Abstract
Mitochondria form a cellular network of organelles, or cellular compartments, that efficiently couple nutrients to energy production in the form of ATP. As cancer cells rely heavily on glycolysis, historically mitochondria and the cellular pathways in place to maintain mitochondrial activities were thought to be more relevant to diseases observed in non-dividing cells such as muscles and neurons. However, more recently it has become clear that cancers rely heavily on mitochondrial activities including lipid, nucleotide and amino acid synthesis, suppression of mitochondria-mediated apoptosis as well as oxidative phosphorylation (OXPHOS) for growth and survival. Considering the variety of conditions and stresses that cancer cell mitochondria may incur such as hypoxia, reactive oxygen species and mitochondrial genome mutagenesis, we examine potential roles for a mitochondrial-protective transcriptional response known as the mitochondrial unfolded protein response (UPRmt) in cancer cell biology.
Keywords: Mitochondria, UPRmt, ATF5, cancer
I. Introduction
Mitochondrial organization and functions
Mitochondria are cellular compartments that form a dynamic network located throughout the cytosol that harbor the tricarboxylic acid (TCA) cycle and oxidative phosphorylation (OXPHOS) machinery. These two processes convert acetyl-CoA generated from carbohydrate, protein and fat catabolism into ATP via the electron transport chain (respiratory chain) complexes and the ATP synthase. Mitochondria are also required for many other essential cellular activities, including amino acid, lipid, nucleotide, and iron-sulfur cluster synthesis as well as calcium homeostasis. Beyond metabolism, mitochondria significantly contribute to multiple signal transduction events including the regulation of cell death, cell differentiation, growth and innate immunity [1–5].
Mitochondria are double membrane bound organelles consisting of about 1200 proteins. Nearly 99% of mitochondrial proteins are encoded by nuclear genes and synthesized on cytosolic ribosomes. These proteins harbor mitochondrial targeting sequences (MTS) that direct them to either the mitochondrial outer membrane, intermembrane space, inner membrane or the mitochondrial matrix. At the outer membrane, proteins destined for the mitochondrial matrix interact with the TOM (translocase of the outer membrane) complex and the TIM (translocase of the inner membrane) complex sequentially to traverse both mitochondrial membranes [6]. In addition to the channels, transportation across the inner membrane requires a proton gradient generated by the respiratory chain as well as molecular chaperones located in the mitochondrial matrix. Once in the matrix, the MTS is typically cleaved and the protein folds and/or assembles which is facilitated by molecular chaperones and complex assembly factors.
The remaining components of the mitochondrial proteome are encoded by the mitochondrial genome (mtDNA). Typically, human cells harbor hundreds, or thousands of mtDNA copies, which require extensive cellular machinery to maintain [7, 8]. Human mtDNA encodes 13 essential components of the OXPHOS complexes as well as 2 rRNAs and 22 tRNAs, which are required for the synthesis of mtDNA-encoded proteins within the mitochondrial matrix [9]. As the OXPHOS system is composed of large multi-subunits complexes encoded by separate genomes, transcription, protein expression and complex assembly must be tightly coordinated to prevent the accumulation of toxic protein folding or assembly intermediates in the mitochondria.
Mitochondria in cancer
Mitochondrial dysfunction caused by lesions associated with nuclear- or mtDNA-encoded genes contributes to a variety of human diseases, which typically present as neuro-muscular disorders [10–13]. Interestingly, similar mutations or lesions have been found in a variety of cancers, suggesting a relationship between mitochondrial dysregulation and cancer cell biology [14, 15]. Many of these mutations reduce OXPHOS efficiency [16–18] and force cells to rely more heavily on glycolysis for ATP production, potentially providing an underlying mechanism for what Warburg observed in cancer cells nearly 100 years ago; that cancer cells rely heavily on glycolysis even in the presence of oxygen [19].
However, a bevy of recent studies have found that cancer cells rely more heavily on mitochondrial functions than previously thought. In contrast to Warburg’s observation that mitochondria are dysfunctional in cancers, mitochondria are likely metabolically altered to support cell proliferation and tumorigenesis, consistent with mitochondrial perturbations impairing tumorigenicity. For example, depletion of mtDNA in glioblastoma and breast carcinoma cells significantly impairs tumor proliferation [20, 21]. Additionally, inhibition of mtDNA replication or mitochondrial biogenesis by suppressing PGC-1α or TFAM also reduces the invasion and metastasis of mammary epithelial cancer cells and lung cancer cells, respectively [22, 23]. These studies suggest that cancer cells rely on mitochondrial activities as well cellular pathways that evolved to maintain and recover mitochondrial function.
Furthermore, considerable evidence suggests that signals emanating from stressed or dysfunctional mitochondria promote cancer cell growth and survival. For example, mutations in the TCA cycle gene encoding isocitrate dehydrogenase (IDH) cause an over-production of the onco-metabolite 2-hydroxyglutarate, which contributes to glioma formation and leukemogensis [24, 25]. And, succinate dehydrogenase (SDH) or fumarate hydratase (FH) mutations also cause accumulation of the TCA cycle metabolic intermediates, succinate and fumarate, that activate hypoxia-inducible factor 1 alpha (HIF-1α), which promotes cancer cell growth and survival [26–29]. Moreover, mutations in genes encoding complex III components of electron transport chain impair apoptosis, thus contributing to tumor progression [30].
Further suggestive of a contribution of dysfunctional mitochondria to tumor progression, several conserved mtDNA mutations, such as mutations in cytochrome c oxidase subunit 1 (MTCO1) and NADH dehydrogenase (ND5), are found in tumors of patients with diverse mtDNA backgrounds [16, 31]. Although the impact on cancer biology remains controversial, several studies have suggested that mtDNA mutations perturbing the respiratory chain cause increased reactive oxygen species (ROS) which engage components of pro-growth or pro-survival pathways such as KRas, Akt and TLR4 [22, 32–36]. While it is well-documented that mtDNA lesions are relatively common in cancer cells, it should be noted that some studies have found no effect of mtDNA mutations on cancer cell biology [37]. Hopefully, future studies will be able to determine if mtDNA mutations are correlative or causative in tumorigenesis.
While mitochondrial lesions may or may not benefit tumor growth and survival, accumulation of mitochondrial stress in cancer has the potential to perturb mitochondrial and cellular activities, and must be dealt with to maintain cellular integrity. Here, we review a mitochondrial stress responsive pathway, the mitochondrial unfolded protein response (UPRmt), that responds to mitochondrial dysfunction by promoting mitochondrial repair, metabolic adaptations as well as survival by inducing anti-apoptotic factors, and its potential role in cancer biology.
II. The mitochondrial unfolded protein response (UPRmt)
Discovery of a mitochondrial stress response: A brief history
Considerable evidence has suggested the existence of adaptive transcriptional responses to mitochondrial dysfunction. Transcript profiling studies comparing affected tissues in patients with mitochondrial diseases demonstrated a variety of potentially adaptive transcriptional alterations [38]. Consistent with the patient studies, mitochondrial dysfunction in cultured cells caused by depletion of mtDNA or the overexpression of a terminally misfolded mitochondrial protein (ΔOTC) causes transcriptional induction of a number of mitochondrial-specific molecular chaperones and proteases [39, 40]. As these findings were conceptually similar to a well-studied response that mediates endoplasmic reticulum protein homeostasis known as the unfolded protein response (UPR), it was dubbed a mitochondrial stress response, and later a UPRmt [41]. While conceptually similar, regulation of the UPRmt is different and completely independent of the UPRER, likely deriving from differences in the two organelles and their respective protein folding compartments. And, the scope of the response is reflective of diverse cellular activities affected by mitochondrial functions.
In line with an organelle-specific response, the UPRmt is specifically activated by mitochondrial perturbations [41]. In addition to mtDNA depletion and mitochondrial unfolded protein accumulation, OXPHOS defects, inhibition of mitochondrial protein synthesis, mtDNA mutations, reactive oxygen species, hypoxia, as well as pathogenic bacteria that target mitochondria as part of a virulence response can trigger the UPRmt [42–49]. As indicated in the previous section, many of the listed mitochondrial defects can be observed in cancer cells suggesting activation of the UPRmt.
UPRmt signaling in C. elegans
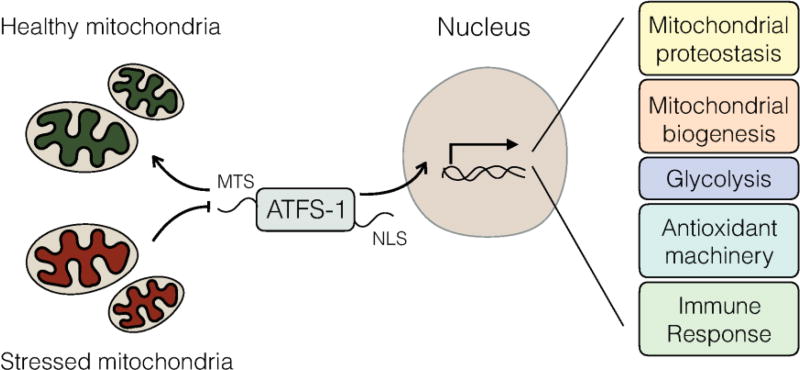
Genetic and biochemical studies in C. elegans have indicated that cells evaluate or monitor mitochondrial protein import efficiency to regulate the UPRmt. Mitochondrial import likely serves as a useful surrogate for mitochondrial function, as multiple activities including OXPHOS and mitochondrial protein homeostasis, are required for efficient mitochondrial import [50]. Several components that regulate UPRmt activation have been discovered via genetic screens. The bZip transcription factor, ATFS-1, directly regulates UPRmt gene promoters during mitochondrial dysfunction and is regulated by organellar compartmentalization (Figure 1). ATFS-1 harbors both a mitochondrial targeting sequence as well as a nuclear localization sequence allowing it to respond to mitochondrial import efficiency [43]. In cells with a healthy mitochondrial network, ATFS-1 is synthesized and rapidly imported into mitochondria where it is degraded. However, during mitochondrial stress or dysfunction, reduced mitochondrial protein import efficiency causes a percentage of mitochondrial-targeted proteins to accumulate in the cytosol. As ATFS-1 harbors a nuclear localization sequence, it traffics to the nucleus to regulate a broad transcriptional response [43, 51]. In addition to transcriptional adaptations, UPRmt activation also requires chromatin rearrangements for a sustained response [52]. Interestingly, UPRmt activation can also be communicated between cells or different tissues presumably to allow for metabolic coordination or to prepare tissues for future conditions that may impact mitochondrial functions, although the signaling mechanism remains to be further defined [49, 53].
Figure 1. UPRmt signaling in C. elegans.
In the absence of mitochondrial stress, ATFS-1 is targeted to mitochondria via an amino-terminal mitochondrial targeting sequence (MTS) and is subsequently degraded in the mitochondrial matrix. During mitochondrial stress or dysfunction, mitochondrial protein import is impaired causing ATFS-1 to accumulate in the cytosol. Subsequently, ATFS-1 traffics to nucleus via its nuclear localization signal (NLS) and regulates transcription of ~500 genes that promote mitochondrial protein homeostasis (proteostasis), mitochondrial recovery or biogenesis, metabolic adaptations such as glycolysis, antioxidants, and genes involved in xenobiotic detoxification to promote survival and the resolution of mitochondrial stress.
The transcriptional responses
Once in the nucleus, ATFS-1 regulates transcription of over 500 genes that orchestrate a coherent mitochondrial stress response including genes that promote mitochondrial protein homeostasis (chaperones, proteases and antioxidant genes). ATFS-1 also regulates diverse metabolic adaptations including an increase of all glycolysis genes while simultaneously limiting transcription of TCA cycle and OXPHOS genes, presumably to reduce mitochondrial metabolic loads and maintain cellular ATP levels via glycolysis, which occurs in the cytosol. And, the UPRmt coordinates a mitochondrial repair and recovery program that includes a mitochondrial biogenesis pathway [54]. Concomitantly, ATFS-1 regulates the expression of xenobiotic detoxifying genes and an innate immune response likely to reduce the effects of toxic metabolic intermediates or environmental toxins, and detect those pathogens that perturb mitochondrial function as part of their virulence responses. For example, the pathogen Pseudomonas aeruginosa, which produces the OXPHOS inhibitor cyanide as a virulence factor, activates the UPRmt, which is required to clear the infection [46, 54].
UPRmt signaling in mammals
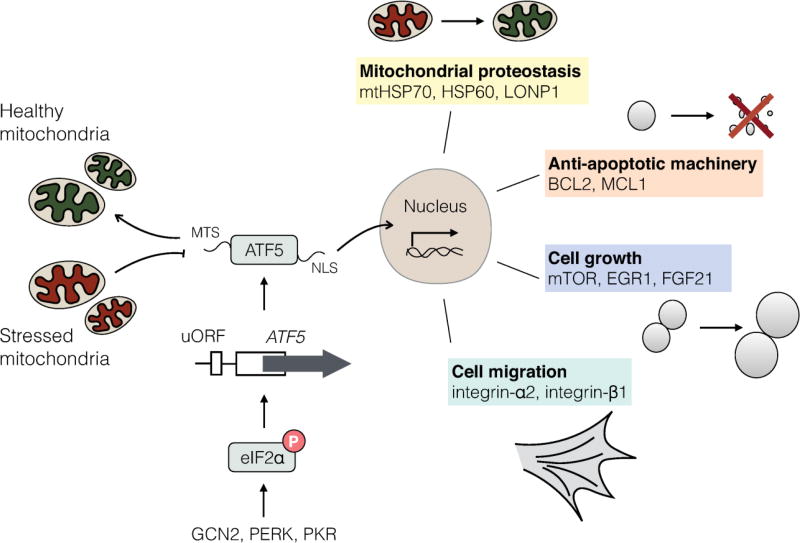
The UPRmt was first discovered in cultured mammalian cells expressing a terminally misfolded protein to the mitochondrial matrix, which yielded the increased transcription of multiple proteostasis genes as well as the bZip transcription factor CHOP [40]. Subsequent studies have demonstrated that UPRmt activation in mammals relies on similar mechanisms and bZip transcription factors to those in, however signaling in mammals is likely more elaborate as multiple transcription factors including CHOP, ATF4 and ATF5, are involved. The interaction or coordination of these three transcription factors remains to be understood, but all three are induced during mitochondrial dysfunction and required for UPRmt induction [55–59]. For example, ATF4 has been shown to respond to mitochondrial dysfunction and induce mitochondrial proteases, components of one carbon metabolism, as well as the hormone FGF21 that coordinates metabolism between cells and tissues [60–62]. And most recently, ATF5 was found to regulate a UPRmt, by mediating a transcription response that includes mitochondrial chaperones and proteases similar to ATFS-1 in C. elegans [59] (Figure 2). Interestingly, ATF5 is transcriptionally induced in several mitochondrial disorders [63–66], and cells with impaired ATF5 are susceptible to mitochondrial stress [59].
Figure 2. UPRmt signaling in mammalian cells.
The mammalian UPRmt is regulated by multiple bZip transcription factors such as ATF5, which is regulated by at least two mechanisms. Expression of ATF5 is regulated by the phosphorylation of the translation initiation factor eIF2α, which is regulated by the kinases GCN2, PERK or PKR. Because the ATF5-encoding mRNA harbors upstream open reading frames (uORFs) in the 5`-untranslated region, its synthesis requires phosphorylated eIF2α which can be stimulated during nutrient deprivation, mitochondria or endoplasmic reticulum dysfunction or the accumulation of double-stranded RNA in the cytosol by the above-mentioned kinases. Once it is expressed, ATF5 is regulated by mitochondrial protein import efficiency. In the absence of mitochondrial stress, ATF5 is targeted to mitochondria via its amino-terminal mitochondrial targeting sequence (MTS). However, during mitochondrial dysfunction, ATF5 fails to be imported into mitochondria and traffics to the nucleus via its nuclear localization signal (NLS) to induce transcription of genes that influence mitochondrial proteostasis, anti-apoptotic machinery, cell growth and migration.
Accumulating evidence indicates that, like in C. elegans, the UPRmt in mammals is regulated at least in part by mitochondrial import efficiency. For example, during mitochondrial dysfunction, a subunit of the TIM23 mitochondrial protein import complex is rapidly degraded, resulting in reduced import efficiency and induction of a UPRmt [67]. Importantly, ATF5 is regulated by mitochondrial protein import efficiency similar to ATFS-1 [59], and potentially responds to degradation of Tim17a or other forms of mitochondrial stress that perturb protein import. Similar to ATFS-1, the steady-state localization of ATF5 is within mitochondria when expressed in healthy cells. However, during mitochondrial stress it accumulates in nuclei and induces the expression of mitochondrial-protective genes.
The UPRmt requires the integrated stress response (ISR)
Interactions between CHOP, ATF4 and ATF5 have been documented by multiple studies. For example, ATF4 and CHOP are capable of forming a heterodimer, however ATF5 does not associate with ATF4 or CHOP at least in vitro [68]. And, CHOP and ATF4 are both required for transcriptional induction of ATF5 [69–71]. Of note, the relationship between the three transcription factors has not been determined during mitochondrial dysfunction. One potential mechanism consistent with current data is simply that CHOP and/or ATF4 are required for basal transcription of ATF5, which can serve as a mitochondrial stress sensor similar to ATFS-1 in C. elegans. This model is consistent with CHOP being activated by multiple stressors, yet contribute specifically to a mitochondrial stress specific transcriptional program via increasing expression of ATF5, a transcription factor capable of responding to mitochondrial dysfunction. Although, this model remains to be tested during mitochondrial dysfunction.
Interestingly, CHOP, ATF4 and ATF5 are preferentially translated during conditions that activate the integrated stress response (ISR) [69, 72–76]. The ISR is comprised of four kinases that are stimulated by different cellular conditions that phosphorylate the translation initiation factor eIF2α. The kinase PERK is activated by endoplasmic reticulum dysfunction, GCN2 is activated by amino acid depletion, PKR is activated by double stranded RNA in the cytosol typically during viral infection, and HRI is activated by heme depletion [77]. Importantly, all four kinases have been shown to be activated during mitochondrial stress, consistent with the pleiotropic nature of mitochondrial dysfunction. Phosphorylation of eIF2α results in reduced synthesis of most proteins, however proteins such as CHOP, ATF4 and ATF5 that are encoded by mRNAs harboring upstream open reading frames (uORFs) in the 5’ untranslated regions are preferentially synthesized [69, 70, 73–75]. The activation or contribution of each eIF2α kinase and CHOP, ATF4 and ATF5 during mitochondrial dysfunction remains to be elucidated. Although, the interactions are potentially cell type-specific as different baseline levels of eIF2α phosphorylation driven by specific kinases exist in different tissues. For example, pancreatic beta cells rely heavily on PERK [78], and liver cells on GCN2 [79]. In tissue culture, it is clear that the ISR responds to mitochondrial stress and is required for UPRmt activation. However, it remains to be determined if the ISR kinases dynamically respond to mitochondrial stress in vivo or simply set the baseline level of eIF2α phosphorylation and cell-specific expression of CHOP, ATF4 and ATF5. Alternatively, ATF5 transcription can also be induced by increased growth factor signaling via ERK and PI3K signaling [80].
III. The UPRmt and cancer
As discussed in the previous sections, mitochondrial dysfunction is well documented in multiple cancers, as is the increased eIF2α phosphorylation that affects CHOP, ATF4 and ATF5 expression, suggesting a role of the UPRmt in cancer cell survival and growth. However, to our knowledge, a specific role for the UPRmt in cancer biology has not been explicitly examined. In principle, the UPRmt could promote cell growth and survival by ensuring mitochondrial function in the presence of mitochondrial stress related to cancer cell physiology or mutation accumulation, or influencing cancer cell metabolism, growth and inflammatory signaling, or responses to therapeutic agents perceived by the cell as xenobiotics [81, 82]. In this section, we review recent findings that suggest functions for UPRmt regulatory components as well as transcriptional outputs in cancer cell growth and survival.
UPRmt signaling components
Many studies indicate that the ISR via PERK, PKR or GCN2 activation is important for tumor progression [83–88], although their relationship with mitochondrial dysfunction remains to be elucidated. However, there is evidence showing that mitochondrial dysfunction enhances chemotherapeutic resistance in tumors via GCN2 [82], demonstrating a relationship between mitochondrial stress and ISR activation. However, the impact of ATF5 in the context of ISR activation in cancer remains to be addressed.
While CHOP, ATF4 and ATF5 have all been reported to contribute to tumor growth and survival [83–85, 89, 90], we focus on ATF5, as it functions downstream of CHOP and ATF4, and recent studies suggest a role for ATF5 in cancer cell survival and growth. ATF5 knockout mice are viable despite the failure of olfactory neuron differentiation [91, 92]. However, many cancers including epithelial ovarian carcinoma, thyroid follicular lymphoma, chronic lymphocytic leukemia, colorectal adenocarcinoma, breast carcinoma, pancreatic cancer and malignant glioma cells require ATF5 for growth and tumor formation [80, 93–100].
Initially, ATF5 was identified as an anti-apoptotic factor as it regulates the expression of the anti-apoptotic components BCL2 and MCL1 [80, 93, 97], and multiple studies demonstrated that ATF5 inhibition leads to cell death in multiple cancer cells. ATF5 also regulates growth and metabolism coordinating factors such as EGR1, mTOR and FGF21, as well as the mitochondrial-protective genes outlined above [59, 101–103]. In addition to pro-growth and anti-apoptotic phenotypes, ATF5 contributes the resistance to radiotherapy [81] and the invasiveness of tumor cells by inducing integrin-α2 and integrin-β1 [96].
In general, the relationship between mitochondrial dysfunction or metabolism and the requirement for ATF5 has not been explicitly examined. However, mitochondrial dysfunction is common in gliomas [104, 105], ovarian cancer [106, 107], breast cancer [108] and leukemia [109, 110], where ATF5 is highly expressed. And, our lab recently demonstrated that ATF5 is required to maintain OXPHOS function and promote growth in multiple cancer cell lines including a line derived from a thyroid oncocytoma harboring high quantities of deleterious mtDNAs [59, 111]. Though further investigation is required, the ATF5-mediated UPRmt potentially contributes to the survival and growth of these cancers by adapting them to various forms of mitochondrial dysfunction, sustaining mitochondrial proteostasis and preventing mitochondrial-induced cell death.
UPRmt transcriptional outputs
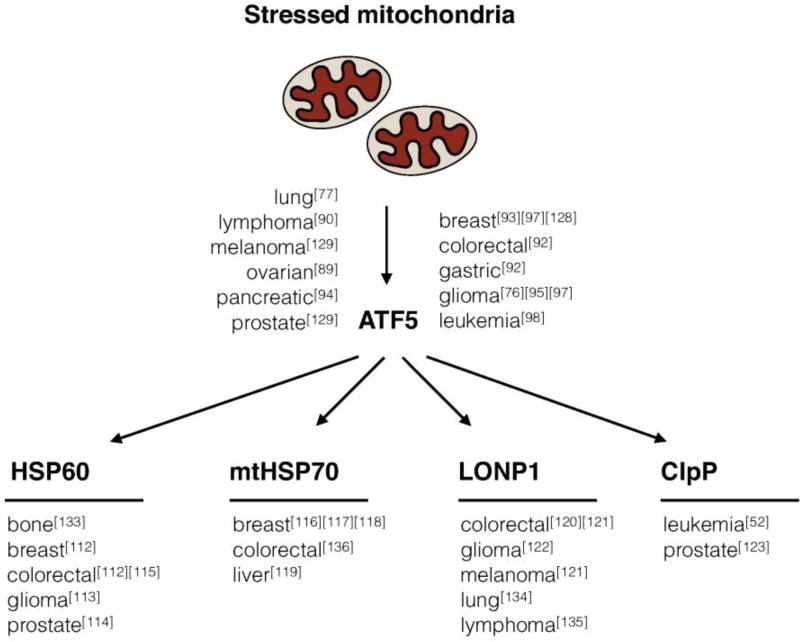
The UPRmt-induced genes such as mitochondrial chaperones and proteases are highly induced in many cancers [112–115], some of which have been shown to be ATF5-dependent [59]. Consistent with promoting protein folding and organelle homeostasis, mitochondrial chaperones contribute to signal transduction, protect against cell death and senescence, and thus are required for cancer cell survival and growth (Figure 3). For example, the mitochondrial chaperone HSP60 protects tumor cells from Bax-dependent cell death and CypD-dependent cell death by regulating mitochondrial permeability transition [116, 117]. HSP60 is also positively associated with tumor progression and hormone resistance in prostate cancer [118], and tumor differentiation in colorectal cancer [119]. mtHSP70, or mortalin, promotes tumor cell survival and epithelial-to-mesenchymal transition (EMT) by regulating the activities of p53 and PI3K–Akt pathways [120, 121], as well as facilitating metastasis of breast carcinoma and hepatocellular carcinoma [122, 123].
Figure 3. Requirements for UPRmt-related components in cancer cell biology.
The transcription factor ATF5 and the UPRmt-induced mitochondrial chaperones (HSP60, mtHSP70) and mitochondrial proteases (LONP1, ClpP) are required for the growth and survival of multiple cancers.
Mitochondrial proteases also promote mitochondrial function by degrading proteins that fail to fold or assemble, or that become damaged over time. LONP1 and ClpP are mitochondrial matrix-localized proteases that can be induced by ATF5. Loss of either LONP1 or ClpP impairs tumor proliferation and metastasis due to the dysregulation of mitochondrial activities [124–127]. Additionally, mitochondrial proteases can contribute to the metabolic remodeling observed in cancer. For example, some cancer cells increase OXPHOS capacity to promote survival and proliferation, a phenomenon known as the “reverse Warburg effect” [128]. Breast cancer, pancreatic cancer, and AML cells have elevated expression of respiratory chain components and increased mitochondrial mass and basal oxygen consumption [129–131]. Perhaps not surprisingly, the UPRmt is required for the maintenance of mitochondrial function in these cancers. Consistent with this idea, ClpP inhibition impairs the growth and viability of multiple AML cell lines [132].
The UPRmt and cancer treatment
Multiple studies in vitro and in vivo have demonstrated that the inhibition of the UPRmt can selectively repress the growth and progression of tumor cells. And, because inhibition of UPRmt components is often toxic to cancer cells, while having relatively modest impact on normal cells, UPRmt-related reagents are being developed as cancer-specific therapies.
Several studies have shown that ATF5 inhibition suppresses the viability of cancer cells. For example, expression of a dominant-negative form of ATF5 specifically increased death in cancer cell lines relative to non-neoplastic cell lines [133]. Interestingly, a cell permeable peptide, CP-d/n-ATF5-S1, has been developed as an ATF5-specific inhibitor. The inhibitory peptide impairs the growth of prostate cancer, glioblastoma, melanoma and triple receptor-negative breast cancer cells in cell culture and xenograft models by inducing apoptosis [134]. Lastly, ATF5 inhibition can selectively kill rat and human glioblastoma cells as well as human pancreatic cancer cells while sparing the neighboring normal cells in vivo [98, 99].
The mitochondrial-localized HSP90 paralog, TRAP-1, is expressed in normal cells at low levels, but enriched in mitochondria of pancreatic and breast adenocarcinoma cells [135]. Repression of TRAP-1 by G-TPP, a derivative of the HSP90 inhibitor 17-AAG that localizes to mitochondria, leads to the apoptosis of patient-derived and cultured glioblastoma cells [136, 137]. Similarly, knocking-down of HSP60 by siRNA causes Bax overexpression and Bax-dependent apoptosis in breast and colon adenocarcinoma cells [116], as well as canine osteosarcoma cells [138], but not in normal cells, providing a potential therapeutic approach for breast and colon cancers.
Targeting UPRmt-regulated mitochondrial proteases has also been shown effective in impairing cancer cell survival and proliferation. For instance, inhibition of mitochondrial protease LONP1 by obtusilactone A or CDDO selectively kills non-small-cell lung carcinoma and lymphoma cells, respectively [139, 140]. Also, inhibition of ClpP can selectively kill human leukemic cells by disturbing the folding of mitochondrial metabolic proteins [132].
V. Conclusion and Perspective
Underlying mechanisms that regulate the UPRmt in mammals are emerging with significant similarities to what has been elucidated in C. elegans. Cells utilize mitochondrial protein import efficiency to determine the function of the cellular pool of mitochondria. And, if import efficiency declines due to damaged OXPHOS, reduced membrane potential or perturbed mitochondrial proteostasis, ATF5 fails to be imported, which allows it to traffic to the nucleus and activate the UPRmt. Studies from worms and cell culture indicate that the UPRmt promotes survival and mitochondrial recovery during a number of mitochondrial stresses, and similar data is emerging from in vivo systems.
Considerable evidence suggests that the UPRmt is active in multiple cancer types, and thus, may provide viable therapeutic targets. While ATF5 can induce a number of mitochondrial chaperone and protease genes during mitochondrial stress, along with anti-apoptotic components, the full transcriptional scope of the response is unclear. Furthermore, it will be important to understand how UPRmt activation affects aspects of cancer cell biology such as metabolic adaptations and xenobiotic detoxification. A number of interesting parameters remain to be elucidated in cancer cells as well. For example, is the UPRmt exclusively activated by mitochondrial dysfunction, or are there other pathways to engage the protective effects of the UPRmt (gene amplification, etc.)? It will also be interesting to determine how the UPRmt interacts with known oncogenes and tumor suppressors to impact mitochondrial physiology.
Recent studies using small molecules and cell permeable peptides to impair both UPRmt signaling component (ATF5) and UPRmt transcriptional outputs (mitochondrial chaperones and proteases) provide optimism that the UPRmt pathway can be manipulated to improve cancer treatment. Presumably, inhibition of either will synthetically interact with cancer-specific mitochondrial stress and thus impair tumor growth or induce cell death.
Acknowledgments
This work was supported by a Frank Lappin Horsfall Jr. Student Fellowship to P. D. and grants from the NIH (R01HL127891 and R01AG047182) to C.M.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no competing financial interests.
References
- 1.Liu X, et al. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86(1):147–57. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 2.Sciacovelli M, et al. The mitochondrial chaperone TRAP1 promotes neoplastic growth by inhibiting succinate dehydrogenase. Cell Metab. 2013;17(6):988–99. doi: 10.1016/j.cmet.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol. 2014;15(4):243–56. doi: 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kasahara A, Scorrano L. Mitochondria: from cell death executioners to regulators of cell differentiation. Trends Cell Biol. 2014;24(12):761–70. doi: 10.1016/j.tcb.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Weinberg SE, Sena LA, Chandel NS. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015;42(3):406–17. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoogenraad NJ, Ward LA, Ryan MT. Import and assembly of proteins into mitochondria of mammalian cells. Biochim Biophys Acta. 2002;1592(1):97–105. doi: 10.1016/s0167-4889(02)00268-9. [DOI] [PubMed] [Google Scholar]
- 7.Miller FJ, et al. Precise determination of mitochondrial DNA copy number in human skeletal and cardiac muscle by a PCR-based assay: lack of change of copy number with age. Nucleic Acids Res. 2003;31(11):e61. doi: 10.1093/nar/gng060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wachsmuth M, et al. Age-Related and Heteroplasmy-Related Variation in Human mtDNA Copy Number. PLoS Genet. 2016;12(3):e1005939. doi: 10.1371/journal.pgen.1005939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta. 1999;1410(2):103–23. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 10.Schwarz K, et al. The breathing heart - mitochondrial respiratory chain dysfunction in cardiac disease. Int J Cardiol. 2014;171(2):134–43. doi: 10.1016/j.ijcard.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 11.Russell AP, et al. Skeletal muscle mitochondria: a major player in exercise, health and disease. Biochim Biophys Acta. 2014;1840(4):1276–84. doi: 10.1016/j.bbagen.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 12.Subramaniam SR, Chesselet MF. Mitochondrial dysfunction and oxidative stress in Parkinson’s disease. Prog Neurobiol. 2013;106–107:17–32. doi: 10.1016/j.pneurobio.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, et al. Oxidative stress and mitochondrial dysfunction in Alzheimer’s disease. Biochim Biophys Acta. 2014;1842(8):1240–7. doi: 10.1016/j.bbadis.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larman TC, et al. Spectrum of somatic mitochondrial mutations in five cancers. Proc Natl Acad Sci U S A. 2012;109(35):14087–91. doi: 10.1073/pnas.1211502109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaude E, Frezza C. Defects in mitochondrial metabolism and cancer. Cancer Metab. 2014;2:10. doi: 10.1186/2049-3002-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petros JA, et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci U S A. 2005;102(3):719–24. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonora E, et al. Defective oxidative phosphorylation in thyroid oncocytic carcinoma is associated with pathogenic mitochondrial DNA mutations affecting complexes I and III. Cancer Res. 2006;66(12):6087–96. doi: 10.1158/0008-5472.CAN-06-0171. [DOI] [PubMed] [Google Scholar]
- 18.Singh KK, et al. Mutations in mitochondrial DNA polymerase-gamma promote breast tumorigenesis. J Hum Genet. 2009;54(9):516–24. doi: 10.1038/jhg.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. J Gen Physiol. 1927;8(6):519–30. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickinson A, et al. The regulation of mitochondrial DNA copy number in glioblastoma cells. Cell Death Differ. 2013;20(12):1644–53. doi: 10.1038/cdd.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan AS, et al. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab. 2015;21(1):81–94. doi: 10.1016/j.cmet.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107(19):8788–93. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LeBleu VS, et al. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16(10):992–1003. doi: 10.1038/ncb3039. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2010;465(7300):966. doi: 10.1038/nature09132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Losman JA, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339(6127):1621–5. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isaacs JS, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8(2):143–53. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 27.Selak MA, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7(1):77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 28.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20(1):51–6. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laukka T, et al. Fumarate and Succinate Regulate Expression of Hypoxia-inducible Genes via TET Enzymes. J Biol Chem. 2016;291(8):4256–65. doi: 10.1074/jbc.M115.688762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dasgupta S, et al. Forced cytochrome B gene mutation expression induces mitochondrial proliferation and prevents apoptosis in human uroepithelial SV-HUC-1 cells. Int J Cancer. 2009;125(12):2829–35. doi: 10.1002/ijc.24701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gasparre G, et al. Clonal expansion of mutated mitochondrial DNA is associated with tumor formation and complex I deficiency in the benign renal oncocytoma. Hum Mol Genet. 2008;17(7):986–95. doi: 10.1093/hmg/ddm371. [DOI] [PubMed] [Google Scholar]
- 32.Sharma LK, et al. Mitochondrial respiratory complex I dysfunction promotes tumorigenesis through ROS alteration and AKT activation. Hum Mol Genet. 2011;20(23):4605–16. doi: 10.1093/hmg/ddr395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, et al. NADPH oxidase 1-dependent ROS is crucial for TLR4 signaling to promote tumor metastasis of non-small cell lung cancer. Tumour Biol. 2015;36(3):1493–502. doi: 10.1007/s13277-014-2639-9. [DOI] [PubMed] [Google Scholar]
- 34.Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17(4):351–9. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sabharwal SS, Schumacker PT. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles’ heel? Nat Rev Cancer. 2014;14(11):709–21. doi: 10.1038/nrc3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishikawa K, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320(5876):661–4. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 37.Deichmann M, et al. Somatic mitochondrial mutations in melanoma resection specimens. Int J Oncol. 2004;24(1):137–41. [PubMed] [Google Scholar]
- 38.Cizkova A, et al. Development of a human mitochondrial oligonucleotide microarray (h-MitoArray) and gene expression analysis of fibroblast cell lines from 13 patients with isolated F1Fo ATP synthase deficiency. BMC Genomics. 2008;9:38. doi: 10.1186/1471-2164-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinus RD, et al. Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. Eur J Biochem. 1996;240(1):98–103. doi: 10.1111/j.1432-1033.1996.0098h.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhao Q, et al. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21(17):4411–9. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoneda T, et al. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci. 2004;117(Pt 18):4055–66. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- 42.Liu L, et al. Hypoxic reactive oxygen species regulate the integrated stress response and cell survival. J Biol Chem. 2008;283(45):31153–62. doi: 10.1074/jbc.M805056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nargund AM, et al. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337(6094):587–90. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Houtkooper RH, et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497(7450):451–7. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cano M, et al. Oxidative stress induces mitochondrial dysfunction and a protective unfolded protein response in RPE cells. Free Radic Biol Med. 2014;69:1–14. doi: 10.1016/j.freeradbiomed.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pellegrino MW, et al. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature. 2014;516(7531):414–7. doi: 10.1038/nature13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jovaisaite V, Auwerx J. The mitochondrial unfolded protein response-synchronizing genomes. Curr Opin Cell Biol. 2015;33:74–81. doi: 10.1016/j.ceb.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rauthan M, et al. A Mutation in Caenorhabditis elegans NDUF-7 Activates the Mitochondrial Stress Response and Prolongs Lifespan via ROS and CED-4. G3 (Bethesda) 2015;5(8):1639–48. doi: 10.1534/g3.115.018598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shao LW, Niu R, Liu Y. Neuropeptide signals cell non-autonomous mitochondrial unfolded protein response. Cell Res. 2016;26(11):1182–1196. doi: 10.1038/cr.2016.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wright G, et al. Oxidative stress inhibits the mitochondrial import of preproteins and leads to their degradation. Exp Cell Res. 2001;263(1):107–17. doi: 10.1006/excr.2000.5096. [DOI] [PubMed] [Google Scholar]
- 51.Haynes CM, et al. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell. 2010;37(4):529–40. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian Y, et al. Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPR(mt) Cell. 2016;165(5):1197–208. doi: 10.1016/j.cell.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berendzen KM, et al. Neuroendocrine Coordination of Mitochondrial Stress Signaling and Proteostasis. Cell. 2016;166(6):1553–1563. doi: 10.1016/j.cell.2016.08.042. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nargund AM, et al. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt) Mol Cell. 2015;58(1):123–33. doi: 10.1016/j.molcel.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Horibe T, Hoogenraad NJ. The chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response. PLoS One. 2007;2(9):e835. doi: 10.1371/journal.pone.0000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Papa L, Germain D. SirT3 regulates the mitochondrial unfolded protein response. Mol Cell Biol. 2014;34(4):699–710. doi: 10.1128/MCB.01337-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chung HK, et al. Growth differentiation factor 15 is a myomitokine governing systemic energy homeostasis. J Cell Biol. 2017;216(1):149–165. doi: 10.1083/jcb.201607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bao XR, et al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. Elife. 2016:5. doi: 10.7554/eLife.10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fiorese CJ, et al. The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Curr Biol. 2016;26(15):2037–43. doi: 10.1016/j.cub.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Munch C, Harper JW. Mitochondrial unfolded protein response controls matrix pre-RNA processing and translation. Nature. 2016;534(7609):710–3. doi: 10.1038/nature18302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim KH, et al. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med. 2013;19(1):83–92. doi: 10.1038/nm.3014. [DOI] [PubMed] [Google Scholar]
- 62.Kim KH, Lee MS. FGF21 as a Stress Hormone: The Roles of FGF21 in Stress Adaptation and the Treatment of Metabolic Diseases. Diabetes Metab J. 2014;38(4):245–51. doi: 10.4093/dmj.2014.38.4.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Endo J, et al. Metabolic remodeling induced by mitochondrial aldehyde stress stimulates tolerance to oxidative stress in the heart. Circ Res. 2009;105(11):1118–27. doi: 10.1161/CIRCRESAHA.109.206607. [DOI] [PubMed] [Google Scholar]
- 64.Tyynismaa H, et al. Mitochondrial myopathy induces a starvation-like response. Hum Mol Genet. 2010;19(20):3948–58. doi: 10.1093/hmg/ddq310. [DOI] [PubMed] [Google Scholar]
- 65.Torres-Peraza JF, et al. Protective neuronal induction of ATF5 in endoplasmic reticulum stress induced by status epilepticus. Brain. 2013;136(Pt 4):1161–76. doi: 10.1093/brain/awt044. [DOI] [PubMed] [Google Scholar]
- 66.Yap YW, et al. Comparative Microarray Analysis Identifies Commonalities in Neuronal Injury: Evidence for Oxidative Stress, Dysfunction of Calcium Signalling, and Inhibition of Autophagy-Lysosomal Pathway. Neurochem Res. 2016;41(3):554–67. doi: 10.1007/s11064-015-1666-2. [DOI] [PubMed] [Google Scholar]
- 67.Rainbolt TK, et al. Stress-regulated translational attenuation adapts mitochondrial protein import through Tim17A degradation. Cell Metab. 2013;18(6):908–19. doi: 10.1016/j.cmet.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Newman JR, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300(5628):2097–101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- 69.Zhou D, et al. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem. 2008;283(11):7064–73. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]
- 70.Teske BF, et al. CHOP induces activating transcription factor 5 (ATF5) to trigger apoptosis in response to perturbations in protein homeostasis. Mol Biol Cell. 2013;24(15):2477–90. doi: 10.1091/mbc.E13-01-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fusakio ME, et al. Transcription factor ATF4 directs basal and stress-induced gene expression in the unfolded protein response and cholesterol metabolism in the liver. Mol Biol Cell. 2016;27(9):1536–51. doi: 10.1091/mbc.E16-01-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harding HP, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6(5):1099–108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 73.Jousse C, et al. Inhibition of CHOP translation by a peptide encoded by an open reading frame localized in the chop 5’UTR. Nucleic Acids Res. 2001;29(21):4341–51. doi: 10.1093/nar/29.21.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167(1):27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A. 2004;101(31):11269–74. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watatani Y, et al. Stress-induced translation of ATF5 mRNA is regulated by the 5’-untranslated region. J Biol Chem. 2008;283(5):2543–53. doi: 10.1074/jbc.M707781200. [DOI] [PubMed] [Google Scholar]
- 77.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34(Pt 1):7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 78.Zhang W, et al. PERK EIF2AK3 control of pancreatic beta cell differentiation and proliferation is required for postnatal glucose homeostasis. Cell Metab. 2006;4(6):491–7. doi: 10.1016/j.cmet.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 79.Guo F, Cavener DR. The GCN2 eIF2alpha kinase regulates fatty-acid homeostasis in the liver during deprivation of an essential amino acid. Cell Metab. 2007;5(2):103–14. doi: 10.1016/j.cmet.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 80.Sheng Z, et al. A genome-wide RNA interference screen reveals an essential CREB3L2-ATF5-MCL1 survival pathway in malignant glioma with therapeutic implications. Nat Med. 2010;16(6):671–7. doi: 10.1038/nm.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ishihara S, Haga H. ATF5: development of oncogenic resistance to radiotherapy. Aging (Albany NY) 2015;7(7):453–4. doi: 10.18632/aging.100775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang SF, et al. Mitochondrial dysfunction enhances cisplatin resistance in human gastric cancer cells via the ROS-activated GCN2-eIF2alpha-ATF4-xCT pathway. Oncotarget. 2016;7(45):74132–74151. doi: 10.18632/oncotarget.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim SH, et al. Human breast cancer cells contain elevated levels and activity of the protein kinase, PKR. Oncogene. 2000;19(27):3086–94. doi: 10.1038/sj.onc.1203632. [DOI] [PubMed] [Google Scholar]
- 84.Kim SH, et al. Neoplastic progression in melanoma and colon cancer is associated with increased expression and activity of the interferon-inducible protein kinase, PKR. Oncogene. 2002;21(57):8741–8. doi: 10.1038/sj.onc.1205987. [DOI] [PubMed] [Google Scholar]
- 85.Ye J, et al. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29(12):2082–96. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bobrovnikova-Marjon E, et al. PERK promotes cancer cell proliferation and tumor growth by limiting oxidative DNA damage. Oncogene. 2010;29(27):3881–95. doi: 10.1038/onc.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nagelkerke A, et al. Hypoxia stimulates migration of breast cancer cells via the PERK/ATF4/LAMP3-arm of the unfolded protein response. Breast Cancer Res. 2013;15(1):R2. doi: 10.1186/bcr3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nagasawa I, et al. BRAF-mutated cells activate GCN2-mediated integrated stress response as a cytoprotective mechanism in response to vemurafenib. Biochem Biophys Res Commun. 2017;482(4):1491–1497. doi: 10.1016/j.bbrc.2016.12.062. [DOI] [PubMed] [Google Scholar]
- 89.Rask K, et al. Increased expression of the transcription factors CCAAT-enhancer binding protein-beta (C/EBBeta) and C/EBzeta (CHOP) correlate with invasiveness of human colorectal cancer. Int J Cancer. 2000;86(3):337–43. doi: 10.1002/(sici)1097-0215(20000501)86:3<337::aid-ijc6>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 90.Greene LA, Lee HY, Angelastro JM. The transcription factor ATF5: role in neurodevelopment and neural tumors. J Neurochem. 2009;108(1):11–22. doi: 10.1111/j.1471-4159.2008.05749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang SZ, et al. Transcription factor ATF5 is required for terminal differentiation and survival of olfactory sensory neurons. Proc Natl Acad Sci U S A. 2012;109(45):18589–94. doi: 10.1073/pnas.1210479109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dalton RP, Lyons DB, Lomvardas S. Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell. 2013;155(2):321–32. doi: 10.1016/j.cell.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen A, et al. ATF5 is overexpressed in epithelial ovarian carcinomas and interference with its function increases apoptosis through the downregulation of Bcl-2 in SKOV-3 cells. Int J Gynecol Pathol. 2012;31(6):532–7. doi: 10.1097/PGP.0b013e31824df26b. [DOI] [PubMed] [Google Scholar]
- 94.Bisikirska B, et al. Elucidation and Pharmacological Targeting of Novel Molecular Drivers of Follicular Lymphoma Progression. Cancer Res. 2016;76(3):664–74. doi: 10.1158/0008-5472.CAN-15-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mittal AK, et al. Molecular basis of aggressive disease in chronic lymphocytic leukemia patients with 11q deletion and trisomy 12 chromosomal abnormalities. Int J Mol Med. 2007;20(4):461–9. [PubMed] [Google Scholar]
- 96.Nukuda A, et al. Role of ATF5 in the invasive potential of diverse human cancer cell lines. Biochem Biophys Res Commun. 2016;474(3):509–14. doi: 10.1016/j.bbrc.2016.04.131. [DOI] [PubMed] [Google Scholar]
- 97.Dluzen D, et al. BCL-2 is a downstream target of ATF5 that mediates the prosurvival function of ATF5 in a cell type-dependent manner. J Biol Chem. 2011;286(9):7705–13. doi: 10.1074/jbc.M110.207639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hu M, et al. Interference with ATF5 function enhances the sensitivity of human pancreatic cancer cells to paclitaxel-induced apoptosis. Anticancer Res. 2012;32(10):4385–94. [PubMed] [Google Scholar]
- 99.Angelastro JM, et al. Selective destruction of glioblastoma cells by interference with the activity or expression of ATF5. Oncogene. 2006;25(6):907–16. doi: 10.1038/sj.onc.1209116. [DOI] [PubMed] [Google Scholar]
- 100.Li G, et al. HSP70 protein promotes survival of C6 and U87 glioma cells by inhibition of ATF5 degradation. J Biol Chem. 2011;286(23):20251–9. doi: 10.1074/jbc.M110.211771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li G, et al. Identification of a novel DNA binding site and a transcriptional target for activating transcription factor 5 in c6 glioma and mcf-7 breast cancer cells. Mol Cancer Res. 2009;7(6):933–43. doi: 10.1158/1541-7786.MCR-08-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sheng Z, et al. BCR-ABL suppresses autophagy through ATF5-mediated regulation of mTOR transcription. Blood. 2011;118(10):2840–8. doi: 10.1182/blood-2010-12-322537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Laeger T, et al. FGF21 is an endocrine signal of protein restriction. J Clin Invest. 2014;124(9):3913–22. doi: 10.1172/JCI74915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhao S, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324(5924):261–5. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Baysal BE, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287(5454):848–51. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- 106.Liu VW, et al. High incidence of somatic mitochondrial DNA mutations in human ovarian carcinomas. Cancer Res. 2001;61(16):5998–6001. [PubMed] [Google Scholar]
- 107.Permuth-Wey J, et al. Inherited variants in mitochondrial biogenesis genes may influence epithelial ovarian cancer risk. Cancer Epidemiol Biomarkers Prev. 2011;20(6):1131–45. doi: 10.1158/1055-9965.EPI-10-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pelicano H, et al. Mitochondrial dysfunction and reactive oxygen species imbalance promote breast cancer cell motility through a CXCL14-mediated mechanism. Cancer Res. 2009;69(6):2375–83. doi: 10.1158/0008-5472.CAN-08-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Linnartz B, Anglmayer R, Zanssen S. Comprehensive scanning of somatic mitochondrial DNA alterations in acute leukemia developing from myelodysplastic syndromes. Cancer Res. 2004;64(6):1966–71. doi: 10.1158/0008-5472.can-03-2956. [DOI] [PubMed] [Google Scholar]
- 110.Yao YG, et al. Mitochondrial DNA sequence variation in single cells from leukemia patients. Blood. 2007;109(2):756–62. doi: 10.1182/blood-2006-01-011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zielke A, et al. Establishment of a highly differentiated thyroid cancer cell line of Hurthle cell origin. Thyroid. 1998;8(6):475–83. doi: 10.1089/thy.1998.8.475. [DOI] [PubMed] [Google Scholar]
- 112.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Goard CA, Schimmer AD. Mitochondrial matrix proteases as novel therapeutic targets in malignancy. Oncogene. 2014;33(21):2690–9. doi: 10.1038/onc.2013.228. [DOI] [PubMed] [Google Scholar]
- 115.Czarnecka AM, et al. Mitochondrial chaperones in cancer: from molecular biology to clinical diagnostics. Cancer Biol Ther. 2006;5(7):714–20. doi: 10.4161/cbt.5.7.2975. [DOI] [PubMed] [Google Scholar]
- 116.Ghosh JC, et al. Hsp60 regulation of tumor cell apoptosis. J Biol Chem. 2008;283(8):5188–94. doi: 10.1074/jbc.M705904200. [DOI] [PubMed] [Google Scholar]
- 117.Ghosh JC, et al. Heat shock protein 60 regulation of the mitochondrial permeability transition pore in tumor cells. Cancer Res. 2010;70(22):8988–93. doi: 10.1158/0008-5472.CAN-10-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Castilla C, et al. Immunohistochemical expression of Hsp60 correlates with tumor progression and hormone resistance in prostate cancer. Urology. 2010;76(4):1017. doi: 10.1016/j.urology.2010.05.045. e1–6. [DOI] [PubMed] [Google Scholar]
- 119.Hamelin C, et al. Identification and verification of heat shock protein 60 as a potential serum marker for colorectal cancer. FEBS J. 2011;278(24):4845–59. doi: 10.1111/j.1742-4658.2011.08385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wadhwa R, et al. Upregulation of mortalin/mthsp70/Grp75 contributes to human carcinogenesis. Int J Cancer. 2006;118(12):2973–80. doi: 10.1002/ijc.21773. [DOI] [PubMed] [Google Scholar]
- 121.Na Y, et al. Stress chaperone mortalin contributes to epithelial-mesenchymal transition and cancer metastasis. Cancer Res. 2016 doi: 10.1158/0008-5472.CAN-15-2704. [DOI] [PubMed] [Google Scholar]
- 122.Ryu J, et al. Identification and functional characterization of nuclear mortalin in human carcinogenesis. J Biol Chem. 2014;289(36):24832–44. doi: 10.1074/jbc.M114.565929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yi X, et al. Association of mortalin (HSPA9) with liver cancer metastasis and prediction for early tumor recurrence. Mol Cell Proteomics. 2008;7(2):315–25. doi: 10.1074/mcp.M700116-MCP200. [DOI] [PubMed] [Google Scholar]
- 124.Gibellini L, et al. Silencing of mitochondrial Lon protease deeply impairs mitochondrial proteome and function in colon cancer cells. FASEB J. 2014;28(12):5122–35. doi: 10.1096/fj.14-255869. [DOI] [PubMed] [Google Scholar]
- 125.Quiros PM, et al. ATP-dependent Lon protease controls tumor bioenergetics by reprogramming mitochondrial activity. Cell Rep. 2014;8(2):542–56. doi: 10.1016/j.celrep.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 126.Di K, et al. Mitochondrial Lon is over-expressed in high-grade gliomas, and mediates hypoxic adaptation: potential role of Lon as a therapeutic target in glioma. Oncotarget. 2016;7(47):77457–77467. doi: 10.18632/oncotarget.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Seo JH, et al. The Mitochondrial Unfoldase-Peptidase Complex ClpXP Controls Bioenergetics Stress and Metastasis. PLoS Biol. 2016;14(7):e1002507. doi: 10.1371/journal.pbio.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pavlides S, et al. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle. 2009;8(23):3984–4001. doi: 10.4161/cc.8.23.10238. [DOI] [PubMed] [Google Scholar]
- 129.Skrtic M, et al. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 2011;20(5):674–88. doi: 10.1016/j.ccr.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sotgia F, et al. Mitochondria “fuel” breast cancer metabolism: fifteen markers of mitochondrial biogenesis label epithelial cancer cells, but are excluded from adjacent stromal cells. Cell Cycle. 2012;11(23):4390–401. doi: 10.4161/cc.22777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Viale A, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514(7524):628–32. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cole A, et al. Inhibition of the Mitochondrial Protease ClpP as a Therapeutic Strategy for Human Acute Myeloid Leukemia. Cancer Cell. 2015;27(6):864–76. doi: 10.1016/j.ccell.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Monaco SE, et al. The transcription factor ATF5 is widely expressed in carcinomas, and interference with its function selectively kills neoplastic, but not nontransformed, breast cell lines. Int J Cancer. 2007;120(9):1883–90. doi: 10.1002/ijc.22469. [DOI] [PubMed] [Google Scholar]
- 134.Karpel-Massler G, et al. A Synthetic Cell-Penetrating Dominant-Negative ATF5 Peptide Exerts Anticancer Activity against a Broad Spectrum of Treatment-Resistant Cancers. Clin Cancer Res. 2016;22(18):4698–711. doi: 10.1158/1078-0432.CCR-15-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kang BH, et al. Regulation of tumor cell mitochondrial homeostasis by an organelle-specific Hsp90 chaperone network. Cell. 2007;131(2):257–70. doi: 10.1016/j.cell.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 136.Kang BH, et al. Combinatorial drug design targeting multiple cancer signaling networks controlled by mitochondrial Hsp90. J Clin Invest. 2009;119(3):454–64. doi: 10.1172/JCI37613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Siegelin MD, et al. Exploiting the mitochondrial unfolded protein response for cancer therapy in mice and human cells. J Clin Invest. 2011;121(4):1349–60. doi: 10.1172/JCI44855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Selvarajah GT, et al. Heat shock protein expression analysis in canine osteosarcoma reveals HSP60 as a potentially relevant therapeutic target. Cell Stress Chaperones. 2013;18(5):607–22. doi: 10.1007/s12192-013-0414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang HM, et al. Obtusilactone A and (−)-sesamin induce apoptosis in human lung cancer cells by inhibiting mitochondrial Lon protease and activating DNA damage checkpoints. Cancer Sci. 2010;101(12):2612–20. doi: 10.1111/j.1349-7006.2010.01701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bernstein SH, et al. The mitochondrial ATP-dependent Lon protease: a novel target in lymphoma death mediated by the synthetic triterpenoid CDDO and its derivatives. Blood. 2012;119(14):3321–9. doi: 10.1182/blood-2011-02-340075. [DOI] [PMC free article] [PubMed] [Google Scholar]