Abstract
Pythium insidiosum is a unique oomycete microorganism, capable of infecting humans and animals. The organism can be phylogenetically categorized into three distinct clades: Clade-I (strains from the Americas); Clade-II (strains from Asia and Australia), and Clade–III (strains from Thailand and the United States). Two draft genomes of the P. insidiosum Clade-I strain CDC-B5653 and Clade-II strain Pi-S are available in the public domain. The genome of P. insidiosum from the distinct Clade-III, which is distantly-related to the other two clades, is lacking. Here, we report the draft genome sequence of the P. insidiosum strain Pi45 (also known as MCC13; isolated from a Thai patient with pythiosis; accession numbers BCFM01000001-BCFM01017277) as a representative strain of the phylogenetically-distinct Clade-III. We also report a genome-scale data set of sequence variants (i.e., SNPs and INDELs) found in P. insidiosum (accessible online at the Mendeley database: http://dx.doi.org/10.17632/r75799jy6c.1).
Keywords: Pythium insidiosum, Pythiosis, Draft genome, Sequence variant
Specifications Table
| Subject area | Biology |
| More specific subject area | Microbiology, Genomics |
| Type of data | Genome sequence, Sequence variants, Phylogenetic relationship |
| How data was acquired | IlluminaHiSeq 2500 Next Generation Sequencing Platform |
| Data format | Assembled genome sequence, Sequence variants [i.e., single-nucleotide polymorphisms (SNPs) and small insertions and deletions (INDELs)], Phylogenetic tree |
| Experimental factors | Genomic DNA was extracted from the Pythium insidiosum strain Pi45, which is categorized in the phylogenetically-distinct clade-III. |
| Experimental features | A rDNA-based phylogenetic tree of P. insidiosum was generated. Genome of the P. insidiosum strain Pi45 was sequenced and assembled. The reference genome sequence of the P. insidiosum strain Pi-S was mapped with sequence reads from the P. insidiosum strain Pi45 to identify SNPs and INDELs. |
| Data source location | The organism was isolated from a patient with pythiosis in Thailand. |
| Data accessibility | The draft genome sequence of the P. insidiosum strain Pi45 (also known as MCC13) has been deposited in the Data Bank of Japan (DDBJ) under the accession numbers: BCFM01000001-BCFM01017277. The sequence variant data (i.e., SNPs and INDELs) of the P. insidiosum strain Pi45 is accessible online at the Mendeley database (http://dx.doi.org/10.17632/r75799jy6c.1). |
Value of the data
-
•
The first draft genome sequence of a P. insidiosum strain from the rDNA-based phylogenetic-distinct clade-III is now available.
-
•
Draft genome data of the P. insidiosum strain Pi45 will be valuable for comparative genomic studies of Pythium species and related oomycetes.
-
•
Sequence variant data (i.e., SNPs and INDELs) will be applicable for identification of the organism, genetic polymorphism analyses, genotype-phenotype association studies, and epidemiological exploration.
1. Data
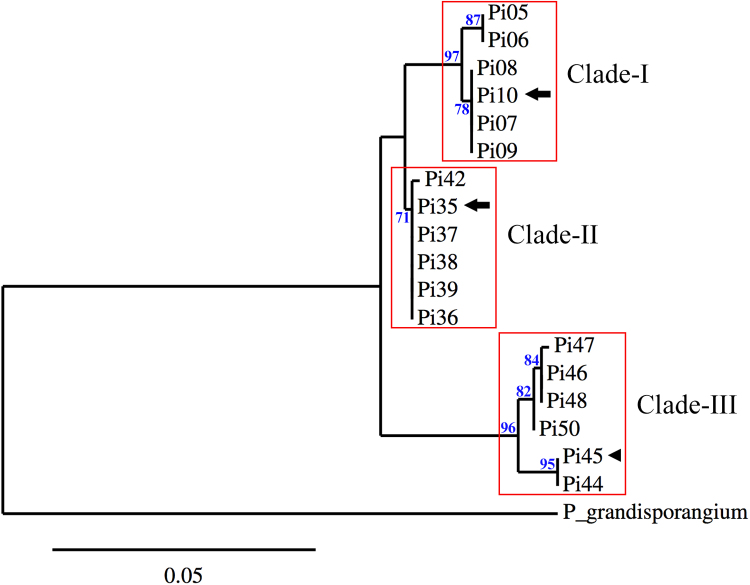
Pythium insidiosum is a member of the oomycetes, a unique group of fungus-like microorganisms belonging to the Kingdom Stramenopiles [1]. P. insidiosum is distinguished from other oomycetes by its capacity to infect humans and animals [1], [2], [3]. The infectious condition called ‘pythiosis’ caused by this organism usually leads to life-long disability or death in affected individuals [2], [3], [4], [5]. Genome sequence is a powerful resource that can be used to explore an organism of interest at the molecular level. Two draft genomes of the P. insidiosum strains CDC-B5653 [6] and Pi-S [7] are available in the public domain. P. insidiosum can be divided into three distinct clades: Clade-I (strains from Americas); Clade-II (strains from Asia and Australia); and Clade–III (strains from Thailand and the United States) (Table 1; Fig. 1). The strain CDC-B5653 (labeled as Pi10) is placed in the Clade-I, whereas the strain Pi-S (labeled as Pi35) is placed in the Clade-II (Fig. 1). The genome of P. insidiosum from the distinct Clade-III, which is distantly-related to the other two clades, is lacking. Here, we report genome data of the P. insidiosum strain Pi45, isolated from a Thai patient and categorized as Clade-III (Fig. 1). We also report a genome-scale data set of sequence variants (i.e., SNPs and INDELs) found in P. insidiosum.
Table 1.
Eighteen strains of Pythium insidiosum used for generation of the rDNA-based phylogenetic tree. Strain identification numbers, reference numbers, host sources, geographic origins, assigned phylogenetically-distinct clades, and accession numbers of the rDNA sequences of all strains are summarized in the table. ‘*’ indicates the strains, including Pi45, where genome sequences are publically available.
| ID | Reference ID | Source | Country of origin | Clade | Accession number |
|---|---|---|---|---|---|
| Pi05 | CBS 575.85 | Equine | Costa Rica | I | AB971178 |
| Pi06 | CBS 574.85 | Equine | Costa Rica | I | AB971179 |
| Pi07 | CBS 573.85 | Equine | Costa Rica | I | AB971180 |
| Pi08 | CBS 580.85 | Equine | Costa Rica | I | AB898107 |
| Pi09 | CBS 101555 | Equine | Brazil | I | AB971181 |
| Pi10* | ATCC 200269 | Human | USA | I | AB898108 |
| Pi35* | Pi-S | Human | Thailand | II | AB898124 |
| Pi36 | ATCC 64221 | Equine | Australia | II | LC199883 |
| Pi37 | ATCC 28251 | Equine | Papua New Guinea | II | LC199884 |
| Pi38 | CBS 101039 | Human | India | II | AB898125 |
| Pi39 | CBS 702.83 | Equine | Japan | II | LC199885 |
| Pi42 | CR02 | Environment | Thailand | II | AB971184 |
| Pi44 | MCC 17 | Human | Thailand | III | AB971185 |
| Pi45* | MCC 13 | Human | Thailand | III | AB971186 |
| Pi46 | SIMI 3306-44 | Human | Thailand | III | AB971187 |
| Pi47 | SIMI 2921-45 | Human | Thailand | III | AB971188 |
| Pi48 | SIMI 4763 | Human | Thailand | III | AB971189 |
| Pi50 | ATCC 90586 | Human | USA | III | AB971190 |
Fig. 1.
Phylogenetic relationship of Pythium insidiosum: the rDNA-based maximum-likelihood phylogenetic tree categorizes 18 strains of Pythium insidiosum into three distinct clades: Clade-I, Clade-II, and Clade-III. Description of each strain of P. insidiosum can be found in Table 1. The arrows indicate the strains [i.e., CDC-B5653 (labeled as Pi10) and Pi-S (labeled as Pi35)] where genome sequences are publically available, while the arrow head indicates the strain Pi45 where genome data is reported here. The rDNA sequence from Pythium granisporangium (accession number: AY151182) is included as an outgroup. Branch support values of greater than 70% are demonstrated at the nodes. Nucleotide substitution per site is shown at the bottom.
2. Experimental design, materials and methods
2.1. rDNA-based phylogenetic tree
rDNA sequences from the strain Pi45 and 17 other strains of P. insidiosum were retrieved from the NCBI database (Table 1). The rDNA sequence from Pythium grandisporangium (accession number: AY151182) served as an outgroup. All rDNA sequences were subjected to phylogenetic analysis, using an array of online tools at www.phylogeny.fr [8], [9], [10], [11], [12], [13].
2.2. Genome sequencing and assembly
Genomic DNA of the P. insidiosum strain Pi45 was extracted [14] and processed to prepare one paired-end library for genome sequencing, using the IlluminaHiSeq 2500 platform (Yourgene Bioscience, Taiwan). Raw reads underwent quality trimming (minimal read length, 35 bases) by CLC Genomics Workbench (http://www.clcbio.com/). Adaptor sequences were removed by Cutadapt 1.8.1 [15]. A total of 33,692,522 adaptor-removed, quality-validated reads, equivalent to 3,488,072,978 total bases, were subjected to genome assembly by Velvet 1.2.10 [16]. The assembled genome consisted of 65,230,783 bases (‘N’ composition, 0.6%) in 17,277 contigs (average length, 3776 bases; range, 300–209,930 bases; N50, 14,374 bases). Assessment of the resulting draft genome sequence by CEGMA [17], [18] showed 78.6% genome completeness. A total of 26,058 open reading frames were predicted by MAKER2 [19].
2.3. Identification of sequence variants
A total of 7,843,910 adaptor-removed quality-validated reads (23.3% of all reads), derived from the P. insidiosum strain Pi45, can be aligned to the reference genome of the P. insidiosum strain Pi-S [7], using the Burrows-Wheeler Alignment tool [20]. A total of 865,332 variants (i.e., SNPs and INDELs) were identified by FreeBayes [21].
2.4. Data accessibility
The draft genome sequence of the P. insidiosum strain Pi45 (also known as MCC13) has been deposited in the Data Bank of Japan (DDBJ) under the accession numbers: BCFM01000001-BCFM01017277. The sequence variant data (i.e., SNPs and INDELs) of the P. insidiosum strain Pi45 can be accessible online at the Mendeley database (http://dx.doi.org/10.17632/r75799jy6c.1).
Acknowledgements
This work was supported by the Faculty of Medicine, Ramathibodi Hospital, Mahidol University; the Thailand Research Fund [Grant number, BRG5980009]; and the Mahidol University [Grant number, ngor-por 09/2560]. We thank Dr. T. Tristan Brandhorst for reviewing the manuscript.
Footnotes
Transparency document associated with this article can be found in the online version at 10.1016/j.dib.2017.10.047.
Transparency document. Supplementary material
Transparency document
.
References
- 1.Kamoun S. Molecular genetics of pathogenic oomycetes. Eukaryot. Cell. 2003;2:191–199. doi: 10.1128/EC.2.2.191-199.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaastra W., Lipman L.J.A., De Cock A.W.A.M., Exel T.K., Pegge R.B.G., Scheurwater J., Vilela R., Mendoza L. Pythium insidiosum: an overview. Vet. Microbiol. 2010;146:1–16. doi: 10.1016/j.vetmic.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 3.Krajaejun T., Sathapatayavongs B., Pracharktam R., Nitiyanant P., Leelachaikul P., Wanachiwanawin W., Chaiprasert A., Assanasen P., Saipetch M., Mootsikapun P., Chetchotisakd P., Lekhakula A., Mitarnun W., Kalnauwakul S., Supparatpinyo K., Chaiwarith R., Chiewchanvit S., Tananuvat N., Srisiri S., Suankratay C., Kulwichit W., Wongsaisuwan M., Somkaew S. Clinical and epidemiological analyses of human pythiosis in Thailand. Clin. Infect. Dis. 2006;43:569–576. doi: 10.1086/506353. [DOI] [PubMed] [Google Scholar]
- 4.Krajaejun T., Pracharktam R., Wongwaisayawan S., Rochanawutinon M., Kunakorn M., Kunavisarut S. Ocular pythiosis: is it under-diagnosed? Am. J. Ophthalmol. 2004;137:370–372. doi: 10.1016/S0002-9394(03)00908-5. [DOI] [PubMed] [Google Scholar]
- 5.Chareonsirisuthigul T., Khositnithikul R., Intaramat A., Inkomlue R., Sriwanichrak K., Piromsontikorn S., Kitiwanwanich S., Lowhnoo T., Yingyong W., Chaiprasert A., Banyong R., Ratanabanangkoon K., Brandhorst T.T., Krajaejun T. Performance comparison of immunodiffusion, enzyme-linked immunosorbent assay, immunochromatography and hemagglutination for serodiagnosis of human pythiosis. Diagn. Microbiol. Infect. Dis. 2013;76:42–45. doi: 10.1016/j.diagmicrobio.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 6.Ascunce M.S., Huguet-Tapia J.C., Braun E.L., Ortiz-Urquiza A., Keyhani N.O., Goss E.M. Whole genome sequence of the emerging oomycete pathogen Pythium insidiosum strain CDC-B5653 isolated from an infected human in the USA. Genom. Data. 2016;7:60–61. doi: 10.1016/j.gdata.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rujirawat T., Patumcharoenpol P., Lohnoo T., Yingyong W., Lerksuthirat T., Tangphatsornruang S., Suriyaphol P., Grenville-Briggs L.J., Garg G., Kittichotirat W., Krajaejun T. Draft genome sequence of the pathogenic oomycete Pythium insidiosum strain Pi-S, isolated from a patient with pythiosis. Genome Announc. 2015;3 doi: 10.1128/genomeA.00574-15. (pii: e00574-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 10.Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 11.Anisimova M., Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 12.Chevenet F., Brun C., Bañuls A.-L., Jacq B., Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinforma. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.-F., Guindon S., Lefort V., Lescot M., Claverie J.-M., Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lohnoo T., Jongruja N., Rujirawat T., Yingyon W., Lerksuthirat T., Nampoon U., Kumsang Y., Onpaew P., Chongtrakool P., Keeratijarut A., Brandhorst T.T., Krajaejun T. Efficiency comparison of three methods for extracting genomic DNA of the pathogenic oomycete Pythium insidiosum. J. Med. Assoc. Thail. 2014;97:342–348. [PubMed] [Google Scholar]
- 15.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10. [Google Scholar]
- 16.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parra G., Bradnam K., Ning Z., Keane T., Korf I. Assessing the gene space in draft genomes. Nucleic Acids Res. 2008;37:289–297. doi: 10.1093/nar/gkn916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parra G., Bradnam K., Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 19.Holt C., Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinforma. 2011;12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.E. Garrison, G. Marth, Haplotype-based variant detection from short-read sequencing, ArXiv PreprarXiv:1207.3907 [q-GN], 2012.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document
Data Availability Statement
The draft genome sequence of the P. insidiosum strain Pi45 (also known as MCC13) has been deposited in the Data Bank of Japan (DDBJ) under the accession numbers: BCFM01000001-BCFM01017277. The sequence variant data (i.e., SNPs and INDELs) of the P. insidiosum strain Pi45 can be accessible online at the Mendeley database (http://dx.doi.org/10.17632/r75799jy6c.1).