Abstract
Purpose of review
When providing accurate clinical diagnosis and genetic counselling in craniosynostosis, the challenge is heightened by knowledge that etiology in any individual case may be entirely genetic, entirely environmental, or anything in between. This review will scope out how recent genetic discoveries from next-generation sequencing have impacted on the clinical genetic evaluation of craniosynostosis.
Recent findings
Survey of a 13-year birth cohort of patients treated at a single craniofacial unit demonstrates that a genetic cause of craniosynostosis can be identified in one quarter of cases. The substantial contributions of mutations in two genes, TCF12 and ERF, is confirmed. Important recent discoveries are mutations of CDC45 and SMO in specific craniosynostosis syndromes, and of SMAD6 in non-syndromic midline synostosis. The added value of exome or whole genome sequencing in the diagnosis of difficult cases is highlighted.
Summary
Strategies to optimise clinical genetic diagnostic pathways by combining both targeted and next-generation sequencing are discussed. As well as improved genetic counselling, recent discoveries spotlight the important roles of signalling through the bone morphogenetic protein and hedgehog pathways in cranial suture biogenesis, as well as a key requirement for adequate cell division in suture maintenance.
Keywords: Exome sequencing, CDC45, SMAD6, SMO
Introduction
Craniosynostosis, the premature fusion of the cranial sutures, has a prevalence of between 1 in 1,400 and 1 in 2,100 children [1, 2*], putting it at the borderline of what constitutes a rare disease. Moreover the frequency of non-syndromic midline synostosis appears to be increasing, for reasons that are not understood [2*]. Clinical management poses multiple challenges, which are best addressed in a specialist multidisciplinary unit.
Etiological assessment of craniosynostosis should start by recognising the substantial heterogeneity in underlying causes. Awareness of both the striking pathological differences between fusion of different cranial sutures, and the complex interplay of potentially causative factors - intrauterine environment, polygenic background, growth and development of the brain, as well as specific monogenic and chromosomal disorders – is essential [3]. Reviews of the clinical approach to diagnosis and associated phenotypic features are covered in several articles [4–6]; here we focus on recent progress in understanding the genetic underpinnings of craniosynostosis.
A current benchmark for diagnosis
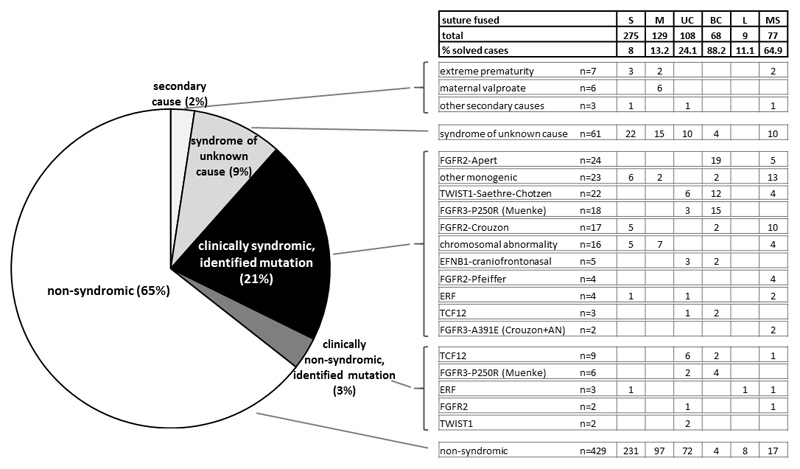
An initial clinical evaluation of a child with craniosynostosis will categorise them according to the cranial vault suture, or sutures, shown to be fused (preferably using 3-dimensional computed tomographic reconstruction); whether any risk factors can be identified in the obstetric or perinatal history; and features suggesting an underlying syndrome, based on positive family history, associated malformations or dysmorphic features, and evidence of significant developmental or cognitive delay. To provide context, Figure 1 shows data (updated from a previously published series [7]), collected from a single specialist unit at Oxford, UK for a 13-year birth cohort (n = 666) with minimum 5-year follow-up. The frequency of different types of craniosynostosis varies widely, with sagittal synostosis being most common (41%) and lambdoid synostosis rarest (1%). Equally striking, the proportion of cases in which a cause is delineated varies widely, from 88% for bicoronal synostosis to only 8% for sagittal synostosis. The high genetic load in coronal synostosis likely reflects the distinct biogenesis of the coronal sutures during embryonic development [8–10].
Figure 1. Classification and causes of craniosynostosis in a prospectively ascertained 13-year cohort.
Data are based on a cohort of patients with craniosynostosis (n = 666) born between 1998 and 2010 inclusive, presenting to a single specialized unit (Oxford, UK) and requiring at least one major craniofacial procedure by the end of 2015. The pie chart on the left shows a broad classification according to presence or absence of syndromic features and identification of a likely secondary or genetic cause. The grid on the right provides a more detailed breakdown according to the pattern of suture involvement and precise diagnosis. Abbreviations for different suture fusions as follows: S, sagittal; M, metopic; UC, unilateral coronal; BC, bilateral coronal; L, unilateral or bilateral lambdoid; MS, multiple suture fusion excluding bilateral coronal or lambdoid. AN, acanthosis nigricans. Data updated from previously published 5-year cohort [7].
Although an adverse intrauterine environment may contribute substantially to the origin of many cases of craniosynostosis [11–13], this is difficult to prove in the individual patient [14]. Overall, only 2.4% of cases were attributed to a likely secondary cause in the Oxford series. Of the remainder, 31% were classified clinically as syndromic and 69% as non-syndromic (Fig. 1). Not surprisingly, a positive genetic diagnosis was obtained in a much higher proportion of the syndromic cases (69%) than those initially classified as non-syndromic (5%). However, as discussed below, the recognition that a growing minority of non-syndromic craniosynostosis may have an underlying monogenic basis has important implications for the approach to genetic testing and genetic counselling.
Syndromic craniosynostosis
As shown in Fig. 1, there are six genes frequently (each >0.5% overall) mutated in craniosynostosis: FGFR2, FGFR3, TWIST1, EFNB1, TCF12 and ERF. Mutations in the first four of these mostly cause recognisable syndromes (FGFR2, predominantly Apert, Crouzon and Pfeiffer; FGFR3, Muenke and Crouzon with acanthosis nigricans; TWIST1, Saethre-Chotzen; and EFNB1, craniofrontonasal syndrome), for which the molecular basis was determined a decade or more ago, so their clinical features and genotype-phenotype correlations are largely well documented [3–6]. Recent clinical updates have been published on prevalence of tracheal cartilaginous sleeve [15] and progressive postnatal pansynostosis [16] in syndromic craniosynostosis, both of which highlight the particularly high burden of complications arising from FGFR2 mutations. Additional FGFR2 mutation-focused phenotype studies have been published on foramen magnum size [17], ophthalmic complications [18] and intestinal malrotation [19]. Complications were found to be differentially enriched in different syndromes: tracheal sleeve, proptosis and exposure keratitis were particularly associated with Pfeiffer syndrome, whereas insidious postnatal pansynostosis was most common in Crouzon syndrome. Recently, useful series have been published on the two most severe - fortunately rare - FGFR2-associated craniosynostosis syndromes, Beare-Stevenson cutis gyrata syndrome (BSS) and bent-bone dysplasia (BBD). BSS is caused by two specific heterozygous missense mutations in the juxtatransmembrane region of FGFR2, encoding p.Ser372Tyr and p.Tyr375Cys. Wenger et al. [20] reviewed 21 previously published cases of BSS and added two new cases, highlighting the substantial mortality in the first year of life (70%) and developmental delay in survivors. BBD was only recognised as a distinct clinical entity in 2012 [21] and Krakow et al. provide a useful overview of the clinical features of 11 cases, including 7 previously unpublished [22*]. Although the missense mutations responsible for BBD, encoding p.Tyr381Asp and p.Met391Arg, lie close to those for BBS, the BBD substitutions localise to the transmembrane region and are associated with a distinct pathophysiology involving enhanced nucleolar rRNA transcripton [23].
Muenke syndrome, defined by a specific 749C>G (p.P250R) mutation in FGFR3 that represents the single most common nucleotide substitution in craniosynostosis, can only be confidently identified by genetic testing. A clinical survey of 106 subjects provides an overview of the natural history of this disorder [24]; 15% of individuals did not have craniosynostosis, and association with isolated hydrocephalus has been separately described [25]. The mechanisms underlying the clinical variability of Muenke syndrome are not understood, indicating that a wide range of possible outcomes should be mentioned when providing preconceptual and prenatal advice. Not surprisingly there is a diversity of attitudes towards prenatal testing, as recently surveyed amongst five adult couples, in each of which one individual was affected with Muenke syndrome [26].
Two other genes mutated in >1% of craniosynostosis, TCF12 and ERF, were first reported in 2013 [27, 28], so description of the associated natural history is less complete. TCF12 is discussed in the section on non-syndromic craniosynostosis. ERF encodes a negative regulator of ERK1/2, the key signal transducer at the base of the pathway from growth factor receptors through RAS-MAP kinase. Clinical presentation of ERF mutations varies from a mild Crouzon-like picture to non-syndromic craniosynostosis. Further published information to augment the original clinical descriptions [28] is still scanty; surprisingly, in view of the relatively high frequency in the Oxford cohort, Paumard-Hernandez et al. [29] did not identify any ERF mutations in a series of 69 undiagnosed craniosynostosis cases. Chaudhry et al. [30] described two subjects with ERF mutations, who had features overlapping those originally reported. Surprisingly - given that the pathogenic mechanism of most ERF mutations appears to be haploinsufficiency - a specific missense substitution, p.Tyr89Cys, located within the DNA-binding ETS domain of ERF, was identified in 4 unrelated patients or families with Chitayat syndrome, which is characterised by a bilateral accessory phalanx resulting in shortening of the index finger, hallux valgus and respiratory compromise [31]. Although facial features were similar to other subjects with ERF mutations, craniosynostosis was not documented. The distinct clinical features might result from altered DNA-binding properties associated with the specific missense substitution, but this has not so far been investigated experimentally.
Recently identified disease genes
Next-generation sequencing has substantially accelerated the discovery of new gene/disease associations in syndromic craniosynostosis. Two examples in 2016 were a mosaic heterozygous mutation of SMO in Curry-Jones syndrome (CJS) [32*] and biallelic mutations of CDC45 in Meier-Gorlin syndrome (MGS) associated with craniosynostosis [33*]. CJS is characteristed by patchy skin lesions, polysyndactyly, diverse cerebral malformations, coronal craniosynostosis, iris colobomas, microphthalmia and intestinal malrotation. SMO encodes smoothened, a G-protein-coupled receptor that transduces signalling by the hedgehog family of proteins; the recurrent, mosaic activating c.1234C>T substitution encoding p.Leu412Phe was identified in eight of ten CJS cases analyzed [32*]. CJS has unusual abdominal symptomatology associated with smooth muscle hamartomas on the mesentery and surface of the bowel; motility disorders and upper gastrointestinal bleeding are frequent [34]. The identical SMO mutation has also been identified in several tumors, particularly involving the skin or brain; these are potentially treatable using hedgehog pathway inhibitors [35]. CDC45 encodes a key component of the machinery of DNA replication, present in all eukaryotes, so the identification of mutations in craniosynostosis may appear surprising. Clinical presentation varied from unicoronal or bicoronal synostosis and mild short stature, to a severe phenotype of MGS (defined by the triad of short stature, microtia and a/hypoplastic patellae), combined with multi-suture synostosis. Mutations were found to reduce protein levels, either by affecting splicing or through protein instability (missense mutations); variation in the amount of residual protein activity probably explains the variability of the phenotype observed [33*].
Non-syndromic craniosynostosis
The contribution of genetic diagnoses has been substantially lower in non-syndromic craniosynostosis, <1% in sagittal and metopic synostosis (Fig. 1 and [36]). However diagnostic success rates are higher in unicoronal (13%), multisuture (15%) and bicoronal synostosis (60%) cases (Fig. 1). The single largest contributor to these diagnoses is TCF12, which encodes a partner protein of TWIST1 particularly critical for coronal suture development [27]. Two follow-up studies have confirmed the importance of TCF12 mutations in coronal craniosynostosis, both in the context of familial mutations [37], and in a more general screen of craniosynostosis [29]. Given the haploinsufficiency mechanism of TCF12 mutations, heterozygous deletions are also expected to be pathogenic and this has been confirmed in two reports [38, 39]. At present, diagnostic labs rely on DNA sequencing to test TCF12, pointing to the need for a dedicated diagnostic method such as multiplex ligation-dependent probe amplification (MLPA) to detect TCF12 deletions. Further analysis of the phenotype associated with TCF12 mutations is awaited: although the clinical outlook in most affected individuals is good (and non-penetrance occurs in 50% or more of mutation-positive individuals [27]), a minority may present with learning disability or autistic spectrum disorder [40]. The reasons for this clinical variability require further investigation.
A recent discovery that is likely to change the previously negative diagnostic picture for the midline synostoses was reported by Timberlake et al. [41**]. In an exome sequencing study of 132 parent-offspring trios and 59 additional probands with either sagittal or metopic synostosis, these authors reported a significant enrichment of mutations in SMAD6, which encodes a negative regulator of signaling through the bone morphogenetic protein (BMP) pathway. Thirteen percent of individuals with metopic, and 3% with sagittal synostosis, were heterozygous for loss-of-function or rare missense variants in SMAD6, and the positive diagnostic rate was higher (24%) in familial cases. Although de novo mutations occurred in 3 of the 13 families identified, in the others, the variant was inherited from a parent who was usually unaffected. Confirming that non-penetrance for SMAD6 mutations is frequent, the Exome Aggregation Consortium [42] did not identify any deficiency of SMAD6 loss-of-function mutations, yielding a pLI (probability of being intolerant to loss-of-function mutations, also termed constraint) value of zero. Timberlake et al. [41**] proposed an ingenious explanation for this paradox. Observing that in the only reported genome-wide association study of non-syndromic sagittal synostosis, the strongest signal (odds ratio = 4.6 for the risk allele) was with a single nucleotide polymorphism (SNP) rs1884302 located 345 kb away from the BMP2 gene [43], these authors genotyped the SMAD6 mutation-positive individuals for the rs1884302 SNP. They found that 14 of 17 affected individuals harbored at least one risk allele (C), whereas all 13 unaffected individuals were homozygous for the non-risk (T) allele, a highly significant difference that appears to support a digenic disease mechanism involving two different components of BMP signalling. This finding could have major implications for molecular diagnostics, as no genetic testing is currently routinely indicated in either non-syndromic sagittal or metopic synostosis, the two most common clinical presentations of craniosynostosis (Fig. 1). However, the paradoxically low pLI score urges caution in interpretating these data; other groups are currently attempting to replicate the findings to reach consensus regarding future diagnostic use. A further caveat is emerging evidence that a similar spectrum of SMAD6 mutations may predispose to cardiac abnormalities, particularly bicuspid aortic valve with ascending aortic dilatation [44, 45]. This raises the question whether echocardiography should be undertaken on all mutation-positive individuals; clinicians need to have a clear, evidence-based care pathway before offering genetic testing.
Molecular diagnostic approach to craniosynostosis
Although mutations in just six genes constitute three-quarters of all genetically diagnosed cases, the etiology of the remaining quarter is very diverse. Fifty-seven genes were classified as validated “craniosynostosis genes” by Twigg and Wilkie [3], based on identification of mutations in two or more independent cases, and some additional potentially causative genes were highlighted by Lattanzi et al. [6]. The long tail of rare genetic diagnoses is apparent in Fig. 1, which shows that in the category of syndromic craniosynostosis with an identified mutation, “other monogenic” (comprising mutations in 20 different genes) is the second leading causal category after FGFR2 mutations in Apert syndrome. These rare diagnoses include potentially treatable conditions for which early recognition is particularly important, such as hypophosphatasia (ALPL) [46], Albright osteodystrophy (GNAS1) [47, 48] and rickets (XLH) [49]. A diverse variety of chromosomal abnormalities also occurs in association with craniosynostosis, probably often through non-specific mechanisms involving suboptimal brain growth. The question arises how to design an optimal diagnostic algorithm that accommodates both the simple and complex aspects of the overall presentation.
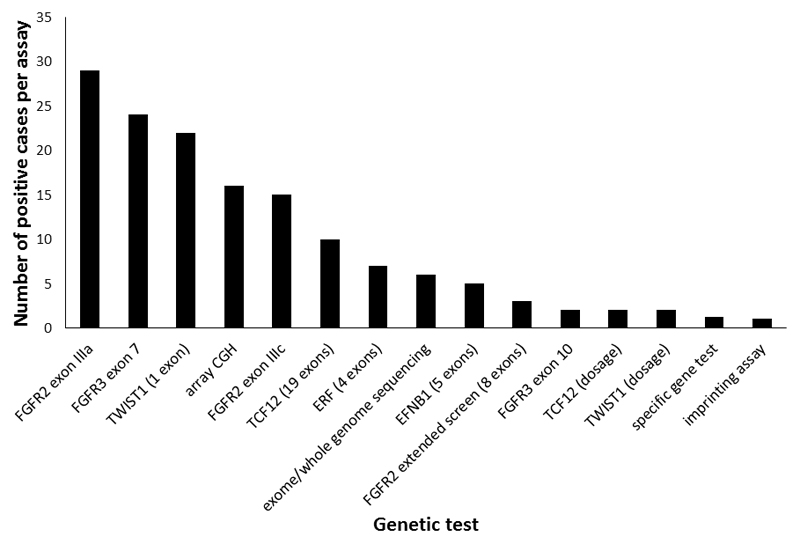
As a guide, Fig. 2 provides a hierarchical summary of the effort required to make each successful diagnosis. Sixty-six percent of all diagnoses were made using just five diagnostic tests – targeted DNA sequencing of FGFR2 exons IIIa and IIIc, FGFR3 exon 7 and TWIST1 exon 1, plus array CGH, so in many clinical situations it will make sense to start with a combination of these investigations. Moving further down the hierarchy, some tests are more complex (for example, TCF12 contains 19 coding exons) and the economic argument for using next-generation sequencing is increasingly strong, although orthogonal technology is currently still required to detect specific copy number variations.
Figure 2. Genetic testing in craniosynostosis.
The cohort described in Fig. 1 (n = 666) was analyzed in terms of the number of successes in achieving a positive diagnosis for different genetic tests. The tests are arranged hierarchically with those yielding the highest number of diagnoses at the left.
Recently the first use of exome and whole genome sequencing for difficult-to-diagnose craniosynostosis was presented [50*]. Of 40 probands studied (previously negative for a wide range of targeted testing), a molecular genetic diagnosis was resolved in 15 (37.5%) of cases. IL11RA [51–53] was the only recurrently mutated gene, further underlining the very substantial genetic heterogeneity in rare causes of craniosynostosis. Mutations were classified according to four categories: commonly mutated craniosynostosis genes with atypical presentation; other core craniosynostosis genes; more rarely associated genes; and known disease genes not known to be associated with craniosynostosis. The genes in which mutations occurred were distributed across all four categories, making an argument for the value of a genome-wide search strategy rather than gene panel. Another important finding from this study [50*] was that in 5 of the 15 positive cases, the novel molecular diagnosis had immediate, actionable consequences for genetic or clinical management, either in terms of reproductive diagnostic options or for the medical management of potential complications revealed by the diagnosis.
Genetic counselling in craniosynsotosis
Aside from the uncertainties that face geneticists when counselling about the reproductive implications for many disorders (such as variable expressivity and gonadal mosaicism), a particular issue in craniosynostosis is that 45% of identified genetic causes pinpoint within the FGFR2 and FGFR3 genes [Fig. 1]. These genes show markedly elevated apparent mutation rates owing to selective advantage of these mutations when they arise spontaneously in the adult male testis (a process termed selfish spermatogonial selection [54]). Direct methods to identify the source of the originating mutations within individual seminiferous tubules of testes (removed because of incidental pathology) were recently described [55, 56], providing further support for the proposed pathophysiological mechanism. The clinical significance of this knowledge is that sibling recurrence risk for de novo FGFR2 and FGFR3 mutations is likely to be exceptionally low, making it justified to reassure parents and mitigate demand for prenatal diagnosis [57].
Conclusion
Several initiatives are under way to undertake wide-scale exome/genome sequencing in craniosynostosis, which are expected to yield further novel gene mutations; however these are likely to be either rare, or associated with substantial non-penetrance (as is the case, for example, with TCF12 and SMAD6). Further genome-wide association studies are also in progress, which might, like the BMP2 SNP, also have possible diagnostic implications.
Partly fuelled by these human genetic studies, fundamental research into the developmental biology of the cranial sutures is continuing to make progress. Maintenance of sutural patency requires a delicate balance between stem cell maintenance, proliferation and osteogenic differentiation [3]; a key goal is to identify the stem cells required to maintain sutural integrity, and delineate their niche (this is likely to involve integration with molecular stress/strain transduction mechanisms, about which very little is currently known). Importantly, markers are now becoming available to mark murine sutural cells at different stages of differentiation including Gli1 [58], Prrx1 [59] and Axin2 [60*]. A detailed understanding of the complex processes underlying normal sutural homeostasis may eventually lead to medical preventions or therapies for craniosynostosis [61]. For the time being, however, surgery continues to be the mainstay of treatment, although lack of consensus about timing and surgical approaches remains a persisting issue in this field [62].
Key points.
The causes of craniosynostosis are very heterogeneous, with monogenic, chromosomal, polygenic and environmental/teratogenic factors all playing an important role
A specific genetic diagnosis can currently be identified in one quarter of patients with craniosynostosis
Five percent of patients initially classified as having a non-syndromic diagnosis are subsequently found to harbour a pathogenic mutation; the TCF12 gene is most frequently implicated (coronal synostosis)
The recent discovery of SMAD6 mutations in midline synostosis may have further implications for diagnostic assessment, but both the proposed digenic inheritance mechanism, and potential implications for cardiovascular risk, require further evaluation before clinical implementation
Craniosynostosis occurs at low frequency in a large number of rare monogenic disorders, many of which have require specific protocols for therapy or screening for additional complications. Accurate and prompt diagnosis requires a combination of careful clinical evaluation and correctly targeted diagnostic testing, proceeding to exome/whole genome sequencing if necessary
Acknowledgements
Financial support and sponsorship
Work in Prof Wilkie’s laboratory is supported by Wellcome (Investigator Award), NIHR Oxford Biomedical Research Centre, National Institutes of Health and Action Medical Research.
Footnotes
Conflicts of interest
None
References
- [1].Lajeunie E, Le Merrer M, Bonaïti-Pellie C, et al. Genetic study of nonsyndromic coronal craniosynostosis. Am J Med Genet. 1995;55:500–504. doi: 10.1002/ajmg.1320550422. [DOI] [PubMed] [Google Scholar]
- [2].Cornelissen M, den Ottelander B, Rizopoulos D, et al. Increase of prevalence of craniosynostosis. J Craniomaxillofac Surg. 2016;44:1273–1279. doi: 10.1016/j.jcms.2016.07.007. [This paper reported a 12.5% increase in the overall prevalence of craniosynostosis in the Netherlands between 1997 and 2013. The increase was most marked in metopic synostosis (+20.5%), as found in other studies. The proportion of cases with identified mutations (7%) was considerably lower than presented in this review.] [DOI] [PubMed] [Google Scholar]
- [3].Twigg SRF, Wilkie AOM. A genetic-pathophysiological framework for craniosynostosis. Am J Hum Genet. 2015;97:359–377. doi: 10.1016/j.ajhg.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Johnson D, Wilkie AOM. Craniosynostosis. Eur J Hum Genet. 2011;19:369–376. doi: 10.1038/ejhg.2010.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Forrest CR, Hopper RA. Craniofacial syndromes and surgery. Plast Reconstr Surg. 2013;131:86e–109e. doi: 10.1097/PRS.0b013e318272c12b. [DOI] [PubMed] [Google Scholar]
- [6].Lattanzi W, Barba M, Di Pietro L, Boyadjiev SA. Genetic advances in craniosynostosis. Am J Med Genet. 2017;173A:1406–1429. doi: 10.1002/ajmg.a.38159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wilkie AOM, Byren JC, Hurst JA, et al. Prevalence and complications of single-gene and chromosomal disorders in craniosynostosis. Pediatrics. 2010;126:e391–400. doi: 10.1542/peds.2009-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jiang X, Iseki S, Maxson RE, et al. Tissue origins and interactions in the mammalian skull vault. Dev Biol. 2002;241:106–116. doi: 10.1006/dbio.2001.0487. [DOI] [PubMed] [Google Scholar]
- [9].Merrill AE, Bochukova EG, Brugger SM, et al. Cell mixing at a neural crest-mesoderm boundary and deficient ephrin-Eph signaling in the pathogenesis of craniosynostosis. Hum Mol Genet. 2006;15:1319–1328. doi: 10.1093/hmg/ddl052. [DOI] [PubMed] [Google Scholar]
- [10].Deckelbaum RA, Holmes G, Zhao Z, et al. Regulation of cranial morphogenesis and cell fate at the neural crest-mesoderm boundary by engrailed 1. Development. 2012;139:1346–1358. doi: 10.1242/dev.076729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sanchez-Lara PA, Carmichael SL, Graham JM., Jr Fetal constraint as a potential risk factor for craniosynostosis. Am J Med Genet. 2010;152A:394–400. doi: 10.1002/ajmg.a.33246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kini U, Hurst JA, Byren JC, et al. Etiological heterogeneity and clinical characteristics of metopic synostosis: Evidence from a tertiary craniofacial unit. Am J Med Genet. 2010;152A:1383–1389. doi: 10.1002/ajmg.a.33435. [DOI] [PubMed] [Google Scholar]
- [13].Lakin GE, Sinkin JC, Chen R, et al. Genetic and epigenetic influences of twins on the pathogenesis of craniosynostosis: a meta-analysis. Plast Reconstr Surg. 2012;129:945–954. doi: 10.1097/PRS.0b013e31824422a8. [DOI] [PubMed] [Google Scholar]
- [14].Johnson D, Wall SA, Mann S, Wilkie AOM. A novel mutation, Ala315Ser, in FGFR2: a gene-environment interaction leading to craniosynostosis? Eur J Hum Genet. 2000;8:571–577. doi: 10.1038/sj.ejhg.5200499. [DOI] [PubMed] [Google Scholar]
- [15].Wenger TL, Dahl J, Bhoj EJ, et al. Tracheal cartilaginous sleeves in children with syndromic craniosynostosis. Genet Med. 2017;19:62–68. doi: 10.1038/gim.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wood BC, Oh AK, Keating RF, et al. Progressive postnatal pansynostosis: an insidious and pernicious form of craniosynostosis. J Neurosurg Pediatr. 2015;16:309–316. doi: 10.3171/2015.1.PEDS14464. [DOI] [PubMed] [Google Scholar]
- [17].Coll G, Arnaud E, Collet C, et al. Skull base morphology in fibroblast growth factor receptor type 2-related faciocraniosynostosis: a descriptive analysis. Neurosurgery. 2015;76:571–583. doi: 10.1227/NEU.0000000000000676. discussion 583. [DOI] [PubMed] [Google Scholar]
- [18].Sharma N, Greenwell T, Hammerton M, et al. The ophthalmic sequelae of Pfeiffer syndrome and the long-term visual outcomes after craniofacial surgery. J AAPOS. 2016;20:315–319. doi: 10.1016/j.jaapos.2016.04.007. [DOI] [PubMed] [Google Scholar]
- [19].Hibberd CE, Bowdin S, Arudchelvan Y, et al. FGFR-associated craniosynostosis syndromes and gastrointestinal defects. Am J Med Genet. 2016;170A:3215–3221. doi: 10.1002/ajmg.a.37862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wenger TL, Bhoj EJ, Wetmore RF, et al. Beare-Stevenson syndrome: two new patients, including a novel finding of tracheal cartilaginous sleeve. Am J Med Genet. 2015;167A:852–857. doi: 10.1002/ajmg.a.36985. [DOI] [PubMed] [Google Scholar]
- [21].Merrill AE, Sarukhanov A, Krejci P, et al. Bent bone dysplasia-FGFR2 type, a distinct skeletal disorder, has deficient canonical FGF signaling. Am J Hum Genet. 2012;90:550–557. doi: 10.1016/j.ajhg.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Krakow D, Cohn DH, Wilcox WR, et al. Clinical and radiographic delineation of bent bone dysplasia-FGFR2 type or bent bone dysplasia with distinctive clavicles and angel-shaped phalanges. Am J Med Genet. 2016;170:2652–2661. doi: 10.1002/ajmg.a.37772. [This paper expands knowledge of bent bone dysplasia. Pathognomonic skeletal features include poor ossification of the calvarium, craniosynostosis, small clavicles, bent appendicular bones, and angel shaped metacarpal/metatarsal phalangeal bones. All three individuals with p.Met391Arg were stillborn, whereas a minority of those with p.Tyr381Arg survived the neonatal period.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Neben CL, Idoni B, Salva JE, et al. Bent bone dysplasia syndrome reveals nucleolar activity for FGFR2 in ribosomal DNA transcription. Hum Mol Genet. 2014;23:5659–5671. doi: 10.1093/hmg/ddu282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kruszka P, Addissie YA, Yarnell CM, et al. Muenke syndrome: An international multicenter natural history study. Am J Med Genet. 2016;170A:918–929. doi: 10.1002/ajmg.a.37528. [DOI] [PubMed] [Google Scholar]
- [25].Gonzalez-Del Angel A, Estandia-Ortega B, Alcantara-Ortigoza MA, et al. Expansion of the variable expression of Muenke syndrome: Hydrocephalus without craniosynostosis. Am J Med Genet. 2016;170A:3189–3196. doi: 10.1002/ajmg.a.37951. [DOI] [PubMed] [Google Scholar]
- [26].Phipps J, Skirton H. A qualitative study to explore the views and attitudes towards prenatal testing in adults who have Muenke syndrome and their partners. J Genet Couns. 2017 doi: 10.1007/s10897-017-0103-x. [DOI] [PubMed] [Google Scholar]
- [27].Sharma VP, Fenwick AL, Brockop MS, et al. Mutations in TCF12, encoding a basic helix-loop-helix partner of TWIST1, are a frequent cause of coronal craniosynostosis. Nat Genet. 2013;45:304–307. doi: 10.1038/ng.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Twigg SRF, Vorgia E, McGowan SJ, et al. Reduced dosage of ERF causes complex craniosynostosis in humans and mice and links ERK1/2 signaling to regulation of osteogenesis. Nat Genet. 2013;45:308–313. doi: 10.1038/ng.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Paumard-Hernandez B, Berges-Soria J, Barroso E, et al. Expanding the mutation spectrum in 182 Spanish probands with craniosynostosis: identification and characterization of novel TCF12 variants. Eur J Hum Genet. 2015;23:907–914. doi: 10.1038/ejhg.2014.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chaudhry A, Sabatini P, Han L, et al. Heterozygous mutations in ERF cause syndromic craniosynostosis with multiple suture involvement. Am J Med Genet. 2015;167A:2544–2547. doi: 10.1002/ajmg.a.37218. [DOI] [PubMed] [Google Scholar]
- [31].Balasubramanian M, Lord H, Levesque S, et al. Chitayat syndrome: hyperphalangism, characteristic facies, hallux valgus and bronchomalacia results from a recurrent c.266A<G p.(Tyr89Cys) variant in the ERF gene. J Med Genet. 2017;54:157–165. doi: 10.1136/jmedgenet-2016-104143. [DOI] [PubMed] [Google Scholar]
- [32].Twigg SRF, Hufnagel RB, Miller KA, et al. A recurrent mosaic mutation in SMO, encoding the hedgehog signal transducer smoothened, is the major cause of Curry-Jones syndrome. Am J Hum Genet. 2016;98:1256–1265. doi: 10.1016/j.ajhg.2016.04.007. [This paper and the next one [33]* identify two new monogenic causes of craniosynostosis. Illustrating the diverse pathophysiological processes underlying this heterogeneous disorder, the new genes classify within separate pathways and processes, respectively involved in hedgehog signalling (SMO) and DNA replication (CDC45).] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fenwick AL, Kliszczak M, Cooper F, et al. Mutations in CDC45, encoding an essential component of the pre-initiation complex, cause Meier-Gorlin syndrome and craniosynostosis. Am J Hum Genet. 2016;99:125–138. doi: 10.1016/j.ajhg.2016.05.019. [This paper and the previous one [32]* identify two new monogenic causes of craniosynostosis. Illustrating the diverse pathophysiological processes underlying this heterogeneous disorder, the new genes classify within separate pathways and processes, respectively involved in hedgehog signalling (SMO) and DNA replication (CDC45).] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wigby K, Twigg SRF, Broderick R, et al. Gastrointestinal disorders in Curry-Jones syndrome: Clinical and molecular insights from an affected newborn. Am J Med Genet. 2017;173A:1586–1592. doi: 10.1002/ajmg.a.38232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Khamaysi Z, Bochner R, Indelman M, et al. Segmental basal cell nevus syndrome caused by an activating mutation in smoothened. Br J Dermatol. 2016;175:178–181. doi: 10.1111/bjd.14425. [DOI] [PubMed] [Google Scholar]
- [36].Ye X, Guilmatre A, Reva B, et al. Mutation screening of candidate genes in patients with nonsyndromic sagittal craniosynostosis. Plast Reconstr Surg. 2016;137:952–961. doi: 10.1097/01.prs.0000479978.75545.ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].di Rocco F, Baujat G, Arnaud E, et al. Clinical spectrum and outcomes in families with coronal synostosis and TCF12 mutations. Eur J Hum Genet. 2014;22:1413–1416. doi: 10.1038/ejhg.2014.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Piard J, Roze V, Czorny A, et al. TCF12 microdeletion in a 72-year-old woman with intellectual disability. Am J Med Genet. 2015;167A:1897–1901. doi: 10.1002/ajmg.a.37083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Goos JA, Fenwick AL, Swagemakers SM, et al. Identification of intragenic exon deletions and duplication of TCF12 by whole genome or targeted sequencing as a cause of TCF12-related craniosynostosis. Hum Mutat. 2016;37:732–736. doi: 10.1002/humu.23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Tammimies K, Marshall CR, Walker S, et al. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. JAMA. 2015;314:895–903. doi: 10.1001/jama.2015.10078. [DOI] [PubMed] [Google Scholar]
- [41].Timberlake AT, Choi J, Zaidi S, et al. Two locus inheritance of non-syndromic midline craniosynostosis via rare SMAD6 and common BMP2 alleles. eLife. 2016;5:e20125. doi: 10.7554/eLife.20125. [The first paper to use exome sequencing to seek monogenic causes of nonsyndromic midline synostosis. Given the previous epidemiological and genetic evidence suggesting a multifactorial basis (polygenes and environment), the identification of three de novo mutations in SMAD6 was unexpected. Whether there is also an increased burden of de novo mutations in additional genes remains unclear.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Justice CM, Yagnik G, Kim Y, et al. A genome-wide association study identifies susceptibility loci for nonsyndromic sagittal craniosynostosis near BMP2 and within BBS9. Nat Genet. 2012;44:1360–1364. doi: 10.1038/ng.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tan HL, Glen E, Topf A, et al. Nonsynonymous variants in the SMAD6 gene predispose to congenital cardiovascular malformation. Hum Mutat. 2012;33:720–727. doi: 10.1002/humu.22030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gillis E, Kumar A, Luyckx I, et al. Candidate gene resequencing in a large bicuspid aortic valve-associated thoracic aortic aneurysm cohort: SMAD6 as an important contributor. Front Physiol. 2017 doi: 10.3389/fphys.2017.00400. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Linglart A, Biosse-Duplan M. Hypophosphatasia. Curr Osteoporos Rep. 2016;14:95–105. doi: 10.1007/s11914-016-0309-0. [DOI] [PubMed] [Google Scholar]
- [47].Mamoei S, Cortnum S. Raised intracranial pressure as a result of pansynostosis in a child with Albright's hereditary osteodystrophy. Childs Nerv Syst. 2017;33:865–868. doi: 10.1007/s00381-016-3330-9. [DOI] [PubMed] [Google Scholar]
- [48].Adetayo OA, Fearon JA. Craniosynostosis and guanine nucleotide-binding protein alpha stimulating mutation: risk of bleeding diathesis and circulatory collapse in patients undergoing cranial vault reconstruction. J Craniofac Surg. 2017 doi: 10.1097/SCS.0000000000003592. [DOI] [PubMed] [Google Scholar]
- [49].Jaszczuk P, Rogers GF, Guzman R, Proctor MR. X-linked hypophosphatemic rickets and sagittal craniosynostosis: three patients requiring operative cranial expansion: case series and literature review. Childs Nerv Syst. 2016;32:887–891. doi: 10.1007/s00381-015-2934-9. [DOI] [PubMed] [Google Scholar]
- [50].Miller KA, Twigg SRF, McGowan SJ, et al. Diagnostic value of exome and whole genome sequencing in craniosynostosis. J Med Genet. 2017;54:260–268. doi: 10.1136/jmedgenet-2016-104215. [The first use of genome-wide sequencing technology applied to craniosynostosis patients refractory to routine testing. Previously unanticipated diagnoses with immediate implications for clinical management included craniofrontonasal, Marfan, Noonan, and STAT3-related hyper IgE syndromes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Nieminen P, Morgan NV, Fenwick AL, et al. Inactivation of IL11 signaling causes craniosynostosis, delayed tooth eruption, and supernumerary teeth. Am J Hum Genet. 2011;89:67–81. doi: 10.1016/j.ajhg.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Keupp K, Li Y, Vargel I, et al. Mutations in the interleukin receptor IL11RA cause autosomal recessive Crouzon-like craniosynostosis. Mol Genet Genomic Med. 2013;1:223–237. doi: 10.1002/mgg3.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Papachristoforou R, Petrou PP, Sawyer H, et al. A novel large deletion encompassing the whole of the galactose-1-phosphate uridyltransferase (GALT) gene and extending into the adjacent interleukin 11 receptor alpha (IL11RA) gene causes classic galactosemia associated with additional phenotypic abnormalities. JIMD Rep. 2014;12:91–98. doi: 10.1007/8904_2013_249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Goriely A, Wilkie AOM. Paternal age effect mutations and selfish spermatogonial selection: causes and consequences for human disease. Am J Hum Genet. 2012;90:175–200. doi: 10.1016/j.ajhg.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Maher GJ, Goriely A, Wilkie AOM. Cellular evidence for selfish spermatogonial selection in aged human testes. Andrology. 2014;2:304–314. doi: 10.1111/j.2047-2927.2013.00175.x. [DOI] [PubMed] [Google Scholar]
- [56].Maher GJ, McGowan SJ, Giannoulatou E, et al. Visualizing the origins of selfish de novo mutations in individual seminiferous tubules of human testes. Proc Natl Acad Sci USA. 2016;113:2454–2459. doi: 10.1073/pnas.1521325113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Wilkie AOM, Goriely A. Gonadal mosaicism and non-invasive prenatal diagnosis for “reassurance” in sporadic paternal age effect (PAE) disorders. Prenat Diagn. 2017 doi: 10.1002/pd.5108. under revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhao H, Feng J, Ho TV, et al. The suture provides a niche for mesenchymal stem cells of craniofacial bones. Nat Cell Biol. 2015;17:386–396. doi: 10.1038/ncb3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Takarada T, Nakazato R, Tsuchikane A, et al. Genetic analysis of Runx2 function during intramembranous ossification. Development. 2016;143:211–218. doi: 10.1242/dev.128793. [DOI] [PubMed] [Google Scholar]
- [60].Maruyama T, Jeong J, Sheu TJ, Hsu W. Stem cells of the suture mesenchyme in craniofacial bone development, repair and regeneration. Nat Commun. 2016;7:10526. doi: 10.1038/ncomms10526. [Axin2, a transcriptional target of β–catenin, the effector of the Wnt signalling pathway, was previously shown to be expressed in cranial sutures. In this paper, Axin2-expressing cells were isolated and shown to be enriched for stem cell markers and self-renewal properties.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kosty J, Vogel TW. Insights into the development of molecular therapies for craniosynostosis. Neurosurg Focus. 2015;38:E2. doi: 10.3171/2015.2.FOCUS155. [DOI] [PubMed] [Google Scholar]
- [62].Doumit GD, Papay FA, Moores N, et al. Opinion leaders and evidence-based medicine in craniofacial surgery. J Craniofac Surg. 2014;25:106–110. doi: 10.1097/SCS.0b013e3182a2ea31. [DOI] [PubMed] [Google Scholar]