Abstract
A full understanding and mediation of nanoparticle–serum protein interactions is key to design nanoparticles with vivid functions within the body, and to solve this problem one needs to differentiate and characterize individual nano-protein conjugates. In this paper, the authors applied capillary electrophoresis combined with inductively coupled plasma mass spectrometry detection to study the behavior of gold nanoparticles of different geometry, size and surface functionalization upon interacting with serum proteins and their mixtures. Due to high-resolution and -sensitivity benefits of this combined technique baseline separations were attained for free nanoparticles (at real-life doses) and different protein conjugates, and the conversion into the protein-bound form was scrutinized in terms of reaction time.
Electronic supplementary material
The online version of this article (doi:10.1007/s10337-017-3387-y) contains supplementary material, which is available to authorized users.
Keywords: Capillary electrophoresis, Mass spectrometry, Gold nanoparticles, Serum proteins
Introduction
Gold nanoparticles (AuNPs) have recently gained much attention due to a wide range of possible biomedical applications, including in vitro and in vivo imaging, drug delivery, photothermal therapy, etc. [1–4]. For therapeutic and diagnostic purposes, AuNPs are mainly administered intravenously and after entering the blood, the nanomaterial surface is instantly coated with plasma proteins. The protein corona provides AuNPs with a biological identity which is different from the original, engineered one and has a consequence for cellular uptake, accumulation, degradation and elimination of nanomaterial from the body [5]. Moreover, protein adsorption onto the nanoparticle surface induces the protein conformational transformations that can influence the biological reactivity of nanomaterial [6, 7]. Therefore, examining the relation between nanomaterial properties (such as the shape, size, type of terminal groups on its surface, etc.) and a tendency to adsorb specific plasma proteins may lead to a better understanding of nanoparticle behavior in vivo. The obtained information can be valuable in terms of developing new theranostic nanomaterials that could reach the desired region in the body without major side effects.
Analytical techniques suitable for characterization of the protein corona can be subdivided into two basic categories: (1) direct methods usually applied to analyze the proteins adsorbed on the nanomaterial surface, e.g., circular dichroism [8] or Fourier transformed infrared spectroscopy [9], and (2) indirect methods with the help of which changes in the nanoparticle properties are scrutinized (e.g. dynamic light scattering [10] or differential centrifugal sedimentation [11]). It should be emphasized that using some indirect techniques, such as fluorescence correlation spectroscopy [12] or nanoparticle tracking and analysis by light scattering [13], makes it possible to examine the protein corona in situ. On the other hand, techniques such as ESI–MS [14–16] and transmission electron microscopy [17] require that the excess of protein is removed prior to or during analysis. Application of the separation-based techniques, such as liquid/nanoliquid chromatography, capillary electrophoresis (CE) and gel electrophoresis, in investigations of the protein-bound nanoparticles has received a great deal of recent research trials [18–32]. Among these methods, CE has been most widely used to explore nano–bio interactions, as follows from a recent literature examination by Aleksenko et al. [33]. In that review, the authors discussed merits and present shortcomings of the CE method in the area, placing special focus on contributions based on using inductively coupled plasma mass spectrometry (ICP-MS) detection. It was concluded that recent advent of ICP-MS made CE a versatile speciation tool for biomedical studies of nanomaterials containing metals. In our original research, we utilized CE–ICP-MS specifically to explore compositional changes of metal-based nanoparticles in human serum environment [34, 35]. However, while the formation of distinct protein conjugates was recorded and characterized both kinetically and thermodynamically, in most of cases they remained unidentified. To cope with this challenge, it was deemed essential to portray the conjugates of the same AuNPs with individual serum proteins, as well as their physiologically-like mixtures, under identical sample preparation/separation conditions. This is the main objective of the present work.
Materials and Methods
Chemicals
Suspensions of spherical AuNPs (sAuNPs; 5, 10, 20, and 50 nm in diameter, functionalized with and stabilized by citric acid) were purchased from British Biocell International (Cardiff, UK) and stored in darkness at 4 °C. Rod-shaped AuNPs (rAuNPs) (6 × 29 nm), functionalized with polyethylene glycol and further modified by introducing amino- or carboxy-groups, were synthesized at the University of Florence. Human serum albumin, transferrin, apo-transferrin, and holo-transferrin, as well as chemicals used to prepare buffer solutions (Na2HPO4, NaH2PO4, 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid or HEPES, piperazine-N,N′-bis(2-ethanesulfonic acid, PIPES), were the products of Sigma-Aldrich (St. Louis, MO, USA).
Instrumentation
Analyses were performed on a HP3DCE system (Agilent Technologies, Waldbronn, Germany) coupled to a 7500a ICP mass spectrometer (Agilent Technologies, Tokyo, Japan) via a microconcentric CEI-100 nebulizer (CETAC, Omaha, NE, USA) and a custom-machined low-dead-volume conical spray chamber. A cross-piece was used to merge the sheath liquid flow. Polyimide-coated fused-silica capillaries (i.d. 75 mm; o.d. 375 mm; length 70 cm) were purchased from CM Scientific Ltd. (Silsden, UK). The197Au, 34S, and 57Fe isotopes were monitored to observe the speciation changes upon binding of nanoparticles with proteins. The signal of 72Ge was recorded to control the stability of CE flow and hyphenation performance as well as the efficiency of nebulization. Instrumental control and data analysis were performed using Agilent ChemStation software. Procedures for capillary initialization and rinsing before and between analyses are described elsewhere [34, 35]. Optimum operational parameters of CE–ICP-MS are presented in Table 1.
Table 1.
CE–ICP-MS operational parameters
| CE system | |
| Capillary | Fused silica capillary, I.D. 75 μm, O.D. 375 μm, length 70 cm |
| Capillary electrolyte | HEPES 40 mM, pH 7.4 (sAuNPs) PIPES 10 mM, pH 7.4 (rAuNPs) |
| Voltage | 15 kV (sAuNPs) 12 or 20 kV (rAuNPs) |
| Temperature | 37 °C |
| Sample injection | Hydrodynamic, 100 mbar × s |
| ICP-MS system | |
| RF power | 1320–1400 W |
| Sample depth | 6.7–7.0 mm |
| Plasma gas | 15.0 L min−1 |
| Nebulizer gas flow | 1.0 L min−1 |
| Monitored isotopes | 197Au, 72Ge, 57Fe, 34S |
Sample Preparation and Analysis
Protein conjugation of AuNPs was performed as described previously [34, 35]. Briefly, the AuNPs diluted in 10 mM phosphate buffer (pH 7.4) containing 100 mM NaCl (final gold concentrations 9.5 and 4.3 or 1.7 mg L−1 for sAuNPs and rAuNPs, respectively) were added to an individual protein solution in simulated physiological buffer (10 mM phosphate buffer, pH 7.4, 100 mM NaCl) and the mixture was incubated at 37 °C for a desired period. Final concentrations of albumin and transferrin were 19 and 3 mg L−1 (sAuNPs samples) or 1.5 and 0.1 g L−1 (rAuNPs samples), respectively. Optimized CE conditions (see Table 1 and Refs. [34, 35] for more detail) ensured satisfactory separation efficiency and quantitative elution of AuNPs and their protein conjugates. Also importantly given the scope of this study, the repeatability of migration times was high enough (≤7% RSD; see ESM Table S-1) for a successful identification of nanoconjugates by this migration parameter. ICP-MS conditions were optimized using a tune-up solution containing 10 µg L−1 Li, Y, Ce, and Bi to obtain the highest signals for yttrium and bismuth and the lowest level of polyatomic, oxide and doubly charged ions.
Results and Discussion
Interaction of Nanoparticles with Albumin
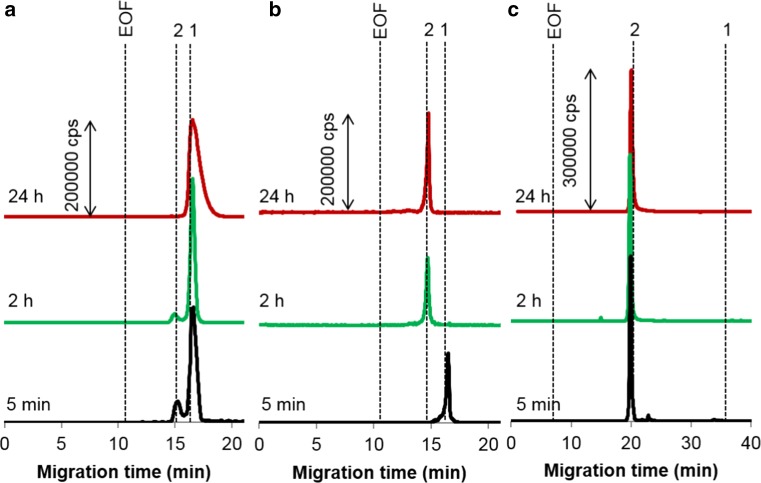
Since the conjugation of sAuNPs with albumin, the most abundant plasma protein, has been in-depth examined earlier [34, 36–38], rAuNPs of different surface functionalization were the target of these binding experiments. As can be expected, the type of terminal groups regulates the rate of the corona formation (cf. traces a and c in Fig. 1), nanoparticles containing amino groups having a higher reactivity toward albumin than carboxyl-modified analogues. In the latter case, albumin molecules initially surround not more than 20% of nanoparticles and upon further incubation the conjugates are underwent decomposition (see also ESM Fig. S-1). Finally, after 24 h, carboxy-rAuNPs return into initial, protein-free form. Increasing the protein-to-metal ratio breaks down this tendency and after 2 h, all rod-shaped nanoparticles are converted in the conjugated form (Fig. 1b). This finding confirms that the applied dose of nanomaterial is a crucial factor governing the trafficking of AuNPs to the targets.
Fig. 1.
197Au electropherograms illustrating interaction between rAuNPs and albumin under simulated extracellular conditions. Particles (molar ratio to the protein): a carboxy-rAuNPs (1:1); b carboxy-rAuNPs (1:2.5); c amino-rAuNPs (1:1). Concentrations: albumin 1.5 g L−1; gold (a) and c 4.3 mg L−1, b 1.7 mg L−1. Peak assignment: 1 free rAuNPs, 2 albumin conjugate, EOF electroosmotic flow. Other conditions, see Table 1
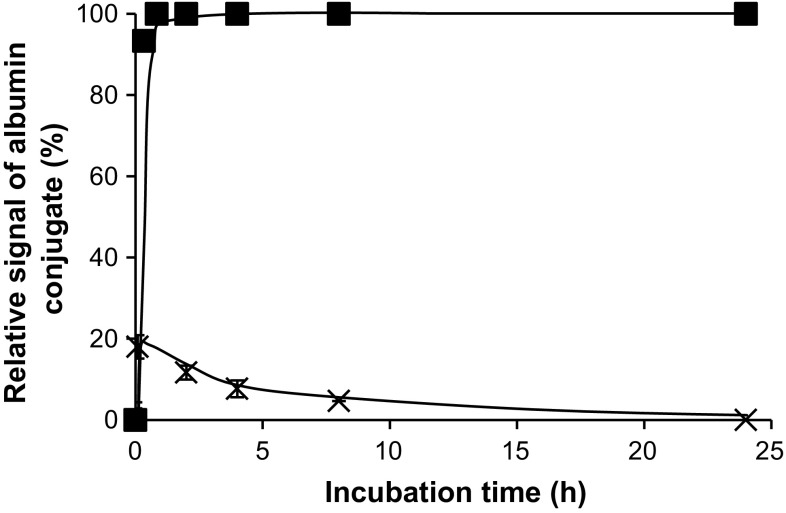
It is important to note that our observations are mostly in accord with the data of Scaletti et al. [39], who found out that the reaction of amino-functionalized rAuNPs with BSA led to greater changes in the intensity of plasmonic extinction band than in case of the carboxylated particles. Using BSA instead of HSA, however, is always an approximation as these proteins are compositionally homogeneous only for 76% [40]. Also interesting was to compare AuNPs of both types of shape at their doses and molar ratios to albumin mimicking circumstances to be encountered in blood (Fig. 2). From this figure, it is obvious that the albumin-binding affinity of sAuNPs is much higher, likely, on the account of more accessible metal centers.
Fig. 2.
Binding of 5-nm sAuNPs (squares) and carboxy-rAuNPs (crosses) to albumin as a function of time. Gold concentration, 9.5 and 4.3 mg L−1, respectively
Interaction of Nanoparticles with Transferrin
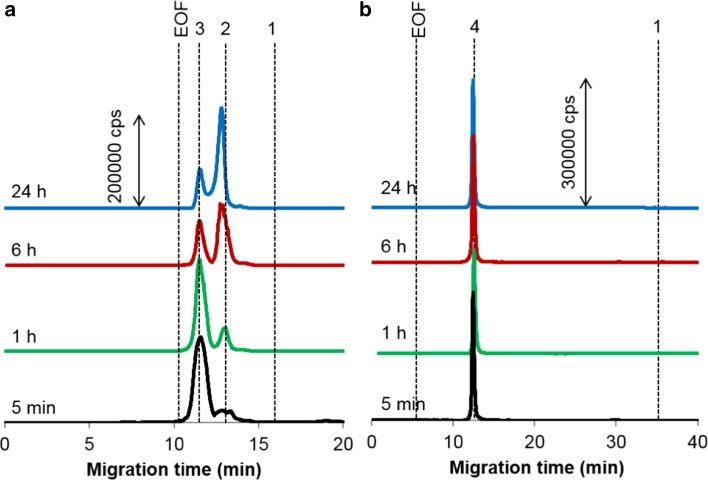
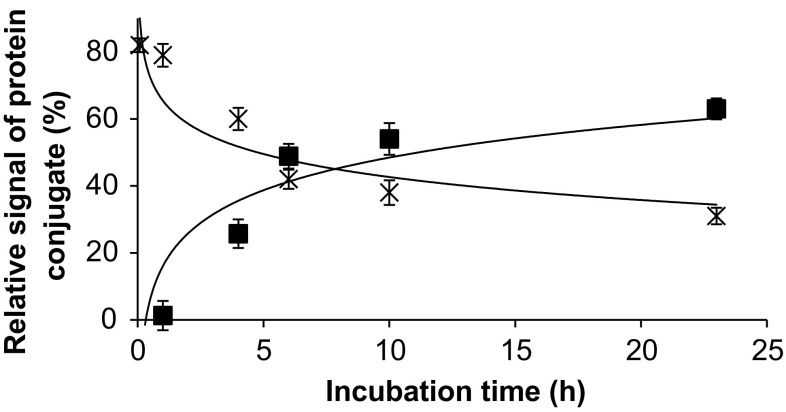
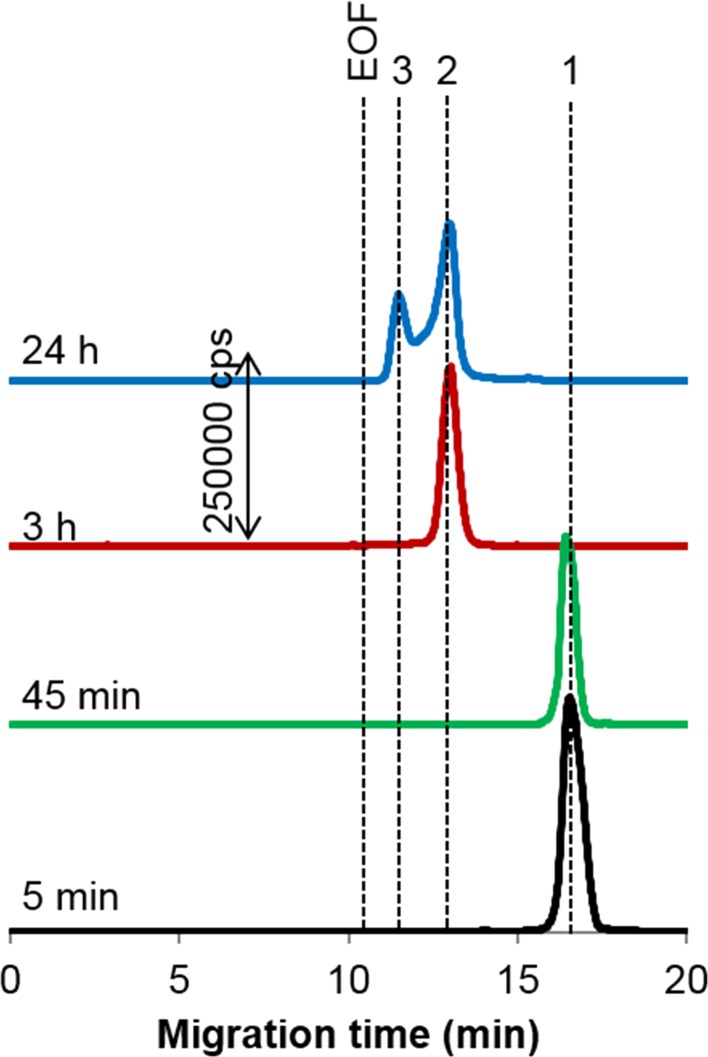
For sAuNPs, binding to transferrin also reaches equilibrium fast (in ca. 5 min), though most of the particles remain uncovered with the protein such as 90% of 50-nm particles [34]. In contrast, rAuNPs became fully conjugated after the same period of incubation (Fig. 3), regardless of the type of functionalization. This implies that for nanoparticles with high affinity to the protein, the nature of terminal groups has lesser influence on the rate of the corona formation. As can also be seen in Fig. 3, for carboxy-rAuNPs, displaying higher electrophoretic mobility than amino-rAuNPs, the signals corresponding to the conjugates of apo- (iron-void form) and holo-transferrin (iron-saturated form) are well resolved. This allowed us to follow the time-dependent changes in relative content of both types of transferrin conjugates, as shown in Fig. 4. Remarkably, after equilibrium is attained (at about 24 h), the shares the transferrin forms take in forming the corona correspond to their physiological ratio, i.e., 70% of apo- and 30% of holo-transferrin (see also below). This observation was verified by analyzing carboxy-rAuNPs incubated for 24 h with a mixture of two protein standards of the same concentration ratio (Fig. S-2; compare to Fig. 3a, upper trace).
Fig. 3.
197Au electropherograms of the conjugates of a carboxy- and b amino-rAuNPs with transferrin. Concentrations: transferrin, 0.1 g L−1; gold, 4.3 mg L−1 after various incubation times. Peaks: 1 free particles, 2 conjugate with apo-transferrin, 3 conjugate with holo-transferrin, 4 conjugate with transferrin. Other conditions, see Table 1
Fig. 4.
Interconversion of carboxy-rAuNPs conjugates with holo- (crosses) and apo-transferrin (squares). Gold concentration, 4.3 mg L−1. For regression parameters, see ESM Table S-2
Interaction with the Physiological Mixture of Albumin and Transferrin
In human blood, the concentration of albumin falls in the range from 35 to 50 g L−1, while the transferrin content is about 15-fold lower (2–4 g L−1) [41, 42]. When sAuNPs were subject to treatment with a mixture of these proteins simulating the physiological situation, only a single signal belonging to the albumin conjugate was recorded, no matter what the particle size was or their dose tested (data not shown). In each case explored, a complete conversion of sAuNPs into the albumin-bound form was faster than 2 min and the protein corona was stable even after 24 h of observation. Quite another binding behavior was characteristic to rAuNPs that demonstrate much slower reactivity toward the proteins under scrutiny. Figure 5 shows no peaks recorded before 2 h, and formed the first was the conjugate of apo-transferrin. Afterward, a two-protein binding scenario was observed, with holo-transferrin contributing at maximum 28% in the equilibrium proteinization. It should be underscored again that the equilibrium state is like that found out when transferrin was the only binding partner or the physiological mixture of two transferrin forms was applied (see above). The fact that albumin does not take part in the speciation changes in the presence of transferrin is quite understandable, considering poor albumin affinity of this sort of nanomaterial (see Fig. 1a).
Fig. 5.
197Au electropherograms showing the proteinization of carboxy-rAuNPs (4.3 mg L−1 Au) in the mixture of human transferrin (0.1 g L−1) and albumin (1.5 g L−1) as a function of the incubation time. Peak assignment as in Fig. 3. Other conditions, see Table 1
Conclusion
In this work, CE equipped with ICP-MS detection was used as an analytical system proved fitting perfectly to carry out metallic nanoparticle bioanalyses under physiological conditions. Alterations of AuNPs in the presence of serum proteins and their mixtures was shown to depend on the nanoparticle geometry and size, surface functionalization and applied dose (nanoparticle-to-protein molar ratio), as well as time for which AuNPs reside in a proteinaceous setting. Perhaps most interestingly, the protein-binding kinetics for differently shaped AuNPs was for the first time compared within a single CE–ICP-MS study. Provided that the results on the characterization of individual nano-protein conjugates would be supported by further studies, using a molecular-type or/and high-resolution ICP-MS detector, the impending task of deciphering the protein-mediated transformations of AuNPs on the way to the target could be completed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Financial support from the National Science Centre, Poland (under Grant number 2015/17/B/ST4/03707), Warsaw university of Technology and the Russian Foundation of Basic Research (Grant 16-03-00492) is gratefully acknowledged. The authors are thankful to Dr. Federica Scaletti and Prof. Luigi Messori, Department of Chemistry, University of Florence, for providing the rod-shaped nanoparticles.
Compliance with Ethical Standards
Funding
This study was funded by the National Science Centre, Poland (Grant number 2015/17/B/ST4/03707), Warsaw University of Technology and the Russian Foundation of Basic Research (Grant number 16-03-00492).
Conflict of Interest
Authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s10337-017-3387-y) contains supplementary material, which is available to authorized users.
An erratum to this article is available at https://doi.org/10.1007/s10337-017-3410-3.
References
- 1.Durr NJ, Larson T, Smith DK, Korgel BA, Sokolov K, Ben-Yakar A. Two-photon luminescence imaging of cancer cells using molecularly targeted gold nanorods. Nano Lett. 2007;7:941–945. doi: 10.1021/nl062962v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keren S, Zavaleta C, Cheng Z, de la Zerda A, Gheysens O, Gambhir SS. Noninvasive molecular imaging of small living subjects using Raman spectroscopy. Proc Natl Acad Sci USA. 2008;105:5844–5849. doi: 10.1073/pnas.0710575105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Visaria RK, Griffi RJ, Williams BW, Ebbini ES, Paciotti GF, Song CW, Bischof JC. Enhancement of tumor thermal therapy using gold nanoparticle-assisted tumor necrosis factor-α delivery. Mol Cancer Ther. 2006;5:1014–1020. doi: 10.1158/1535-7163.MCT-05-0381. [DOI] [PubMed] [Google Scholar]
- 4.Huang X, El-Sayed IH, Qian W, El-Sayed MA. Cancer cell imaging and photothermal therapy in the near-infrared region by using gold nanorods. J Am Chem Soc. 2006;128:2115–2120. doi: 10.1021/ja057254a. [DOI] [PubMed] [Google Scholar]
- 5.Caracciolo G, Farokhzad OC, Mahmoudi M. Biological identity of nanoparticles in vivo: clinical implications of the protein corona. Trends Biotechnol. 2017;35:257–264. doi: 10.1016/j.tibtech.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 6.Walkey CD, Chan WC. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem Soc Rev. 2012;41:2780–2799. doi: 10.1039/C1CS15233E. [DOI] [PubMed] [Google Scholar]
- 7.Saptarshi SR, Duschl A, Lopata AL. Interaction of nanoparticles with proteins: relation to bio-reactivity of the nanoparticle. J Nanobiotechnol. 2013;11:26–38. doi: 10.1186/1477-3155-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S, Peng Z, Leblanc RM. Method to determine protein concentration in the protein–nanoparticle conjugates aqueous solution using circular dichroism spectroscopy. Anal Chem. 2015;87:6455–6459. doi: 10.1021/acs.analchem.5b01451. [DOI] [PubMed] [Google Scholar]
- 9.Wang M, Fu C, Liu X, Lin Z, Yang N, Yu S. Probing the mechanism of plasma protein adsorption on Au and Ag nanoparticles with FT-IR spectroscopy. Nanoscale. 2015;7:15191–15196. doi: 10.1039/C5NR04498G. [DOI] [PubMed] [Google Scholar]
- 10.Fischer K, Schmidt M. Pitfalls and novel applications of particle sizing by dynamic light scattering. Biomaterials. 2016;98:79–91. doi: 10.1016/j.biomaterials.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Maiolo D, Bergese P, Mahon E, Dawson KA, Monopoli MP. Surfactant titration of nanoparticle–protein corona. Anal Chem. 2014;24:12055–12063. doi: 10.1021/ac5027176. [DOI] [PubMed] [Google Scholar]
- 12.Shang L, Nienhaus GU. In situ characterization of protein adsorption onto nanoparticles by fluorescence correlation spectroscopy. Acc Chem Res. 2017;50:387–395. doi: 10.1021/acs.accounts.6b00579. [DOI] [PubMed] [Google Scholar]
- 13.Montes-Burgos I, Walczyk D, Hole P, Smith J, Lynch I, Dawson K. Characterisation of nanoparticle size and state prior to nanotoxicological studies. J Nanopart Res. 2010;12:47–53. doi: 10.1007/s11051-009-9774-z. [DOI] [Google Scholar]
- 14.Capriotti AL, Caracciolo G, Cavaliere C, Colapicchioni V, Piovesana S, Pozzi D, Laganà A. Analytical methods for characterizing the nanoparticle–protein corona. Chromatographia. 2014;77:755–769. doi: 10.1007/s10337-014-2677-x. [DOI] [Google Scholar]
- 15.Capriotti AL, Caracciolo G, Caruso G, Foglia P, Pozzi D, Samperi R, Laganà A. Differential analysis of “protein corona” profile adsorbed onto different nonviral gene delivery systems. Anal Biochem. 2011;419:180–189. doi: 10.1016/j.ab.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Capriotti AL, Caracciolo G, Caruso G, Cavaliere C, Pozzi D, Samperi R, Laganà A. Label-free quantitative analysis for studying the interactions between nanoparticles and plasma proteins. Anal Bioanal Chem. 2013;405:635–645. doi: 10.1007/s00216-011-5691-y. [DOI] [PubMed] [Google Scholar]
- 17.Carrillo-Carrion C, Carril M, Parak WJ. Techniques for the experimental investigation of the protein corona. Curr Opin Biotechnol. 2017;46:106–113. doi: 10.1016/j.copbio.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Caracciolo G, Pozzi D, Capriotti AL, Marianecci C, Carafa M, Marchini C, Montani M, Amici A, Amenitsch H, Digman MA, Gratton E, Sanchez SS, Laganà A. Factors determining the superior performance of lipid/DNA/protammine nanoparticles over lipoplexes. J Med Chem. 2011;54:4160–4171. doi: 10.1021/jm200237p. [DOI] [PubMed] [Google Scholar]
- 19.Bewersdorff T, Vonnemann J, Kanik A, Haag R, Haase A. The influence of surface charge on serum protein interaction and cellular uptake: studies with dendritic polyglycerols and dendritic polyglycerol-coated gold nanoparticles. Int J Nanomed. 2017;12:2001–2019. doi: 10.2147/IJN.S124295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Shang L, Maffre P, Hohmann S, Kirschhöfer F, Brenner-Weiß G, Nienhaus GU. The nature of a hard protein corona forming on quantum dots exposed to human blood serum. Small. 2016;12:5836–5844. doi: 10.1002/smll.201602283. [DOI] [PubMed] [Google Scholar]
- 21.Mirsadeghi S, Dinarvand R, Ghahremani MH, Hormozi-Nezhad MR, Mahmoudi Z, Hajipour MJ, Atyabi F, Ghavamia M, Mahmoudi M. Protein corona composition of gold nanoparticles/nanorods affects amyloid beta fibrillation process. Nanoscale. 2015;7:5004–5013. doi: 10.1039/C4NR06009A. [DOI] [PubMed] [Google Scholar]
- 22.Fernández-Iglesias N, Bettmer J. Complementary mass spectrometric techniques for the quantification of the protein corona: a case study on gold nanoparticles and human serum proteins. Nanoscale. 2015;7:14324–14331. doi: 10.1039/C5NR02625C. [DOI] [PubMed] [Google Scholar]
- 23.Caracciolo G, Palchetti S, Colapicchioni V, Digiacomo L, Pozzi D, Capriotti AL, La Barbera G, Laganà A. Stealth effect of biomolecular corona on nanoparticles uptake by immune cells. Langmuir. 2015;31:10764–10773. doi: 10.1021/acs.langmuir.5b02158. [DOI] [PubMed] [Google Scholar]
- 24.Shim KH, Hulme J, Maeng EH, Kim MK, An SSA. Analysis of SiO2 nanoparticles binding proteins in rat blood and brain homogenate. Int J Nanomed. 2014;9:207–215. doi: 10.2217/nnm.13.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walkey CD, Olsen JB, Song F, Liu R, Guo H, Olsen DWH, Cohen Y, Emili A, Chan WCW. Protein corona fingerprinting predicts the cellular interaction of gold and silver nanoparticles. ACS Nano. 2014;8:2439–2455. doi: 10.1021/nn406018q. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Li J, Chen Y, Teng Y, Wang C, Li J, Liu L, Dong B, Qiu L, Jiang P. Capillary electrophoretic studies on quantum dots and histidine appended peptides self-assembly. Electrophoresis. 2015;36:2419–2424. doi: 10.1002/elps.201500205. [DOI] [PubMed] [Google Scholar]
- 27.Matczuk M, Legat J, Timerbaev AR, Jarosz M. A sensitive and versatile method for characterization of protein-mediated transformations of quantum dots. Analyst. 2016;141:2574–2580. doi: 10.1039/C6AN00276E. [DOI] [PubMed] [Google Scholar]
- 28.Boulos SP, Davis TA, Yang JA, Lohse SE, Alkilany AM, Holland LA, Murphy CJ. Nanoparticle–protein interactions: a thermodynamic and kinetic study of the adsorption of bovine serum albumin to gold nanoparticle surfaces. Langmuir. 2013;29:14984–14996. doi: 10.1021/la402920f. [DOI] [PubMed] [Google Scholar]
- 29.Deng Z, Hu K, Bi L, Yuan H, Chen Y, Zhao S, Du H, Yuan X, Huang Y, Zhang S. Selective removal of IgG from the urine of patients with proteinuria using a polymer coated core–shell magnetic nanoparticle. RSC Adv. 2016;6:107732–107738. doi: 10.1039/C6RA24560A. [DOI] [Google Scholar]
- 30.Wang J, Fan J, Li J, Liu L, Wang J, Jiang P, Liu X, Qiu L. In-capillary probing of quantum dots and fluorescent protein self-assembly and displacement using Förster resonance energy transfer. J Sep Sci. 2017;40:933–939. doi: 10.1002/jssc.201600937. [DOI] [PubMed] [Google Scholar]
- 31.Brambilla D, Verpillot R, Taverna M, De Kimpe L, Le Droumaguet B, Nicolas J, Canovi M, Gobbi M, Mantegazza F, Salmona M, Nicolas V, Scheper W, Couvreur P, Andrieux K. New method based on capillary electrophoresis with laser-induced fluorescence detection (CE–LIF) to monitor interaction between nanoparticles and the amyloid-β peptide. Anal Chem. 2010;82:10083–10089. doi: 10.1021/ac102045x. [DOI] [PubMed] [Google Scholar]
- 32.Qin H, Jiang X, Fan J, Wang J, Liu L, Qiu L, Wang J, Jiang P. Investigation of the weak binding of a tetrahistidine-tagged peptide to quantum dots by using capillary electrophoresis with fluorescence detection. J Sep Sci. 2017;40:567–573. doi: 10.1002/jssc.201601183. [DOI] [PubMed] [Google Scholar]
- 33.Aleksenko SS, Matczuk M, Timerbaev AR. Characterization of interactions of metal-containing nanoparticles with biomolecules by CE: an update (2012–2016) Electrophoresis. 2017;38:1661–1668. doi: 10.1002/elps.201700132. [DOI] [PubMed] [Google Scholar]
- 34.Matczuk M, Anecka K, Scaletti F, Messori L, Keppler BK, Timerbaev AR, Jarosz M. Speciation of metal-based nanomaterials in human serum characterized by capillary electrophoresis coupled to ICP-MS: a case study of gold nanoparticles. Metallomics. 2015;7:1364–1370. doi: 10.1039/C5MT00109A. [DOI] [PubMed] [Google Scholar]
- 35.Matczuk M, Legat J, Scaletti F, Messori L, Timerbaev AR, Jarosz M. The fate of differently functionalized gold nanorods in human serum: a response from CE–ICP-MS. J Chromatogr A. 2017;1499:222–225. doi: 10.1016/j.chroma.2017.03.081. [DOI] [PubMed] [Google Scholar]
- 36.Matczuk M, Legat J, Shtykov SN, Jarosz M, Timerbaev AR. Characterization of the protein corona of gold nanoparticles by an advanced treatment of CE–ICP-MS data. Electrophoresis. 2016;37:2257–2259. doi: 10.1002/elps.201600152. [DOI] [PubMed] [Google Scholar]
- 37.Khan S, Gupta A, Verma NC, Nandi CK. Kinetics of protein adsorption on gold nanoparticle with variable protein structure and nanoparticle size. J Chem Phys. 2015;143:164709. doi: 10.1063/1.4934605. [DOI] [PubMed] [Google Scholar]
- 38.Naveenraj S, Anandan S, Kathiravan A, Renganathan R, Ashokkumar M. The interaction of sonochemically synthesized gold nanoparticles with serum albumins. J Pharm Biomed Anal. 2010;53:804–810. doi: 10.1016/j.jpba.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 39.Scaletti F, Feis A, Centi S, Pini R, Rotello VM, Messori L. Tuning the interactions of PEG-coated gold nanorods with BSA and model proteins through insertion of amino or carboxylate groups. J Inorg Biochem. 2015;150:120–125. doi: 10.1016/j.jinorgbio.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 40.Bujacz A. Structures of bovine, equine and leporine serum albumin. Acta Cryst. 2012;68:1278–1289. doi: 10.1107/S0907444912027047. [DOI] [PubMed] [Google Scholar]
- 41.Anderson NL, Anderson NG. The human plasma proteome. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.R200007-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Sadler PJ, Sun H. Unexpectedly strong binding of a large metal ion (Bi3+) to human serum transferrin. J Biol Chem. 1996;271:9483–9489. doi: 10.1074/jbc.271.16.9483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.