Abstract
Diabetes affects millions of people worldwide, and β-cell replacement is one of the promising new strategies for treatment. Induced pluripotent stem cells (iPSCs) can differentiate into any cell type, including pancreatic β cells, providing a potential treatment for diabetes. However, the molecular mechanisms underlying the differentiation of iPSC-derived β cells have not yet been fully elucidated. Here, we generated pancreatic β-like cells from mouse iPSCs using a 3-step protocol and performed deep RNA sequencing to get a transcriptional landscape of iPSC-derived pancreatic β-like cells during the selective differentiation period. We then focused on the differentially expressed genes (DEGs) during the time course of the differentiation period, and these genes underwent Gene Ontology annotation and Kyoto Encyclopedia of Genes and Genomes pathway analysis. In addition, gene-act networks were constructed for these DEGs, and the expression of pivotal genes detected by quantitative real-time polymerase chain reaction was well correlated with RNA sequence (RNA-seq). Overall, our study provides valuable information regarding the transcriptome changes in β cells derived from iPSCs during differentiation, elucidates the biological process and pathways underlying β-cell differentiation, and promotes the identification and functional analysis of potential genes that could be used for improving functional β-cell generation from iPSCs.
Keywords: iPSCs, RNA sequencing, β-cell differentiation
Introduction
Diabetes is a major health-care problem affecting millions of people all over the world, and it is now widely accepted that both type 1 and type 2 diabetes mellitus may lead to the loss of β cells.1–3 β-cell replacement is a promising strategy for diabetes treatment. Although such replacements achieved by whole pancreatic organ or islet transplants are effective, they are limited by a shortage of donor tissues and immune rejection.4 Pluripotent stem cells (PSCs), including embryonic stem cells (ESCs) and induced PSCs (iPSCs), have an unlimited capacity for self-renewal and can differentiate into any cell type in the body, offering potential for clinical cell therapies.5,6 The generation of functional human or mouse pancreatic β cells in vitro from iPSCs has been reported.1,7,8 However, current protocols are not yet optimized for a number of reasons including the complexity of signaling pathways involved.9 The molecular mechanisms underlying the differentiation of iPSCs into β cells have not yet been fully elucidated. A better understanding of the processes and intracellular signaling pathways during β-cell differentiation from iPSCs may facilitate the identification of pivotal molecules and pathways, promoting the establishment of efficient and reproducible protocols for iPSC-derived β-cell generation.
RNA sequencing (RNA-seq) is the direct sequencing of transcripts by high-throughput sequencing technologies. Unlike microarrays, which are based on a reference transcriptome for prior probe selection and have limitations such as hybridization artifacts and measurement accuracy, RNA-seq has considerable advantages for whole-genome transcriptome profiling.10 RNA-seq directly detects transcripts, avoiding hybridization biases of microarrays and providing more precise information on transcript levels. Due to its nearly unlimited sensitivity, RNA-seq technology has now been widely applied for the investigation of novel transcripts, transcriptome analyses, and alternative splicing identification in diverse biological processes and diseases, including iPSC-derived cells.11–14 However, until now, there has been little research about the transcriptome changes during the differentiation of iPSCs into pancreatic β-like cells.
In this study, we used a 3-step protocol to generate pancreatic β-like cells from mouse iPSCs and then conducted RNA-seq to get a transcriptional landscape of iPSC-derived pancreatic β-like cells during the selective differentiation period. We focused on the differentially expressed genes (DEGs) at different time points and performed bioinformatic analyses to find important processes and pathways involved in iPSC differentiation. Key genes were also identified and validated by quantitative real-time polymerase chain reaction (qRT-PCR). Thus, by RNA-seq, our study illustrated the dynamic gene alterations and the related processes and pathways during the selective differentiation of mouse iPSCs into pancreatic β-like cells, which might facilitate the improvement of the generation of pancreatic β-like cells from iPSCs.
Materials and Methods
Ethical Statement
All of the experimental procedures involving animals were conducted in accordance with the Institutional Animal Care guidelines of Nantong University, China and approved by the Administration Committee of Experimental Animals, Jiangsu Province, China.
In Vitro Differentiation of Mouse Green Fluorescent Protein Positive (GFP+)-iPSCs Into Pancreatic β-like Cells
Mouse green fluorescent protein positive (GFP+)-iPSCs (Innovative Cellular Therapeutics Ltd., Shanghai, China) were induced to differentiate into pancreatic β-like cells using a 3-step protocol as previously described.15,16 Briefly, at step 1, embryoid bodies (EBs) were formed from GFP+-iPSCs. GFP+-iPSCs were detached into single cells using 0.25% trypsin (Thermo Fisher Scientific, Carlsbad, CA, USA) and plated onto a 10-cm culture dish for 1 h to remove the feeder cells. The feeder-depleted cells were then collected and centrifuged at 400 g for 5 min. The cell pellet was resuspended in EB medium composed of knockout Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific), 15% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA), 2 mM l-glutamine (Thermo Fisher Scientific), 1 × 10−4 M nonessential amino acids (Thermo Fisher Scientific), 1×10−4 M 2-mercaptoethanol (Sigma-Aldrich), and 1× penicillin–streptomycin (Thermo Fisher Scientific) at 5,000 cells/mL and plated on ultralow attachment plates (Corning Inc., Corning, NY, USA). Cells were then incubated at 37 °C for 4 d. At step 2, EBs were induced to multilineage progenitors (MPs). The EBs were collected and transferred to 10 cm plates coated with 0.1% gelatin (Sigma-Aldrich). Each plate contained 8 to 12 EBs and was incubated for another 9 d with EB medium, which was replaced every 3 d. At step 3, EBs were induced to β-like cells. EB medium was replaced with β-cell selective differentiation medium containing DMEM: nutrient mixture F-12 (DMEM/F12) (Corning, Inc.); 15% FBS, 20 nM progesterone (Sigma-Aldrich); 100 μM putrescine (Sigma-Aldrich); 1 μg/mL laminin (Sigma-Aldrich); 10 mM nicotinamide (Sigma-Aldrich); 1× ITS premix containing insulin, transferrin, and selenic acid (Thermo Fisher Scientific); B27 media supplement (Thermo Fisher Scientific); and 1× penicillin–streptomycin (Thermo Fisher Scientific). On the sixth day, cells were trypsinized and transferred into unused culture dishes throughout step 3 with medium changed every 3 d.
Characterization of Pancreatic β-like Cells
The mouse GFP+-iPSC-derived β-like cells were characterized by immunofluorescence and glucose-stimulated insulin secretion. For immunofluorescence, cells were fixed by 4% paraformaldehyde (PFA) (Solarbio, Beijing, China) for 20 min, permeabilized by 0.1% Triton X-100 (Solarbio) for 10 min, and blocked with 5% bovine serum albumin (BSA) (Solarbio) for 30 min. After that, the cells were incubated with rabbit anti-insulin 1:100 (Abcam, Cambridge, MA, USA) or rabbit anti-C-peptide 1:100 (Abcam) overnight at 4 °C. The next day, the cells were washed 3 times with phosphate-buffered saline (PBS) and incubated with Alexa Flour 594-conjugated goat anti-rabbit secondary antibody 1:500 (Abcam) at room temperature for 1 h. Subsequently, the cells were stained with Hoechst (Sigma-Aldrich) for 15 min and washed with PBS 3 times. The cells were visualized by an Olympus fluorescence microscope (Olympus, Tokyo, Japan).
For a glucose-stimulated insulin secretion assay, the iPSC-derived β-like cells were exposed to a glucose gradient (0, 5, 15, 30, and 45 mM), and insulin secretion was measured by an ultrasensitive mouse insulin enzyme-linked immunosorbent assay (ELISA) kit (Mercodia, Uppsala, Sweden).
RNA Extraction and Transcriptome Sequencing
Total RNA was isolated from iPSCs at different time points during the differentiation using Trizol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions and then purified with RNeasy spin columns (Qiagen, Valencia, CA, USA) to remove contaminating DNA. The quality of isolated RNA samples was evaluated with an Agilent Bioanalyzer 2200 (Agilent Technologies, Santa Clara, CA, USA). Samples with an RNA Integrity Number >8.0 were used for complementary DNA (cDNA) library preparation. A cDNA library was generated using an Ion Total RNA-Seq Kit v2.0 (Thermo Fisher Scientific) and sequenced on Proton Sequencers according to Ion PI Sequencing 200 Kit v2.0 (Thermo Fisher Scientific).
Before mapping of single-end reads, raw reads were subjected to quality control (QC) to remove dirty raw reads, the reads that contain the sequence of the adapter, reads with >5% ambiguous bases (noted as N), and low-quality reads containing more than 20% of bases with qualities <13. After QC, the filtered reads (clean reads) were aligned to the reference sequences with the Map Splice program (v2.1.8; University of Kentucky, Bioinformatics Lab, http://www.netlab.uky.edu/p/bioinfo/MapSplice/).
Screening of DEGs
The gene expression level was calculated by the reads per kilobase transcriptome per million mapped reads (RPKM method), eliminating the influence of different gene lengths and sequencing discrepancies in the calculation of gene expression. Therefore, the calculated gene expression can be directly used for comparing the difference in gene expression among samples.
We then applied the EB-seq algorithm which was a package that can be downloaded from Bioconductor (http://www.bioconductor.org/packages/devel/bioc/html/EBSeq.html) to filter the DEGs, after the analyses of significance and false discovery rate (FDR), with the following criteria: (i) fold change >2 or <0.5 and (ii) FDR <0.05.
Analysis of Gene Ontology (GO), Pathway, Pathway-Act Network, and Gene-Act Network
GO analysis was applied to elucidate the biological implications of the differential expression of genes.17 We downloaded the GO annotations from NCBI (http://www.ncbi.nlm.nih.gov/), UniProt (http://www.uniprot.org/), and GO (http://www.geneontology.org/). Fisher’s exact test was applied to identify significant GO categories, and FDR was used to correct the P values. Pathway analysis was used to elucidate the significant pathways of the differential genes according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/).18 Fisher’s exact test was used to find the significant enrichment pathway. The resulting P values were adjusted using the Benjamini–Hochberg FDR algorithm and pathway categories, and a P value <0.05 was considered significant (indicated significant pathways). These pathway terms (P < 0.05) were then selected to build a pathway-act network based on the relationships among these pathways in the KEGG database. The KEGG database was also used to build a network of genes according to the relationships among genes in the database. Gene degree centrality was defined as the number of links of one node to others.19
qRT-PCR
The same RNA samples described in RNA extraction and transcriptome sequencing were subjected to qRT-PCR to validate the results of transcriptome sequencing. RNA samples were reverse transcribed to cDNA using the Prime-Script reagent kit (TaKaRa, Dalian, China). qRT-PCR was performed with Synergy Brands (SYBR) Premix Ex Taq (TaKaRa) on an ABI Step One system (Applied Biosystems, Foster City, CA, USA) according to standard protocols; each sample was run in triplicate. Relative quantification of messenger RNA was conducted using the comparative 2−ΔΔCt method and normalized against glyceraldehyde 3-phosphate dehydrogenase (GAPDH) level. Primers used were listed in Supplementary Table S1 (Supplementary Materials available at http://pan.baidu.com/s/1o8RaUUY using the password zxy4). We then created a line graph comparing the expression of genes detected by qRT-PCR and RNA-seq, and the correlation coefficient between qRT-PCR and RNA-Seq was calculated automatically by Excel software (Microsoft Corporation, Redmond, WA, USA).
Statistical Analysis
Statistical analyses were performed using Prism 5 software (GraphPad, San Diego, CA, USA). Student’s t tests were used for comparison between groups. P < 0.05 was considered statistically significant. All data were expressed as means ± standard deviation (SD).
Results
Selective Differentiation of Mouse GFP+-iPSCs Into Pancreatic β-like Cells
Mouse GFP+-iPSCs were differentiated into β-like cells in 3 steps (Fig. 1A). At step 1, GFP+-iPSCs were detached into single cells, resuspended in EB medium, and plated on ultralow attachment plates for 4 d, forming EBs (Fig. 1B, columns 1 and 2). At step 2, the EBs were transferred to an adherent culture and differentiated into MPs for 9 d. At step 3, which lasted for 21 d, the cells were treated with fresh β-cell selective medium. On day 4 of step 3, cells that migrated from EBs were growing by adherence (early stage; Fig. 1B, column 3). After being trypsinized and resuspended in selective medium, these cells began to form clusters on day 14 of step 3 (midstage; Fig. 1B, column 4), and on day 21, large clusters were evident (late stage; Fig. 1B, column 5).
Figure 1.
Generation of pancreatic β-like cells from mouse green fluorescent protein positive (GFP+)-induced pluripotent stem cells (iPSCs). (A) Schematic diagram depicting a 3-step protocol for iPSC-derived β-like cells. EB, embryoid body; MP, multilineage progenitor. (B) Morphologies of differentiating GFP+-iPSCs into β-like cells. Images of iPSCs at different time points during differentiation into β-like cells in vitro were shown. GFP continues to be expressed throughout all steps. BF, bright field image.
Characterization of iPSC-Derived Pancreatic β-like Cells
Since pancreatic β cells are characterized by insulin secretion and C-peptide is a by-product of insulin synthesis, we examined these 2 β-cell markers in the mouse GFP+-iPSC-derived β-like cells to evaluate the efficiency of β-like cell differentiation. The results show that, at day 21 of step 3 (late stage), the majority of the differentiated cells were positive for insulin and C-peptide by immunofluorescence (Fig. 2A). To detect whether the differentiated cells were glucose responsive, these iPSC-derived β-like cells were exposed to glucose at different concentrations (0, 5, 15, 30, and 45 mM), and insulin secretion was measured. As shown in Figure 2B, insulin was hardly detected at 0 mM. Following glucose treatment, insulin secretion in these β-like cells increased with a peak at the 30 mM glucose concentration. No insulin was induced when glucose was washed out, suggesting these β-like cells could only produce insulin in response to glucose.
Figure 2.
Characterization of induced pluripotent stem cell (iPSC)-derived β-like cells. (A) Immunofluorescent staining of iPSCs at step 3 on day 21. In vitro-derived β-like cells were stained with antibodies to green fluorescent protein (GFP), insulin, and C-peptide on day 21 of step 3. Cells were counterstained with Hoechst. Scale bar: 50 µm. (B) Glucose-stimulated insulin secretion in vitro. iPSC-derived β-like cells on day 21 of step 3 were first exposed to different glucose concentrations (0, 5, 15, 30, and 45 mM), and the insulin levels in their supernatants were determined. Glucose was then removed from the medium, and insulin levels were again measured in supernatants from the same cells.
RNA-Seq of iPSC-Derived β-like Cells in Different Time Courses
In order to get a global view of transcriptome changes during the differentiation of β cells, we collected iPSC-derived β-like cells at different time points, including the undifferentiated iPSCs and iPSC-derived β-like cells at day 4 (early stage), day 14 (midstage), and day 21 (late stage) during step 3, and performed deep sequencing (Supplementary Figure S1). Total raw reads among the 4 samples ranged from 14 to 17 million. The average guanine/cytosine (G/C) base content was approximately 50% for each sample. After QC, about 95% of the reads were obtained as clean reads (Supplementary Table S2). We then mapped the filtered clean reads to the reference genome. It showed that, for each sample, more than 89% of reads could be mapped to the reference genome with uniquely mapped reads accounting for more than 78% of reads (Table 1), indicating good reliability of the data.
Table 1.
Summary of the Sequencing Reads Alignment to the Reference Genome..
| Statistics | S0 | S9_4 | S9_14 | S9_21 |
|---|---|---|---|---|
| Clean Reads | 15,212,293 | 15,972,975 | 16,642,840 | 13,880,227 |
| Unmapped Reads | 1,573,251 | 1,128,714 | 1,023,309 | 1,138,379 |
| Mapped (Rate) | 13,639,042 (0.897) | 14,844,261 (0.929) | 15,619,531 (0.939) | 12,741,848 (0.918) |
| Unique Mapped (Rate) | 11,916,424 (0.783) | 13,720,153 (0.859) | 14,059,435 (0.845) | 11,665,377 (0.84) |
| Repeat Mapped | 1,722,618 | 1,124,108 | 1,560,096 | 1,076,471 |
Abbreviation: iPSCs, induced pluripotent stem cells.
The value within parenthesis were the percentage of Mapped or Unique mapped reads in clean reads.
DEGs Following iPSC Differentiation
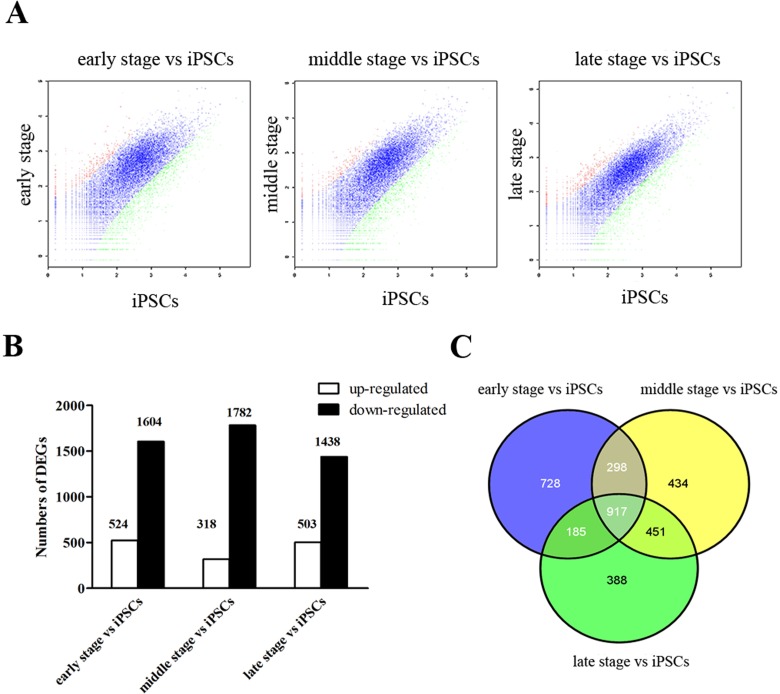
By RNA-Seq, we identified 26,547 genes during iPSC differentiation. The EB-seq algorithm was then used to find DEGs between time points. Under the threshold of 2-fold change and FDR <0.05, we identified 2,128, 2,100, and 1,941 DEGs compared to iPSCs at early stage, middle stage, and late stage in step 3, respectively. Most of these DEGs were downregulated (Fig. 3A and B). A Venn diagram shows that DEGs in the middle stage versus iPSCs and DEGs in the late stage versus iPSCs have fifty percent overlap of genes (Fig. 3C).
Figure 3.
Differentially expressed genes (DEGs) in the time course during the step 3 differentiation period. The (A) charts and (B) bar graph showed that a huge number of genes were differentially expressed in induced pluripotent stem cell (iPSC)-derived β-like cells at early, middle, and late stages of step 3 compared to iPSCs. Red and green dots indicate upregulation and downregulation, respectively, while blue dots indicate no significant difference. (C) Venn diagram showing separate and overlapping expression between DEGs in iPSC-derived β-like cells at early, middle, and late stages of step 3 compared to iPSCs.
Functional Enrichment Analysis of DEGs in Different Time Courses
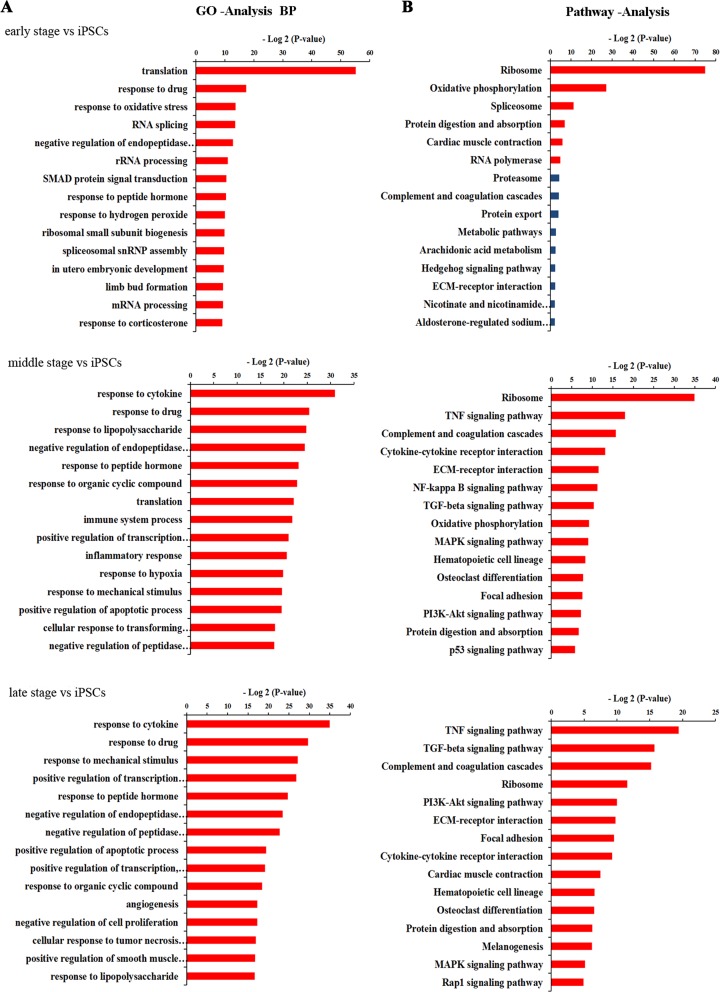
We then performed GO and KEGG pathway analyses on DEGs in early stage versus iPSCs, middle stage versus iPSCs, and late stage versus iPSCs, respectively, to find possible biological processes and pathways involved in the differentiation from iPSCs to pancreatic β-like cells. The top enriched categories of biological processes in each group were shown in Figure 4A and Supplementary Table S3. We found that translation was significantly affected in early stage of step 3, while biological processes such as response to cytokines, regulation of transcription, and regulation of apoptotic process were enriched in the later period of step 3. Analysis of pathways during the iPSC differentiation indicated that the ribosome signaling pathway was enriched in early stage versus iPSCs, consisting of the GO category analyses of biological processes (Fig. 4B; Supplementary Table S3). On the other hand, signaling pathways such as the tumor necrosis factor (TNF) signaling pathway, transforming growth factor-β (TGF-β) signaling pathway, phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)-protein kinase B (Akt) signaling pathway, mitogen-activated protein kinase (MAPK) signaling pathway, extracellular matrix (ECM)–receptor interaction, and focal adhesion exhibited significant enrichment in middle and late stages versus iPSCs. We then built pathway interaction networks of the pathway terms that exhibited significant enrichment for deep analysis. As shown in Supplementary Figure S2, most pathways in middle and late phases versus iPSCs were associated with MAPK signaling pathway, which was in the center of the pathway-act network. In addition, the PI3K-Akt and TGF-β signaling pathways interacted with a set of their surrounding signaling pathways in the pathway-act network of middle and late stages versus iPSCs, indicating that these pathways might be the secondary core pathways and the DEGs involved in these pathways may play important roles in iPSC differentiation.
Figure 4.
Enriched Gene Ontology (GO) categories and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of differentially expressed genes (DEGs) in different time courses during pancreatic β-like cell differentiation. (A) Enrichment analysis of biological process GO terms of DEGs during β-like cell differentiation. The horizontal axis represents the −log2 (P value) of the significant GO terms. (B) Significant pathways (excluding diseases) involved in DEGs during β-like cell differentiation. The horizontal axis represents the −log2 (P_value) of the significant pathways.
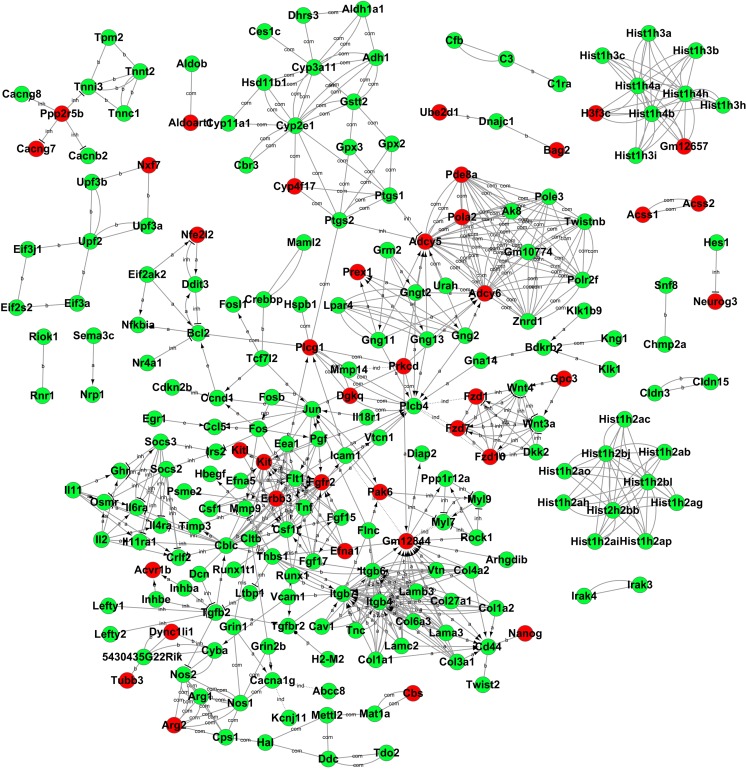
Gene-Act Network Analysis and qRT-PCR Validation
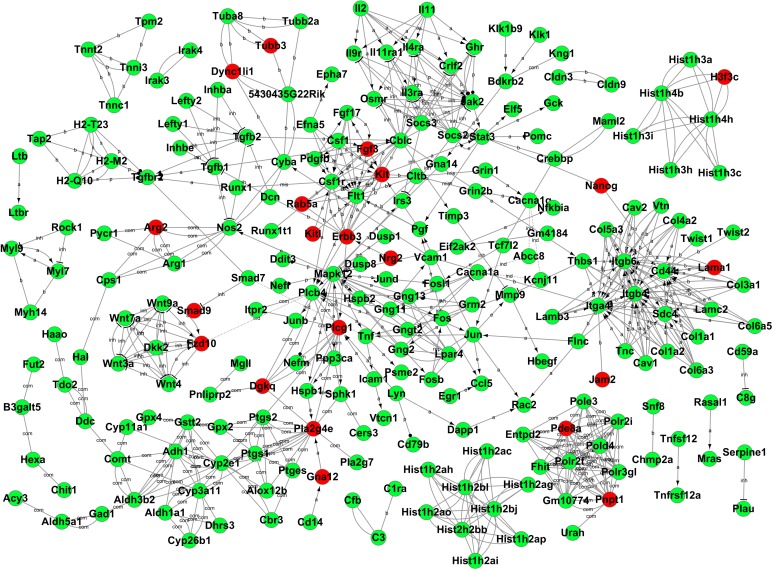
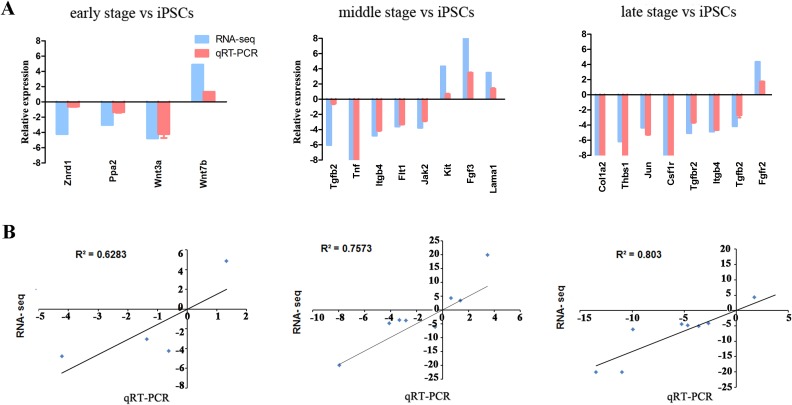
After functional analysis, we then constructed a gene-act network based on the KEGG database to explore relationships between DEGs in different time points and to identify pivotal genes during the differentiation from iPSCs to pancreatic β-like cells. The gene-act networks were shown in Figs. 5, 6, and 7. Red and green circle nodes represent upregulated and downregulated genes, respectively. Edges between 2 nodes represent interactions between genes, including activation, binding/association, repression, compound, expression, inhibition, indirect effect, phosphorylation, and state change. The more genes a single gene interacted with, the more centrally the gene in the network, which was represented as gene degree. Referring to the interaction degree of genes and the KEGG pathway they involved, we selected several key genes in each time point to validate their expression during the differentiation of iPSCs into pancreatic β-like cells by qRT-PCR (Supplementary Table S4). As shown in Fig. 8, qRT-PCR and RNA-seq data were well correlated with each other.
Figure 5.
Gene-act network of differentially expressed genes (DEGs) in induced pluripotent stem cell (iPSC)-derived β-like cells at early stage of step 3 compared to iPSCs. Cycle nodes represent genes and the line between 2 nodes represents interaction between genes. Related abbreviations: a, activation; b, binding/association; com, compound; inh, inhibition.
Figure 6.
Gene-act network of differentially expressed genes (DEGs) in induced pluripotent stem cell (iPSC)-derived β-like cells at midstage of step 3 compared to iPSCs. Cycle nodes represent genes and the line between 2 nodes represents interaction between genes. Related abbreviations: a, activation; b, binding/association; com, compound; inh, inhibition.
Figure 8.
Quantitative real-time polymerase chain reaction (qRT-PCR) validation of messenger RNA (mRNA) expression of the key genes in each time point. (A) Relative mRNA expression of genes in indicated time points compared to induced pluripotent stem cells (iPSCs) by RNA-seq (blue bar) and qRT-PCR (red bar). (B) The correlation coefficient between qRT-PCR and RNA-seq.
Figure 7.
Gene-act network of differentially expressed genes (DEGs) in induced pluripotent stem cell (iPSC)-derived β-like cells at late stage of step 3 compared to iPSCs. Cycle nodes represent genes and the line between 2 nodes represents interaction between genes. Related abbreviations: activation (a); binding/association (b); compound (com); inhibition (inh).
Discussion
Although pancreatic β-cell differentiation could be achieved with cells not only from healthy individuals or mice but also from iPSCs of diabetic patients and mouse models using different iPSC generation technologies,20,21 the information of precise transcriptional changes during the selective differentiation period is limited. Here, we used a 3-step method to generate pancreatic β-like cells from mouse iPSCs and performed RNA-seq of the iPSC-derived β-like cells at different time points to get a transcriptional landscape in iPSC-derived β-cell differentiation.
Since the first mouse iPSCs were generated from terminally differentiated murine fibroblasts using retrovirus-mediated forced expression of the transcription factors octamer-binding transcription factor 4 (Oct4), sex determining region Y (SRY)-box 2 (Sox2), c-Myc, and Krüppel-like factor 4 (Klf4) in 2006, various mammalian iPSCs were successfully generated.22–24 These iPSCs have the capacity to differentiate into any cell type, including insulin-secreting pancreatic β cells, providing a potential treatment option for diabetes mellitus. Various techniques, requiring several weeks and multiple steps, for the differentiation from iPSCs to pancreatic β-like cells have been reported. They are accompanied with a number of transcription factors, small molecules, and specific differentiation markers.7,16,25 Among them, a 3-step differentiation protocol into insulin-producing β-like cells, modified from a previously published protocol for ESCs,15 was concise and effective. This protocol is comprised of (i) EB formation from iPSCs, (ii) spontaneous differentiation of EBs into multilineage progenitors, and (iii) the induction of differentiation into insulin-producing β-like cells. In this article, we followed this protocol to generate pancreatic β-like cells from mouse iPSCs, and morphological changes were observed in iPSCs during the differentiation procedure. MPs were generated and migrated from the iPSC-derived EBs during step 2, and numerous clusters were formed during step 3 differentiation. We then performed immunofluorescence and glucose-stimulated insulin secretion tests to characterize phenotypic and functional properties of the iPSC-derived pancreatic β-like cells. The results show that a majority of the differentiated cells were positive for insulin and C-peptide, 2 markers of insulin-producing β cells. These cells were responsive to glucose stimulation, indicating a successful generation of functional β-like cells from iPSCs.
Following validation of the iPSC-derived pancreatic β-like cells, whole transcriptome RNA sequencing was used to find transcriptional alterations during the step 3 differentiation period. Compared with the undifferentiated state iPSCs, a number of DEGs have been identified and most of them were downregulated. By GO and KEGG pathway analyses, we gained a comprehensive view of the related functions and pathways of the molecular changes (Supplementary Figure S3). It was observed that most DEGs in early stage in step 3 were mainly involved in translation, which suggested that protein synthesis activity might be affected to prepare the MPs to differentiate into β cells following selective media treatment. Then on day 14 of step 3 (midstage), the DEGs were enriched in biological processes related to response (response to cytokines, drugs, etc.), while on day 21 of step 3 (late stage), biological processes were enriched in regulation including regulation of transcription, apoptotic process, cell proliferation, and so on. Pathways, such as the TGF-β, MAPK, and PI3K-Akt signaling pathways, played important roles during the later period of differentiation. As it has been reported, the TGF-β signaling pathway is a major regulator during pancreatic development, especially for β-cell differentiation.26 This signaling pathway is complex and includes almost 30 different growth and differentiation factors, such as activin and TGF-β1.27 A protocol including activin A in the culture media has successfully generated functional pancreatic β cells in vitro from human PSCs.7 The PI3K signaling pathway is a negative regulator of cellular differentiation in human and murine ESCs.9 Inhibitors of the PI3K pathway, such as LY 294002 and AKT1-II, could efficiently promote differentiation of human embryonic stem cells (hESCs) into mesendoderm and then definitive endoderm under certain conditions, acting as late stage maturation factor.28 This might suggest a regulatory action of PI3K at a later stage in β-cell differentiation, which was consistent with our pathway analysis. The MAPK signaling pathway controls many cellular events, including cell proliferation, migration, terminal differentiation, and cell death.29 It has been reported that MAPK cascades such asERK1/2, p38, and c-Jun N-terminal kinase 1/2 (JNK1/2) have a crucial role in osteogenic differentiation and are activated by both bone morphogenetic protein-2 and TNF-α/interleukin-1β.30 In our research, during the middle and late stage vs iPSCs, the MAPK signaling pathway was also enriched and palyed a central role in the pathway-act network, during the later stage of iPSC-derived β-cell differentiation, though the detailed mechanism was not clear and deserved our further investigation. In addition, the Hedgehog signaling pathway, which has an important role in early pancreas development, was enriched in the differentiation process of iPSC-derived β cells.
After GO and KEGG pathway analyses, we constructed gene-act networks of DEGs, which focused on gene relationships and described potential modules of coordinately expressed genes,31 providing a functional framework for understanding gene expression patterns during iPSC-derived β-cell differentiation. The pivotal genes identified by the gene-act network were coincident with the biological process and pathways. The key genes we selected according to their degree in the gene-act network and their related pathway were validated by qRT-PCR. Among them, Tnf, Tgfb2, Tgfbr2, and Thbs1 were involved in TGF-β signaling pathway; Itgb4, Lama1, Fgf3, Kit, Jak2, Flt1, Thbs1, Col1a2, Fgfr2, and Csf1r were involved in PI3K signaling pathway, while Tnf, Fgf3, Tgfb2, Tgfbr2, Fgfr2, and Jun were involved in MAPK signaling pathway. The role of some of these genes in β-cell differentiation has been described previously, such as Fgf3 and Fgfr2, which could regulate expression and differentiation of the pancreatic epithelium.26 c-Kit was reported to be critical in β-cell function and could increase insulin secretion.32 Tnf and c-Jun could control apoptosis in differentiating pancreatic β cells.33 On the other hand, the exact mechanisms of genes, such as Flt1 and Thbs1, in β-cell differentiation need our further exploration, which will provide new molecules to improve iPSC-derived β-cell differentiation. In addition, there are still some limitations in our study. In our research, we just performed immunofluorescence and glucose-stimulated insulin secretion tests to confirm the differentiation of iPSCs into β cells. However, to make the data more reliable, it is better to detect the rate of iPSCs’ differentiation into β cell by fluorescent-activated cell sorting. In our study, we compared only the third step (though including early, middle, late stages) to that of iPSCs. It is known that EB and MP phases are also very important for β-like cells differentiation, involving certain genes and pathways. In our future research, we might also focus on these factors to make our knowledge of the iPSC differentiation into β-like cells more comprehensive.
In conclusion, we used a 3-step protocol to generate pancreatic β-like cells from mouse GFP+-iPSCs and got an integrated global view of gene expression patterns during iPSC-derived β-cell differentiation by deep RNA sequencing. GO and KEGG pathway analyses provided a comprehensive view of the biological function and signaling pathways during these periods. A gene-act network further enabled us to identify pivotal genes in different time courses of β-cell differentiation. Overall, our results may help to elucidate the molecular mechanisms underlying iPSC-derived β-cell differentiation and to facilitate protocol improvement for generating β-like cells from iPSCs.
Footnotes
Authors’ Note: Yan Huang and Jian Wan contributed equally to this work.
Ethical Approval: This study was approved by the Administration Committee of Experimental Animals, Jiangsu Province, China.
Statement of Human and Animal Rights: All of the experimental procedures involving animals were conducted in accordance with the Institutional Animal Care guidelines of Nantong University, China and approved by the Administration Committee of Experimental Animals, Jiangsu Province, China.
Statement of Informed Consent: There are no human subjects in this article and informed consent is not applicable.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by National Natural Science Foundation of China (81471801, 81672903, 81502569), Medical Innovation Team Program of Jiangsu Province, Science and technology project of Nantong City (MS12015017, M12016018, MS22015063).
Supplemental Material: The supplements for the article are available online.
References
- 1. Atkinson MA, Eisenbarth GS, Michels AW. Type 1 diabetes. Lancet. 2014;383(9911):69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nolan CJ, Damm P, Prentki M. Type 2 diabetes across generations: from pathophysiology to prevention and management. Lancet. 2011;378(9786):169–181. [DOI] [PubMed] [Google Scholar]
- 3. Zhang Z, Yu B, Gu Y, Zhou S, Qian T, Wang Y, Ding G, Ding F, Gu X. Fibroblast-derived tenascin-C promotes Schwann cell migration through beta1-integrin dependent pathway during peripheral nerve regeneration. Glia. 2016;64(3):374–385. [DOI] [PubMed] [Google Scholar]
- 4. Schroeder IS. Potential of pluripotent stem cells for diabetes therapy. Curr Diab Rep. 2012;12(5):490–498. [DOI] [PubMed] [Google Scholar]
- 5. Sancho-Martinez I, Li M, Izpisua Belmonte JC. Disease correction the iPSC way: advances in iPSC-based therapy. Clin Pharmacol Ther. 2011;89(5):746–749. [DOI] [PubMed] [Google Scholar]
- 6. Kim C. Disease modeling and cell based therapy with iPSC: future therapeutic option with fast and safe application. Blood Res. 2014;49(1):7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pagliuca FW, Millman JR, Gurtler M, Segel M, Van Dervort A, Ryu JH, Peterson QP, Greiner D, Melton DA. Generation of functional human pancreatic beta cells in vitro. Cell. 2014;159(2):428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang D, Jiang W, Liu M, Sui X, Yin X, Chen S, Shi Y, Deng H. Highly efficient differentiation of human ES cells and iPS cells into mature pancreatic insulin-producing cells. Cell Res. 2009;19(4):429–438. [DOI] [PubMed] [Google Scholar]
- 9. Tsaniras SC, Jones PM. Generating pancreatic beta-cells from embryonic stem cells by manipulating signaling pathways. J Endocrinol. 2010;206(1):13–26. [DOI] [PubMed] [Google Scholar]
- 10. Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12(2):87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Klerk E, ‘t Hoen PAC. Alternative mRNA transcription, processing, and translation: insights from RNA sequencing. Trends Genet. 2015;31(3):128–139. [DOI] [PubMed] [Google Scholar]
- 12. Voellenkle C, Garcia-Manteiga JM, Pedrotti S, Perfetti A, De Toma I, Da Silva D, Maimone B, Greco S, Fasanaro P, Creo P, et al. Implication of long noncoding RNAs in the endothelial cell response to hypoxia revealed by RNA-sequencing. Sci Rep. 2016;6:24141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aggarwal P, Turner A, Matter A, Kattman SJ, Stoddard A, Lorier R, Swanson BJ, Arnett DK, Broeckel U. RNA expression profiling of human iPSC-derived cardiomyocytes in a cardiac hypertrophy model. Plos One. 2014;9(9):e108051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Woodard CM, Campos BA, Kuo SH, Nirenberg MJ, Nestor MW, Zimmer M, Mosharov EV, Sulzer D, Zhou H, Paull D, et al. iPSC-derived dopamine neurons reveal differences between monozygotic twins discordant for Parkinson’s disease. Cell Rep. 2014;9(4):1173–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schroeder IS, Rolletschek A, Blyszczuk P, Kania G, Wobus AM. Differentiation of mouse embryonic stem cells to insulin-producing cells. Nat Protoc. 2006;1(2):495–507. [DOI] [PubMed] [Google Scholar]
- 16. Alipio Z, Liao W, Roemer EJ, Waner M, Fink LM, Ward DC, Ma Y. Reversal of hyperglycemia in diabetic mouse models using induced-pluripotent stem (iPS)-derived pancreatic beta-like cells. Proc Natl Acad Sci USA. 2010;107(30):13426–13431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17(10):1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5(2):101–113. [DOI] [PubMed] [Google Scholar]
- 20. Quiskamp N, Bruin JE, Kieffer TJ. Differentiation of human pluripotent stem cells into beta-cells: potential and challenges. Best Pract Res Clin Endocrinol Metab. 2015;29(6):833–847. [DOI] [PubMed] [Google Scholar]
- 21. Stepniewski J, Kachamakova-Trojanowska N, Ogrocki D, Szopa M, Matlok M, Beilharz M, Dyduch G, Malecki MT, Jozkowicz A, Dulak J. Induced pluripotent stem cells as a model for diabetes investigation. Sci Rep. 2015;5:8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. [DOI] [PubMed] [Google Scholar]
- 23. Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007;318(5858):1917–1920. [DOI] [PubMed] [Google Scholar]
- 24. Liao J, Cui C, Chen S, Ren J, Chen J, Gao Y, Li H, Jia N, Cheng L, Xiao H, et al. Generation of induced pluripotent stem cell Lines from adult rat cells. Cell Stem Cell. 2009;4(1):11–15. [DOI] [PubMed] [Google Scholar]
- 25. Tateishi K, He J, Taranova O, Liang G, D’Alessio AC, Zhang Y. Generation of insulin-secreting islet-like clusters from human skin fibroblasts. Biol Chem. 2008;283(46):31601–31607. [DOI] [PubMed] [Google Scholar]
- 26. Oliver-Krasinski JM, Stoffers DA. On the origin of the beta cell. Genes Dev. 2008;22(15):1998–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim SK, Hebrok M. Intercellular signals regulating pancreas development and function. Genes Dev. 2001;15(2):111–127. [DOI] [PubMed] [Google Scholar]
- 28. McLean AB, D’Amour KA, Jones KL, Krishnamoorthy M, Kulik MJ, Reynolds DM, Sheppard AM, Liu H, Xu Y, Baetge EE. Activin A efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells. 2007;25(1):29–38. [DOI] [PubMed] [Google Scholar]
- 29. Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410(6824):37–40. [DOI] [PubMed] [Google Scholar]
- 30. Huang RL, Yuan Y, Tu J, Zou GM, Li Q. Opposing TNF-alpha/IL-1 beta- and BMP-2-activated MAPK signaling pathways converge on Runx2 to regulate BMP-2-induced osteoblastic differentiation. Cell Death Dis. 2014;5:e1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. [DOI] [PubMed] [Google Scholar]
- 32. Feng ZC, Li J, Turco BA, Riopel M, Yee SP, Wang R. Critical role of c-Kit in beta cell function: increased insulin secretion and protection against diabetes in a mouse model. Diabetologia. 2012;55(8):2214–2225. [DOI] [PubMed] [Google Scholar]
- 33. Ammendrup A, Maillard A, Nielsen K, Aabenhus Andersen N, Serup P, Dragsbaek Madsen O, Mandrup-Poulsen T, Bonny C. The c-Jun amino-terminal kinase pathway is preferentially activated by interleukin-1 and controls apoptosis in differentiating pancreatic beta-cells. Diabetes. 2000;49(9):1468–1476. [DOI] [PubMed] [Google Scholar]