Abstract
We conducted genome-wide (GWAS) and candidate gene association studies of maternal mitochondrial DNA copy number. Maternal peripheral blood was collected during labor and delivery admission from 471 participants of a placental abruption case-control study conducted in Lima, Peru. SNP genotyping was performed using the Illumina Cardio-Metabo Chip. Whole blood mtDNA copy number was measured using qRT-PCR techniques. We evaluated 119,629 SNPs in the GWAS and 161 SNPs (in 29 mitochondrial biogenesis and oxidative phosphorylation genes) in the candidate association study. Top hits from GWAS and the candidate gene study were selected to compute weighted genetic risk scores (wGRS). Linear regression models were used to calculate effect size estimates and related nominal p-values. The top hit in our GWAS was chr19:51063065 in FOXA3 (empirical p-value=2.20e-6). A total of 134 SNPs had p-values<0.001 including rs17111633 in CNNM1 (p-value = 6.32e-6) and chr19:51083059 in MYPOP (p-value = 3.23e-5). In the candidate association study, several SNPs in PPARG, PRKCA, SP1, and THRB were associated with mtDNA copy number (p-values<0.05). mtDNA copy number was significantly associated with wGRS based on top GWAS hits (β =0.49, 95% CI:0.38–0.60, p<0.001). Variations in nuclear DNA are potentially associated with maternal mtDNA copy number.
Keywords: Maternal Mitochondrial DNA copy number, genetic variation, oxidative phosphorylation, mitochondrial biogenesis, pregnancy
Introduction
Mitochondrial dysfunction underlies cardiometabolic disorders, neurodegenerative disease and cancer (1–3). Accumulating evidence, including work from our group, supports the role of mitochondrial dysfunction in adverse pregnancy complications (4–8). Approximately 1,500 proteins encoded by nuclear DNA regulate mitochondrial biogenesis and maintain mitochondrial structure and function by regulating mitochondrial DNA replication, transcription, and translation, as well as pathways such as oxidative phosphorylation and apoptosis (9). Mitochondrial DNA copy number (mtDNA) abundance in various tissues (including whole blood and placenta) has emerged as a possible biomarker of mitochondrial dysfunction (10) and related pathophysiologic disturbances, such as oxidative stress (6, 11). Cells under oxidative stress, such as aging tissues, have higher replication of the mitochondria (12), which may be reflected by higher mtDNA copy number (10, 13). On the other hand, severely damaged cells or tissues could sustain mitochondrial replication failure leading to lower mtDNA copy number (14).
Genetic factors that are related to mitochondrial dysfunction in general, and mtDNA copy number in particular, have not been fully described. While previous work on genetic variations and mitochondrial dysfunction has focused on variations in mtDNA, recently, investigators have noted that single nucleotide polymorphisms (SNPs) in nuclear DNA may be associated with mitochondrial dysfunction (15). Only one previous study investigated genome-wide nuclear DNA variation and mtDNA copy number associations (15). The genome-wide association study (GWAS) was conducted among 386 subjects belonging to 21 Spanish families in the Genetic Analysis of Idiopathic Thrombophilia (GAIT) Project. To our knowledge, no prior study has investigated variations in nuclear DNA in relation to maternal mtDNA copy number during pregnancy. We conducted a GWAS and a candidate gene association study (genes participating in mitochondrial biogenesis and oxidative phosphorylation) of maternal mtDNA copy number.
Methods
Study setting and study population
This study was conducted among participants enrolled in the Peruvian Abruptio Placentae Epidemiology (PAPE). This case-control study is designed to investigate risk factors of placental abruption, and has been described previously (16). Recruitment was conducted at the Hospital Nacional Dos de Mayo, Instituto Especializado Materno Perinatal, and Hospital Madre-Niño San Bartolomé in Lima, Peru, between August 2002 and May 2004. Hospital admission and delivery logs were monitored daily to identify placental abruption cases and controls among pregnant women who delivered at participating institutes. Placental abruption cases were selected based on evidence of retroplacental bleeding (fresh blood entrapped between the decidua and the placenta) or blood clots behind the placenta and two of the following: (i) vaginal bleeding in late pregnancy not due to placenta previa or cervical lesions; (ii) uterine tenderness and/or abdominal pain; (iii) fetal distress or death. Controls were selected from eligible pregnant women who delivered at participating hospitals during the study period and who did not have a diagnosis of placental abruption in the current pregnancy. We included both placental abruption cases and controls in the current analyses, described below. Institutional Review Boards of participating institutions approved the project protocol. All participants provided written informed consent.
Data collection
Participants were interviewed by trained research personnel using standardized structured questionnaires. Information was collected on socio-demographic, behavorial and medical characteristics including maternal age, marital status, employment status during pregnancy, smoking and medical history. Blood specimens from 233 placental abruption cases and 238 controls were processed and analyzed for the current GWAS.
DNA extraction and genotyping
DNA extraction and genotyping procedures were previously described (16). Briefly, DNA was extracted from collected blood specimens using the Gentra PureGene Cell kit for DNA preparations (Qiagen, Hilden, Germany). Variants in cardiovascular and metabolism genes were characterized using 217,697 Single Nucleotide Polymorphisms (SNPs) represented on the Illumina Cardio-Metabo Chip (Illumina Inc, San Diego, CA). The Cardio-Metabo Chip is a high-density custom array that captures DNA variation at regions identified by well-powered GWAS meta-analyses for diseases and traits relevant to metabolic and atherosclerotic-cardiovascular endpoints, constructed using the Illumina iSelect technology. Our analyses involved genome-wide and candidate gene components. For the candidate gene study, we evaluated 161 SNPs in 29 genes that participate in mitochondrial biogenesis (CAMK2B, CAMK2D CAMK4, CREB1, NR1H3, PPARG, PPARGC1B, PRKCA, SP1, THRB, and TRNT1) and oxidative phosphorylation (COX5A, COX10, COX4I1, COX7A2, COX7B2, LRPPRC, NDUFA10, NDUFA12, NDUFA3, NDUFB1, NDUFC1, NDUFC2, NDUFS3, NDUFS4, PPA2, SDHB, TSFM, and TUFM).
Maternal mtDNA copy number measurement
Maternal peripheral blood was used for measurement of mtDNA copy number. Real-time qPCR analyses were performed to measure mtDNA copy number using the NovaQUANT™ Human Mitochondrial to Nuclear DNA Ratio Kit (catalog #72620 EMD Millipore, Billerica, MA) according to the manufacturer's instructions (21). This kit provides plates pre-aliquoted with the primers to compare the levels of two nuclear genes (BECN1 and NEB) to two mitochondrial genes (ND1 and ND6) in a RT-qPCR. 2ng of DNA was added to 80uL of SYBR Select Master Mix (catalog #4472908 Life Technologies, Carlsbad, CA) and 20uL of the mixture was applied to each of the four wells of pre-aliquoted primers. Plates were run in the ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA) with the cycling conditions: 95°C for 10 minutes followed by 40 Cycles of: 95°C for 15 seconds 64°C for 1 minute. Analyses were done using the ABI Prism 7000 SDS software RQ Study Application version 1.1. Relative copy number method was used to calculate the mtDNA copy number. Ct values were defined as the cycle number in which fluorescence first crosses the threshold, obtained for each of the two target genes and two reference genes. This is accomplished by averaging the copy numbers calculated from the ND1/BECN1 pair and the ND6/NEB pair. To calculate copy number, we determined N = 2-ΔCt where ΔCt1 = CtND1 − CtBECN1 and ΔCt2 = CtND6 − CtNEB, then averaged 2-ΔCt1 and 2-ΔCt2. All assays were performed without knowledge of pregnancy outcome.
Data quality control and preprocessing
SNP quality control procedures were applied before analyses (17). Participants were excluded if they had genotyping failure for more than 10% of the sites. SNPs were excluded if the minor allele frequency was less than 1%, if the SNP failed to be genotyped in more than 10% of the study participants, or if the SNP was not in Hardy-Weinberg equilibrium among controls (critical p-value=0.00001). We evaluated 119,629 SNPs in the GWAS and 161 SNPs (in 29 mitochondrial biogenesis and oxidative phosphorylation genes) in the candidate association study. Principal components were computed to adjust for large-scale differences in ancestry (17). To determine how many principal components to include in our set of adjustment variables, we used the scree plot from the principal components analysis. We found that after the first two principal components, there is no clear breakpoint in the scree plot. Thus, the first two principal components were identified to explain ancestral variability and were included in the analyses. The final analytic sample (N=471) included participants with complete mtDNA copy number measurement, genotype data, and covariate data.
Statistical analyses
Study participant characteristics were described using mean (standard deviation) and number (percent) for continuous and categorical variables, respectively, for strata defined by placental abruption case or control status. Simple linear regression models were fit to evaluate genetic associations between SNPs and mtDNA copy number in all participants adjusting for placental abruption status and the first two principal components. The Bonferroni corrected threshold (p-value < 4.18e-7) was used to identify significant associations after accounting for multiple testing in overall genome-wide level analyses.
In the candidate gene study, associations of mtDNA copy number with genetic variations (N=161 SNPs) in genes involved in mitochondrial biogenesis and oxidative phosphorylation were evaluated using similar independent linear regression models adjusted for placental abruption case control status and the first two principal components. For these analyses, the nominal significance level threshold (alpha=0.05) was used.
In secondary analyses, top hits from GWAS and the candidate gene study were selected to compute weighted genetic risk scores (GRS) (18). SNPs were included in the final set if they were common (MAF > 0.10) and they were not in linkage disequilibrium (R2<0.64) with other SNPs in our set of selected SNPs. Each SNP was weighted according to its relative effect size estimated from the simple linear regression models. The range of scores (addition of betas multiplied by number of risk alleles) was from 40.1 to 659.9 when using the 9 selected top GWAS SNPs and 25.2 to 182.8 when using the selected 4 candidate SNPs. We then evaluated associations of the GWAS- and candidate gene-based scores with mtDNA copy number using independent linear regression models adjusted for placental abruption case control status and first two principal components. In these analyses, we used P<0.05 as a cut-off to determine statistical significance. In pathway analysis, we examined the functional relationships of genes represented by the top GWAS hits using Ingenuity Pathway Analysis. Analyses were conducted using PLINKv1.07 (19), R (R Foundation), SAS version 9.4 (SAS Institute) and Ingenuity Pathway Analysis (IPA, Ingenuity, Redwood City, CA, www.qiagen.com/ingenuity).
Results
Study participant characteristics are summarized in Table 1. Among placental abruption cases (N=233), mean ± SD maternal age at delivery, gestational age at delivery and whole blood mtDNA copy number were 27.3±6.5 years, 35.5±4.3 weeks and 289.7±153.5, respectively. Among controls (N=238), mean ± SD maternal age at delivery, gestational age at delivery and whole blood mtDNA copy number were 27.2±6.9 years, 37.9±3.4 weeks and 271.5 ± 125.4, respectively.
Table 1.
Baseline Characteristics of Study Population
| Characteristics | Placental Abruption Casesa |
Controlsa |
|---|---|---|
| (N=233) | (N=238) | |
| Maternal age at delivery (years) | 27.3 ± 6.5 | 27.2 ± 6.9 |
| Gestational age at delivery (weeks) | 35.5 ± 4.3 | 37.9 ± 3.4 |
| Whole blood mtDNA copy number | 289.7 ± 153.5 | 271.5 ± 125.4 |
| Pre-pregnancy body mass index (kg/m2) | 23.5 ± 3.5 | 23.9 ± 4.2 |
| Nulliparous | 23 (10.0%) | 8 (3.4%) |
| Maternal education ≤ high school | 186 (80.0%) | 202 (84.9%) |
| Single marital status | 43 (18.0%) | 35 (14.7%) |
| Received prenatal care | 194 (83.0%) | 215 (90.3%) |
| Smoked during pregnancy | 46 (20.0%) | 39 (16.4%) |
| Chronic hypertension | 42 (18.0%) | 18 (8.0%) |
| Preeclampsia | 70 (31.1%) | 28 (12.0%) |
| Weighted GRSb (selected candidate genes) | 100.5 ± 25.7 | 108.2 ± 24.0 |
| Weighted GRS (selected top GWASc) | 289.3 ± 106.1 | 289.8 ± 118.1 |
mean ± standard deviation (SD), otherwise N (%);
Genetic Risk Score;
Genome-Wide Association Study
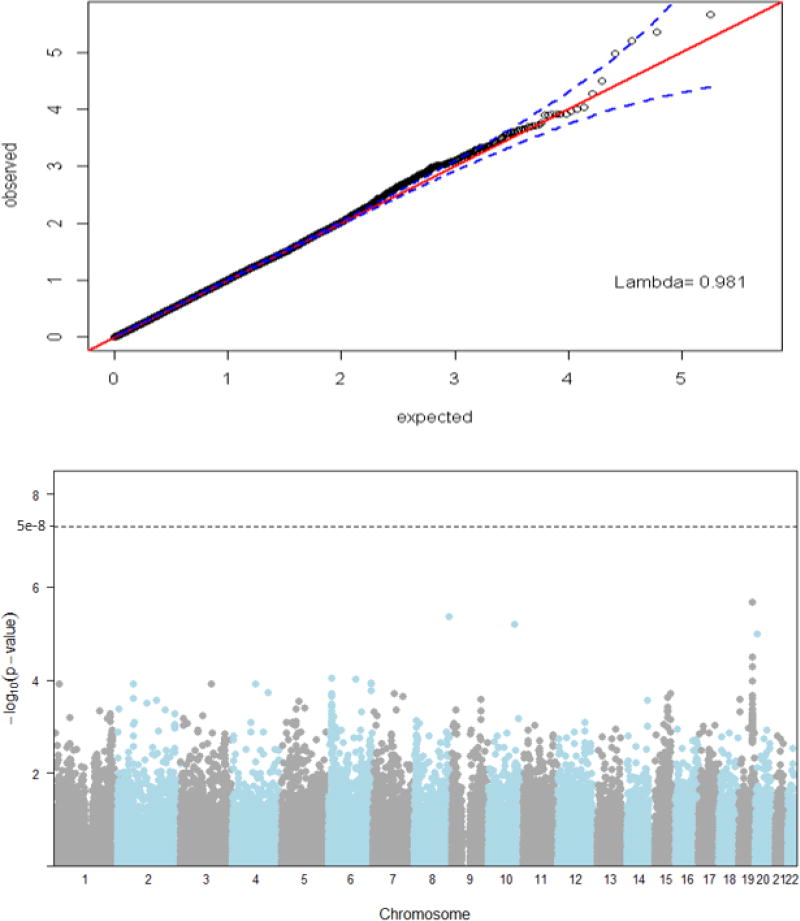
In the GWAS, we did not observe significant genomic inflation (λ = 0.98) as shown in Figure 1. Identified associations were not statistically significant at the genome-wide level (p-value <5.00e-8) (Figure 1). However, several signals that suggest associations of SNPs in chromosome 6 and chromosome 19 with mtDNA copy number were observed. The top hit in our genome-wide analysis was chr19:51063065 in FOXA3 (empirical p-value = 2.20e-6). A total of 134 SNPs had p-values<0.001. SNPs represented by the top 20 hits are shown in Table 2. These included rs17111633 in CNNM1 (p-value = 6.32e-6) and chr19:51083059 in MYPOP (p-value = 3.23e-5). The top network enriched by the GWAS hits (FOXA3, CNNM1, MYPOP, RSPH6A, PHACTR1, LOC105377914, PACRG, PHACTR1) included a network involved in cellular development, growth, proliferation, death and survival (score=20) (Supplementary Figure 1). In the candidate association study, SNPs in PPARG (chr3:12340308), PRKCA (chr17:61731170), SP1 (rs7131938), and THRB (rs17787379) were associated with mtDNA copy number (p-values<0.05) (Supplementary Table 1).
Figure 1.
Quantile-quantile plot with 95% confidence intervals for p-values examining genome-wide genetic variations and mtDNA copy number. Inflation factor, Lambda = 0.98, and Manhattan plot for p-values examining genome-wide genetic variations and mtDNA copy number.
Table 2.
Top mitochondrial DNA copy number GWAS and selected candidate gene association hits
| SNP a | Chr | Position a | rsID | Gene Abbriviation |
Gene Nomenclature |
Location | Effect Size b |
MAF c | P-value b |
|---|---|---|---|---|---|---|---|---|---|
| Top GWAS hits | |||||||||
| chr19:51063065 d | 19 | 51063065 | rs59547719 | FOXA3 | Foxhead Box transcript | Intronic (non-coding, mRNA) | 62.2 | 0.15 | 2.2E-06 |
| rs11989281 | 8 | 134731887 | rs11989281 | NA | NA | NA | 83.1 | 0.07 | 4.4E-06 |
| rs17111633 | 10 | 101134457 | rs17111633 | CNNM1 | Cyclin mediator 1 | Intronic (non-coding, mRNA) | 151.6 | 0.02 | 6.3E-06 |
| rs16991150 | 20 | 5588410 | rs16991150 | NA | NA | NA | 201.1 | 0.01 | 1.1E-05 |
| chr19:51083059 | 19 | 51083059 | NA | MYPOP | Myb-related transcription | 3’ downstream, mRNA | 65.5 | 0.10 | 3.2E-05 |
| chr19:51005665 d | 19 | 51005665 | rs10407137 | RSPH6A | Radial spoke head 6 homolog A | Intronic (non-coding, mRNA) | 40.5 | 0.31 | 5.2E-05 |
| chr6:13154215 | 6 | 13154215 | rs11961962 | PHACTR1 | Phosphatase and actin regulator 1 | Intronic (non-coding, mRNA) | 82.1 | 0.05 | 9.3E-05 |
| rs6939784 d | 6 | 103603097 | rs6939784 | LOC105377914 | Contig mRNA transcript | NA | −40.7 | 0.30 | 9.9E-05 |
| chr19:51059105 d | 19 | 51059105 | rs10410870 | FOXA3 | Foxhead Box transcript | 5’ upstream, mRNA | 38.5 | 0.32 | 1.1E-04 |
| rs6941513 d | 6 | 163950790 | rs6941513 | NA | NA | NA | 35.9 | 0.40 | 1.2E-04 |
| rs9855301 | 3 | 119416106 | rs9855301 | NA | NA | NA | 69.7 | 0.08 | 1.2E-04 |
| rs12513009 d | 4 | 90774543 | rs12513009 | LOC105377329 | NA | Intronic (non-coding, mRNA) | −41.1 | 0.27 | 1.2E-04 |
| rs17184588 d | 1 | 24238099 | rs17184588 | NA | NA | NA | 36.4 | 0.31 | 1.2E-04 |
| rs13017599 | 2 | 61017835 | rs13017599 | NA | NA | NA | −59.1 | 0.10 | 1.3E-04 |
| rs7745161 d | 6 | 163949460 | rs7745161 | NA | NA | NA | 36.6 | 0.41 | 1.3E-04 |
| rs1008295 d | 6 | 163530086 | rs1008295 | PACRG | Parkin coregulated gene | Intronic (non-coding, mRNA) | 35.0 | 0.37 | 1.7E-04 |
| rs10519453 | 4 | 139432369 | rs10519453 | SLC7A11 | Solute carrier family 7 (anionic amino acid transporter light chain, xc-system) | Intronic (non-coding, mRNA) | 52.0 | 0.13 | 1.9E-04 |
| rs6467991 | 7 | 83792673 | rs6467991 | SEMA3A | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A | Intronic (non-coding, mRNA) | 75.0 | 0.06 | 2.0E-04 |
| chr6:13108480 | 6 | 13108480 | ENSSNP8230549 | PHACTR1 | NA | Intronic (non-coding, mRNA) | 82.7 | 0.05 | 2.0E-04 |
| chr6:13095627 | 6 | 13095627 | rs4715043 | PHACTR1 | Phosphatase and actin regulator 1 | Intronic (non-coding, mRNA) | 62.0 | 0.10 | 2.0E-04 |
| Selected MB and OP Candidate SNPs | |||||||||
| rs2279238 | 11 | 47238600 | rs2279238 | NR1H3 | Liver X receptor, alpha|liver X receptor-alpha | Coding mRNA | 17.7 | 0.32 | 7.2E-02 |
| rs627297 | 11 | 77441437 | rs627297 | NDUFC2_KCTD14 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa | Intronic (non-coding, mRNA) | −23.8 | 0.17 | 5.2E-02 |
| chr3:12340308 | 3 | 12340308 | NA | PPARG | Peroxisome proliferator-activated receptor gamma | Intronic (non-coding, mRNA) | −24.8 | 0.26 | 2.6E-02 |
| rs7131938 | 12 | 52059194 | rs7131938 | SP1 | Sp1 transcription factor | 5’ upstream mRNA | −25.2 | 0.21 | 3.7E-02 |
NCBI build 36.1 hg18 assembly;
Linear regression estimates of SNPs in relation to mtDNA copy number adjusted for placental abruption status and first two principal components;
Minor Allele Frequency;
Selected top GWAS SNPs for wGRS analysis
Two weighted GRSs were computed using nine selected top GWAS SNPs and four selected MB and OP candidate SNPs. mtDNA copy number was significantly associated with the wGRS based on top GWAS SNPs (β = 0.49, 95% CI: 0.38 – 0.60, p < 0.001). mtDNA copy number was marginally associated with the candidate gene-wGRS (β = 0.46, 95% CI: −0.07 – 0.99, p=0.09).
Discussion
In this study we identified nuclear DNA variants that are potentially associated with mtDNA copy number, although associations were not statistically significant at the genome-wide level (p-value<5.00e-8). The top hits in our GWAS analysis included SNPs in FOXA3, CNNM1, and MYPOP. In the candidate association study, several SNPs in PPARG, PRKCA, SP1, and THRB were associated with mtDNA copy number (p-values<0.05). In addition, we observed significant association between wGRS based on our top GWAS hits and mtDNA copy number, and marginal association between wGRS based on our candidate gene analysis hits and mtDNA copy number.
The significance of diseases with mitochondrial basis, including adverse pregnancy complications, such as placental abruption and preeclampsia (6, 20), motivate the need to understand common genetic variations that underlie mtDNA copy number, a trait that reflects mitochondrial dysfunction (1–3). The first GWAS study in 2012 that identified nuclear DNA variation that may influence mtDNA levels (measured as the ratio of mtDNA quantity to nuclear DNA quantity, mtDNA/nDNA) was conducted among 386 Spanish families of the GAIT (21). This study highlighted the association between mtDNA levels and rs10888838 in MRPL37, a gene that is involved in mitochondrial protein translation, among male participants (p-value=4.0e-06). The study analyzed only 485 microsatellite markers distributed through the autosomal genome. In 2014, the same investigators analyzed 283,437 SNPs in the same participants and reported rs4708928 as their top GWAS hit (p-value=1.7e-6). Among females, rs4708928 in PARK2, Parkinson protein 2, E3 ubiquitin protein ligase, may be associated with mtDNA levels (p-value=3.3e-6) (15). Of note, in that study, investigators did a sex-stratified analysis to detect sex-specific SNPs that could influence mtDNA. Although the GAIT study did not identify genome-wide significant SNPs, the findings provided the first evidence from GWAS study that suggests nuclear genetic variations in the control of mtDNA levels. None of the top 23 SNPs reported in the study were included in our metabochip samples.
In the present GWAS analysis of 119,629 maternal nuclear DNA variants from 471 participants, we identified some SNPs that may be associated with mtDNA copy number. Among our top 20 hits, SNPs that warrant further investigation include chr19:51063065 (p-value=2.2e-06) and chr19:51059105 (p-value=1.1e-04) in FOXA3, rs17111633 (p-value=6.3e-06) in chromosome 10 in CNNM1, chr19:51005665 (p-value=5.2e-05) in RSPH6A, and rs1008295 (p-value=1.7e-04) in chromosome 6 in PACRG. In addition, a number of SNPs that participate in mitochondrial biogenesis and oxidative phosphorylation were potentially associated with mtDNA copy number in our study. These include chr3:12340308 in PPARG (p=0.03) and rs7131938 in chromosome 12 in SP1 (p=0.04). Notably wGRS based on the selected top GWAS hits was associated with mtDNA copy number (β=0.49, 95% CI: 0.38 – 0.60) while the candidate gene based wGRS was marginally associated with mtDNA copy number.
Previous studies that examined the expression of FOXA (forkhead box transcription factors, including FOXA3) family, which play important roles in metabolism and homeostasis, highlighted that FOXA1 is essential in regulating mitochondrial citrate carrier (CIC) gene expression and hence the activity of CIC, the major route for citrate export from mitochondria (22). In a mouse study, postnatal hepatocyte overexpression of the FOXA2 transcription factor in transthyretin-FOXA2 transgenic has been shown to cause mitochondrial damage while maintaining serum levels of glucose sufficient for survival without increased levels of ketone bodies and free fatty acids (23). CNNM1, cyclin mediator 1 is one of the genes implicated in mitochondrial magnesium homeostasis (24). A haplotype analysis study of 7,287 African American women showed that carriers of each additional copy of rs6584273 SNP in CNNM1 had 16% lower risk of type 2 diabetes (24). MYPOP, Myb-related transcription factor gene and its functions are largely unknown. No prior study reported association of genetic variations in FOXA3, CNNM1 or MYPOP in relation to mtDNA copy number.
In pathway analysis of genes represented by top GWAS hits related to mtDNA copy number, a network involved in cellular development, growth, proliferation, death and survival was identified. Our finding highlights the strong relationship between mitochondrial function and fundamental cellular processes and the potential mechanisms linking mitochondrial dysfunction and pathogenesis of adverse pregnancy complications. For instance, in a recent GWAS conducted by our group, we reported that a network involved in cell cycle, growth and proliferation was associated with placental abruption, a disorder that has been associated with mitochondrial dysfunction (4, 16).
Our study is among the few that examined genetic variations related to maternal mtDNA copy number. In addition, there are several strengths of our current study. Participants were genotyped using the metabochip platform that is designed to support genotyping of SNPs selected based on evidence for association in GWA meta-analysis of diseases and traits relevant to metabolic and atherosclerotic-cardiovascular endpoints relevant to adverse complications of pregnancy. We also conducted a candidate gene study of genetic variations in Mitochondrial Biogenesis and Oxidative Phosphorylate, relevant to mitochondrial function and dysfunction. Finally, our study population is relatively homogenous minimizing issues of potential population stratification.
Several limitations of this study deserve mention, and our findings should be interpreted with caution. Investigators have previously reported inverse U-shaped relationships between mtDNA copy number and adverse outcomes (e.g. colorectal cancer risk) (25), suggesting that lower mtDNA copy number can be observed in cases of failing organ/tissues or in patients with underlying lipid disorders as a feedback response to compensate defective mitochondria (6, 11). Our analyses evaluated linear relationships and did not examine such types of relationships. Our study population comprised of both placental abruption cases and controls. Although we adjusted for case-control status in analyses, findings need to be replicated in other cohorts. We conducted a sensitivity analyses to assess whether findings differ when analyses populations are limited to controls and found no significant differences in findings to those we report in this manuscript. Our study may be underpowered to detect associations between variants in nuclear DNA and mtDNA copy number, particularly since several of the identified SNPs have low MAF. The candidate SNP association findings were not adjusted for multiple testing, although some priors for our candidate genes exist in other studies. Finally, several of the identified top hits in the GWAS and candidate gene study are located in non-coding regions and have not been well-investigated before. Therefore, additional studies (including larger studies in diverse cohorts as well as functional studies) are needed to characterize functional relevance of identified genetic variations.
Findings from our current study contribute to the understanding of genetic determinants of mitochondrial dysfunction, an important pathophysiologic process in a number of pregnancy complications. Similar studies can have potential roles in risk stratification and public health/clinical applications in prenatal care.
Supplementary Material
Acknowledgments
This study was supported by grants from the National Institute of Health, the Eunice Kennedy Shriver National Institute of Child Health and Human Development (R01HD059827 and T32HD052462) and the National Heart Lung and Blood Institute (K01HL10374).
Footnotes
Conflicts of Interest
The authors have no conflicts of interest to disclose.
References
- 1.DiMauro S, Garone C, Naini A. Metabolic myopathies. Current Rheumatology Reports. 2010;12(5):386–93. doi: 10.1007/s11926-010-0119-9. [DOI] [PubMed] [Google Scholar]
- 2.Ježek P, Plecitá-Hlavatá L, Smolková K, Rossignol R. Distinctions and similarities of cell bioenergetics and the role of mitochondria in hypoxia, cancer, and embryonic development. The International Journal of Biochemistry & Cell Biology. 2010;42(5):604–22. doi: 10.1016/j.biocel.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Stark R, Roden M. Mitochondrial function and endocrine diseases. European Journal of Clinical Investigation. 2007;37(4):236–48. doi: 10.1111/j.1365-2362.2007.01773.x. [DOI] [PubMed] [Google Scholar]
- 4.Denis M, Enquobahrie DA, Tadesse MG, Gelaye B, Sanchez SE, Salazar M, et al. Placental genome and maternal-placental genetic interactions: a genome-wide and candidate gene association study of placental abruption. PloS One. 2014;9(12):e116346. doi: 10.1371/journal.pone.0116346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiu C, Sanchez SE, Hevner K, Enquobahrie DA, Williams MA. Placental mitochondrial DNA content and placental abruption: a pilot study. BMC Research Notes. 2015;8(1):447. doi: 10.1186/s13104-015-1340-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams MA, Sanchez SE, Ananth CV, Hevner K, Qiu C, Enquobahrie DA. Maternal blood mitochondrial DNA copy number and placental abruption risk: results from a preliminary study. Int J Mol Epidemiol Genet. 2013;4(2):120–7. [PMC free article] [PubMed] [Google Scholar]
- 7.Workalemahu T, Enquobahrie DA, Moore A, Sanchez SE, Ananth CV, Pacora PN, et al. Genome-wide and candidate gene association studies of placental abruption. International Journal of Molecular Epidemiology and Genetics. 2013;4(3):128–39. [PMC free article] [PubMed] [Google Scholar]
- 8.Workalemahu T, Enquobahrie DA, Yohannes E, Sanchez SE, Gelaye B, Qiu C, et al. Placental telomere length and risk of placental abruption. The Journal of Maternal-Fetal & Neonatal Medicine. 2015:1–6. doi: 10.3109/14767058.2015.1103224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Modica-Napolitano JS, Kulawiec M, Singh KK. Mitochondria and human cancer. Current Molecular Medicine. 2007;7(1):121–31. doi: 10.2174/156652407779940495. [DOI] [PubMed] [Google Scholar]
- 10.Malik AN, Czajka A. Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction? Mitochondrion. 2013;13(5):481–92. doi: 10.1016/j.mito.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 11.Tan D, Goerlitz DS, Dumitrescu RG, Han D, Seillier-Moiseiwitsch F, Spernak SM, et al. Associations between cigarette smoking and mitochondrial DNA abnormalities in buccal cells. Carcinogenesis. 2008;29(6):1170–7. doi: 10.1093/carcin/bgn034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee H-C, Wei Y-H. Mitochondrial role in life and death of the cell. Journal of Biomedical Science. 2000;7(1):2–15. doi: 10.1007/BF02255913. [DOI] [PubMed] [Google Scholar]
- 13.Malik AN, Shahni R, Iqbal MM. Increased peripheral blood mitochondrial DNA in type 2 diabetic patients with nephropathy. Diabetes Research and Clinical Practice. 2009;86(2):e22–e4. doi: 10.1016/j.diabres.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Torregrosa-Muñumer R, Goffart S, Haikonen JA, Pohjoismäki JLO. Low doses of ultraviolet radiation and oxidative damage induce dramatic accumulation of mitochondrial DNA replication intermediates, fork regression, and replication initiation shift. Molecular Biology of the Cell. 2015;26(23):4197–208. doi: 10.1091/mbc.E15-06-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.López S, Buil A, Souto JC, Casademont J, Martinez-Perez A, Almasy L, et al. A genome-wide association study in the genetic analysis of idiopathic thrombophilia project suggests sex-specific regulation of mitochondrial DNA levels. Mitochondrion. 2014;18:34–40. doi: 10.1016/j.mito.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Moore A, Enquobahrie DA, Sanchez SE, Ananth CV, Pacora PN, Williams MA. A genome-wide association study of variations in maternal cardiometabolic genes and risk of placental abruption. Int J Mol Epidemiol Genet. 2012;3(4):305–13. [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nature protocols. 2010;5(9):1564–73. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horne B, Anderson J, Carlquist J, Muhlestein J, Renlund D, Bair T, et al. Generating genetic risk scores from intermediate phenotypes for use in association studies of clinically significant endpoints. Annals of Human Genetics. 2005;69(2):176–86. doi: 10.1046/j.1529-8817.2005.00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Walsh S. Placental mitochondria as a source of oxidative stress in pre-eclampsia. Placenta. 1998;19(8):581–6. doi: 10.1016/s0143-4004(98)90018-2. [DOI] [PubMed] [Google Scholar]
- 21.López S, Buil A, Souto JC, Casademont J, Blangero J, Martinez-Perez A, et al. Sex-specific regulation of mitochondrial DNA levels: genome-wide linkage analysis to identify quantitative trait loci. PloS One. 2012;7(8):e42711. doi: 10.1371/journal.pone.0042711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iacobazzi V, Infantino V, Bisaccia F, Castegna A, Palmieri F. Role of FOXA in mitochondrial citrate carrier gene expression and insulin secretion. Biochemical and Biophysical Research Communications. 2009;385(2):220–4. doi: 10.1016/j.bbrc.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 23.Hughes DE, Stolz DB, Yu S, Tan Y, Reddy JK, Watkins SC, et al. Elevated hepatocyte levels of the forkhead box A2 (HNF-3β) transcription factor cause postnatal steatosis and mitochondrial damage. Hepatology. 2003;37(6):1414–24. doi: 10.1053/jhep.2003.50253. [DOI] [PubMed] [Google Scholar]
- 24.Chan KHK, Chacko SA, Song Y, Cho M, Eaton CB, Wu W-CH, et al. Genetic variations in magnesium-related ion channels may affect diabetes risk among African American and Hispanic American women. The Journal of Nutrition. 2015;145(3):418–24. doi: 10.3945/jn.114.203489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thyagarajan B, Wang R, Barcelo H, Koh W-P, Yuan J-M. Mitochondrial copy number is associated with colorectal cancer risk. Cancer Epidemiology Biomarkers & Prevention. 2012;21(9):1574–81. doi: 10.1158/1055-9965.EPI-12-0138-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.