Abstract
Background
Our previous studies showed that in ischemic and nonischemic heart failure (HF), the voltage-gated cardiac Na+ channel α subunit (SCN5A) mRNA is abnormally spliced to produce two truncated transcript variants (E28C and D) that activate the unfolded protein response (UPR). We tested whether SCN5A post-transcriptional regulation was abnormal in hypertrophic cardiomyopathy (HCM).
Material and Methods
Human heart tissue was obtained from HCM patients. The changes in relative abundances of SCN5A, its variants, splicing factors RBM25 and LUC7A, and PERK, a major effector of the UPR, were analyzed by real time RT-PCR and the expression changes were confirmed by Western Blot.
Results
We found reduced full-length transcript, increased SCN5A truncation variants and activation of UPR in HCM when compared to control hearts. In these patients, real time RT-PCR revealed that HCM patients had decreased SCN5A mRNA to 27.8 ± 4.07% of control (P<0.01) and an increased abundance of E28C and E28D (3.4 ± 0.3 and 2.8 ± 0.3-fold, respectively, P<0.05). PERK mRNA increased 8.2 ± 3.1 fold (P<0.01) in HCM patients. Western blot confirmed a significant increase of PERK.
Conclusions
These data suggested that the full-length SCN5A was reduced in patients with HCM. This reduction was accompanied by abnormal SCN5A pre-mRNA splicing and UPR activation. These changes may contribute to the arrhythmic risk in HCM.
Keywords: sodium channels, hypertrophic cardiomyopathy, sudden cardiac death, human, SCN5A, PERK, splicing variants
1 Introduction
Hypertrophic cardiomyopathy (HCM) leading to sustained ventricular tachyarrhythmias is the most common cause of sudden cardiac death (SCD) in young patients. The cause of arrhythmias in HCM is unclear, however. Previous reports show that, in ischemic or nonischemic systolic heart failure (HF), SCN5A mRNA that encodes the cardiac Na+ channel is abnormally spliced to produce two truncated transcript variants (E28C and D) that encode nonfunctional Na+ channels that are trapped in the endoplasmic reticulum (ER), leading to activation of the unfolded protein response (UPR) 1–3.
The activity of SCN5A-encoded sodium channel determines cardiac depolarization and electrical conduction. Mutations in SCN5A genes, encoding the α-subunit of ion channels that conduct Na+ current (INa), can be responsible for arrhythmogenic disorders. Sodium channel defects have been linked to the development of cardiac arrhythmias and to SCD 4. In this work, we tested whether SCN5A post-transcriptional regulation was abnormal in HCM and whether the presence of truncated Na+ channels was associated with activation of the UPR, potentially contributing to electrical remodeling and arrhythmias in HCM.
2 Material and Methods
2.1 Human heart tissue samples
De-identified human heart tissue was obtained from myectomy samples from 7 patients with HCM, and 6 patient samples of normal human ventricular tissue obtained from unused transplant donor hearts were provided by Dr J. Andrew Wasserstrom (Northwestern University, Chicago, IL). The control tissue samples were obtained from mid left ventricular free wall using a transmural section. All tissues were obtained with informed consent in accordance with the stipulations in the World Medical Association Declaration of Helsinki.
HCM tissue sample numbers obtained were 26, 32, 38, 46, 48, 61, 83. All patients had cardiac myosin binding protein C (MYBPC3) truncating mutations, except 46, which had a cardiac regulatory myosin light chain (MYL2) mutation. The control patients had no family history of SCD, arrhythmia or syncope. HCM, including specific gene mutations, and control patient demographics are reported in Table 1.
Table 1.
Demographics of study patients
| Subject | Sex | Age | Race | Mutation | Family History of SCD | Prior Ventricular Arrhythmia* | Prior Syncope | NYHA Class | LVOT Resting (mmHg) |
|---|---|---|---|---|---|---|---|---|---|
| HCM | F | 48 | C | MYBPC3 c.2905+1 G>A | Yes | Yes | Yes | II | 163 |
| HCM | F | 43 | C | MYBPC3 c.3330+2 T>G | Yes | Yes | No | III | 130 |
| HCM | F | 44 | C | MYBPC3 c.772G>A | No | Yes | No | I | 92 |
| HCM | F | 19 | C | MYL2 c.482 A>G | No | Yes | No | III | 55 |
| HCM | M | 35 | C | MYBPC3 c.2308 G>A | Yes | Yes | No | I | 10 |
| HCM | F | 57 | C | MYBPC3 c.1928-2 A>G | Yes | No | Yes | II | 94 |
| HCM | M | 60 | C | MYBPC3 c.927-9 G>A | No | Yes | No | III | 44 |
| CON | F | 56 | C | N/A | No | No | No | N/A | N/A |
| CON | M | 58 | AA | N/A | No | No | No | N/A | N/A |
| CON | F | 76 | C | N/A | No | No | No | N/A | N/A |
| CON | M | 61 | C | N/A | No | No | No | N/A | N/A |
| CON | M | 65 | C | N/A | No | No | No | N/A | N/A |
| CON | M | 55 | C | N/A | No | No | No | N/A | N/A |
AA: African American; C: Caucasian; CON: control; HCM: Hypertrophic Cardiomyopathy; MYBPC: Myosin-Binding Protein C; MYL: Myosin Light Chain; LVOT: Left Ventricular Outflow Tract; N/A: Not Applicable; NYHA: New York Heart Association; SCD: Sudden Cardiac Death
Ventricular arrhythmic events were defined as more than 3 consecutive beats of ventricular ectopy at a heart rate equal to or above 120 beats per minute detected by ambulatory monitoring.
2.2 Real-Time PCR quantification
Total RNA was isolated from human ventricular tissue using the RNeasy Lipid Tissue Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s instruction. Total RNA was reverse transcribed into complementary DNA (cDNA) using random primer and SuperScript III (Termor Scientific, Waltham, MA) following the manufacturer’s protocol. Quantitative real-time reverse-transcriptase polymerase chain reaction (qRT-PCR) was carried out using gene-specific primers described previously 1, 2, 5 Fast SYBR® Green Master Mix (Thermo Scientific, Waltham, MA) and 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). The qRT-PCR reaction was activated with an initial denaturation step at 95°C for 20 seconds, followed by cycles of denaturation at 95°C for 3 seconds, and annealing and extension at 60°C for 30 seconds. Samples were run in triplicate and averaged. Gene expression levels were normalized to the level of β-actin.
2.3 Antibodies and Western Blot
Anti-RBM25 and anti-LUC7L3 antibodies were purchased from Novus Biologicals (Littleton, CO). Anti-PERK antibody was purchased from Cell Signaling Technologies (Danvers, MA). Anti pan-actin antibody was purchased from Thermo Fisher Scientific (Waltham, MA). Frozen cardiac samples were homogenized in triple-detergent lysis buffer (50 mmol/L Tris-HCl, 150 mmol/L NaCl, 0.1% SDS, 1% NP-40, and 1x Halt® Protease & Phosphatase Inhibitor Cocktail (Thermo Scientific, Waltham, MA), pH 7.5). The homogenates were centrifuged at 10,000g at 4°C for 20 minutes and supernatants were collected for Western blot analysis. Equal amounts (30 μg) of proteins were separated on 4–20% mini-PROTEAN® TGX™ gels (Biorad, Hercules, CA) and were transferred on to PVDF membrane (EMD Millipore, Billerica, MA). Protein expression levels were detected by using specific primary antibodies and Clarity™ Western ECL Blotting Substrate (Biorad, Hercules, CA). Band intensities were be detected by ChemiDoc MP imaging System and analyzed by Image Lab software (Biorad, Hercules, CA).
2.4 Statistics
All data are presented as means and SD. Means were compared using unpaired Student’s t test. A probability value P <0.05 was considered statistically significant.
3 Results
3.1 Cardiac Na+ channel SCN5A pre-mRNA was abnormally spliced in HCM
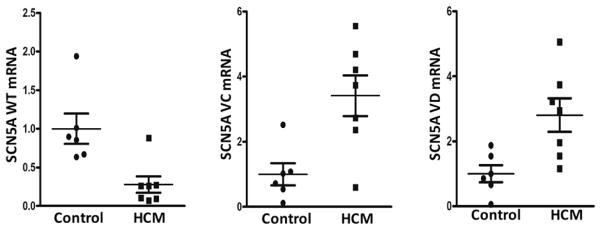
We have shown previously that the pre-mRNA of SCN5A exon 28 is abnormally spliced in ischemic or nonischemic, acquired cardiomyopathy 3, 4. Here, we investigated whether the pre-mRNA splicing of SCN5A exon 28 was altered in HCM. Compared to that in the normal control human hearts, the full-length SCN5A transcript was substantially decreased in HCM heart tissues. The mRNA level of the full-length SCN5A in HCM patient group was 27.8% ± 4.1% (Figure 1, Panel A, P<0.01) of that in the normal control group. Consistent with an inverse relationship of the full-length transcripts to the SCN5A splice variants, the expression levels of the truncated variants E28C and E28D were increased significantly in HCM hearts. SCN5A variants E28C and E28D increased 3.4 ± 0.3 fold (Figure 1, Panel B, P<0.05) and 2.8 ± 0.3 fold, (Figure 1, Panel C, P<0.05) in HCM patients, respectively.
Figure 1.
mRNA expression levels of SCN5A splice variants in HCM heart tissues. Panels A, B, and C show tissue level of full length SCN5A mRNA, SCN5A variant E28C mRNA in HCM, and SCN5A variant E28D mRNA in HCM. All mRNA levels were normalized with β–actin mRNA. *P< 0.05 vs. control (n=13).
3.2 Splicing factors RBM25 and Luc7A were upregulated in HCM
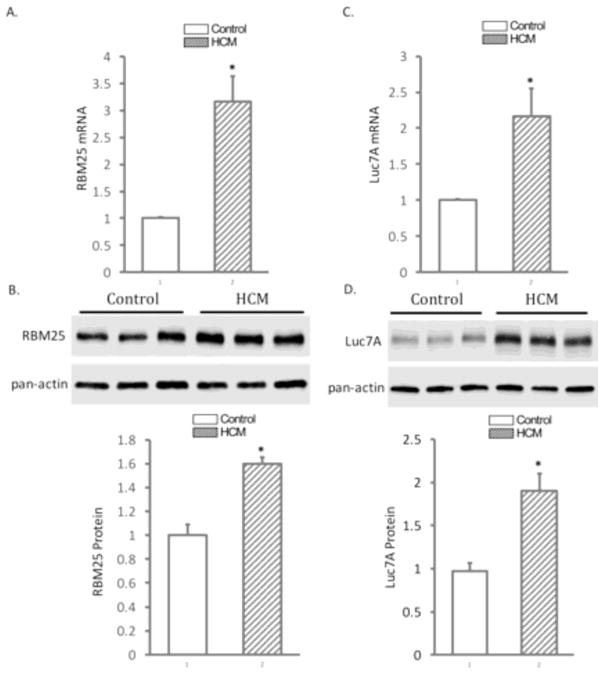
In dilated or ischemic cardiomyopathy, abnormal splicing of SCN5A pre-mRNA is mediated by splicing factors RBM25 and Luc7A. In HCM, the expression of RBM25 and Luc7A increased at both the mRNA and protein levels. Compared to that in the normal human heart tissue, RBM25 increased 3.17 ± 0.47 fold (Figure 2, Panel A, P< 0.05) and Western blot confirmed an increase of RBM25 protein of 1.6 ± 0.05 fold (Figure 2, Panel B, p<0.05). The expression of Luc7A mRNA increased 2.16 ± 0.40 fold (Figure 2, Panel C, P < 0.05) fold. Western blot confirmed the expression of Luc7A protein increased 1.90 ± 0.20 fold (Figure 2, Panel D, P<0.05) in patients with HCM.
Figure 2.
The expression levels of splicing factors, RBM25 and Luc7A, in HCM hearts. Panel A shows tissue level of RBM25 mRNA in HCM. Panel B: Western blot (top) and quantification (bottom) confirms the upregulation of RBM25 in HCM heart tissues. Panel C: The expression levels of Luc7A mRNA in HCM. Panel D: Western blot (top) and quantification (bottom) confirms the upregulation of Luc7A in HCM heart tissues. The mRNA expression was normalized with β-actin and the protein expression was normalized with pan-actin. *P< 0.05 vs control (n=13).
3.3 UPR component PERK was upregulated in human HCM
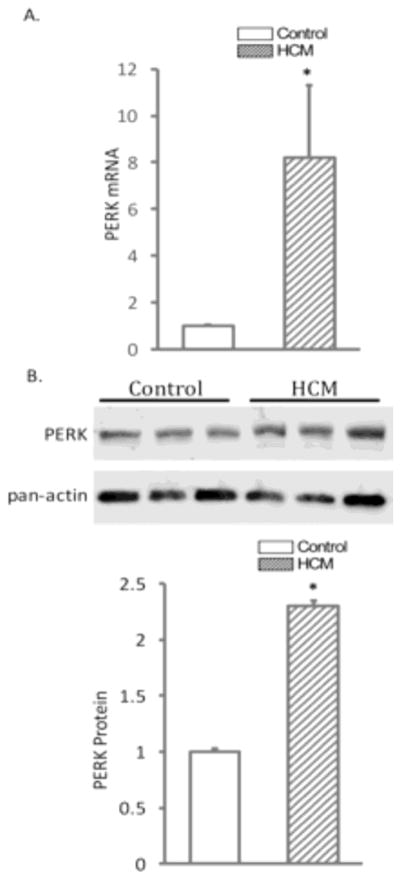
In ischemic or dilated cardiomyopathy, abnormal SCN5A splicing variants are known to activate the UPR, and this activation further reduces SCN5A full-length mRNA transcript abundance 1. To investigate whether increased SCN5A variants promote UPR activation in HCM, an UPR major component PERK was measured at both mRNA and protein levels by qRT-PCR and Western blotting. Real time RT-PCR revealed that PERK mRNA increased 8.2 ± 3.1 fold (Figure 3, Panel A, P<0.05) in HCM patients compared to controls. Western blotting confirmed a significant increase of PERK protein of 2.3 ± 0.01 fold (Figure 3, Panel B, P<0.05) in HCM.
Figure 3.
The expression of PERK in HCM. Panel A shows tissue level of PERK mRNA in HCM. Panel B: Western blot (top) and quantification (bottom) confirms the upregulation of PERK in HCM heart tissues. The mRNA expression was normalized with β-actin and the protein expression was normalized with pan-actin. *P< 0.05 vs control (n=13).
4 Discussion
Modification in cardiac sodium current alters cardiac conduction and is associated with arrhythmogenesis 6. Hypertrophic cardiomyopathy is frequently associated with arrhythmic SCD 7, 8, though the pathophysiological triggers remain poorly understood 9. Ventricular arrhythmia is common in patients with HCM 10 with ventricular tachycardia proving to be a significant independent risk factor for SCD in HCM, especially in the young 11.
Previously, we have shown that ischemic and nonischemic heart failure are associated with increased abnormal SCN5A mRNA splicing, that the expression of SCN5A cardiac Na+ channel mRNA splice variants in white blood cells (WBCs) correlates with levels in the heart, and that normalized WBC splicing levels predict increased risk of ventricular arrhythmias 12. In dilated or ischemic cardiomyopathy, the two SCN5A mRNA splicing variants reach greater that >50% of the total SCN5A mRNA and do not produce functional Na+ channels 2, 5. The expression of these variants in cells stably expressing the full-length Na+ channel causes a dose-dependent reduction in full-length SCN5A transcripts, as well as the reduction in functional Na+ current expression 2, 5. This dominant negative effect is mediated by the UPR 1. Here, we were able to show that heart tissue from patients with HCM showed a decreased full-length SCN5A mRNA transcript abundance and increased abundances of abnormal SCN5A mRNA splice variants, similar to the previous results in other forms of cardiomyopathy 1, 2, 12. This abnormal SCN5A mRNA splicing was accompanied by elevations in the splicing factors Luc7a and RBM25 which are known to cause abnormal SCN5A mRNA splicing 2. In the case of ischemic or dilated cardiomyopathy, the increases in these two splicing factors were thought to result from excess angiotensin II or hypoxia 13. The cause of the increase in HCM is unclear. Moreover, it is unclear if the presence of the HCM mutation also contributed to the UPR activation.
Activation of the UPR has been observed in cardiac hypertrophy with mouse models demonstrating elevated CHOP, a downstream pro-apoptotic effector of the PERK branch of the UPR 14. Here, we observed an increase in PERK in HCM. UPR was responsible for a dominant negative effect on full-length SCN5A transcript abundance in a mouse model expressing an SCN5A truncation mRNA 5. This effect was thought to contribute to the arrhythmic risk induced by abnormal SCN5A splicing and may play a role in the reduced full-length SCN5A mRNA abundance seen in human HCM.
Limitations
Cardiac tissue was obtained from a small number of patients who were referred for septal myectomy, most having cardiac MYBPC3 mutations. Therefore, it is not clear if the results would be similar in patients with mutations in other HCM genes or without obstruction. Nevertheless, the degree of the left ventricular-aortic pressure gradient did not appear to influence the results significantly. The reduction in full-length sodium channel mRNA is consistent with that seen in the arrhythmic condition known as Brugada Syndrome 15. Nevertheless, we were not able to establish if the reduction in SCN5A mRNA was associated with reduced sodium current, but we have shown previously that a reduction in full-length sodium channel mRNA abundance is correlated with a reduction in channel protein and current in other forms of human myopathy 5. From the small sample size, we were unable to assess fully whether the degree of hypertrophy or the HCM genotype affected abnormal SCN5A mRNA splicing or if the degree of alteration in SCN5A mRNA abundance correlated with arrhythmic outcomes. Nevertheless, all but one of our patients studied had suffered previous arrhythmic events.
5 Conclusion
These results show that there is a reduction in sodium channel full-length mRNA during human HCM that likely results in part from abnormal SCN5A mRNA splicing and from activation of the UPR. The data suggest that SCN5A may be reduced in patients with HCM. A reduction in SCN5A may contribute to the arrhythmic risk in HCM patients.
Acknowledgments
Acknowledgements and Funding
This work was supported by National Institute of Health (NIH R01HL106592, R01HL104025 and R01HL119095).
Footnotes
Disclosures
Dr. Dudley is the inventor of on U.S. Application No. 13/291,826, entitled “SCN5A Splicing Factors and Splice Variants for Use in Diagnostic and Prognostic Methods.”
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gao G, Xie A, Zhang J, Herman AM, Jeong EM, Gu L, Liu M, Yang KC, Kamp TJ, Dudley SC. Unfolded protein response regulates cardiac sodium current in systolic human heart failure. Circ Arrhythm Electrophysiol. 2013;6:1018–1024. doi: 10.1161/CIRCEP.113.000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao G, Xie A, Huang SC, Zhou A, Zhang J, Herman AM, Ghassemzadeh S, Jeong EM, Kasturirangan S, Raicu M, Sobieski MA, Bhat G, Tatooles A, Benz EJ, Jr, Kamp TJ, Dudley SC., Jr Role of rbm25/luc7l3 in abnormal cardiac sodium channel splicing regulation in human heart failure. Circulation. 2011;124:1124–1131. doi: 10.1161/CIRCULATIONAHA.111.044495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsiao PY, Tien HC, Lo CP, Juang JM, Wang YH, Sung RJ. Gene mutations in cardiac arrhythmias: A review of recent evidence in ion channelopathies. Appl Clin Genet. 2013;6:1–13. doi: 10.2147/TACG.S29676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu M, Yang KC, Dudley SC., Jr Cardiac sodium channel mutations: Why so many phenotypes? Nat Rev Cardiol. 2014 doi: 10.1038/nrcardio.2014.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shang LL, Pfahnl AE, Sanyal S, Jiao Z, Allen J, Banach K, Fahrenbach J, Weiss D, Taylor WR, Zafari AM, Dudley SC., Jr Human heart failure is associated with abnormal c-terminal splicing variants in the cardiac sodium channel. Circ Res. 2007;101:1146–1154. doi: 10.1161/CIRCRESAHA.107.152918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Remme CA, Bezzina CR. Sodium channel (dys)function and cardiac arrhythmias. Cardiovasc Ther. 2010;28:287–294. doi: 10.1111/j.1755-5922.2010.00210.x. [DOI] [PubMed] [Google Scholar]
- 7.Maron BJ, McKenna WJ, Danielson GK, Kappenberger LJ, Kuhn HJ, Seidman CE, Shah PM, Spencer WH, 3rd, Spirito P, Ten Cate FJ, Wigle ED American College of Cardiology Foundation Task Force on Clinical Expert Consensus D, European Society of Cardiology Committee for Practice G. American college of cardiology/european society of cardiology clinical expert consensus document on hypertrophic cardiomyopathy. A report of the american college of cardiology foundation task force on clinical expert consensus documents and the european society of cardiology committee for practice guidelines. Eur Heart J. 2003;24:1965–1991. doi: 10.1016/s0195-668x(03)00479-2. [DOI] [PubMed] [Google Scholar]
- 8.Maron BJ, Shirani J, Poliac LC, Mathenge R, Roberts WC, Mueller FO. Sudden death in young competitive athletes. Clinical, demographic, and pathological profiles. JAMA. 1996;276:199–204. [PubMed] [Google Scholar]
- 9.Elliott P, Spirito P. Prevention of hypertrophic cardiomyopathy-related deaths: Theory and practice. Heart. 2008;94:1269–1275. doi: 10.1136/hrt.2008.154385. [DOI] [PubMed] [Google Scholar]
- 10.Gao XJ, Kang LM, Zhang J, Dou KF, Yuan JS, Yang YJ. Mid-ventricular obstructive hypertrophic cardiomyopathy with apical aneurysm and sustained ventricular tachycardia: A case report and literature review. Chin Med J (Engl) 2011;124:1754–1757. [PubMed] [Google Scholar]
- 11.Christiaans I, van Engelen K, van Langen IM, Birnie E, Bonsel GJ, Elliott PM, Wilde AA. Risk stratification for sudden cardiac death in hypertrophic cardiomyopathy: Systematic review of clinical risk markers. Europace. 2010;12:313–321. doi: 10.1093/europace/eup431. [DOI] [PubMed] [Google Scholar]
- 12.Gao G, Brahmanandam V, Raicu M, Gu L, Zhou L, Kasturirangan S, Shah A, Negi SI, Wood MR, Desai AA, Tatooles A, Schwartz A, Dudley SC., Jr Enhanced risk profiling of implanted defibrillator shocks with circulating scn5a mrna splicing variants: A pilot trial. J Am Coll Cardiol. 2014;63:2261–2269. doi: 10.1016/j.jacc.2014.02.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao G, Dudley SC., Jr Rbm25/luc7l3 function in cardiac sodium channel splicing regulation of human heart failure. Trends Cardiovasc Med. 2013;23:5–8. doi: 10.1016/j.tcm.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu M, Dudley SC., Jr Role for the unfolded protein response in heart disease and cardiac arrhythmias. Int J Mol Sci. 2016:17. doi: 10.3390/ijms17010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antzelevitch C, Patocskai B. Brugada syndrome: Clinical, genetic, molecular, cellular, and ionic aspects. Curr Probl Cardiol. 2016;41:7–57. doi: 10.1016/j.cpcardiol.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]