Abstract
Point mutations in the peripheral myelin protein 22 (PMP22) gene have been identified to cause demyelinating Charcot-Marie-Tooth disease (CMT) and hereditary neuropathy with liability to pressure palsy (HNPP). To investigate the mutation spectrum of PMP22 in Han-Chinese population residing in Taiwan, 53 patients with molecularly unassigned demyelinating CMT and 52 patients with HNPP-like neuropathy of unknown genetic causes were screened for PMP22 mutations by Sanger sequencing. Three point mutations were identified in four patients with demyelinating CMT, including c.256 C > T (p.Q86X) in two, and c.310delA (p.I104FfsX7) and c.319 + 1G > A in one each. One PMP22 missense mutation, c.124 T > C (p.C42R), was identified in a patient with HNPP-like neuropathy. The clinical presentations of these mutations vary from mild HNPP-like syndrome to severe infantile-onset demyelinating CMT. In vitro analyses revealed that both PMP22 p.Q86X and p.I104FfsX7 mutations result in truncated PMP22 proteins that are almost totally retained within cytosol, whereas the p.C42R mutation partially impairs cell membrane localization of PMP22 protein. In conclusion, PMP22 point mutations account for 7.5% and 1.9% of demyelinating CMT and HNPP patients with unknown genetic causes, respectively. This study delineates the clinical and molecular features of PMP22 point mutations in Taiwan, and emphasizes their roles in demyelinating CMT or HNPP-like neuropathy.
Introduction
Mutations in the peripheral myelin protein 22 (PMP22) gene are the most common cause of inherited neuropathies, and different types of PMP22 mutations lead to diverse phenotypes. Duplication of a 1.5-Mb DNA segment on chromosome 17p11.2-12 encompassing the PMP22 gene causes Charcot-Marie-Tooth disease type 1 A (CMT1A)1,2, which is an autosomal dominant demyelinating neuropathy and the most common subtype of CMT. The same 1.5-Mb DNA segment triplication causes a more severe demyelinating polyneuropathy3, whereas large deletion at the same segment results in hereditary neuropathy with liability to pressure palsies (HNPP) characterized by episodic, recurrent sensory and motor mono-neuropathies at nerve entrapment sites4,5. In addition, a variety of point mutations in the PMP22 gene has been identified in patients with a broad continuum of inherited neuropathies ranging from HNPP, CMT1E, infantile-onset severe dysmyelinating neuropathies similar to Dejerine-Sottas syndrome, to congenital hypomyelinating neuropathy6–10.
The protein encoded by the PMP22 gene is a glycoprotein of 160 amino acids and constitutes 2–5% of overall peripheral myelin proteins11,12. PMP22 protein forms a predicted structure of four transmembrane domains, two extracellular loops, and cytoplasmic N- and C-terminal tails11,12. The homophilic adhesion between two PMP22 proteins and the heterophilic interaction between PMP22 and myelin protein zero (P0) protein on the opposing membranes of two Schwann cells are essential for the compactness and stability of peripheral myelin13,14. Besides, PMP22 protein also modulates the proliferation and apoptosis of Schwann cells15–18. More than 60 different mutations in the PMP22 gene have been reported to date6; however, a majority of them have not been functionally validated by in vitro analyses and studies about PMP22 point mutations in Chinese populations remain sparse. The aim of present study is to investigate the frequency and spectrum of PMP22 point mutations in cohorts of Taiwanese patients with CMT or HNPP-like neuropathy. The clinical and molecular features of the identified PMP22 mutations were also characterized.
Results
Identification of the PMP22 point mutations
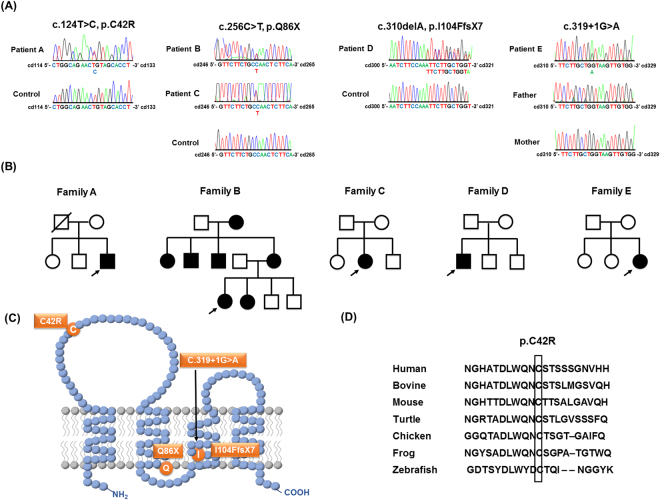
Fifty-three patients of unknown genetic diagnosis were selected from 265 unrelated individuals with demyelinating CMT in whom 175 have PMP22 duplication, 26 have a mutation in the gap junction protein beta 1 (GJB1) gene and 11 have a mutation in the myelin protein zero (MPZ) gene. Another group of 52 molecularly unassigned patients were recruited from 138 index patients with a HNPP-like phenotype, after PMP22 deletion was excluded. Mutational analyses of PMP22 in the 53 patients with demyelinating CMT revealed three point mutations, including c.256 C > T (p.Q86X) in two patients, and c.310delA (p.I104FfsX7) and c.319 + 1G > A in one patient each (Fig. 1A and B). One of the 52 patients with HNPP-like phenotype was found to carry a PMP22 missense mutation, c.124 T > C (p.C42R). All the five patients harboring a PMP22 point mutation are heterozygous for the mutation.
Figure 1.
The PMP22 mutations and the pedigrees harboring the PMP22 mutations in this study. (A) Sanger sequencing traces demonstrating the PMP22 c.124 T > C (p.C42R), c.256 C > T (p.Q86X), c.310delA (p.I104FfsX7) and c.319 + 1G > A mutations. (B) The five pedigrees harboring the PMP22 mutations. Open symbol: unaffected; filled symbol: affected; symbol with a diagonal line: deceased; arrow: proband. (C) Schematic representation of the PMP22 protein structure and the positions of the four PMP22 mutations. (D) The p.C42R mutation resides in an evolutionarily conserved region, as shown by the alignment of multiple PMP22 orthologs from various species.
Among these four mutations, the p.C42R mutation alters the amino acid residue residing in the extracellular loop of PMP22 protein and the other three mutations affect the amino acid sequence of PMP22 transmembrane domains (Fig. 1C). The c.256 C > T (p.Q86X) and c.319 + 1G > A mutations have been recognized to be pathogenic for CMT1 before19,20, whereas the c.310delA (p.I104FfsX7) mutation is novel. The c.124 T > C (p.C42R) mutation was lately reported by a study analyzing patients with inherited neuropathies by a targeted gene panel without clear phenotype information and functional validation21. The pathogenicity of the c.310delA (p.I104FfsX7) is evident by the fact that PMP22 c.312delT (p.I104IfsX7) mutation, of which putative protein product is almost identical to that produced by the c.310delA mutation, has been shown to be a pathogenic mutation for CMT120. The c.124 T > C (p.C42R) was found in an apparently sporadic patient (Fig. 1B), whose families’ sample was unavailable for co-segregation analysis. This mutation is not present in the 141,352 individuals with diverse ethnicities in the genome Aggregation Database (gnomAD) that includes approximately 9300 individuals from East Asian populations. It is also absent in the dbSNP database and 1497 ethnically matched control individuals. The 42th amino acid residue of human PMP22 protein is highly evolutionarily conserved (Fig. 1D). Additionally, the pathogenicity of the c.124 T > C (p.C42R) mutation is supported by in silico analyses using three different programs—Polyphen2, Mutation Taster and CADD. Polyphen2 predicts PMP22 c.124 T > C (p.C42R) to be probably damaging with a score 0.999 and false positive rate 0.0122. Mutation Taster predicts this variant to be disease causing with a very strong probability value, 0.9999, which indicates a high “security” of the prediction23. The CADD v1.2 PHRED score for PMP22 c.124 T > C (p.C42R) is 25.7, which places this variant in the top 0.27% most deleterious variants in the genome24.
Clinical features of patients harboring the PMP22 point mutations
The clinical characteristics of the five patients with PMP22 point mutations are summarized in Table 1. Patient A, who is heterozygous for the PMP22 p.C42R mutation, developed episodic numbness in his palms while holding things since age 20 years. Labored work evoked recurrent, transient numbness and soreness in his legs since age 35 years. Neurological examinations at age 40 years demonstrated diffusely diminished deep tendon reflexes, but there was neither weakness nor atrophy over limb muscles. All modalities of sensation were preserved. The nerve conduction studies (NCS) showed generalized prolonged distal latencies and F-wave latencies of the motor or sensory nerves and mildly decreased motor nerve conduction velocities (Table 2).
Table 1.
Clinical manifestations of the patients with PMP22 point mutations.
| Patient | A | B | C | D | E |
|---|---|---|---|---|---|
| PMP22 mutation | c.124 T > C, p.C42R | c.256 C > T, p.Q86X | c.256 C > T, p.Q86X | c.310delA, p.I104FfsX7 | c.319 + 1G > A |
| Sex | Male | Female | Female | Male | Female |
| Age at onset (y) | 20 | Teenage | 20 | < 1 y | 19 |
| Age at exam (y) | 40 | 28 | 29 | 26 | 20 |
| Clinical diagnosis | HNPP | Demyelinating CMT | Demyelinating CMT | Demyelinating CMT | Demyelinating CMT |
| Inheritance | Apparently sporadic | Autosomal dominant | Apparently sporadic | Apparently sporadic | de novo |
| First symptom | Left hand numbness | Foot drop | Foot drop | Delayed walking | Left hand numbness |
| Muscle strength (MRC scale) | |||||
| Dorsiflexion | 5 | 0 | 0 | 0 | 5 |
| Plantar flexion | 5 | 2 | 2 | 1 | 5 |
| Knee flexion | 5 | 4 | 4 | 4 | 5 |
| Thumb abduction | 5 | 3 | 4 | 3 | 5 |
| Wrist extension | 5 | 4 | 5 | 4 | 5 |
| Muscle atrophy | Nil | Distal UL + LL | Distal UL + LL | Distal UL + LL | Distal LL |
| Knee DTR (Rt/Lt) | +/+ | −/− | −/− | −/− | −/− |
| Ankle DTR (Rt/Lt) | −/+ | −/− | −/− | −/− | −/− |
| Sensory loss | Nil | Distal to ankles | Toes and distal fingers | Distal to ankles and wrists | Nil |
| References | Laššuthová et al.21 | Numakura et al.19 | Numakura et al.19 | This study | Nelis et al.20 |
Abbreviation: HNPP = hereditary neuropathy with liability to pressure palsies; CMT = Charcot-Marie-Tooth disease; MRC = Medical Research Council; LL = lower limbs; UL = upper limbs; DTR = deep tendon reflex; Rt = right; Lt = left.
Table 2.
Nerve conduction studies of patients with PMP22 point mutations.
| Patient | A | B | C | D | E |
|---|---|---|---|---|---|
| PMP22 mutation | c.124 T > C, p.C42R | c.256 C > T, p.Q86X | c.256 C > T, p.Q86X | c.310delA, p.I104FfsX7 | c.319 + 1G > A |
| Median nerve | |||||
| DML (ms) | 8.3 | NR | NR | 18.2 | 10.8 |
| MNCV (m/s) | 42.0 | NR | NR | 4 | 21.6 |
| CMAP (mV) | 10.2 | NR | NR | 0.4 | 4.4 |
| F-wave L (ms) | 36.3 | NR | NR | NR | NR |
| DSL (ms) | 3.9 | NR | NR | NR | NR |
| SNAP (uV) | 10 | NR | NR | NR | NR |
| Ulnar nerve | |||||
| DML (ms) | 5.8 | 18.2 | 22.1 | NR | 7.0 |
| MNCV (m/s) | 48.9 | 5.9 | 7.6 | NR | 20.5 |
| CMAP (mV) | 7.5 | 0.4 | 0.2 | NR | 6.1 |
| F-wave L (ms) | 37.6 | NR | NR | NR | NR |
| DSL (ms) | 3.9 | NR | NR | NR | NR |
| SNAP (uV) | 10 | NR | NR | NR | NR |
| Tibial nerve | |||||
| DML (ms) | 5.4 | NR | NR | NR | NR |
| MNCV (m/s) | 34.9 | NR | NR | NR | NR |
| CMAP (mV) | 11.5 | NR | NR | NR | NR |
| F-wave L (ms) | 61.8 | NR | NR | NR | NR |
| Sural nerve | |||||
| DSL (ms) | 4.5 | NR | NR | NR | NR |
| SNAP (uV) | 12.6 | NR | NR | NR | NR |
Abbreviation:
DML = distal motor latency; MNCV = motor nerve conduction velocity; CMAP = compound motor action potential amplitude; F-wave L = F-wave latency; DSL = distal sensory latency; SNAP = sensory nerve action potential (antidromic); NR = no response.
Normal values:
Median nerve: DML ≤ 4.4 ms; MNCV ≥ 51.9 m/s; CMAP ≥ 6.4 mV; F-wave L ≤ 29 ms; DSL ≤ 3.2 ms; SNAP ≥ 17μV.
Ulnar nerve: DML ≤ 3.5 ms; MNCV ≥ 56.1 m/s; CMAP ≥ 7.0 mV; F-wave L ≤ 29 ms; DSL ≤ 3.0 ms; SNAP ≥ 8 μV.
Tibial nerve: DML ≤ 6.4 ms; MNCV ≥ 42.9 m/s; CMAP ≥ 4.1 mV; F-wave L ≤ 53 ms;
Sural nerve: DSL ≤ 3.5 ms; SNAP ≥ 12 μV.
Patient B, who has the PMP22 p.Q86X mutation, suffered from a slowly progressive weakness and atrophy in the distal limbs since teenage. She had a normal developmental milestone on walking. Her sister, mother and another four of her maternal relatives also had a similar clinical phenotype. Physical examinations at age 28 revealed generalized weakness over four limbs with predominant involvement of the lower limbs and distal limb muscles, severe atrophy of the muscles in the legs and feet, generalized areflexia, and stock and glove pattern of sensory loss in the regions below ankles. Patient C, also carrying the PMP22 p.Q86X mutation, presented with foot drop as the initial manifestation and then developed slowly progressive weakness and atrophy of the distal limb muscles since age 20 years. She had a normal onset of walking and denied any family history of neuromuscular diseases. Neurological examinations at age 29 demonstrated severe atrophy and weakness of the muscles in the legs and feet (score 2/5 and 0/5 on the Medic Research Council scale, respectively), mild weakness of the thigh muscles and intrinsic hand muscles (score 4/5), generalized areflexia, and sensory loss in the toes and distal fingers. For both patients carrying the PMP22 p.Q86X mutation, the NCS revealed a severe demyelinating polyneuropathy with a single digit ulnar motor nerve conduction velocity (Table 2).
Patient D is heterozygous for the PMP22 p.I104FfsX7 mutation. The patient had delayed motor milestones and was not able to walk by himself till 3 years of age. He could never run, jump or walk well. He had difficulties in buttoning and opening a jar at age 7 years and experienced slowly progressive weakness, atrophy and sensory loss in the distal limbs since age 18 years. Neurological examinations at age 26 years revealed hammer toes, severe atrophy and weakness of tibialis anterior, gastrocnemius and foot muscles (score 0–1/5), mild weakness of the thigh muscles (score 4/5), atrophy and weakness of the intrinsic hand muscles (score 3/5), generalized areflexia, and sensory loss in the regions distal to ankles and wrists with a positive Romberg’s test (see Supplementary video and Supplementary Fig. S1). The NCS showed a severe demyelinating polyneuropathy with a median motor nerve conduction velocity of 4 m/s (Table 2). He denied any relevant family history.
Patient E, who harbored a de novo PMP22 c.319 + 1G > A mutation, experienced two episodes of transient numbness over left 4, 5th digits lasting for 2 months each at age 19 years. She could still play tennis well at age 20 years. Physical examinations at age 20 revealed pes cavus, hammer toes, generalized areflexia, mild weakness of bilateral extensor digitorum brevis and no sensory loss. The NCS revealed a demyelinating polyneuropathy with a median motor nerve conduction velocity of 21.6 m/s (Table 2). Her parents were healthy and did not carry the PMP22 mutation.
In vitro analyses of PMP22 expression
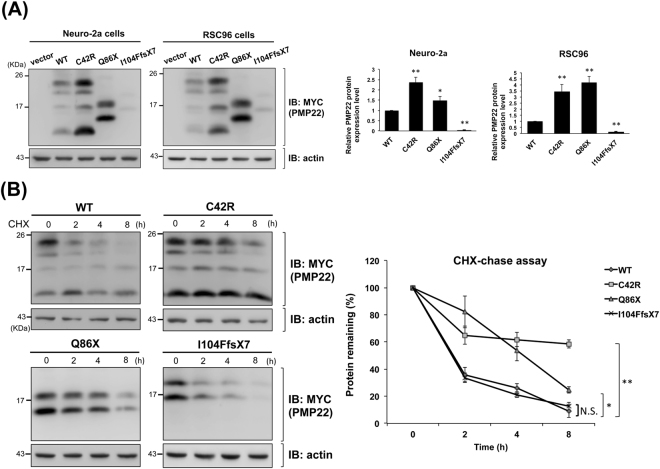
To investigate the molecular consequences of the PMP22 mutations identified in this study, we first cloned the wide-type (WT) PMP22 cDNA into pcDNA3.1/myc-His vector and then introduced the p.C42R, p.I104FfsX7, and p.Q86X mutations into WT PMP22 expression plasmids separately. Mouse neuroblastoma cell line (Neuro-2a) and two kinds of rat neuronal Schwannoma cell lines (RSC96 and RT4-D6P2T) were used in the in vitro functional studies. Cells were transfected with WT plasmids or either one of the mutant constructs to investigate functional consequences of these mutant PMP22 proteins. The expression levels of WT or mutant PMP22 protein at steady-state were measured by Western blotting. The p.Q86X and p.I104FfsX7 mutations resulted in a smaller, truncated protein product. The I104FfsX7 mutant protein was expressed at a significantly lower level than achieved with WT PMP22 protein (Fig. 2A, p < 0.001 for both Neuro-2a cells and RSC96 cells). Interestingly, we also observed substantial protein signal of the I104FfsX7 mutant at the interface between stacking gel and separating gel (see Supplementary Fig. S2), suggesting that the mutant protein got aggregated and thus tended to be stuck at the gel interface. Additionally, in both Neuro-2a and RSC96 cell lines, the steady-state protein levels of C42R and Q86X mutant PMP22 were significantly greater than that of WT PMP22 (Fig. 2A, all p < 0.05). We next evaluated the influence of these mutations on PMP22 degradation by a cycloheximide (CHX)-chase assay. The p.C42R and p.Q86X mutations attenuated PMP22 protein degradation (Fig. 2B and Supplementary Fig. S3), which might be responsible for the higher steady-state levels of these two mutant proteins.
Figure 2.
In vitro characterization of the wide-type (WT), C42R, Q86X, and I104FfsX7 mutant PMP22 proteins. (A) Representative Western blot analysis and densitometric quantification of steady-state PMP22 expression in the Neuro-2a cells and RSC96 cells. Cells were transfected with WT or mutant PMP22 expression plasmids. Actin was used as a loading control. The error bars indicate standard error of the mean (SEM) from 3 independent experiments. The asterisks indicate statistically significant differences (* p < 0.05, ** p < 0.01). (B) Analyses of the stability of the WT and mutant PMP22 proteins. Neuro-2a cells were transfected with WT or mutant PMP22 expression plasmids for 48 hours and then subjected to cycloheximide (CHX)-chase assays. Representive Western blots are shown. All values are shown as means ± SEM (n = 3). The asterisks indicate statistically significant differences (* p < 0.05, ** p < 0.01, NS = not significant).
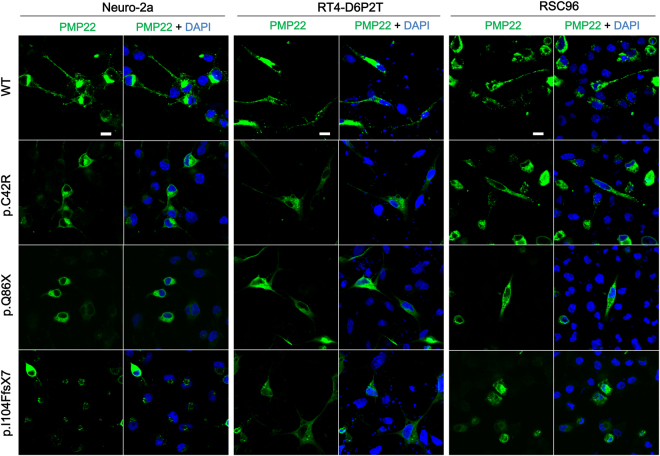
PMP22 is a transmembrane protein and a proper cell membrane localization is critical for PMP22 protein to maintain myelin integrity; therefore, we conducted immunofluorescence analyses to visualize the myc-tagged WT or mutant PMP22 proteins in three cell lines with neuronal background. Cells expressing WT or C42R PMP22 had a clear PMP22-specific staining on the cell membrane, but cells expressing C42R PMP22 had a much greater portion of PMP22-specific staining in the cytoplasm than cell membrane (Fig. 3). For cells expressing Q86X or I104FfsX7 PMP22, the PMP22-specific reticular staining was abundant throughout the cytosol but scarcely present on the cell membrane (Fig. 3). And not surprisingly, numerous small aggregates were found in Neuro-2a cells expressing I104FfsX7 PMP22. Similar results were also found in both RSC96 and RT4-D6P2T cell lines expressing the I104FfsX7 mutant, although the aggregates tended to be smaller and fewer in these two cell lines. These results are consistent with the western blot detection of I104FfsX7 PMP22 aggregates.
Figure 3.
Immunofluorescence analyses of wide-type (WT) and mutant PMP22 proteins in the transfected Neuro-2a, RT4-D6P2T, and RSC96 cells. Confocal fluorescence images of transfected cells labeled with an Alexa Flour 488-conjugated anti-c-Myc antibody for detecting exogenous myc-tagged PMP22 proteins (green). Cells expressing WT or C42R PMP22 had a clear PMP22-specific staining on the cell membrane, but cells expressing C42R PMP22 had a much greater portion of PMP22-specific staining in the cytoplasm than cell membrane. Cells expressing Q86X or I104FfsX7 PMP22 showed an abundant PMP22-specific reticular staining throughout the cytosol but scarcely present on the cell membrane. Cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, blue). Scale bar = 10 um.
Discussion
To understand the contribution of PMP22 point mutations to inherited neuropathy in Taiwan and their clinical and genetic features, we screened patients with demyelinating CMT or HNPP-like phenotype for mutations in the PMP22 gene using Sanger sequencing. We identified four disparate PMP22 point mutations in five unrelated patients with variable clinical presentations from mild HNPP-like syndrome to severe infantile-onset demyelinating CMT. Among these mutations, the p.C42R mutation was associated with a mild HNPP phenotype and partially affected intracellular trafficking of PMP22 protein to cell membrane in in vitro studies. The p.Q86X mutation, which exhibited a more severe phenotype, resulted in a truncated protein product and significantly impaired cell membrane localization of PMP22 protein. The p.Q86X mutation was found in two unrelated patients with similar clinical manifestations, presenting as a young adulthood-onset, progressive and disable polyneuropathy. In addition, the p.I104FfsX7 mutation linked to an infantile-onset severe polyneuropathy led to a truncated PMP22 with a very low expression level, insoluble cytoplasmic aggregate formations, and the failure of cell membrane localization. The c.319 + 1G > A mutation was identified in a 20-year-old lady with a very mild symptom but severe electrophysiological abnormalities. The wide clinical spectrum in the patients with PMP22 point mutations indicates that PMP22 sequencing should be considered in all of the molecularly unassigned patients with inherited demyelinating neuropathy or HNPP-like syndrome, regardless of their clinical severities.
Among the four PMP22 mutations identified in this study, the pathogenicity of the p.Q86X, p.I104FfsX7 and c.319 + 1G > A mutations in CMT have been supported by previous reports19,20. However, the causal role of p.C42R mutation in inherited neuropathies has not been clearly demonstrated yet. The present study further supported the pathogenicity of PMP22 p.C42R mutation by the following evidences. First, this mutation was identified in one of 52 unrelated patients with HNPP-like phenotype, but not in any of the 1,497 Taiwanese control individuals. The significant discrepancy in the prevalence of this mutation between patients and controls sustains the association between the PMP22 p.C42R mutation and HNPP (Fisher’s exact test, p value = 0.034). Second, this mutation is absent in the gnomAD, the large database containing 126,216 exome sequences and 15,136 whole-genome sequences from different ethnic groups. Third, the p.C42R mutation occurs at an evolutionarily conserved amino acid residue of the human PMP22 protein and multiple computational predictive programs support its pathogenic effect. Forth, multiple PMP22 missense mutations have also been reported to cause HNPP. Accordingly, the p.C42R mutation is classified as a likely pathogenic variant based on the guidelines for the interpretation of sequence variants recommended by the American College of Medical Genetics and Genomics and the Association for Molecular Pathology25. PMP22 point mutation is a rare cause of inherited neuropathy. Previous studies showed that PMP22 point mutations account for 0.6–1.4% of total CMT cases in Caucasian populations26–29. According to our findings, the prevalence of PMP22 point mutations in Taiwanese CMT is also within this range. We identified PMP22 point mutations in four out of 53 molecularly unassigned demyelinating CMT patients selected from 265 demyelinating CMT patients or 382 patients with all kinds of CMT, indicating that PMP22 point mutations are responsible for 1.5% (4/265) of demyelinating CMT and 1.0% (4/382) of total CMT in Taiwan. Despite with a low prevalence, the PMP22 point mutations remain to be an important cause of demyelinating CMT without common mutations because they explain for 7.5% (4/53) of such cases.
Only 68–84% patients with a HNPP-like neuropathy are attributed by a 1.5-Mb DNA segment deletion on PMP22 30–32. Frameshift, nonsense, or missense mutations on the PMP22 gene are also well-known causes of HNPP6; however, clinical manifestations and electrophysiological features are not useful to distinguish patients with PMP22 point mutation from those with PMP22 deletion31. We found a PMP22 point mutation in one out of 52 HNPP-like neuropathy patients with unclear causes, whom were chosen from 138 patients with HNPP-like phenotype after excluding PMP22 deletion. Therefore, PMP22 point mutations only accounted for 0.7% (1/138) of overall HNPP-like neuropathy and 1.9% (1/52) of HNPP-like neuropathy with unknown genetic diagnosis in Taiwan. Additional genetic contributions of HNPP-like neuropathy in Han-Chinese population await further investigation. The other possible causes of HNPP-like neuropathy include mutations on the septin 9 (SEPT9) gene that causes hereditary neuralgic amyotrophy33, a heterozygous MPZ p.Y145X mutation34, and a novel genetic locus on chromosome 21q21 implicated in a hereditary recurrent neuropathy35.
The molecular pathogenic mechanism of PMP22 point mutations might differ from those of CMT1A with a PMP22 duplication and HNPP with a PMP22 deletion. The gene dosage effect of PMP22 has been proposed to explain the phenotypic differences between CMT1A and HNPP, based on the facts that PMP22 mRNA and protein levels in the peripheral nerve system are increased in CMT1A but decreased in HNPP. In the cellular transfection studies, we demonstrated that the PMP22 p.Q86X and p.I104FfsX7 mutations, which are separately associated with young adulthood-onset and infantile-onset demyelinating CMT, result in almost total cytoplasmic retention and loss of cell membrane localization of PMP22. These findings are different from the increased PMP22 expression in CMT1A. We also revealed that the HNPP-causing PMP22 p.C42R mutation had a mildly defective intracellular trafficking of PMP22 to cell membrane. Similar to previous studies, our findings affirm that cytoplasmic retention and impaired cell membrane localization of PMP22 are common mechanisms underlying the pathology of CMT1A/HNPP due to PMP22 point mutations36,37.
In conclusion, PMP22 point mutations are uncommon causes of demyelinating CMT and HNPP-like neuropathy. However, it still accounts for 7.5% of molecularly unassigned demyelinating CMT in the absence of PMP22 duplication, GJB1 mutations or MPZ mutations, as well as 1.9% of HNPP-like neuropathy of unknown genetic causes in Taiwan. This study delineates the clinical and molecular features of PMP22 point mutations in Taiwan, expands the spectrum of PMP22 point mutations, and emphasizes its role in demyelinating CMT and HNPP-like neuropathy.
Methods
Patients
Fifty-three individuals with demyelinating CMT of unknown genetic diagnosis were enrolled into this study. These patients were selected from a consecutive series of 382 unrelated patients with CMT at the Neurology Clinics of Taipei Veterans General Hospital, of whom 265 patients have demyelinating polyneuropathy ascertained by standard clinical and electrophysiological evaluations. Median or ulnar nerve motor nerve conduction velocity with a cutoff value of 38 m/s is used to distinguish between demyelinating and axonal CMT38.
Another group of 52 molecularly unassigned patients was recruited from 138 index patients with a HNPP-like phenotype, after PMP22 deletion was excluded by a RT-qPCR-based assay. The HNPP-like phenotype was defined by clinical manifestations compatible with the diagnostic guideline of HNPP regardless of genetic causes39. Sequencing analysis of the PMP22 gene was applied to the 53 selected patients with demyelinating CMT and 52 cases with HNPP-like neuropathy. All the participants are of Han Chinese origin. Peripheral blood samples were collected after written informed consent was obtained from the participants or their parents on behalf of them for those younger than 18 years. This study conformed to the tenets of the Declaration of Helsinki, and all protocols of this study were approved by the Institutional Review Board of Taipei Veterans General Hospital. Written informed consent was obtained from all of the participants. Informed consent for publication of the images/video have been obtained from patient D.
Mutation Analyses
Genomic DNA was extracted from peripheral blood cells using a standard protocol. The coding and flanking sequences of PMP22 were amplified by PCR with intronic primers, and Sanger sequencing was then performed using the Big Dye 3.1 dideoxy terminator method (Applied Biosystems, Foster City, CA) on an ABI Prism 3700 Genetic Analyzer (Applied Biosystems). Amplicon sequences were compared with the reference PMP22 coding genome (GRCh38, NM_000304.3). The pathogenicity of the identified variants was further ascertained by their absence in 500 neurologically healthy individuals of Han-Chinese origin recruited at our hospital and whole genome sequencing data of 997 Taiwanese controls available from Taiwan Biobank database (https://taiwanview.twbiobank.org.tw). The dbSNP databases (Build 149; https://www.ncbi.nlm.nih.gov/snp) and gnomAD (http://gnomad.broadinstitute.org) were also queried for the putative pathogenic variants40. Functional impacts of the PMP22 variants were predicted in silico using PolyPhen-2 (http://genetics.bwh.harvard.edu)22, Mutation Taster (http://www.mutationtaster.org)23, and Combined Annotation Dependent Depletion (CADD) (http://cadd.gs.washington.edu)24. Evolutionary conservation of the mutation sites was analyzed by aligning amino-acid sequences of PMP22 orthologs from multiple species using the UniProt website (http://www.uniprot.org)41.
In vitro analyses of PMP22 expression
Expression plasmids, cell culture and transfection
The WT cDNA clone of human PMP22 (MGC 4588473) was purchased from Invitrogen (Carlsbad, CA). The full-length coding region of PMP22 was cloned into pcDNA3.1/myc-His vector (Invitrogen) to generate the PMP22 expression plasmid, which could produce a C-terminal myc-tagged WT PMP22 protein. To generate the mutant construct expressing myc-tagged Q86X PMP22 protein, the 5′-terminal 255 base pairs fragment of the WT PMP22 coding sequence was subcloned in-frame into pcDNA3.1/myc-His vector. The mutations, c.124 T > C (p.C42R) and c.310delA (p.I104FfsX7), were separately introduced into the WT construct using QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). Subsequently, the first 330 nucleotides of the c.310delA construct was subcloned in-frame into pcDNA3.1/myc-His vector to create the myc-tagged I104FfsX7 PMP22 expression plasmid.
Mouse neuroblastoma cell line Neuro-2a (ATCC® CCL-131TM) were maintained in Eagle’s Minimum Essential Medium (ATCC no.30-2003) containing 10% fetal bovine serum. Two kinds of rat neuronal Schwannoma cell lines, RSC96 (ATCC® CRL-2765TM) and RT4-D6P2T (ATCC® CRL-2768TM), were maintained in Dulbecco’s Modified Eagle’s Medium (ATCC no. 30-2002) containing 10% fetal bovine serum. All cells were cultured in a humidified 5% CO2 incubator at 37 °C. Transient transfections were performed using Lipofectamine 2000 (Invitrogen).
Western blot analyses and cycloheximide (CHX)-chase assays
Neuro-2a cells and RSC96 cells were transfected with WT PMP22 or either one of the mutant PMP22 expression plasmids (C42R, Q86X, or I104FfsX7). Forty-eight hours post-transfection, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with protease inhibitor cocktail (Merck Millipore, Darmstadt, Germany). The protein concentration was determined using a Bradford protein assay kit (Bio-Rad, Hercules, CA), and 50 μg of proteins from each lysate sample were used for Western blotting with c-Myc antibody (Santa Cruz Biotechnology, Dallas, TX). The protein bands were detected using a standard enhanced chemiluminescence method, and the densitometric analyses were performed using NIH ImageJ software.
To determine the stability of WT and mutant PMP22 proteins, CHX-chase assays were conducted with cells transfected with different PMP22 constructs. Twenty-four hours after transfection, cells were trypsinized and re-seeded into 6-well culture plates. After additional 24 hours, CHX (Sigma-Aldrich, St. Louis, MO) was added to a final concentration of 0.1 mg/ml. Cell lysates were harvested at the indicated time points and subjected to Western blotting with the c-Myc antibody. Actin was used as a loading control. The ratios of PMP22 to actin were calculated densitometrically.
Immunofluorescence analyses
Forty-eight hours after transfection with the WT or mutant PMP22 plasmids, Neuro-2a cells, RSC96 cells and RT4-D6P2T cells were fixed with 4% paraformaldehyde and then permeabilized in 0.2% tween-20. After blocking of non-specific binding with 5% bovine serum albumin, the cells were stained for PMP22 using the c-Myc antibody conjugated to Alexa Fluor 488 (Invitrogen) together with 4′,6-diamidino-2-phenylindole (DAPI) counter staining of cell nuclei. Immunofluorescent staining was examined under an Olympus FluoView FV10i confocal laser scanning fluorescence microscopy system with a 60X oil immersion objective (Olympus, Tokyo, Japan).
Electronic supplementary material
Acknowledgements
We would like to thank the patients who participated in this study. This work was supported by the grants from Ministry of Science and Technology, Taiwan (105-2628-B-075-002-MY3, 104-2314-B-075-045-MY4, 103-2314-B-075-076-MY3), Taipei Veterans General Hospital (V105C-027, V105C-118) and the High-throughput Genome Analysis Core Facility of National Core Facility Program for Biotechnology, Taiwan (NSC-101-2319-B-010-001).
Author Contributions
Y.C.Liao and Y.C.Lee conceived the idea and supervised the research. P.C.T. and N.C.C. conducted the experiments. T.S.L., C.T.H., K.P.L. and Y.C.Lee recruited and conducted clinical assessment of patients. Y.C.Liao and Y.C.Lee analyzed the data and drafted the manuscript. All authors read and approved the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14771-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Birouk N, et al. Charcot-Marie-Tooth disease type 1A with 17p11.2 duplication. Clinical and electrophysiological phenotype study and factors influencing disease severity in 119 cases. Brain. 1997;120(Pt 5):813–823. doi: 10.1093/brain/120.5.813. [DOI] [PubMed] [Google Scholar]
- 2.Lupski JR, et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66:219–232. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- 3.Liu P, et al. Mechanism, prevalence, and more severe neuropathy phenotype of the Charcot-Marie-Tooth type 1A triplication. Am. J. Hum. Genet. 2014;94:462–469. doi: 10.1016/j.ajhg.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chance PF, et al. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72:143–151. doi: 10.1016/0092-8674(93)90058-X. [DOI] [PubMed] [Google Scholar]
- 5.Mouton P, et al. Spectrum of clinical and electrophysiologic features in HNPP patients with the 17p11.2 deletion. Neurology. 1999;52:1440–1446. doi: 10.1212/WNL.52.7.1440. [DOI] [PubMed] [Google Scholar]
- 6.Li J, Parker B, Martyn C, Natarajan C, Guo J. The PMP22 gene and its related diseases. Mol. Neurobiol. 2013;47:673–698. doi: 10.1007/s12035-012-8370-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicholson GA, et al. A frame shift mutation in the PMP22 gene in hereditary neuropathy with liability to pressure palsies. Nat. Genet. 1994;6:263–266. doi: 10.1038/ng0394-263. [DOI] [PubMed] [Google Scholar]
- 8.Russo M, et al. Variable phenotypes are associated with PMP22 missense mutations. Neuromuscul. Disord. 2011;21:106–114. doi: 10.1016/j.nmd.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Simonati A, et al. Congenital hypomyelination neuropathy with Ser72Leu substitution in PMP22. Neuromuscul. Disord. 1999;9:257–261. doi: 10.1016/S0960-8966(99)00008-5. [DOI] [PubMed] [Google Scholar]
- 10.Roa BB, Dyck PJ, Marks HG, Chance PF, Lupski JR. Dejerine-Sottas syndrome associated with point mutation in the peripheral myelin protein 22 (PMP22) gene. Nat. Genet. 1993;5:269–273. doi: 10.1038/ng1193-269. [DOI] [PubMed] [Google Scholar]
- 11.D’Urso D, Schmalenbach C, Zoidl G, Prior R, Muller HW. Studies on the effects of altered PMP22 expression during myelination in vitro. J. Neurosci. Res. 1997;48:31–42. doi: 10.1002/(SICI)1097-4547(19970401)48:1<31::AID-JNR3>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 12.Suter U, Snipes GJ. Peripheral myelin protein 22: facts and hypotheses. J. Neurosci. Res. 1995;40:145–151. doi: 10.1002/jnr.490400202. [DOI] [PubMed] [Google Scholar]
- 13.D’Urso D, Ehrhardt P, Muller HW. Peripheral myelin protein 22 and protein zero: a novel association in peripheral nervous system myelin. J. Neurosci. 1999;19:3396–3403. doi: 10.1523/JNEUROSCI.19-09-03396.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasse B, Bosse F, Hanenberg H, Muller HW. Peripheral myelin protein 22 kDa and protein zero: domain specific trans-interactions. Mol. Cell. Neurosci. 2004;27:370–378. doi: 10.1016/j.mcn.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Brancolini C, Edomi P, Marzinotto S, Schneider C. Exposure at the cell surface is required for gas3/PMP22 To regulate both cell death and cell spreading: implication for the Charcot-Marie-Tooth type 1A and Dejerine-Sottas diseases. Mol. Biol. Cell. 2000;11:2901–2914. doi: 10.1091/mbc.11.9.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erdem S, Mendell JR, Sahenk Z. Fate of Schwann cells in CMT1A and HNPP: evidence for apoptosis. J. Neuropathol. Exp. Neurol. 1998;57:635–642. doi: 10.1097/00005072-199806000-00009. [DOI] [PubMed] [Google Scholar]
- 17.Nobbio L, et al. Impairment of PMP22 transgenic Schwann cells differentiation in culture: implications for Charcot-Marie-Tooth type 1A disease. Neurobiol. Dis. 2004;16:263–273. doi: 10.1016/j.nbd.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Suh JG, et al. An in-frame deletion in peripheral myelin protein-22 gene causes hypomyelination and cell death of the Schwann cells in the new Trembler mutant mice. Neuroscience. 1997;79:735–744. doi: 10.1016/S0306-4522(96)00692-6. [DOI] [PubMed] [Google Scholar]
- 19.Nelis E, Timmerman V, De Jonghe P, Van Broeckhoven C. Identification of a 5′ splice site mutation in the PMP-22 gene in autosomal dominant Charcot-Marie-Tooth disease type 1. Hum. Mol. Genet. 1994;3:515–516. doi: 10.1093/hmg/3.3.515. [DOI] [PubMed] [Google Scholar]
- 20.Numakura C, Lin C, Ikegami T, Guldberg P, Hayasaka K. Molecular analysis in Japanese patients with Charcot-Marie-Tooth disease: DGGE analysis for PMP22, MPZ, and Cx32/GJB1 mutations. Hum. Mutat. 2002;20:392–398. doi: 10.1002/humu.10134. [DOI] [PubMed] [Google Scholar]
- 21.Lassuthova P, et al. Improving diagnosis of inherited peripheral neuropathies through gene panel analysis. Orphanet J. Rare Dis. 2016;11:118. doi: 10.1186/s13023-016-0500-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 24.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fridman V, et al. CMT subtypes and disease burden in patients enrolled in the Inherited Neuropathies Consortium natural history study: a cross-sectional analysis. J. Neurol. Neurosurg. Psychiatry. 2015;86:873–878. doi: 10.1136/jnnp-2014-308826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gess B, Schirmacher A, Boentert M, Young P. Charcot-Marie-Tooth disease: frequency of genetic subtypes in a German neuromuscular center population. Neuromuscul. Disord. 2013;23:647–651. doi: 10.1016/j.nmd.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Murphy SM, et al. Charcot-Marie-Tooth disease: frequency of genetic subtypes and guidelines for genetic testing. J. Neurol. Neurosurg. Psychiatry. 2012;83:706–710. doi: 10.1136/jnnp-2012-302451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saporta AS, et al. Charcot-Marie-Tooth disease subtypes and genetic testing strategies. Ann. Neurol. 2011;69:22–33. doi: 10.1002/ana.22166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mariman EC, et al. Evidence for genetic heterogeneity underlying hereditary neuropathy with liability to pressure palsies. Hum. Genet. 1994;93:151–156. doi: 10.1007/BF00210601. [DOI] [PubMed] [Google Scholar]
- 31.Mariman EC, et al. Prevalence of the 1.5-Mb 17p deletion in families with hereditary neuropathy with liability to pressure palsies. Ann. Neurol. 1994;36:650–655. doi: 10.1002/ana.410360415. [DOI] [PubMed] [Google Scholar]
- 32.Nelis E, et al. Estimation of the mutation frequencies in Charcot-Marie-Tooth disease type 1 and hereditary neuropathy with liability to pressure palsies: a European collaborative study. Eur. J. Hum. Genet. 1996;4:25–33. doi: 10.1159/000472166. [DOI] [PubMed] [Google Scholar]
- 33.Kuhlenbaumer G, et al. Mutations in SEPT9 cause hereditary neuralgic amyotrophy. Nat. Genet. 2005;37:1044–1046. doi: 10.1038/ng1649. [DOI] [PubMed] [Google Scholar]
- 34.Magot A, et al. A new MPZ mutation associated with a mild CMT1 phenotype presenting with recurrent nerve compression. Muscle Nerve. 2008;38:1055–1059. doi: 10.1002/mus.21050. [DOI] [PubMed] [Google Scholar]
- 35.Calpena E, et al. A novel locus for a hereditary recurrent neuropathy on chromosome 21q21. Neuromuscul. Disord. 2014;24:660–665. doi: 10.1016/j.nmd.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 36.Naef R, Suter U. Impaired intracellular trafficking is a common disease mechanism of PMP22 point mutations in peripheral neuropathies. Neurobiol. Dis. 1999;6:1–14. doi: 10.1006/nbdi.1998.0227. [DOI] [PubMed] [Google Scholar]
- 37.Shames I, Fraser A, Colby J, Orfali W, Snipes GJ. Phenotypic differences between peripheral myelin protein-22 (PMP22) and myelin protein zero (P0) mutations associated with Charcot-Marie-Tooth-related diseases. J. Neuropathol. Exp. Neurol. 2003;62:751–764. doi: 10.1093/jnen/62.7.751. [DOI] [PubMed] [Google Scholar]
- 38.Harding AE, Thomas PK. Genetic aspects of hereditary motor and sensory neuropathy (types I and II) J. Med. Genet. 1980;17:329–336. doi: 10.1136/jmg.17.5.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dubourg O, Mouton P, Brice A, LeGuern E, Bouche P. Guidelines for diagnosis of hereditary neuropathy with liability to pressure palsies. Neuromuscul. Disord. 2000;10:206–208. doi: 10.1016/S0960-8966(99)00103-0. [DOI] [PubMed] [Google Scholar]
- 40.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.UniProt C. Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42:D191–198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.