ABSTRACT
Circular RNAs (circRNAs) are highly abundant and evolutionarily conserved non-coding RNAs produced by circularization of specific exons. Since their re-discovery as potential regulators of gene expression, thousands of circRNAs were detected in different tissues and cell types across most organisms. Accumulating data suggest key roles for them in the central nervous system. Neuronal-expressed RNAs are diverted to yield highly enriched CircRNAs in human, mouse, pig and flies, with many of them enriched in neuronal tissues. CircRNA levels are dynamically modulated in neurons, both during differentiation and following bursts of electrical activity, and accumulate with age, and many of them are enriched in synapses. Together, available data suggest that circRNAs have important roles in synaptic plasticity and neuronal function. This review covers current advances in the field and lays out hypotheses regarding functions of circRNAs in the brain as well as their putative involvement in initiation and progression of neurodegenerative processes.
KEYWORDS: Brain, circRNA, development, neurodegeneration, neuronal-function, synapse
The importance of RNA metabolism for brain function
Normal brain function requires tightly orchestrated control of gene expression at many levels, including regulation of transcription, mRNA processing and alternative splicing, as well as mRNA transport and regulation of translation.1,2,3 Moreover, the brain displays the highest degree of alternative splicing and editing, which might stand behind the morphological and functional diversity of neurons yielding high complexity of neurons.1 Numerous rapidly expanding technologies enable the detection and quantification of whole transcriptomes, and have identified strong links between RNA miss-regulation and several neurodegenerative diseases like Amyotrophic Lateral Sclerosis (ALS) and frontotemporal dementia (FTP,4). Specifically, mutations in several RNA binding proteins (RBPs), including TDP-43, FUS, ataxin2 and others are associated with disrupted RNP granule function and protein aggregates in these and other neurodegenerative diseases (e.g. Alzheimer disease).5–9 In addition, nucleotide repeat expansions (NRE) within the chromosome 9 open reading frame 72 (C9orf72) gene were identified in a substantial fraction of ALS and FTP patients.8 The neurotoxic impact of these repeats is still under debate, but loss of C9orf72 function, and the gain-of-function mechanisms of C9orf72 driven by toxic RNA and protein species encoded by the NRE were reported.4,10 Therefore, dysfunction of RNA metabolism is hence considered as another important factor important for neurodegenerative disease etiology.11,12
Although mRNAs are mainly translated in the cytoplasm (either in free or endoplasmic reticulum-associated ribosomes), some neuronal mRNAs are locally translated in distal compartments including dendrites and synapses.13,14 Proper transport and translation of those mRNAs dictates synaptic or dendritic content, affects neuronal activity and plasticity and is essential for adaptive responses to different stimuli.15 Importantly, disruption of the proper control of mRNA localization and translation is associated with diverse brain diseases such as spinal muscular atrophy (SMA).16 Taken together, these reports suggest that dysfunction of RNA metabolism and/or intracellular transport might explain certain aspects of the mechanisms underlying neuronal dysfunction in these diseases.
CircRNAs: A new regulation layer of gene expression
Circular RNAs (circRNAs) are highly abundant, tissue-specific and evolutionarily conserved RNAs, and are widely expressed across the animal kingdom.17–19 It has been argued that circRNAs are a by-product of splicing.20 However, numerous reports argue against this view. First, circRNAs are extremely abundant. Indeed, in many cases circRNA expression is significantly higher than its linear host transcript and generally the expression levels of the circRNAs and linear molecules originated from the same locus are rather weakly correlated, suggesting specific regulation.17,21 Second, pioneering studies have now showed functionality of specific circRNAs in both normal physiology and disease.18,22–25 Third, circRNAs are cytoplasmic, whereas most (if not all) undesired splicing products accumulate at the site of transcription.26 Fourth, circRNAs are evolutionarily conserved at both their sequence level and their pattern of expression, in particular in the brain.27 Also, the nucleotide sequences of circRNAs are significantly better conserved than their flanking exons, strongly suggesting functional importance.18,21,28 In addition, hundreds of mouse/human/rat circRNAs are highly and specifically expressed in brain tissues. This expression pattern is highly conserved- 80% of all efficiently expressed mouse neuronal circRNAs are also detected in the human brain, with very similar expression patterns across neuronal tissues.27
CircRNAs are mostly derived from protein-coding genes, tend to be upregulated during neuronal differentiation and span complete exons.19,20,27,29 They are generated co-transcriptionally via a special splicing event termed ‘back-splicing’, which is mediated by the spliceosome.19,30 Back splicing is the process in which the splice donor splices to an upstream rather than a downstream acceptor. Importantly, we showed that the circularization event competes with the splicing of the linearly spliced form of the same transcript19 (Fig. 1). In general exon circularization is driven by interactions between the introns flanking the exons that will form the circRNA. In this context, RNA-binding proteins and sequence complementarity can both promote exon circularization19,28,31,32 (Fig. 1). These base-pairings form RNA duplexes that collocate with the splicing sites and facilitate circRNA formation. Importantly, RNA pairing may promote or counteract circRNA formation, as base-pairing within individual introns inhibits circRNAs biogenesis.2 Therefore, dynamic changes in base-pairing (within individual introns or between surrounding introns) regulate circRNA or linear mRNA expression. Recent reports by others and us found that circRNAs are enriched in neuronal tissues19,27,29 and accumulate with age.33 In addition, a subset of circRNAs is highly enriched in synapses.27,29
Figure 1.
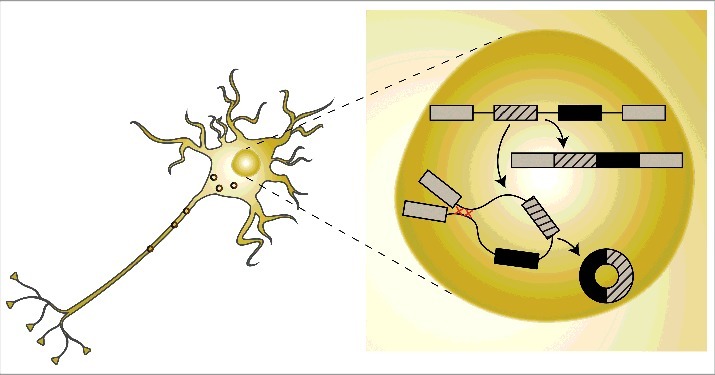
A schematic drawing of circRNA biogenesis. CircRNAs are generated co-transcriptionally via a special splicing event termed ‘back-splicing’. The circularization event is facilitated by base-pairing of intronic sequences that include highly complementary regions and produce stable circular molecules that are transported to the cytoplasm and other cellular compartments.
Notably, functional roles were so far found for only a small fraction of the thousands of identified circRNAs. Two of these RNAs, CDR1as/ciRS-7 and Sry, can act as miRNA sponges. CDR1as/ciRS-7 is the best-characterized and most expressed mammalian circRNA, which is also highly abundant in neurons. The CDR1as circRNA originates from the antisense transcript of the Cerebellar degeneration-related protein 1 (CDR1) gene harbors 74 binding sites for miR-7 and binds the miRNA-effector protein Argonaute in a miR-7-dependent manner.18,25 Therefore, changes in its levels may bear conspicuous effects on miRNA metabolism. Interestingly, downregulation of CDR1as in mammalian cell culture modifies cell migration and overexpression in the zebrafish brain leads to brain developmental defects.18 Other functions of circRNAs have been demonstrated for circFOXO3 and circHIPK3 in cell cycle and growth processes. CircFOXO3 inhibits cell cycle progression by forming a ternary complex with cyclin-dependent kinase 2 (CDK2) and cyclin-dependent kinase inhibitor 1 (p21).23 CircHIPK3 directly binds and inhibits miR-124 activity, and silencing it results in arrested cell growth.22 In addition, we recently reported that the splicing factor muscleblind (mbl) hosts a circRNA which is involved in the auto-regulation of this RNA-binding protein. MBL binds to its own pre-mRNA and promotes the circularization of its own 2nd exon. This mechanism enables tight regulation over circRNA production; when the level of the MBL protein is high, it facilitates circularization of circMBL at the expense of the linear transcript. When the MBL protein level is low, most of the pre-mRNA transcript will be spliced into the linear protein-coding mRNA. Interestingly, the MBL protein strongly binds to circMbl, suggesting the existence of a feedback regulation involving the sequestration and/or transport of this protein.19 Other factors may contribute to the production of circRNAs, including heteronuclear ribo-nuclear proteins (hnRNPs), serine-arginine rich SR proteins and RNA-binding proteins such as the splicing factor Quaking.31,34 These diverse roles indicate regulatory functions for at least part of the brain circRNAs.
Roles for circRNAs in brain function, development and disease
Function
Importantly, circRNAs are highly enriched in neuronal tissues in flies and mammals.17–19 This could be partially explained by the fact that many more neuronal genes encode for circRNAs in comparison to other tissues.19 More precisely, 20 percent of the protein-coding genes in the brain produce circRNAs. CircRNAs are generated at the expense of canonical mRNA isoforms, suggesting that circRNA biogenesis per se may be an important regulator of mRNA production and that their production could function as “mRNA traps”.19,35 Indeed, in many cases the levels of the circular RNA form is higher than that of the linear mRNA “host.”19,21 However, this relationship does not seem to be universal, and in several reported cases, there is no correlation between the expression levels of the circRNA and the linear form.19,27,36 This lack of correlation may be due post-transcriptional events leading to high stability of the circRNAs or low half-life of the linear host mRNA.19,21 Alternatively, this could be achieved by active transcription and dynamic modulation of circularization efficiencies by co-expression of RNA-binding proteins promoting or inhibiting circularization. Further studies will be required to explore these diverse theories and validate or exclude them.
Many of the circRNA-producing genes are exclusively expressed in the brain. Furthermore, in most cases when the host gene is also expressed in other tissues, the percent of transcripts that produce circRNAs is significantly higher in the brain,29 suggesting a neural-specific regulation of circRNA production. Examples of genes which host brain-specific circRNAs include circRims2, circTulp4, circElf2, circPhf21a and circMyst4, most of which originate from genes with pivotal regulatory functions in neurons and during brain development.27,29 Correspondingly, genes that host circRNAs are enriched in gene ontology terms of biological processes related to synaptic function such as nervous system development, neurogenesis, neuron differentiation and include cellular components such as synapse, presynaptic active zone, presynaptic membrane and postsynaptic density.19,27,29,33 When comparing different brain regions, most circRNAs show expression patterns that are compatible with the expression levels of their linear host genes, but some specific circRNAs are highly expressed in specific brain areas in an unrelated manner to the levels of their linear gene products. Such cases include circRims2, circEl2 and circDym, which are enriched in the cerebellum, whereas circPlxnd1 is enriched in the cortex.27 Compatible with the notion of their enrichment in neurons, many circRNAs seem to be particularly enriched in the cerebellum, a brain area very dense with neuronal spines, synapses and neurons in general.27,29
CircRNAs are highly conserved between mouse, rat and human. This conservation includes at least one out of the two specific splice-sites that generate the circRNA (9049 out of 15849 mouse circRNAs were conserved to human by using this criterion).27,29 Thus, the evolutionary conservation of circRNAs in terms of brain region location and developmental time is compatible with the notion of pivotal brain roles.
CircRNAs are mostly found in the cytoplasm, but many of them are localized to and enriched in synapses.27,29 This is supported by enrichment of circRNAs in murine synaptoneurosome fractions and in micro-dissected neuropil from hippocampal slices in comparison to cytoplasmic RNA and total brain RNA. Moreover, those findings are compatible with the detection of circRNAs in high-resolution in-situ hybridization assays of cultured hippocampal neurons and hippocampal slices.27,29 CircStau2a is an example of this type of circRNAs. It is localized to the synaptic space, unlike the linear mRNA produced from the same gene which is almost exclusively found in the cytoplasm. Another example is circRMST, which shows a distinct subcellular localization from its linear lncRNA form that is predominantly nuclear.27 The functional significance of the synaptic localization of circRNAs opens intriguing possibilities. For example, circRNAs could be packaged into vesicles and then act as messaging or trafficking molecules between or within cells; in addition, they could serve as carrier molecules that deliver specific cargoes to the synapse (Fig. 2). Another option is that circRNAs serve as templates for translation, either in the cytoplasm or within the synaptic space, possibly enabling the regulation of translation in response to stimuli. Indeed, it has been recently shown that artificial circRNAs carrying IRES sequences can be translated in insect and mammalian cells.31,37,38 Additional studies will be required to test these hypotheses and validate their physiological significance (see Fig. 2).
Figure 2.
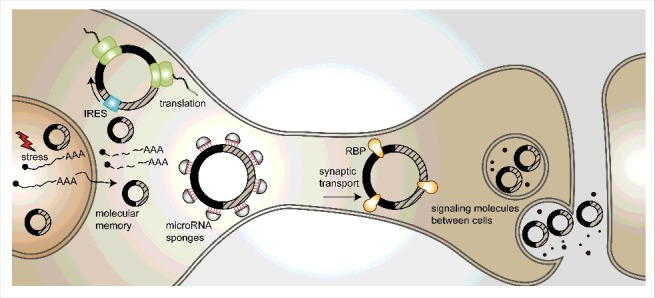
Possible functions of circRNAs in the brain. (1) CircRNAs may be transcribed upon stressful insults and serve as “memory” molecules, as they are extremely stable. (2) CircRNAs can serve as templates for translation into proteins, in the cytoplasm or in synapses. (3) CircRNAs can act as miRNA sponges. (4) Active transport to the synapse or dendrites may utilize circRNAs as transport-hubs for RNA-binding proteins. (5) CircRNAs may encode information that can be stored or even passed on between cells.
Recently, it was reported that neuronal activity changes circRNA expression. Induction of homeostatic plasticity by the use of bicuculline, an antagonist of the GABA-A receptor caused dynamic changes in circRNA expression in mouse primary neurons. Dozens of circRNAs were upregulated (including circHomer, circNell2 and circMpped1), and several others were downregulated, while most of the linear transcripts remained stable following the pharmacological treatment. For example, circHomer is significantly upregulated in both the neuronal somata and in dendrites of murine primary neuron cultures as well as in both the stratum pyramidale (somatic layer) and stratum radiatum (neuropil layer) of mouse brains.29 This regulation implies that circRNAs are either produced or transported in response to changes in neuronal activity, suggesting specific functions of those molecules in synapses.
Another level of circRNA regulation involves RNA editing. The RNA-editing enzyme ADAR, which plays key roles in development and function of the nervous system11,39 is a strong modulator of circRNA production.28 ADAR editing enzymes can efficiently bind and edit reverse complementary sequences, leading to the disruption of RNA secondary structures, which in some cases are key for the modulation of circRNAs production.2,28 Indeed, intronic complementary sequences surrounding circRNAs are enriched in A-to-I editing cites, including hyper-editing.28 Therefore, it is not surprising that ADAR1 activity antagonizes with circRNAs production by inhibiting inter-intron interactions.27,28 More specifically, ADAR1 knockdown increases circRNAs production.28 These observations were also extended to Drosophila, suggesting that this regulation is universal.27 In flies, dADAR levels and activity are strongly regulated by temperature, with more editing events occurring at 18°C than at 29°C. Not surprisingly, we recently reported that more circRNAs are produced in flies after their transfer for 3 days to 29°C.27 Thus, given that circRNA expression occurs at the expense of the host mRNA, the modulation of circRNA expression by ADAR might serve as another layer of regulation of the linear host gene expression, independent of putative functions of the circRNA in trans.
CircRNAs are extremely stable, mostly because of their resistance to exonucleases; they consequently display half-lives longer than 48 hours.19,35 It is therefore possible that circRNAs might serve as “memory” molecules, and encode information that may be stored or even passed on between cells (Fig. 2). For example, one could imagine that upon stressful insults, many mRNAs and circRNAs will be produced. As the system goes back to cellular homeostasis, the transcriptome would come back to its normal steady-state, but the extremely stable circRNA molecules may persist for most of the host's lifetime, especially in organisms with short lifespan such as flies or worms.
CircRNAs in development
Neuronal differentiation and development reflect a complex sequence of events that are tightly regulated and are not yet fully understood.40 Different types of coding and noncoding RNAs such as miRNAs and long non-coding RNAs (lncRNAs) are involved in these regulatory events.41,42 Recently, several reports described important features of circRNA regulation in neuronal development, indicating possible regulatory involvement of circRNAs in these processes.27,29,36 In general, circRNAs are up-regulated during neuronal differentiation in human and murine cell culture systems as well as in mouse primary neurons and porcine brains.27,29,36 In addition, the expression of circRNAs is temporally and spatially changed during neural development, with no apparent correlation to their linear host gene expression. This could indicate a possible requirement for circRNAs in different neuronal tissues and developmental processes.
During differentiation, the number of detected circRNAs increases significantly.27,29 These include highly abundant circRNAs such as CDR1as/CiRS-7, circRTN4, circTULP4 and circRIMS2.27,29 This increase in circRNAs levels suggests that the circularization events are important for neuronal function. Thus, many circRNAs are expressed at different time-points of neuronal differentiation and maturation, and especially during postnatal development. This timeline is compatible with the establishment of neuronal networks and synaptic connections. Intriguingly, many of these circRNAs are the circular products of genes that encode for proteins with synapse-related functions, perhaps indicating a homeostatic role of circRNAs in ensuring a functional balance during neural differentiation.27 In many cases, circRNAs that are differentially expressed during synaptogenesis show no correlation with the expression level of their linear host genes, indicating that their expression is due to specific regulation of the circularization events and/or turnover, but not due to changes in the overall overexpression of those genes. Examples for these circRNAs are circMyst4, circKlhl2 and circAagab, whose circular forms are significantly upregulated during differentiation whereas their linear transcripts are downregulated.29 In addition, many circRNAs that were deregulated during neuronal differentiation in murine cell lines or in mouse brains showed similar expression patterns to circRNAs in differentiating human SH-SY cells, indicating a conserved role of circRNAs expression during neuronal differentiation.27
Unlike the developing murine brain and differentiated cell culture systems, the developing porcine cortex demonstrates a different pattern of expression and accumulation of circRNAs. The number of circRNAs detected increases in the first cortical development stages (E23, E42 and peaked at E60), but drastically decreases later in brain development (E80, E100 and E115).36 The timing of the observed peak in the porcine cortex correlates with processes of gyration, and therefore may indicate a regulatory role for circRNAs specifically during this period. Also, the dynamic expression of a large number of circRNAs is significantly higher in comparison to their host linear transcripts (at least in one of the measured developmental stages). The spatial and temporal expression patterns of circRNAs in the developing porcine brain are also noticeably different from those of mice; brain regions such as the hippocampus, brain stem and cerebellum exhibit distinct patterns of expression, with the highest accumulation of circRNAs in newborn pigs, the last developmental stage measured (E115).36 This may indicate a precise requirement for specific circRNAs in different regions within the brain and at different developmental stages. Enriched pathways associated with the host genes of circRNAs peaking during development include axon guidance, Wnt signaling and TGF-β signaling, 3 pathways that play key roles during embryonic brain development, and are involved in processes such as neuronal migration, specification of axons and dendrites during embryonic development and neuroprotection.36 The issue of circRNA localization within the cell was also addressed for specific candidates and interestingly, one specific circRNA changes its subcellular localization during development: circZEB1 is mainly localized in the cytoplasm in E60 but at E80 it is exclusively localized in the nucleus.36 The developmental and species-specific expression patterns of circRNAs indicate evolutionary changes in their embryonic roles within the mammalian brain, opening novel routes for future interrogations.
CircRNAs in neurodegeneration
Several reports link circRNAs to brain degeneration and disease, and in particular CDR1as/ciRS-7, which is the best-characterized and most highly expressed mammalian circRNA.25 As mentioned above, The human CDR1as/ciRS-7 harbors 74 binding sites for miR-7 and is associated to the Argonaute protein in a miR-7-dependent manner.18,25 Intriguingly, miR-7 has been implicated as key regulator of both various cancers24 and neurological diseases such as Parkinson disease (PD). Several studies suggest a neuro-protective role for miR-7 in dopaminergic neurons: miR-7 can target the PD-related α-synuclein protein and is involved in protecting neurons against cell death induced by the dopaminergic neurotoxic compound 1-Methyl-4-Phenyl-Pyridinium (MPP (+)) via up-regulating the mTOR pathway.43–46 CDR1as/ciRS-7 is also strongly miss-regulated in the hippocampal CA1 region of Alzheimer disease (AD) patients.47 Moreover, neural overexpression of CDR1as/ciRS-7 in zebrafish leads to brain developmental defects and impairment in midbrain development, which resembles the phenotype of miR-7 knock-down.18 The sponge function of circRNAs, and specifically CDR1as/ciRS-7 may hence take an active part in the initiation and progression of PD.
In fly heads, circRNAs accumulate with age, revealed by a significant age-related increase in circRNA abundance when comparing flies of 1, 4 or 20 days33 Indeed, RNA purified from heads of aged Drosophila yielded the highest number of circRNAs detected in comparison to any other Drosophila tissue or developmental stage, even when compared to the linear host transcripts. The expression of 262 circRNAs is significantly higher at the age of 20 days in comparison to day 1 heads, and these host genes are enriched for functional annotation pathways that are related to synaptic function, signaling and developmental processes.33 However, it is not yet known whether accumulation of circRNAs during aging is an evolutionarily conserved feature that is also manifested in other metazoans, and in particular in mammals. Further research should aim at finding the answers to these key questions and be focused on the possible function of circRNAs in the aging brain and the link to other neurodegenerative diseases and cellular senescence in the brain. To conclude, the relevance of circRNAs to neurodegenerative and generally to brain diseases may be twofold. In addition to putative roles of these molecules in the disease itself, they are great candidates to serve as biomarkers, thanks to their extreme stability and abundance in blood.48 Indeed, circRNAs have already been shown to be good biomarkers for different types of cancers.23,24,49
Future prospects and concluding remarks
Many neuronal genes generate multiple circRNAs. These “hot-spots” for circRNA production usually involve splicing with one specific exon, either at the splice donor or splice acceptor. This suggests that the cis or trans-acting elements provide specific exons with circularization capability. This also implies that circRNA levels are tightly regulated.
It is possible that the high degree of exon circularization in the brain is a mere byproduct of the high degree of alternative splicing in the brain,1,3 after all circRNA biogenesis can be viewed as a particular type of alternative splicing, mediated by the spliceosome.19 However, as circRNAs are not produced from exons that are usually alternative spliced, their production is not the sub-product of other alternative splicing events,17,21 possibly indicating a distinct process with particular neuronal roles.
Previous data showed rapid accumulation of circRNAs under induction of neural differentiation in murine and human cell culture models and accumulation in heads of aging Drosophila. These reports raised the question if circRNA expression is a mere byproduct of transcription, especially since neurons are non-dividing cells and would accumulate stable molecules. However, this is not always the case in the porcine brain, a system in which circRNA expression is dynamically modulated during development. There, circRNA expression is highly dynamic and seems to be tightly regulated in development. Understanding the full repertoire of modulators of circularization events is needed in order to elucidate the regulatory potential of these new molecules and more specifically, their roles in neuronal differentiation and maturation. At present, there is no information on the cell-type specificity of circRNAs. Therefore, it would be very useful to profile circRNAs originating from different brain cell types, particularly specific neurons and glia and additionally, map the circRNA populations in different neuronal cell types involved in diverse transmission processes (dopaminergic, cholinergic etc.), and determine the ability of those cells to potentially release and transport circRNAs within the brain's network. In summary, although the functional importance of circRNAs is still not fully understood, it is clear that these molecules are important, as they are evolutionarily conserved, highly expressed and dynamically modulated throughout the brain. Their study has brought a lot of excitement, as they seem to constitute an uncharacterized and unique layer of gene expression regulation.
Funding
This study was supported by the MJF Foundation (grant 11183, to S.K. and H.S.) and The Teva NNE program, which awarded M.H. a pre-doctoral fellowship.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Raj B, Blencowe BJ. Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles. Neuron 2015; 87:14-27; PMID:26139367; https://doi.org/ 10.1016/j.neuron.2015.05.004 [DOI] [PubMed] [Google Scholar]
- 2.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell 2014; 159:134-47; PMID:25242744; https://doi.org/ 10.1016/j.cell.2014.09.001 [DOI] [PubMed] [Google Scholar]
- 3.Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci 2007; 8:819-31; PMID:17895907; https://doi.org/ 10.1038/nrn2237 [DOI] [PubMed] [Google Scholar]
- 4.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al.. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011; 72:257-68; PMID:21944779; https://doi.org/ 10.1016/j.neuron.2011.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berson A, Barbash S, Shaltiel G, Goll Y, Hanin G, Greenberg DS, Ketzef M, Becker AJ, Friedman A, Soreq H. Cholinergic-associated loss of hnRNP-A/B in Alzheimer's disease impairs cortical splicing and cognitive function in mice. EMBO Mol Med 2012; 4:730-42; PMID:22628224; https://doi.org/ 10.1002/emmm.201100995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 2013; 79:416-38; PMID:23931993; https://doi.org/ 10.1016/j.neuron.2013.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orr HT. FTD and ALS: genetic ties that bind. Neuron 2011; 72:189-90; PMID:22017980; https://doi.org/ 10.1016/j.neuron.2011.10.001 [DOI] [PubMed] [Google Scholar]
- 8.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al.. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011; 72:245-56; PMID:21944778; https://doi.org/ 10.1016/j.neuron.2011.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW 3rd, Rademakers R, et al.. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 2013; 77:639-46; PMID:23415312; https://doi.org/ 10.1016/j.neuron.2013.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fratta P, Polke JM, Newcombe J, Mizielinska S, Lashley T, Poulter M, Beck J, Preza E, Devoy A, Sidle K, et al.. Screening a UK amyotrophic lateral sclerosis cohort provides evidence of multiple origins of the C9orf72 expansion. Neurobiol Aging 2015; 36:546 e1-7; PMID:25085785; https://doi.org/ 10.1016/j.neurobiolaging.2014.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behm M, Ohman M. RNA Editing: A Contributor to Neuronal Dynamics in the Mammalian Brain. Trends Genet 2016; 32:165-75; PMID:26803450; https://doi.org/ 10.1016/j.tig.2015.12.005 [DOI] [PubMed] [Google Scholar]
- 12.Szafranski K, Abraham KJ, Mekhail K. Non-coding RNA in neural function, disease, and aging. Front Genet 2015; 6:87; PMID:25806046; https://doi.org/ 10.3389/fgene.2015.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tom Dieck S, Hanus C, Schuman EM. SnapShot: local protein translation in dendrites. Neuron 2014; 81:958–e1; PMID:24559682; https://doi.org/ 10.1016/j.neuron.2014.02.009 [DOI] [PubMed] [Google Scholar]
- 14.Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, Wolozin B. Local RNA translation at the synapse and in disease. J Neurosci 2011; 31:16086-93; PMID:22072660; https://doi.org/ 10.1523/JNEUROSCI.4105-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holt CE, Schuman EM. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 2013; 80:648-57; PMID:24183017; https://doi.org/ 10.1016/j.neuron.2013.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donlin-Asp PG, Bassell GJ, Rossoll W. A role for the survival of motor neuron protein in mRNP assembly and transport. Curr Opin Neurobiol 2016; 39:53-61; PMID:27131421; https://doi.org/ 10.1016/j.conb.2016.04.004 [DOI] [PubMed] [Google Scholar]
- 17.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012; 7:e30733; PMID:22319583; https://doi.org/ 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al.. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495:333-8; PMID:23446348; https://doi.org/ 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 19.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014; 56:55-66; PMID:25242144; https://doi.org/ 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- 20.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014; 15:409; PMID:25070500; https://doi.org/ 10.1186/s13059-014-0409-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19:141-57; PMID:23249747; https://doi.org/ 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al.. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016; 7:11215; PMID:27050392; https://doi.org/ 10.1038/ncomms11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 2016; 44:2846-58; PMID:26861625; https://doi.org/ 10.1093/nar/gkw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res 2013; 73:5609-12; PMID:24014594; https://doi.org/ 10.1158/0008-5472.CAN-13-1568 [DOI] [PubMed] [Google Scholar]
- 25.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384-8; PMID:23446346; https://doi.org/ 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 26.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 2009; 136:688-700; PMID:19239889; https://doi.org/ 10.1016/j.cell.2009.02.001 [DOI] [PubMed] [Google Scholar]
- 27.Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al.. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 2015; 58:870-85; PMID:25921068; https://doi.org/ 10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]
- 28.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, et al.. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 2015; 10:170-7; PMID:25558066; https://doi.org/ 10.1016/j.celrep.2014.12.019 [DOI] [PubMed] [Google Scholar]
- 29.You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al.. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nature neuroscience 2015; 18:603-10; PMID:25714049; https://doi.org/ 10.1038/nn.3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell Rep 2015; 10:103-11; PMID:25543144; https://doi.org/ 10.1016/j.celrep.2014.12.002 [DOI] [PubMed] [Google Scholar]
- 31.Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, Wilusz JE. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev 2015; 29:2168-82; PMID:26450910; https://doi.org/ 10.1101/gad.270421.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 2014; 28:2233-47; PMID:25281217; https://doi.org/ 10.1101/gad.251926.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 2014; 9:1966-80; PMID:25544350; https://doi.org/ 10.1016/j.celrep.2014.10.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA-binding protein quaking regulates formation of circRNAs. Cell 2015; 160:1125-34; PMID:25768908; https://doi.org/ 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- 35.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014; 32:453-61; PMID:24811520; https://doi.org/ 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veno MT, Hansen TB, Veno ST, Clausen BH, Grebing M, Finsen B, Holm IE, Kjems J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol 2015; 16:245; PMID:26541409; https://doi.org/ 10.1186/s13059-015-0801-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA 2015; 21:172-9; PMID:25449546; https://doi.org/ 10.1261/rna.048272.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y, et al.. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci Rep 2015; 5:16435; PMID:26553571; https://doi.org/ 10.1038/srep16435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Overton IM, Baines RA, Keegan LP, O'Connell MA. The ADAR RNA editing enzyme controls neuronal excitability in Drosophila melanogaster. Nucleic Acids Res 2014; 42:1139-51; PMID:24137011; https://doi.org/ 10.1093/nar/gkt909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams ME, de Wit J, Ghosh A. Molecular mechanisms of synaptic specificity in developing neural circuits. Neuron 2010; 68:9-18; PMID:20920787; https://doi.org/ 10.1016/j.neuron.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stappert L, Roese-Koerner B, Brustle O. The role of microRNAs in human neural stem cells, neuronal differentiation and subtype specification. Cell Tissue Res 2015; 359:47-64; PMID:25172833; https://doi.org/ 10.1007/s00441-014-1981-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hart RP, Goff LA. Long noncoding RNAs: Central to nervous system development. Int J Dev Neurosci 2016; PMID:27296516; https://doi.org/ 10.1016/J.Ijdevneu.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi DC, Chae YJ, Kabaria S, Chaudhuri AD, Jain MR, Li H, Mouradian MM, Junn E. MicroRNA-7 protects against 1-methyl-4-phenylpyridinium-induced cell death by targeting RelA. J Neurosci 2014; 34:12725-37; PMID:25232110; https://doi.org/ 10.1523/JNEUROSCI.0985-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fragkouli A, Doxakis E. miR-7 and miR-153 protect neurons against MPP(+)-induced cell death via upregulation of mTOR pathway. Front Cell Neurosci 2014; 8:182; PMID:25071443; https://doi.org/ 10.3389/fncel.2014.00182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doxakis E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J Biol Chem 2010; 285:12726-34; PMID:20106983; https://doi.org/ 10.1074/jbc.M109.086827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A 2009; 106:13052-7; PMID:19628698; https://doi.org/ 10.1073/pnas.0906277106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lukiw WJ. Circular RNA (circRNA) in Alzheimer's disease (AD). Front Genet 2013; 4:307; PMID:24427167; https://doi.org/ 10.3389/fgene.2013.00307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS One 2015; 10:e0141214; PMID:26485708; https://doi.org/ 10.1371/journal.pone.0141214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Yang J, Zhou P, Le Y, Zhou C, Wang S, Xu D, Lin HK, Gong Z. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res 2015; 5:472-80; PMID:25973291 [PMC free article] [PubMed] [Google Scholar]


