ABSTRACT
With a minimal (250–400 nt), non-protein-coding, circular RNA genome, viroids rely on sequence/structural motifs for replication and colonization of their host plants. These motifs are embedded in a compact secondary structure whose elucidation is crucial to understand how they function. Viroid RNA structure has been tackled in silico with algorithms searching for the conformation of minimal free energy, and in vitro by probing in solution with RNases, dimethyl sulphate and bisulphite, and with selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE), which interrogates the RNA backbone at single-nucleotide resolution. However, in vivo approaches at that resolution have not been assayed. Here, after confirming by 3 termodynamics-based predictions and by in vitro SHAPE that the secondary structure adopted by the infectious monomeric circular (+) RNA of potato spindle tuber viroid (PSTVd) is a rod-like conformation with double-stranded segments flanked by loops, we have probed it in vivo with a SHAPE modification. We provide direct evidence that a similar, but not identical, rod-like conformation exists in PSTVd-infected leaves of Nicotiana benthamiana, verifying the long-standing view that this RNA accumulates in planta as a “naked” form rather than tightly associated with protecting host proteins. However, certain nucleotides of the central conserved region, including some of the loop E involved in key functions such as replication, are more SHAPE-reactive in vitro than in vivo. This difference is most likely due to interactions with proteins mediating some of these functions, or to structural changes promoted by other factors of the in vivo habitat.
KEYWORDS: Circular RNAs, non-coding RNAs, RNA secondary structure, SHAPE in vivo, viroids
Abbreviations
- mc and ml
monomeric circular and linear PSTVd RNA, respectively
- PAGE
polyacrylamide gel electrophoresis
- PSTVd
potato spindle tuber viroid
- SHAPE
selective 2′-hydroxyl acylation analyzed by primer extension
Introduction
In contrast to viruses that encode some of the proteins they need to facilitate their infectious cycle, viroids are just composed by a non-protein-coding, small (250–400 nt), circular RNA.1-6 Consequently, these minimal genomes are extremely and specifically dependent on sequence/structural motifs for hijacking components of the transcription, processing and trafficking machineries of their host plants in order to be replicated and invade them systemically, overcoming the defensive barriers they raise.7-11 Some of such motifs have an intrinsically associated catalytic role, as illustrated by the hammerhead ribozymes,12-13 adopted transiently by the strands of both polarities of plastid-replicating viroids (family Avsunviroidae) to mediate self-cleavage of the oligomeric RNAs generated during replication through a symmetric rolling-circle mechanism.6 Yet, certain host proteins enhance the catalytic efficiency of these ribozymes.14
Most other motifs are less complex, seem involved in replication/trafficking, and are embedded in a rod-like secondary structure proposed as an almost universal feature of nuclear-replicating viroids (family Pospiviroidae, type member potato spindle tuber viroid, PSTVd).15-17 An example is the RY motif —located close to the terminal right hairpin loop of the rod-like secondary structure of the monomeric circular (mc) PSTVd (+) RNA— which is responsible for the specific interaction of this RNA in vitro and in vivo with the tomato viroid RNA binding protein 1 (Virp1).18-20 The evidence for the rod-like conformation of the mc PSTVd (+) RNA is strong in silico, with algorithms that retrieve the secondary structure of minimal free energy,21-23 and in vitro, by biochemical approaches that include probing in solution with RNases, bisulphite and dimethyl sulphate,17,24 and more lately by selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE),25-27 which provides data on local backbone RNA flexibility at the single-nucleotide level.28-30 Additionally, electron microscopy31,32 and several biophysical approaches in solution,33-35 also support the aforementioned rod-like secondary structure.
However, direct extrapolation of the existence of a similar structure in planta is questionable for reasons discussed previously,9,36 prominent among which are the initial thermal denaturation/renaturation applied to RNA for SHAPE analysis in vitro, the ionic composition that differs from that of the physiological context, and the interaction of the RNA with different host proteins that may impact significantly on its structure. Nevertheless, there is evidence, albeit indirect and fragmentary, for such a structure in vivo. In brief: i) duplications and deletions observed in natural and artificial variants of several members of the family Pospiviroidae preserve the rod-like structure,37-40 and ii) replication and directional PSTVd trafficking through specific cell boundaries is regulated by RNA motifs, particularly loops, of the rod-like secondary structure (see below). The high genetic variability observed in members of the family Avsunviroidae, a likely consequence of their high mutation rate,41 makes it feasible to validate the in silico and in vitro conformations by the presence in natural sequence variants of co-variations and conversions of canonical into wobble base-pairs, or vice versa, that preserve the pairing of most double-stranded stems and even tertiary interactions.36,42,43 Unfortunately, the lower genetic variability of members of the family Pospiviroidae precludes a similar analysis and leaves open an important issue.
Because novel reagents have been recently developed to extend SHAPE methodology to in vivo conditions,44-45 we decided to apply such approach to dissect the conformation of the mc PSTVd (+) RNA in infected leaves of the experimental host Nicotiana benthamiana. Our results confirm the existence of a rod-like “naked” conformation in vivo showing minor, but significant, changes with that obtained in silico and in vitro. Intriguingly, some of the differences map at nucleotides of the lower strand of the central conserved region (CCR),46 particularly at loop E with a crucial role in replication.47-49 Such differences most likely result from interactions with host proteins or with other factors present in the in vivo habitat.
Results
In silico predictions lead to a rod-like conformations for the mc PSTVd (+) strand
Although other PSTVd variants, particularly the so-called intermediate strain from potato (GenBank accession number NC_002030),15,17 have been subjected to multiple thermodynamics- and kinetics-based predictions,33-35,50 this is neither the case for the NB variant (GenBank accession number AJ634596.1)51,52 used in the present study, nor for the dahlia variant (GenBank accession number AB623143.1)53 used in SHAPE in vitro.26 Moreover, we were specifically interested in the secondary structure of the mc PSTVd (+) RNA, which predominates in infected tissue and is the most relevant viroid form for priming the asymmetric rolling-circle replication and for trafficking in vivo.48,54-59 As in a recent report dealing with another viroid,36 we tackled this issue with 3 independent softwares: Mfold,21 RNAfold22 and RNAstructure,23,60 the first 2 with the version for circular RNAs not available for the third one. The three softwares generated essentially the same rod-like conformation of minimal free energy for the NB variant, except 2 alternative motifs predicted by RNAfold and one small alternative motif predicted by Mfold (in the 3 instances with a relatively low probability) (Fig. 1), thus indicating a high degree of coherence between them.
Figure 1.
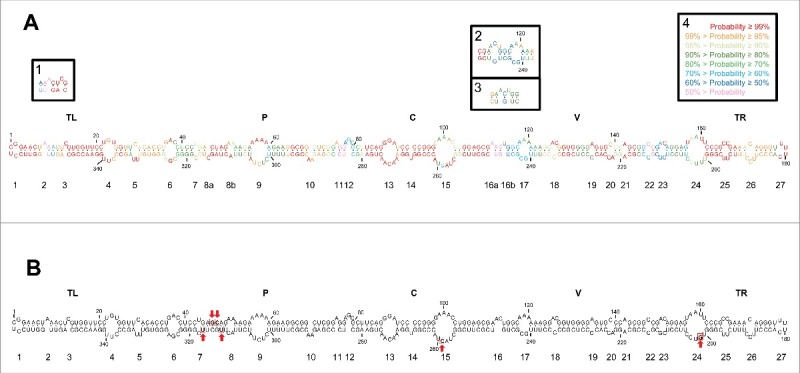
(A) Rod-like secondary structure of minimum free energy predicted by RNAstructure for the mc PSTVd (+) RNA of the NB variant. Insets (1) and (2), alternative motifs predicted by RNAfold and inset (3), alternative motif predicted by Mfold. Colors denote the probability (see inset 4) of a nucleotide being double- or single-stranded as predicted by RNAstructure. (B) Rod-like secondary structure of minimum free energy proposed for the mc PSTVd (+) RNA of the intermediate variant,82 which differs from NB in 6 substitutions: C46→G, U47→C, U201→G, U259→C, A313→U, and C317→U (within squares and marked with arrows). The structure of the intermediate variant, which is presented here for comparative purposes and as a reference for loop numbering,48 has been edited to remove an non-existing extra G-C base pair adjacent to the terminal left loop.48 Loops 8 and 16 in the intermediate variant are split in the NB variant into 8a and 8b, and 16a and 16b, respectively. TL (terminal left), P (pathogenicity), C (central), V (variable) and TR (terminal right) are structural domains defined previously.82
When the conformation of the NB variant in silico was compared with those of the intermediate and dahlia variants, in all cases obtained using RNAstructure that is the software usually implemented in SHAPE in vitro and in vivo,60 additional alternative motifs were observed in the rod-like structure, particularly with the dahlia variant (Fig. S1). Such differences most likely result from changes in the nucleotide sequence: 6 substitutions between the NB and the intermediate variants, and 9 substitutions plus 3 indels between the NB and dahlia variants (Fig. S1).
Non-denaturing page shows that the mc PSTVd (+) RNA adopts a predominant secondary structure in vitro
Then we moved to analyze if, according to in silico predictions, the mc PSTVd (+) form folds into a major conformation in solution. For such purpose, this RNA was isolated and purified from infected tissue and then examined by non-denaturing PAGE. Before electrophoresis, 2 aliquots of the purified RNA resuspended in water were heated at 95°C for 2 min and either gradually-cooled at 25°C along 15 min or snap-cooled on ice, serving as control a third non-treated aliquot. The samples were then incubated at 37 °C for 5 min in the folding buffer.29 Three additional aliquots were similarly processed, but the folding buffer was supplemented with 6 mM Mg2+. The mobility of the mc PSTVd (+) RNA was unaffected by the treatments (Fig. 2), as also occurred when the Mg2+ was added before heating at 95°C for 2 min, although in this latter case the band intensity became attenuated as a likely consequence of the RNA hydrolysis promoted by this metal (data not shown). Importantly, a prominent single band was observed in all instances, consistent with the view that the mc PSTVd (+) form adopts in vitro a single secondary structure, without excluding the presence of others with minor modifications indiscernible by non-denaturing PAGE.
Figure 2.

The mobility of the mc PSTVd (+) RNA in non-denaturing 5% PAGE remains unaffected after different thermal treatments. Before electrophoresis, aliquots of the gel-eluted RNA were heated at 95°C for 2 min and snap-cooled on ice (lane 2 and 5), gradually-cooled at 25°C along 15 min (lane 3 and 6), or left without thermal treatment (NT) (lane 4 and 7). The RNAs were subsequently renatured at 37 °C for 5 min in the folding buffer (with or without 6 mM Mg2+). M refers to DNA markers with their size (in base pairs) indicated on the left (lane 1). The gel was stained with ethidium bromide.
In vitro SHAPE with N-methylisatoic anhydride (NMIA) confirms the rod-like conformation inferred in silico for the mc PSTVd (+) RNA
The observation that the mc PSTVd (+) RNA adopts in vitro a main conformation, made it possible to probe such secondary structure at single-nucleotide resolution with SHAPE in vitro using NMIA29 coupled to computer-assisted prediction using RNAstructure.60 In order to cover entirely the whole molecule, 2 independent primers were used. The analysis with these 2 primers led to consistent results that were also in good agreement with those predicted in silico: the overall conformation adopted in vitro by the mc PSTVd (+) RNA is a rod-like secondary structure with, excepting minor rearrangements, similar double-stranded segments interspersed with small loops (Fig. 3A).
Figure 3.
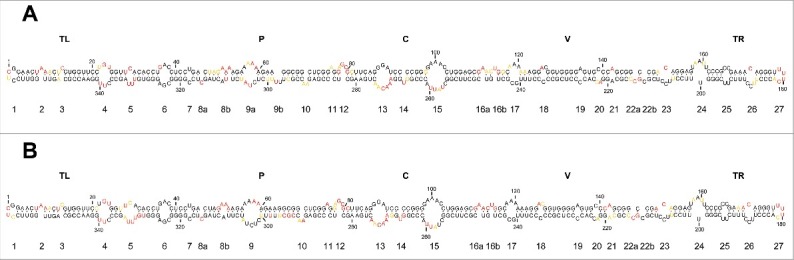
In vitro SHAPE analysis supports a rod-like conformation for the mc PSTVd (+)` RNA. (A) and (B) Results obtained with N-methylisatoic anhydride (NMIA) and 2-methylnicotinic acid imidazolide (NAI), respectively, coupled to computer-assisted prediction using RNAstructure. Nucleotides in red, yellow and black displayed high (more that 0.85), intermediate (0.85–0.40) and low (less than 0.40) SHAPE-reactivity. Other details regarding domains and loops as in the legend to Fig. 1.
Importantly, of the motifs for which the physical and/or biological evidence supporting their existence and involvement in replication/trafficking is strongest —loops 6, 7, 15 (loop E) and 24 (numbering according to;48 see below for a more detailed account of their functions)— all except loop 24 were preserved. We do not know the cause for such discrepancy but the evidence sustaining this loop is only biological and not structural.51 Other motifs, including loops 13 and 14 (located in the CCR and, therefore, most likely relevant from a functional perspective), and loops 23 and 26 (the internal and external RY motifs involved in binding VirP1),18 were also preserved.
It is worth noting that this is the first SHAPE analysis in vitro performed with the mc PSTVd (+) RNA, which apart from being the form that triggers infection and mediates trafficking in vivo (see above), should adopt its intramolecular cooperative folding more easily than the monomeric linear (ml) counterparts used in previous SHAPE studies in vitro.26,27
In vitro SHAPE with 2-methylnicotinic acid imidazolide (NAI) further supports the rod-like conformation predicted for the mc PSTVd (+) RNA
Because we were planning to use NAI as the acylating reagent for SHAPE in vivo,44,45 we first examined with the same 2 primers used before how consistent was the NAI-derived conformation in vitro for the mc PSTVd (+) RNA with that determined with NMIA. In both instances a similar rod-like secondary structure was obtained, and what is even more significant, with essentially the same loops (Fig. 3A and B).
Altogether, the excellent concordance of in vitro SHAPE results with NMIA and NAI provided us with the confidence to take the next step and analyze the conformation of the mc PSTVd (+) RNA in planta with the latter reagent, which due to its higher t1/2 hydrolysis,45 is more appropriate for SHAPE studies in vivo.
SHAPE analysis of the mc PSTVd (+) RNA in infected leaves of Nicotiana benthamiana reveals a “naked” rod-like conformation with minor but significant differences with that observed in vitro
We essentially followed a NAI-based procedure described previously replacing radioactive by fluorescent labeling.44 The other most significant modification was that, following NAI infiltration of PSTVd-infected leaves and RNA extraction and clarification, the mc PSTVd (+) RNA was purified according to a double PAGE approach (see Materials and Methods).36 Such modification ensured that the stops observed during reverse transcription occurred in the mc PSTVd (+) RNA. Moreover, to confirm that acylation of this RNA took place in planta and not during the ensuing isolation, we performed an additional control:44 a preparation of ml forms of eggplant latent viroid (+) RNA obtained by in vitro transcription36 was incorporated immediately after homogenization of the PSTVd-infected tissue infiltrated with NAI. Subsequent SHAPE analysis of this externally-added RNA control failed to reveal the reactivity observed previously in vitro36 (data not shown). We also discarded that the grinding procedure might influence the final outcome because homogenization of NAI-infiltrated PSTVd-infected tissue, with either a mechanical device or with liquid nitrogen, led essentially to the same results (data not shown).
Examination of the data obtained showed that this RNA indeed adopts a rod-like conformation in vivo —an observation relevant per se because it indicates that this RNA accumulates in planta essentially as a “naked” form rather than in tight association with protecting host proteins— in which, as a general trend, loops were more reactive than double-stranded segments. A closer inspection, however, revealed minor but significant differences when this conformation in vivo was compared with that derived from SHAPE in vitro using NAI and the same 2 primers (Fig. 4). The most significant differences are as follows. First, the structure of loop 6 [CGA(36–38)/GAC(323–325) flanked by cis Watson-Crick G/C and G/U wobble base-pairs] mediating trafficking from palisade mesophyll to spongy mesophyll,59 and of loop 7 (U43/C318) critical for systemic trafficking,58 remained essentially unchanged in vitro and in vivo. Notably, except A325 and G326, the corresponding nucleotides displayed no SHAPE reactivity, thus indicating that these 2 loops fold into compact structures not accessible to the acylating reagent. Second, some nucleotides of the lower strand of loop 13 [AACAA(270–275)] forming part of the CCR, were more reactive in vitro than in vivo, consistent with the idea that they are somewhat protected under the latter conditions. Third, on the basis of UV cross-linking studies in vitro61 and in vivo,62,63 nuclease and chemical mapping in solution,24 and sequence comparison, isostericity matrices and mutagenic analyses,57 it has been inferred that a loop E (sarcin/ricin) motif exists in PSTVd (loop 15). This loop, which also forms part of the CCR and encompasses 5 core non-Watson-Crick base pairs57 and a bulged nucleotide (U259 in variant NB),57,64 has been involved in transcription/accumulation,57 ligation,47,65 host specificity64 and pathogenesis,66 with some of these functions being possibly interdependent. Some nucleotides of the lower strand of loop E were more reactive in vitro than in vivo, resembling the situation observed in loop 13 and leading to a similar inference. However, no difference was detected in the intervening loop 14 (U267), with this bulged nucleotide showing similar SHAPE reactivity in vitro and in vivo. Fourth, loop 24 containing U201 as predicted in silico (Fig. 1A), is proposed to form part of a bipartite RNA motif mediating trafficking from the bundle sheath to mesophyll in young tobacco leaves;51 this loop is reorganized according to in vitro and in vivo SHAPE (Fig. 3 and 4), with the critical U20151 now found in an adjacent and basically non-reactive double-stranded segment. And fifth, some nucleotides of the lower strand of loop 23 [C150/UCC(206–208)] and loop 26 [C172/UCC(187–189)], forming part of the VirP1-binding RY-motifs,18 displayed lower SHAPE reactivity in vitro than in vivo, which might indicate some sort of rearrangement in the physiological context.
Figure 4.
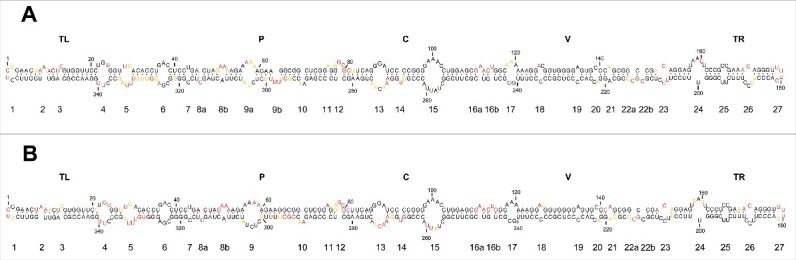
In vivo SHAPE analysis corroborates a rod-like conformation for the mc PSTVd (+) RNA. (A) Results obtained with NAI in vivo coupled to computer-assisted prediction using RNAstructure. (B) Results obtained with the same acylating agent in vitro (see Fig. 3B) are included here to facilitate a direct comparison. Other details as in the legends to Figs. 1 and 3.
Discussion
Recent work has shown that SHAPE chemistry (with NAI) permits structural probing of RNAs of Arabidopsis thaliana,44 extending to the plant kingdom previous studies in bacteria, yeast, fly and mammalian cells.45 Moreover, among the RNAs of A. thaliana examined is the U12 small nuclear RNA,44 indicating that NAI can gain access not only to the cytoplasm but also to organelles —like the nucleus— surrounded by a membrane. This finding is important for the present study because most viroids, including PSTVd, have a nuclear replication and accumulation.
Due to their peculiar properties, viroids must express their associated phenotype without resorting to protein intermediation, mostly relying on sequence/structural motifs embedded in the genomic RNA. Therefore, to dissect finely the conformation of this RNA in vivo is of crucial importance but, unfortunately, no viroid has been reported to infect A. thaliana.56 To address such issue we have selected the system PSTVd/N. benthamiana because this viroid was the first identified and sequenced,15-17 and because the available data about its physical and biological properties exceeds by far those on any other viroid. And N. benthamiana because it is an experimental host in which PSTVd accumulates to a relatively high titer,51,52 and because this virological model plant is very convenient for infiltration studies, particularly with Agrobacterium tumefaciens;67 hence, we presumed an easy and quick penetration of NAI, the acylating reagent commonly used for in vivo SHAPE.
But first we re-examined the structure of minimal free energy predicted for the mc PSTVd (+) RNA (NB variant) by 3 softwares, which generated an almost identical rod-like conformation similar to that proposed previously for this51,52 and other PSTVd variants including the intermediate and that from dahlia.17,48,53 On the other hand, non-denaturing PAGE of the mc PSTVd (+) RNA from NB variant showed a predominant band consistent with a single major folding amenable to in vitro SHAPE, which indeed provided a well-resolved and essentially coincidental structure with 2 acylating reagents (NMIA and NAI). Moreover, this conformation is similar to that obtained previously for the PSTVd intermediate and dahlia variants by in vitro SHAPE with benzoyl cyanide (BzCN) (Fig. S2).26,27 The small differences observed could arise from using in the in vitro SHAPE analyses distinct PSTVd variants and forms (mc and ml), acylating agents (NMIA and NAI on the one side, and BzCN on the other) and folding conditions. In any case, a key point in the present study is the use of the same acylating reagent (NAI) for the in vitro and in vivo SHAPE of the same variant (NB), making the corresponding results directly comparable.
Globally, the rod-like conformation adopted by the mc PSTVd (+) RNA in vivo resembles that predicted in silico and inferred in vitro in a protein-free milieu, thus showing that it is not covered by tightly-associated host protein(s) mimicking the protecting role of coat proteins in viruses (although we cannot exclude the existence of loose RNA-protein interactions or of proteins interacting with double-stranded RNA motifs, which may be undetectable by SHAPE). In other words, our finding affords the first direct evidence supporting the long-held tenet15,68 that such RNA is essentially “naked” in its major subcellular habitat, the nucleolus,69,70 wherein a significant fraction of the accumulating mc PSTVd (+) RNA may function as a reservoir or storage molecule.11,71 During replication, synthesis of (+) and (−) strands of PSTVd and closely-related viroids occurs in the nucleoplasm,69 catalyzed by RNA polymerase II72-75 in association with transcription factor IIIA.76,77 Subsequent processing of oligomeric (+) strands most likely takes place in the nucleolus:56,69 cleavage directed by the upper CCR strand and catalyzed by a class III RNase, and ligation directed by both CCR strands —particularly by loop E (loop 15) possibly assisted by proximal loops 13 and 14— and catalyzed by DNA ligase 1.47,49 Loop E, conserved in the rod-like conformation of all members of the genus Pospiviroid to which PSTVd belongs, positions the termini to be joined in close proximity and proper orientation.47,78 Being ligation the last step of the replication cycle, DNA ligase 1 —or some auxiliary factor— might remain attached to its substrate, thus accounting for the differences observed in loop 13 and loop E between in vitro and in vivo SHAPE; other explanations, however, cannot be ruled out.
In summary, we provide direct evidence at single-nucleotide resolution that the mc PSTVd (+) RNA indeed adopts a rod-like conformation in vivo. However, within this peculiar conformation, some nucleotides forming part of key functional motifs, e.g. loop E, are more SHAPE-reactive in vitro than in vivo, suggesting that they may locally interact with host proteins and through them exert their physiological role. On the other hand, being a “naked” RNA, the mc PSTVd (+) RNA must have evolved to become resistant to degradation through an RNA-based rather than through a protein-based strategy: the lack of free termini in the rod-like conformation makes it resistant against exonucleases, while its compact folding —to a large extent stabilized by double-stranded segments— plays a similar role against endonucleases acting upon either single-stranded regions (like most RNases), or double-stranded segments above a certain length (like the Dicer-like enzymes involved in RNA silencing). The loops, besides limiting that length and providing additional stability, would mediate several key roles including replication and trafficking. Consistent with the intrinsic resistance of the mc PSTVd (+) RNA, the subgenomic viroid RNAs characterized in some PSTVd-infected hosts do not seem to result from cleavage of this form but of replication intermediates.71 It will be interesting to extend the approach developed here to additional viroid/host combinations, although limitations posed by the low viroid accumulation and/or penetration of the acylating reagent will have to be previously overcome.
Materials and methods
Extraction, purification and analysis of the mc PSTVd (+) RNA
Total nucleic acids from young leaves of PSTVd-infected N. benthamiana were extracted with phenol-saturated buffer79 and then fractionated on non-ionic cellulose (CF11; Whatman) with STE (50 mM Tris-HCl, pH 7.2, 100 mM NaCl, 1 mM EDTA) containing 35% ethanol. The viroid-enriched preparations were treated with methoxyethanol for removing polysaccharides.80 Following PAGE in a 5% gel under non-denaturing conditions with 1X TAE (Tris-Acetate-EDTA), a segment delimited by the DNA markers of 300 and 400 bp was cut, applied on top of a single-well 5% polyacrylamide gel, and electrophoresed under denaturing conditions with 0.25X TBE (Tris-Borate-EDTA) and 8 M urea. The second gel was stained with ethidium bromide and the band of interest, identified with appropriate markers, was excised and the corresponding mc PSTVd (+) RNA eluted overnight with 10 mM Tris-HCl, pH 7.5 containing 1 mM EDTA and 0.1% SDS, and recovered by ethanol precipitation. This RNA, after being treated as indicated in Results, was examined by non-denaturing PAGE.
Thermodynamics-based prediction of RNA structure
The structures of minimal free energy for the mc PSTVd (+) RNA were determined with 3 softwares: Mfold version 4.721 and RNAfold included in the ViennaRNA package version 2.2.522 using the circular version and default parameters, and RNAstructure version 5.8 (Shapeknots),23,60 using the default parameters and beginning the sequence of the linear RNA at position 1.
In vitro SHAPE
The substrate for this analysis was the mc PSTVd (+) RNA purified from infected N. benthamiana. SHAPE in vitro with N-methylisatoic anhydride (NMIA) was performed essentially as reported.29 The RNA (3 pmol) was heated at 95°C for 2 min, put on ice for 15 min, and renatured at 37°C for 5 min in the folding buffer (100 mM HEPES-NaOH pH 8.0, 100 mM NaCl, 6 mM MgCl2). Acylation started by adding NMIA (6 mM in dimethyl-sulfoxide), while only dimethyl-sulfoxide was incorporated to the control reaction. To generate the 2′-O-adducts, reaction mixtures were incubated at 37°C for 15 min and ended by adding 3 volumes of ethanol. The RNA was recovered by centrifugation, washed 3 times with 70% ethanol, and subjected to primer extension. SHAPE in vitro with 2-methylnicotinic acid imidazolide (NAI, 50 mM in dimethyl-sulfoxide) was performed similarly, with NAI being synthesized as reported previously.45 The data obtained were coupled to computer-assisted prediction.60
VIC-labeled fluorescent DNA primers (Applied Biosystems) RF-1242 (5’-AAACCCTGTTTCGGCGGGAATTAC-3’) and RF-1367 (5’-GGAGGTCAGGTGTGAACCACAGG-3’), complementary to positions 156–179 and 20–42, respectively, were purified by denaturing PAGE in 20% gels. After adding 4 pmol of the primer, the RNAs with and without the acylating reagent were heated to 95°C for 2 min and snap-cooled on ice for 15 min. Extensions were performed at 52°C for 1 h (in a 20 μl reaction volume) with 100 U SuperScript III RT (Invitrogen) and 0.5 mM dNTPs in the buffer recommended by the supplier. Sequencing reactions used to identify the peaks were prepared in a similar way but adding 10 mM of ddGTP to the primer extension mix and using the same primers, tagged with the NED fluorophore, and non-modified RNA. The resulting cDNAs were ethanol-precipitated, recovered by centrifugation, washed 3 times with 70% ethanol, resuspended in deionized formamide and resolved by capillary electrophoresis in an ABI 3130 XL Genetic Analyzer (Applied Biosystems) as previously described.30 Electrophoregrams were analyzed using the QuShape software,81 which also normalized the reactivity data. At least 4 in vitro SHAPE replicas were performed with each primer and acylating reagent, and the mean and standard deviation of the reactivity of each nucleotide was calculated.
In vivo SHAPE
Young leaves (15 g) of PSTVd-infected N. benthamiana were mixed with 200 ml of the reaction buffer (40 mM HEPES-NaOH pH 7.5, 100 mM KCl, 0.5 mM MgCl2) and gently shaken for 5 min. In planta acylation was performed by adding NAI (100 mM in dimethylsulfoxide and just dimethylsulfoxide to the control reaction) and shaking the mixture at room temperature for 15 min. After adding β-mercaptoethanol (500 mM) and shaking for another 10 min to stop the reaction, the leaves were drained and washed 4 times with water (250 ml). Extraction, purification, and primer-extension of the mc PSTVd (+) RNA was as with in vitro SHAPE. At least 3 in vivo SHAPE replicas were performed with each primer, and the mean and standard deviation of the reactivity of each nucleotide was calculated.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest disclosed.
Acknowledgments
We thank Drs. Cristina Romero and Alicia Barroso for their valuable help with the initial SHAPE experiments, Drs. Sonia Delgado and Pedro Serra for helpful advise, and A. Ahuir for excellent technical assistance.
Funding
This work was supported by grant BFU2014-56812-P (to R.F.) from the Ministerio de Economía y Competitividad of Spain. A.L.C. was the recipient of a predoctoral fellowship from the same organism.
References
- 1.Diener TO. Discovering viroids—a personal perspective. Nature Rev Microbiol 2003; 1:75-80; https://doi.org/ 10.1038/nrmicro736 [DOI] [PubMed] [Google Scholar]
- 2.Flores R, Hernández C, Martínez de Alba E, Daròs JA, Di Serio F. Viroids and viroid-host interactions. Annu Rev Phytopathol 2005; 43:117-39; PMID:16078879; https://doi.org/ 10.1146/annurev.phyto.43.040204.140243 [DOI] [PubMed] [Google Scholar]
- 3.Tsagris EM, Martínez de Alba AE, Gozmanova M, Kalantidis K. Viroids. Cell Microbiol 2008; 10:2168-79; PMID:18764915; https://doi.org/ 10.1111/j.1462-5822.2008.01231.x [DOI] [PubMed] [Google Scholar]
- 4.Kovalskaya N, Hammond RW. Molecular biology of viroid-host interactions and disease control strategies. Plant Sci 2014; 228:48-60; PMID:25438785; https://doi.org/ 10.1016/j.plantsci.2014.05.006 [DOI] [PubMed] [Google Scholar]
- 5.Palukaitis P. What has been happening with viroids? Virus Genes 2014; 49:175-84; PMID:25164861; https://doi.org/ 10.1007/s11262-014-1110-8 [DOI] [PubMed] [Google Scholar]
- 6.Flores R, Minoia S, Carbonell A, Gisel A, Delgado S, López-Carrasco A, Navarro B, Di Serio F. Viroids, the simplest RNA replicons: how they manipulate their hosts for being propagated and how their hosts react for containing the infection. Virus Res 2015; 209:136-45; PMID:25738582; https://doi.org/ 10.1016/j.virusres.2015.02.027 [DOI] [PubMed] [Google Scholar]
- 7.Ding B. The biology of viroid-host interactions. Annu Rev Phytopathol 2009; 47:105-31; PMID:19400635; https://doi.org/ 10.1146/annurev-phyto-080508-081927 [DOI] [PubMed] [Google Scholar]
- 8.Ding B. Viroids: self-replicating, mobile, and fast-evolving noncoding regulatory RNAs. Wiley Interdiscip Rev RNA 2010; 1:362-75; PMID:21956936; https://doi.org/ 10.1002/wrna.22 [DOI] [PubMed] [Google Scholar]
- 9.Flores R, Serra P, Minoia S, Di Serio F, Navarro B. Viroids: from genotype to phenotype just relying on RNA sequence and structural motifs. Front Microbiol 2012; 3:217; PMID:22719735; https://doi.org/ 10.3389/fmicb.2012.00217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao AL, Kalantidis K. Virus-associated small satellite RNAs and viroids display similarities in their replication strategies. Virology 2015; 479–480:627-36; PMID:25731957; https://doi.org/ 10.1016/j.virol.2015.02.018 [DOI] [PubMed] [Google Scholar]
- 11.Steger G, Perreault JP. Structure and associated biological functions of viroids. Adv Virus Res 2016; 94:141-72; PMID:26997592; https://doi.org/ 10.1016/bs.aivir.2015.11.002 [DOI] [PubMed] [Google Scholar]
- 12.Hutchins C, Rathjen PD, Forster AC, Symons RH. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res 1986; 14:3627-40; PMID:3714492; https://doi.org/10.1093/nar/14.9.3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G. Autolytic processing of dimeric plant virus satellite RNA. Science 1986; 231:1577-80; PMID:17833317; https://doi.org/ 10.1126/science.231.4745.1577 [DOI] [PubMed] [Google Scholar]
- 14.Daròs JA, Flores R. A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead-mediated self-cleavage. EMBO J 2002; 21:749-59; https://doi.org/ 10.1093/emboj/21.4.749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diener TO. Potato spindle tuber “virus:” IV. A replicating, low molecular weight RNA. Virology 1971; 45:411-28; PMID:5095900; https://doi.org/ 10.1016/0042-6822(71)90342-4 [DOI] [PubMed] [Google Scholar]
- 16.Diener TO. Potato spindle tuber viroid VIII. Correlation of infectivity with a UV-absorbing component and thermal denaturation properties of the RNA. Virology 1972; 50:606-9; PMID:4636118; https://doi.org/ 10.1016/0042-6822(72)90412-6 [DOI] [PubMed] [Google Scholar]
- 17.Gross HJ, Domdey H, Lossow C, Jank P, Raba M, Alberty H, Sänger HL. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature 1978; 273:203-8; PMID:643081; https://doi.org/ 10.1038/273203a0 [DOI] [PubMed] [Google Scholar]
- 18.Gozmanova M, Denti MA, Minkov IN, Tsagris M, Tabler M. Characterization of the RNA motif responsible for the specific interaction of potato spindle tuber viroid RNA (PSTVd) and the tomato protein Virp1. Nucleic Acids Res 2003; 31:5534-43; PMID:14500815; https://doi.org/ 10.1093/nar/gkg777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martínez de Alba AE, Sägesser R, Tabler M, Tsagris M. A bromodomain-containing protein from tomato binds specifically potato spindle tuber viroid RNA in vitro and in vivo. J Virol 2003; 77:9685-94; PMID:12915580; https://doi.org/ 10.1128/JVI.77.17.9685-9694.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalantidis K, Denti MA, Tzortzakaki S, Marinou E, Tabler M, Tsagris M. Virp1 is a host protein with a major role in potato spindle tuber viroid infection in Nicotiana plants. J Virol 2007; 81:12872-80; PMID:17898061; https://doi.org/ 10.1128/JVI.00974-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31:3406-15; PMID:12824337; https://doi.org/ 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorenz R, Bernhart SH, Hoener zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA Package 2.0. Algorith Mol Biol 2011; 6:26; https://doi.org/ 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinform 2010; 11:129; https://doi.org/ 10.1186/1471-2105-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gast FU, Kempe D, Spieker RL, Sänger HL. Secondary structure probing of potato spindle tuber viroid (PSTVd) and sequence comparison with other small pathogenic RNA replicons provides evidence for central non-canonical base-pairs, large A-rich loops, and a terminal branch. J Mol Biol 1996; 262:652-70; PMID:8876645; https://doi.org/ 10.1006/jmbi.1996.0543 [DOI] [PubMed] [Google Scholar]
- 25.Xu W, Bolduc F, Hong N, Perreault JP. The use of a combination of computer-assisted structure prediction and SHAPE probing to elucidate the secondary structures of five viroids. Mol Plant Pathol 2012; 13:666-76; PMID:22243942; https://doi.org/ 10.1111/j.1364-3703.2011.00776.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giguère T, Raj Adkar-Purushothama C, Perreault JP. Comprehensive secondary structure elucidation of four genera of the family Pospiviroidae. PLoS ONE 2014; 9:e98655; https://doi.org/ 10.1371/journal.pone.0098655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adkar-Purushothama CR, Brosseau C, Giguère T, Sano T, Moffett P, Perreault JP. Small RNA derived from the virulence modulating region of the potato spindle tuber viroid silences callose synthase genes of tomato plants. Plant Cell 2015; 27:2178-94; PMID:26290537; https://doi.org/ 10.1105/tpc.15.00523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2'-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 2005; 127:4223-31; PMID:15783204; https://doi.org/ 10.1021/ja043822v [DOI] [PubMed] [Google Scholar]
- 29.Wilkinson KA, Merino EJ, Weeks KM. Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nature Protoc 2006; 1:1610-6; https://doi.org/ 10.1038/nprot.2006.249 [DOI] [PubMed] [Google Scholar]
- 30.Mortimer SA, Weeks KM. Time-resolved RNA SHAPE chemistry: quantitative RNA structure analysis in one second snapshots and at single nucleotide resolution. Nature Protoc 2009; 4:1413-21; https://doi.org/ 10.1038/nprot.2009.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sogo JM, Koller T, Diener TO. Potato spindle tuber viroid X. Visualization and size determination by electron microscopy. Virology 1973; 55:70-80; PMID:4728831; https://doi.org/ 10.1016/S0042-6822(73)81009-8 [DOI] [PubMed] [Google Scholar]
- 32.Sänger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt A. Viroids are single-stranded covalently-closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976; 73:3852-6; https://doi.org/ 10.1073/pnas.73.11.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riesner D, Henco K, Rokohl U, Klotz G, Kleinschmidt AK, Domdey H, Jank P, Gross HJ, Sänger HL. Structure and structure formation of viroids. J Mol Biol 1979; 133:85-115; PMID:529284; https://doi.org/ 10.1016/0022-2836(79)90252-3 [DOI] [PubMed] [Google Scholar]
- 34.Riesner D, Kaper JM, Randles JW. Stiffness of viroids and viroid-like RNA in solution. Nucleic Acids Res 1982; 10:5587-98; PMID:7145708; https://doi.org/ 10.1093/nar/10.18.5587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dingley AJ, Steger G, Esters B, Riesner D, Grzesiek S. Structural characterization of the 69 nucleotide potato spindle tuber viroid left-terminal domain by NMR and thermodynamic analysis. J Mol Biol 2003; 334:751-67; PMID:14636600; https://doi.org/ 10.1016/j.jmb.2003.10.015 [DOI] [PubMed] [Google Scholar]
- 36.López-Carrasco A, Gago-Zachert S, Mileti G, Minoia S, Flores R, Delgado S. The transcription initiation sites of eggplant latent viroid strands map within distinct motifs in their in vivo RNA conformations. RNA Biol 2016; 13:83-97; https://doi.org/ 10.1080/15476286.2015.1119365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haseloff J, Mohamed NA, Symons RH. Viroid RNAs of the cadang-cadang disease of coconuts. Nature 1982; 229:316-21; https://doi.org/ 10.1038/299316a0 [DOI] [Google Scholar]
- 38.Semancik JS, Szychowski JA, Rakowski AG, Symons RH. A stable 463 nucleotide variant of citrus exocortis viroid produced by terminal repeats. J Gen Virol 1994; 75:727-32; PMID:8151291; https://doi.org/ 10.1099/0022-1317-75-4-727 [DOI] [PubMed] [Google Scholar]
- 39.Wassenegger M, Heimes S, Sänger HL. An infectious viroid RNA replicon evolved from an in vitro-generated non-infectious viroid deletion mutant via a complementary deletion in vivo. EMBO J 1994; 13:6172-7; PMID:7813454; https://doi.org/ 10.1002/wrna.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fadda Z, Daròs JA, Flores R, Durán-Vila N. Identification in eggplant of a variant of citrus exocortis viroid (CEVd) with a 96 nucleotide duplication in the right terminal region of the rod-like secondary structure. Virus Res 2003; 97:145-9; PMID:14602207; https://doi.org/ 10.1016/j.virusres.2003.08.002 [DOI] [PubMed] [Google Scholar]
- 41.Gago S, Elena SF, Flores R, Sanjuán R. Extremely high mutation rate of a hammerhead viroid. Science 2009; 323:1308; PMID:19265013; https://doi.org/ 10.1126/science.1169202 [DOI] [PubMed] [Google Scholar]
- 42.Ambrós S, Hernández C, Desvignes JC, Flores R. Genomic structure of three phenotypically different isolates of peach latent mosaic viroid: implications of the existence of constraints limiting the heterogeneity of viroid quasi-species. J Virol 1998; 72:7397-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gago S, De la Peña M, Flores R. A kissing-loop interaction in a hammerhead viroid RNA critical for its in vitro folding and in vivo viability. RNA 2005; 11:1073-83; PMID:15928342; https://doi.org/ 10.1261/rna.2230605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwok CK, Ding Y, Tang Y, Assmann SM, Bevilacqua PC. Determination of in vivo RNA structure in low-abundance transcripts. Nat Commun 2013; 4:2971; PMID:24336128; https://doi.org/ 10.1038/ncomms3971 [DOI] [PubMed] [Google Scholar]
- 45.Spitale RC, Crisalli P, Flynn RA, Torre EA, Kool ET, Chang HY. RNA SHAPE analysis in living cells. Nat Chem Biol. 2013; 9:18-20; PMID:23178934; https://doi.org/ 10.1038/nchembio.1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McInnes JL, Symons RH. Comparative structure of viroids and their rapid detection using radioactive and nonradioactive nucleic acid probes In Viroids and Satellites: Molecular Parasites at the Frontier of Life (Maramorosch K. ed.) p. 21-58, CRC Press, Boca Raton, Fl: 1991 [Google Scholar]
- 47.Gas ME, Hernández C, Flores R, Daròs JA. Processing of nuclear viroids in vivo: an interplay between RNA conformations. PLoS Pathog 2007; 3:1813-26; https://doi.org/ 10.1371/journal.ppat.0030182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong X, Archual AJ, Amin AA, Ding B. A genomic map of viroid RNA motifs critical for replication and systemic trafficking. Plant Cell 2008; 20:35-47; PMID:18178767; https://doi.org/ 10.1105/tpc.107.056606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nohales MA, Flores R, Daròs JA. A viroid RNA redirects host DNA ligase 1 to act as an RNA ligase. Proc Natl Acad Sci U S A 2012; 109:13805-10; PMID:22869737; https://doi.org/ 10.1073/pnas.1206187109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Repsilber D, Wiese S, Rachen M, Schröder AW, Riesner D, Steger G. Formation of metastable RNA structures by sequential folding during transcription: time-resolved structural analysis of potato spindle tuber viroid (−)-stranded RNA by temperature-gradient gel electrophoresis. RNA 1999; 5:574-84; PMID:10199573; https://doi.org/ 10.1017/S1355838299982018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qi Y, Pelissier T, Itaya A, Hunt E, Wassenegger M, Ding B. Direct role of a viroid RNA motif in mediating directional RNA trafficking across a specific cellular boundary. Plant Cell 2004; 16:1741-52; PMID:15194818; https://doi.org/ 10.1105/tpc.021980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Serio F, Martínez de Alba AE, Navarro B, Gisel A, Flores R. RNA-dependent RNA polymerase 6 delays accumulation and precludes meristem invasion of a nuclear-replicating viroid. J Virol 2010; 84:2477-89; PMID:20015979; https://doi.org/ 10.1128/JVI.02336-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsushima T, Murakami S, Ito H, He Y-H, Raj APC, Sano T. Molecular characterization of potato spindle tuber viroid in dahlia. J Gen Plant Pathol 2011; 77:253-6; https://doi.org/ 10.1007/s10327-011-0316-z [DOI] [Google Scholar]
- 54.Branch AD, Benenfeld BJ, Robertson HD. Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc Natl Acad Sci U S A 1988; 85:9128-32; PMID:16594003; https://doi.org/ 10.1073/pnas.85.23.9128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feldstein PA, Hu Y, Owens RA. Precisely full length, circularizable, complementary RNA: an infectious form of potato spindle tuber viroid. Proc Natl Acad Sci U S A 1998; 95:6560-65; PMID:9601006; https://doi.org/ 10.1073/pnas.95.11.6560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daròs JA, Flores R. Arabidopsis thaliana has the enzymatic machinery for replicating representative viroid species of the family Pospiviroidae. Proc Natl Acad Sci U S A 2004; 101:6792-97; https://doi.org/ 10.1073/pnas.0401090101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhong X, Leontis N, Qiang S, Itaya A, Qi Y, Boris-Lawrie K, Ding B. Tertiary structural and functional analysis of a viroid RNA motif by isostericity matrix and mutagenesis reveal its essential role in replication. J Virol 2006; 80:8566-81; PMID:16912306; https://doi.org/ 10.1128/JVI.00837-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhong X, Tao X, Stombaugh J, Leontis N, Ding B. Tertiary structure and function of an RNA motif required for plant vascular entry to initiate systemic trafficking. EMBO J 2007; 26:3836-46; PMID:17660743; https://doi.org/ 10.1038/sj.emboj.7601812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takeda R, Petrov AI, Leontis NB, Ding B. A three-dimensional RNA motif in potato spindle tuber viroid mediates trafficking from palisade mesophyll to spongy mesophyll in Nicotiana benthamiana. Plant Cell 2011; 23:258-72; PMID:21258006; https://doi.org/ 10.1105/tpc.110.081414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hajdin CE, Bellaousov S, Huggins W, Leonard CW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc Natl Acad Sci U S A 2013; 110:5498-503; PMID:23503844; https://doi.org/ 10.1073/pnas.1219988110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Branch AD, Benenfeld BJ, Robertson HD. Ultraviolet light-induced crosslinking reveals a unique region of local tertiary structure in potato spindle tuber viroid and HeLa 5S RNA. Proc Natl Acad Sci U S A 1985; 82:6590-4; PMID:3863116; https://doi.org/ 10.1073/pnas.82.19.6590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eiras M, Kitajima EW, Flores R, Daròs JA. Existence in vivo of the loop E motif in potato spindle tuber viroid RNA. Arch Virol 2007; 152:1389-93; PMID:17370107; https://doi.org/ 10.1007/s00705-007-0952-y [DOI] [PubMed] [Google Scholar]
- 63.Wang Y, Zhong X, Itaya A, Ding B. Evidence for the existence of the loop E motif of potato spindle tuber viroid in vivo. J Virol 2007; 81:2074-7; PMID:17135317; https://doi.org/ 10.1128/JVI.01781-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wassenegger M, Spieker RL, Thalmeir S, Gast FU, Riedel L, Sänger HL. A single nucleotide substitution converts potato spindle tuber viroid (PSTVd) from a noninfectious to an infectious RNA for Nicotiana tabacum. Virology 1996; 226:191-7; PMID:8955038; https://doi.org/ 10.1006/viro.1996.0646 [DOI] [PubMed] [Google Scholar]
- 65.Gas ME, Molina-Serrano D, Hernández C, Flores R, Daròs JA. Monomeric linear RNA of citrus exocortis viroid resulting from processing in vivo has 5’-phosphomonoester and 3’-hydroxyl termini: implications for the ribonuclease and RNA ligase involved in replication. J Virol 2008; 82:10321-5; PMID:18701598; https://doi.org/ 10.1128/JVI.01229-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qi Y, Ding B. Inhibition of cell growth and shoot development by a specific nucleotide sequence in a noncoding viroid RNA. Plant Cell 2003; 15:1360-74; PMID:12782729; https://doi.org/5543290 10.1105/tpc.011585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goodin MM, Zaitlin D, Naidu RA, Lommel SA. Nicotiana benthamiana: its history and future as a model for plant-pathogen interactions. Mol Plant Microbe Interact 2008; 2:1015-26; https://doi.org/ 10.1094/MPMI-21-8-1015 [DOI] [PubMed] [Google Scholar]
- 68.Diener TO. Potato spindle tuber virus: a plant virus with properties of a free nucleic acid III. Subcellular location of PSTV-RNA and the question of whether virions exist in extracts or in situ. Virology 1971; 43:75-98; PMID:5543290; https://doi.org/ 10.1016/0042-6822(71)90226-1 [DOI] [PubMed] [Google Scholar]
- 69.Qi Y, Ding B. Differential subnuclear localization of RNA strands of opposite polarity derived from an autonomously replicating viroid. Plant Cell 2003; 15:2566-77; PMID:14555700; https://doi.org/2591366 10.1105/tpc.016576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harders J, Lukacs N, Robert-Nicoud M, Jovin JM, Riesner D. Imaging of viroids in nuclei from tomato leaf tissue by in situ hybridization and confocal laser scanning microscopy. EMBO J 1989; 8:3941-49; PMID:2591366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Minoia S, Navarro B, Delgado S, Di Serio F, Flores R. Viroid RNA turnover: characterization of the subgenomic RNAs of potato spindle tuber viroid accumulating in infected tissues provides insights into decay pathways operating in vivo. Nucleic Acids Res 2015; 43:2313-25; PMID:25662219; https://doi.org/ 10.1093/nar/gkv034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mühlbach H, Sänger HL. Viroid replication is inhibited by α-amanitin. Nature 1979; 278:185-8; PMID:763366; https://doi.org/ 10.1038/278185a0 [DOI] [PubMed] [Google Scholar]
- 73.Flores R, Semancik JS. Properties of a cell-free system for synthesis of citrus exocortis viroid. Proc Natl Acad Sci U S A 1982; 79:6285-8; PMID:16593239; https://doi.org/ 10.1073/pnas.79.20.6285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schindler IM, Mühlbach HP. Involvement of nuclear DNA-dependent RNA polymerases in potato spindle tuber viroid replication: a reevaluation. Plant Sci. 1992; 84:221-9; https://doi.org/ 10.1016/0168-9452(92)90138-C [DOI] [Google Scholar]
- 75.Warrilow D, Symons RH. Citrus exocortis viroid RNA is associated with the largest subunit of RNA polymerase II in tomato in vivo. Arch Virol 1999; 144:2367-75; PMID:10664390; https://doi.org/ 10.1007/s007050050650 [DOI] [PubMed] [Google Scholar]
- 76.Eiras M, Nohales MA, Kitajima EW, Flores R, Daròs JA. Ribosomal protein L5 and transcription factor IIIA from Arabidopsis thaliana bind in vitro specifically potato spindle tuber viroid RNA. Arch Virol 2011; 156:529-33; PMID:21153748; https://doi.org/ 10.1007/s00705-010-0867-x [DOI] [PubMed] [Google Scholar]
- 77.Wang Y, Qu J, Ji S, Wallace AJ, Wu J, Li Y, Gopalan V, Ding B. A land plant-specific transcription factor directly enhances transcription of a pathogenic noncoding RNA template by DNA-dependent RNA polymerase II. Plant Cell 2016; 28:1094-107; PMID:27113774; https://doi.org/ 10.1105/tpc.16.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baumstark T, Schröder AR, Riesner D. Viroid processing: switch from cleavage to ligation is driven by a change from a tetraloop to a loop E conformation. EMBO J 1997; 16:599-610; PMID:9034342; https://doi.org/ 10.1093/emboj/16.3.599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pallás V, Navarro A, Flores R. Isolation of a viroid-like RNA from hop different from hop stunt viroid. J Gen Virol 1987; 68:3201-5; https://doi.org/ 10.1099/0022-1317-68-12-3201 [DOI] [Google Scholar]
- 80.Bellamy AR, Ralph RK. Recovery and purification of nucleic acids by means of cetyltrimethylammonium bromide. Methods Enzymol 1968; XII:156-60; https://doi.org/ 10.1016/0076-6879(67)12125-3 [DOI] [Google Scholar]
- 81.Karabiber F, McGinnis JL, Favorov OV, Weeks KM. QuShape: Rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA 2013; 19:63-73; PMID:23188808; https://doi.org/ 10.1261/rna.036327.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Keese P, Symons RH. Domains in viroids: evidence of intermolecular RNA rearrangements and their contribution to viroid evolution. Proc Natl Acad Sci U S A 1985; 82:4582-86; PMID:3860809; https://doi.org/ 10.1073/pnas.82.14.4582 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


