 2013; 58: 629–641. [DOI] [PubMed] [Google Scholar]
- Liu K, Liu S, Zhang W, Jia B, Tan L, Jin Z et al. miR-494 promotes cell proliferation, migration and invasion, and increased sorafenib resistance in hepatocellular carcinoma by targeting PTEN. Oncol Rep 2015; 34: 1003–1010. [DOI] [PubMed] [Google Scholar]
- Tang S, Tan G, Jiang X, Han P, Zhai B, Dong X et al. An artificial lncRNA targeting multiple miRNAs overcomes sorafenib resistance in hepatocellular carcinoma cells. Oncotarget 2016; 7: 73257–73269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Sun B, Lin X, Zhao X, Ji W, He M et al. Therapeutic strategy with artificially-designed i-lncRNA targeting multiple oncogenic microRNAs exhibits effective antitumor activity in diffuse large B-cell lymphoma. Oncotarget 2016; 7: 49143–49155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawar K, Hanisch C, Palma Vera SE, Einspanier R, Sharbati S. Down regulated lncRNA MEG3 eliminates mycobacteria in macrophages via autophagy. Sci Rep 2016; 6: 19416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying L, Huang Y, Chen H, Wang Y, Xia L, Chen Y et al. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol Biosyst 2013; 9: 407–411. [DOI] [PubMed] [Google Scholar]
- Ma B, Gao Z, Lou J, Zhang H, Yuan Z, Wu Q et al. Long noncoding RNA MEG3 contributes to cisplatininduced apoptosis via inhibition of autophagy in human glioma cells. Mol Med Rep 2017; 16: 2946–2952. [DOI] [PubMed] [Google Scholar]
- Xiu YL, Sun KX, Chen X, Chen S, Zhao Y, Guo QG et al. Upregulation of the lncRNA Meg3 induces autophagy to inhibit tumorigenesis and progression of epithelial ovarian carcinoma by regulating activity of ATG3. Oncotarget 2017; 8: 31714–31725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakson P, Bjoras M, Boe SO, Simonsen A. Autophagy contributes to therapy-induced degradation of the PML/RARA oncoprotein. Blood 2010; 116: 2324–2331. [DOI] [PubMed] [Google Scholar]
- Wang Z, Cao L, Kang R, Yang M, Liu L, Zhao Y et al. Autophagy regulates myeloid cell differentiation by p62/SQSTM1-mediated degradation of PML-RARalpha oncoprotein. Autophagy 2011; 7: 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZH, Wang WT, Huang W, Fang K, Sun YM, Liu SR et al. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ 2016; 24: 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polager S, Ofir M, Ginsberg D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene 2008; 27: 4860–4864. [DOI] [PubMed] [Google Scholar]
- Eriksen AB, Torgersen ML, Holm KL, Abrahamsen G, Spurkland A, Moskaug JO et al. Retinoic acid-induced IgG production in TLR-activated human primary B cells involves ULK1-mediated autophagy. Autophagy 2015; 11: 460–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Lee HM, Park KS, Shin DM, Kim TS, Kim YS et al. MIR144* inhibits antimicrobial responses against Mycobacterium tuberculosis in human monocytes and macrophages by targeting the autophagy protein DRAM2. Autophagy 2017; 13: 423–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Yao W, Gumireddy K, Li A, Wang J, Xiao W et al. Pseudogene PTENP1 functions as a competing endogenous RNA to suppress clear-cell renal cell carcinoma progression. Mol Cancer Ther 2014; 13: 3086–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Tseng YW, Wu JC, Chen GY, Lin KC, Hwang SM et al. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and microRNA regulation. Biomaterials 2015; 44: 71–81. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010; 465: 1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol 2014; 15: 65–74. [DOI] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011; 331: 456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Jeong YT, Oh H, Kim SH, Cho JM, Kim YN et al. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med 2013; 19: 83–92. [DOI] [PubMed] [Google Scholar]
- He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 2012; 481: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YN, Cai MY, Xu S, Meng M, Ren X, Yang JW et al. Identification of the lncRNA, AK156230, as a novel regulator of cellular senescence in mouse embryonic fibroblasts. Oncotarget 2016; 7: 52673–52684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamini E. Autophagy: a cell repair mechanism that retards ageing and age-associated diseases and can be intensified pharmacologically. Mol Aspects Med 2006; 27: 403–410. [DOI] [PubMed] [Google Scholar]
- Li L, Chen H, Gao Y, Wang YW, Zhang GQ, Pan SH et al. Long noncoding RNA MALAT1 promotes aggressive pancreatic cancer proliferation and metastasis via the stimulation of autophagy. Mol Cancer Ther 2016; 15: 2232–2243. [DOI] [PubMed] [Google Scholar]
- Yuan P, Cao W, Zang Q, Li G, Guo X, Fan J. The HIF-2alpha-MALAT1-miR-216b axis regulates multi-drug resistance of hepatocellular carcinoma cells via modulating autophagy. Biochem Biophys Res Commun 2016; 478: 1067–1073. [DOI] [PubMed] [Google Scholar]
- Chen K, Shi W. Autophagy regulates resistance of non-small cell lung cancer cells to paclitaxel. Tumour Biol 2016; 37: 10539–10544. [DOI] [PubMed] [Google Scholar]
- Gao D, Xiao Z, Li HP, Han DH, Zhang YP. LncRNA MALAT-1 elevates HMGB1 to promote autophagy resulting in inhibition of tumor cell apoptosis in multiple myeloma. J Cell Biochem 2017; 118: 3341–3348. [DOI] [PubMed] [Google Scholar]
- Li LJ, Chai Y, Guo XJ, Chu SL, Zhang LS. The effects of the long non-coding RNA MALAT-1 regulated autophagy-related signaling pathway on chemotherapy resistance in diffuse large B-cell lymphoma. Biomed Pharmacother 2017; 89: 939–948. [DOI] [PubMed] [Google Scholar]
- Li Z, Li J, Tang N. Long noncoding RNA Malat1 is a potent autophagy inducer protecting brain microvascular endothelial cells against oxygen-glucose deprivation/reoxygenation-induced injury by sponging miR-26b and upregulating ULK2 expression. Neuroscience 2017; 354: 1–10. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hu Y, Sun C, Zhuo S, He Z, Wang H et al. Down-regulation of Risa improves insulin sensitivity by enhancing autophagy. FASEB J 2016; 30: 3133–3145. [DOI] [PubMed] [Google Scholar]
- He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol 2010; 22: 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell 2010; 40: 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 2009; 10: 458–467. [DOI] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008; 182: 685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci 2013; 126: 2534–2544. [DOI] [PubMed] [Google Scholar]
- Lindqvist LM, Vaux DL. BCL2 and related prosurvival proteins require BAK1 and BAX to affect autophagy. Autophagy 2014; 10: 1474–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Yue Z. Atg14L and Rubicon: Yin and Yang of Beclin 1-mediated autophagy control. Autophagy 2009; 5: 890–891. [DOI] [PubMed] [Google Scholar]
- Li Y, Jiang B, Zhu H, Qu X, Zhao L, Tan Y et al. Inhibition of long non-coding RNA ROR reverses resistance to Tamoxifen by inducing autophagy in breast cancer. Tumour Biol 2017; 39: 1010428317705790. [DOI] [PubMed] [Google Scholar]
- Chen YM, Liu Y, Wei HY, Lv KZ, Fu PF. Large intergenic non-coding RNA-ROR reverses gemcitabine-induced autophagy and apoptosis in breast cancer cells. Oncotarget 2016; 7: 59604–59617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng N, Ching T, Wang Y, Liu B, Lin H, Shi O et al. Analysis of microarray data on gene expression and methylation to identify long non-coding RNAs in non-small cell lung cancer. Sci Rep 2016; 6: 37233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Feng N, Zheng M, Ye X, Lin H, Yu X et al. PM2.5 exposure-induced autophagy is mediated by lncRNA loc146880 which also promotes the migration and invasion of lung cancer cells. Biochim Biophys Acta 2017; 1861: 112–125. [DOI] [PubMed] [Google Scholar]
- Ma B, Yuan Z, Zhang L, Lv P, Yang T, Gao J et al. Long non-coding RNA AC023115.3 suppresses chemoresistance of glioblastoma by reducing autophagy. Biochim Biophys Acta 2017; 1864: 1393–1404. [DOI] [PubMed] [Google Scholar]
- Wakatsuki S, Tokunaga S, Shibata M, Araki T. GSK3B-mediated phosphorylation of MCL1 regulates axonal autophagy to promote Wallerian degeneration. J Cell Biol 2017; 216: 477–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y, Noda NN, Nakatogawa H, Ohsumi Y, Inagaki F. Dimeric coiled-coil structure of Saccharomyces cerevisiae Atg16 and its functional significance in autophagy. J Biol Chem 2010; 285: 1508–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y, Noda NN, Fujii K, Yoshimoto K, Ohsumi Y, Inagaki F. In vitro reconstitution of plant Atg8 and Atg12 conjugation systems essential for autophagy. J Biol Chem 2008; 283: 1921–1928. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2001; 2: 211–216. [DOI] [PubMed] [Google Scholar]
- Huang S, Lu W, Ge D, Meng N, Li Y, Su L et al. A new microRNA signal pathway regulated by long noncoding RNA TGFB2-OT1 in autophagy and inflammation of vascular endothelial cells. Autophagy 2015; 11: 2172–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge D, Han L, Huang S, Peng N, Wang P, Jiang Z et al. Identification of a novel MTOR activator and discovery of a competing endogenous RNA regulating autophagy in vascular endothelial cells. Autophagy 2014; 10: 957–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentelle RD, Senkal CE, Jiang W, Ponnusamy S, Gencer S, Selvam SP et al. Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nat Chem Biol 2012; 8: 831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature 2011; 469: 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Yang GQ, Shao XM, Wei L. GAS5 modulated autophagy is a mechanism modulating cisplatin sensitivity in NSCLC cells. Eur Rev Med Pharmacol Sci 2016; 20: 2271–2277. [PubMed] [Google Scholar]
- Song J, Ahn C, Chun CH, Jin EJ. A long non-coding RNA, GAS5, plays a critical role in the regulation of miR-21 during osteoarthritis. J Orthop Res 2014; 32: 1628–1635. [DOI] [PubMed] [Google Scholar]
- Liu Z, Wei X, Zhang A, Li C, Bai J, Dong J. Long non-coding RNA HNF1A-AS1 functioned as an oncogene and autophagy promoter in hepatocellular carcinoma through sponging hsa-miR-30b-5p. Biochem Biophys Res Commun 2016; 473: 1268–1275. [DOI] [PubMed] [Google Scholar]
- Tang B, Li N, Gu J, Zhuang Y, Li Q, Wang HG et al. Compromised autophagy by MIR30B benefits the intracellular survival of Helicobacter pylori. Autophagy 2012; 8: 1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Cui J, Yang XM, Liu ZY, Song F, Li L et al. Cytoplasmic fragment of CD147 generated by regulated intramembrane proteolysis contributes to HCC by promoting autophagy. Cell Death Dis 2017; 8: e2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YF, Shi YH, Ding ZB, Ke AW, Gu CY, Hui B et al. Autophagy inhibition suppresses pulmonary metastasis of HCC in mice via impairing anoikis resistance and colonization of HCC cells. Autophagy 2013; 9: 2056–2068. [DOI] [PubMed] [Google Scholar]
- Li P, Du Q, Cao Z, Guo Z, Evankovich J, Yan W et al. Interferon-gamma induces autophagy with growth inhibition and cell death in human hepatocellular carcinoma (HCC) cells through interferon-regulatory factor-1 (IRF-1). Cancer Lett 2012; 314: 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Song J, Kim D, Ahn C, Park S, Chun CH et al. PCGEM1 stimulates proliferation of osteoarthritic synoviocytes by acting as a sponge for miR-770. J Orthop Res 2016; 34: 412–418. [DOI] [PubMed] [Google Scholar]
- Fu X, Ravindranath L, Tran N, Petrovics G, Srivastava S. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol 2006; 25: 135–141. [DOI] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 2005; 37: 766–770. [DOI] [PubMed] [Google Scholar]
- Matouk IJ, Abbasi I, Hochberg A, Galun E, Dweik H, Akkawi M. Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur J Gastroenterol Hepatol 2009; 21: 688–692. [DOI] [PubMed] [Google Scholar]
- Xiong H, Ni Z, He J, Jiang S, Li X, He J et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene 2017; 36: 3528–3540. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep 2014; 31: 358–364. [DOI] [PubMed] [Google Scholar]
- Wang K, Liu CY, Zhou LY, Wang JX, Wang M, Zhao B et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun 2015; 6: 6779. [DOI] [PubMed] [Google Scholar]
- Zhang ZZ, Shen ZY, Shen YY, Zhao EH, Wang M, Wang CJ et al. HOTAIR long noncoding RNA promotes gastric cancer metastasis through suppression of poly r(C)-binding protein (PCBP) 1. Mol Cancer Ther 2015; 14: 1162–1170. [DOI] [PubMed] [Google Scholar]
- Bao X, Ren T, Huang Y, Sun K, Wang S, Liu K et al. Knockdown of long non-coding RNA HOTAIR increases miR-454-3p by targeting Stat3 and Atg12 to inhibit chondrosarcoma growth. Cell Death Dis 2017; 8: e2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zhang X, Li H, Liu J. The long noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in hepatocellular carcinoma. Mol Biosyst 2016; 12: 2605–2612. [DOI] [PubMed] [Google Scholar]
- Liu YW, Sun M, Xia R, Zhang EB, Liu XH, Zhang ZH et al. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis 2015; 6: e1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer 2014; 13: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma MZ, Li CX, Zhang Y, Weng MZ, Zhang MD, Qin YY et al. Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Mol Cancer 2014; 13: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhao J, Tang J. [miR-34a may regulate sensitivity of breast cancer cells to adriamycin via targeting Notch1]. Zhonghua Zhong Liu Za Zhi 2014; 36: 892–896. [PubMed] [Google Scholar]
- Nguyen HT, Dalmasso G, Muller S, Carriere J, Seibold F, Darfeuille-Michaud A. Crohn's disease-associated adherent invasive Escherichia coli modulate levels of microRNAs in intestinal epithelial cells to reduce autophagy. Gastroenterology 2014; 146: 508–519. [DOI] [PubMed] [Google Scholar]
- Noda T, Fujita N, Yoshimori T. The late stages of autophagy: how does the end begin? Cell Death Differ 2009; 16: 984–990. [DOI] [PubMed] [Google Scholar]
- Balderhaar HJ, Ungermann C. CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. J Cell Sci 2013; 126: 1307–1316. [DOI] [PubMed] [Google Scholar]
- Huynh KK, Eskelinen EL, Scott CC, Malevanets A, Saftig P, Grinstein S. LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J 2007; 26: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saftig P, Beertsen W, Eskelinen EL. LAMP-2: a control step for phagosome and autophagosome maturation. Autophagy 2008; 4: 510–512. [DOI] [PubMed] [Google Scholar]
- McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell 2015; 57: 39–54. [DOI] [PubMed] [Google Scholar]
- Viereck J, Kumarswamy R, Foinquinos A, Xiao K, Avramopoulos P, Kunz M et al. Long noncoding RNA Chast promotes cardiac remodeling. Sci Transl Med 2016; 8: 326ra322. [DOI] [PubMed] [Google Scholar]
- Abdelmohsen K, Panda AC, Kang MJ, Guo R, Kim J, Grammatikakis I et al. 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Res 2014; 42: 10099–10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Guo Q, Zhao Y, Chen J, Wang S, Hu J et al. BRAF-activated long non-coding RNA contributes to cell proliferation and activates autophagy in papillary thyroid carcinoma. Oncol Lett 2014; 8: 1947–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JH, Li BX, Han ZP, Zou MX, Wang L, Lv YB et al. Snail-activated long non-coding RNA PCA3 up-regulates PRKD3 expression by miR-1261 sponging, thereby promotes invasion and migration of prostate cancer cells. Tumour Biol 2016; 37: 16163–16176. [DOI] [PubMed] [Google Scholar]
- Beaver LM, Kuintzle R, Buchanan A, Wiley MW, Glasser ST, Wong CP et al. Long noncoding RNAs and sulforaphane: a target for chemoprevention and suppression of prostate cancer. J Nutr Biochem 2017; 42: 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Liu Z, Wu S. Long non-coding RNA CTA sensitizes osteosarcoma cells to doxorubicin through inhibition of autophagy. Oncotarget 2017; 8: 31465–31477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel LB, Lubas M, Lund AH. Emerging connections between RNA and autophagy. Autophagy 2016; 13: 3–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hao S, Yin H, Gao J, Yang Z. Autophagy ameliorates cognitive impairment through activation of PVT1 and apoptosis in diabetes mice. Behav Brain Res 2016; 305: 265–277. [DOI] [PubMed] [Google Scholar]
- Alvarez ML, DiStefano JK. Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS ONE 2011; 6: e18671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millis MP, Bowen D, Kingsley C, Watanabe RM, Wolford JK. Variants in the plasmacytoma variant translocation gene (PVT1) are associated with end-stage renal disease attributed to type 1 diabetes. Diabetes 2007; 56: 3027–3032. [DOI] [PubMed] [Google Scholar]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014; 505: 344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol 2014; 34: 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst C, Morton CC. Identification and function of long non-coding RNA. Front Cell Neurosci 2013; 7: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa I, Fradet Y, Beaudry G, Hovington H, Beaudry G, Tetu B. Identification of PCA3 (DD3) in prostatic carcinoma by in situ hybridization. Mod Pathol 2007; 20: 1121–1127. [DOI] [PubMed] [Google Scholar]
- Wu H, Wang F, Hu S, Yin C, Li X, Zhao S et al. MiR-20a and miR-106b negatively regulate autophagy induced by leucine deprivation via suppression of ULK1 expression in C2C12 myoblasts. Cell Signal 2012; 24: 2179–2186. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 2009; 113: 2526–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cao B, Han D, Sun M, Feng J. Long non-coding RNA H19 induces cerebral ischemia reperfusion injury via activation of autophagy. Aging Dis 2017; 8: 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao WT, Ye YP, Zhang NJ, Li TT, Wang SY, Cui YM et al. MicroRNA-30b functions as a tumour suppressor in human colorectal cancer by targeting KRAS, PIK3CD and BCL2. J Pathol 2014; 232: 415–427. [DOI] [PubMed] [Google Scholar]