Abstract
Gut microbial communities are key mediators of health and disease and have the capacity to drive the pathogenesis of diverse complex diseases including metabolic and chronic inflammatory diseases as well as aging. Host genetics is also a major determinant of disease phenotypes, whereby two different genomes play a role, the nuclear (nDNA)- and mitochondrial genome (mtDNA). We investigated the impact of mutations in mtDNA on the gut microbiota using conplastic mouse strains exhibiting distinct mutations in their mtDNA on an identical nDNA. Each of three strain tested harbors a distinct gut microbiota, ranging from differences at the phylum- to operational taxonomic units level. The C57BL/6J-mt FVB/NJ strain, carrying a mutation in the mitochondrial ATP8 synthase gene, exhibits higher Firmicutes abundance than Bacteroidetes, indicating a possible indicative for metabolic dysfunctions. In line with this, the C57BL/6J-mt FVB/NJ displays a variety of different phenotypes, including increased susceptibility to metabolic-related and inflammatory disorders. Furthermore, we discuss the cross-talk between mitochondrial genome/mitochondria and commensal microbiota in relation to clinical phenotypes. In summary, we demonstrate that mutations in mtDNA lead to significant differences in the composition of gut microbial communities in mice. Such differences may facilitate the emergence of metabolic disease and therefore constitute potential therapeutic targets.
Introduction
In recent years a pivotal role of the host-associated microbiota in the emergence of chronic diseases has become apparent, largely due to the realization that these communities are much more variable among individuals than previously anticipated1. Chronic diseases, including metabolic diseases such as obesity and type 2 diabetes (T2D)2,3, inflammatory disorders such as inflammatory bowel disease4 as well as aging5 are in most cases assumed to be driven by a multitude of factors, including the host’s individual traits as well as environmental factors. The relative importance of host genetics, both nuclear DNA (nDNA)2 and mitochondrial DNA (mtDNA)6,7, on such conditions is undisputed, although it is conceivable that some host genes may modulate diseases via way of their influence on the microbiota.
Mammalian mtDNA is circular, approximately 16 kilobases in length and encodes 13 protein-coding genes, 22 transfer RNA genes and 2 ribosomal RNA genes. Herein, the proteins encoded are part of the complexes I, III, IV and V of the mitochondrial oxidative phosphorylation (OXPHOS) subunits6,8. Consequently, mutations in the mtDNA impact OXPHOS functions, including adenosine triphosphate (ATP) production, reactive oxygen species (ROS) production and mitochondrial membrane potential. These all contribute to pivotal cellular activities, such as cell activation, proliferation and cell death9.
It is therefore not surprising that mutations in mtDNA can have severe consequences for the entire organism, which is well exemplified by a number of mitochondrially inherited disorders such as Leber’s hereditary optic neuritis (LHON), mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episode (MELAS)6.
Mitochondrial mutations are also associated with chronic diseases including T2DM10, serum lipid levels11, obesity12, cardiovascular diseases13 and neurodegenerative diseases14, as well as autoimmune diseases such as rheumatoid arthritis15 and systemic lupus erythematosus (SLE)16. Given that these same chronic diseases are also associated with shifts in the gut microbiota17–22, it is possible that mtDNA mutations may be linked to such changes.
Ma et al. recently reported that certain mitochondrial haplogroups are associated with the relative abundance of Bacteroides and Prevotella, two prevalent taxa of the gut microbiota23. While differences are apparent according to region-defined haplogroups (Europeans, Asian, and Mexican Americans), non-haplogroup defining single nucleotide polymorphism (SNP) could not be clearly associated to variation in microbial communities. This limitation is in part due to the parallel impact of the highly variable nuclear genome on the composition of the gut microbiota24, making it difficult to reveal the effect of mitochondrial polymorphism.
This obstacle can be circumvented by the use of conplastic mouse strains, which all share the same nuclear genome but differ in their mtDNA. Since common inbred mouse strains demonstrate unique mitochondrial genomes25–27 and the mitochondria are strictly maternally inherited, we systematically generated conplastic strains carrying mostly single mutations in mtDNA with a C57BL/6J nuclear background by repeatedly backcrossing female mitochondrial-donor strains with male C57BL/6J mice over 12 generations27. This unique resource allowed us to investigate the impact of defined mtDNA mutations on gut microbiota composition in the absence of confounding variation in the nuclear genome. For this purpose, we selected two conplastic strains, C57BL/6J-mt FVB/NJ, which carries a mutation (m.7778 G > T) in the mitochondrial ATP synthase 8 gene (mt-Atp8) as a representative of a conplastic strain carrying a single mtDNA mutation, and the C57BL/6J-mt NZB/BlnJ strain, which carries multiple mutations in the mtDNA, and compared the composition of their gut microbial communities to those of the native C57BL/6J strain.
Accordingly, this strategy unveils a number of strain-specific differences in gut bacterial communities with respect to mtDNA mutations. Intriguingly, C57BL/6J-mt FVB/NJ, with their single mutation in the mt-Atp8 gene, display the greatest level of differentiation in their gut microbiota compared to C57BL/6J and C57BL/6J-mt NZB/BlnJ, pointing towards a major role of mitochondrial ATP synthase 8 in shaping the gut microbiota.
Results
High strain-dependent diversity of the gut microbiota diverse in conplastic mouse strains
Mitochondrial mutations impact on gut bacterial communities at both the phylum and family levels
To evaluate the impact of mtDNA mutations on the gut microbiota, we analyzed 48 individuals from three conplastic mouse strains (C57BL/6J, C57BL/6J-mt FVB/NJ and C57BL/6J-mt NZB/BlnJ, N = 16/strain) using 16S rRNA gene amplicon sequencing (V1–V2 region on the Illumina MiSeq). For each individual, 12,500 contigs were randomly selected and clustering sequences into operational taxonomic units (OTUs) yielded 4,853 OTUs after excluding singletons (see Material and methods).
At the level of phylum abundances, the gut microbiota of conplastic mouse strains was similar to previously described studies of C57BL/6J mice, with Bacteroidetes being most abundant, followed by Firmicutes and Proteobacteria (Table 1)28. However, significant strain-dependent differences were also apparent, characterized by C57BL/6J mice differing from C57BL/6J-mt FVB/NJ mice in the abundance of Bacteroidetes, Deferribacteres, Firmicutes, Proteobacteria, and Tenericutes (Mann-Whitney U test, P adj < 0.05) and from C57BL/6J-mt NZB/BlnJ mice in the abundance of Deferribacteres and Tenericutes (P adj < 0.05). The C57BL/6J-mt FVB/NJ and C57BL/6J-mt NZB/BlnJ strains on the other hand did not show any significant differences in phylum abundances. C57BL/6J and C57BL/6J-mt FVB/NJ mice also significantly differed in the ratio of Firmicutes to Bacteroidetes (P = 0.0230, Kruskal-Wallis test; Dunn test, P adj = 0.0111), which was higher in C57BL/6J-mt FVB/NJ mice than in C57BL/6J mice (Fig. 1). This finding is intriguing given the association of Firmicutes to Bacteroidetes ratios being associated with obesity17,29.
Table 1.
Taxonomic analysis at the phylum level in conplastic strains.
| Phylum | C57BL/6J (%) | C57BL/6J-mt FVB/NJ (%) | C57BL/6J-mt NZB/BlnJ (%) |
|---|---|---|---|
| Actinobacteria | 0.09 | 0.08 | 0.16 |
| Bacteroidetes | 80.86 | 66.44* | 70.51 |
| Cyanobacteria | 0.22 | 0.26 | 0.44 |
| Deferribacteres | 0.05 | 0.47* | 0.21# |
| Firmicutes | 13.11 | 22.20* | 20.43 |
| Proteobacteria | 5.32 | 9.10* | 7.81 |
| Tenericutes | 0.03 | 0.27* | 0.18# |
| Unclassified | 0.19 | 0.35 | 0.21 |
| Verrucomicrobia | 0.13 | 0.83 | 0.04 |
*P adj < 0.05 (compared with C57BL/6J); # P adj < 0.05 (compared with C57BL/6J).
Figure 1.
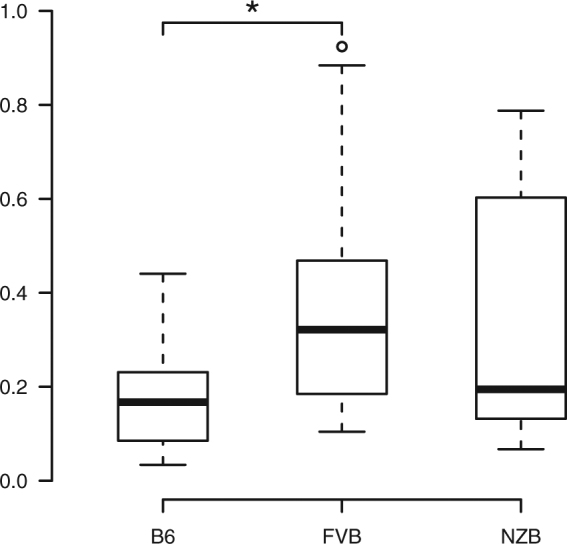
The ratio of Firmicutes and Bacteroidetes significantly differs between conplastic strains. *P = 0.0230 (Kruskal-Wallis test), *P < 0.05 (Dunn test). B6, C57BL/6J; FVB, C57BL/6J-mt FVB/NJ; NZB, C57BL/6J-mt NZB/BlnJ.
Next, we compared the gut microbiota of the conplastic strains at the family level. Bacteroidales S24-7 group, Lachnospiraceae, and Helicobacteraceae were in general the most abundant families (Supplementary Table S1), and there were significant differences in the abundance between the individual conplastic strains. C57BL/6J mice significantly differed from C57BL/6J-mt FVB/NJ mice in the abundance of Anaeroplasmataceae, Bacteroidales S24-7 group, Deferribacteraceae, Desulfovibrionaceae and Helicobacteraceae (MWU-test, P adj < 0.05) and from C57BL/6J-mt NZB/BlnJ mice in Anaeroplasmataceae (P adj = 0.0468) (Supplementary Table S1). C57BL/6J-mt FVB/NJ and C57BL/6J-mt NZB/BlnJ mice showed significant differences in the abundance of Christensenellaceae, Desulfovibrionaceae, and Family XIII (P adj < 0.05).
The impact of mtDNA mutations on alpha diversity and beta diversity
To characterize the level of diversity within strains, alpha diversity was analyzed by applying Mann-Whitney U tests for the Chao1, Shannon and Simpson index. None of the three strains significantly differ in alpha diversity (P adj > 0.05; Mann-Whitney U tests).
Next, to characterize diversity between strains, beta diversity was analyzed using the unweighted UniFrac distance metric, revealing significant differences between strains (Kruskal-Wallis test, P = 0.0252). Applying a post-hoc test identified significant differences in beta diversity between C57BL/6J and C57BL/6J-mt FVB/NJ as well as between C57BL/6J-mt FVB/NJ and C57BL/6J-mt NZB/BlnJ mice (Dunn test, P = 0.0144 and P = 0.0276, respectively). To explore the distance between the communities and the effect of sex on differences in beta diversity, we applied the multivariate analysis of variance (adonis) on the UniFrac distance. Using this approach, we discovered a significant difference in beta diversity between strains (adonis: P = 0.001, R2 = 0.1168) and sexes (P = 0.001, R2 = 0.0376). We also verified an interaction between strain and sex (P = 0.002, R2 = 0.0614).
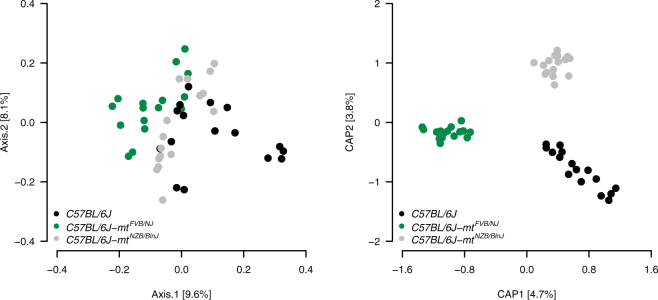
To further detail the differences in the microbiota of the three strains, we conducted a principle coordinate analysis (PCoA). The microbiota of the strains is distinguishable along the first principle coordinate, which explains 9.6% of the variance (Fig. 2A). To investigate this pattern further, we applied a constrained analysis of principal coordinates on the data with strain as an explanatory variable with a correction for sex effects, which reveals a highly significant effect (ANOVA: P = 0.001, F2,44 = 2.030) with R2 = 0.0819 (R2 adj = 0.0425) (Fig. 2B).
Figure 2.
Beta diversity of gut microbiota in conplastic strains. (A) Principal coordinate analysis (PCoA) on unweighted UniFrac distance. (B) Capscale plot on unweighted UniFrac distance. Strain was taken as constraint, and correction for sex effect was applied. The CAP1 and CAP2 were found to be significant with respect to strain with 999 permutations (P = 0.001 for CAP1 and CAP2).
Specific gut microbiota are associated with conplastic mouse strains
To identify bacterial species-level OTUs that are significantly associated with a specific cohort, we applied indicator species analysis using the method of Dufrêne and Legendre30. This revealed four OTUs associated with C57BL/6J, 14 with C57BL/6J-mt FVB/NJ and two with the C57BL/6J-mt NZB/BlnJ strain (Supplementary Table S2).
Basal phenotypes of conplastic strains carrying a mutation in mt-Atp8
Given the marked differences of microbiota between C57BL/6J-mt FVB/NJ and the two other mouse strains, we next characterized C57BL/6J-mt FVB/NJ mice, which carry a mutation in the mitochondrially-encoded ATP synthase 8 gene (mt-Atp8), with respect to their general and cellular metabolic phenotypes.
Using a separate cohort of mice than those included in the gut microbiota analysis, we first weighed C57BL/6J-mt FVB/NJ and C57BL/6J mice at 3, 6, 12, 18 and 24 months of age. In males of both strains body weight increased with age until 18 months and declined without any significant difference between the two strains (Supplementary Fig. S1,A). In contrast, in females body weight increased with time throughout the observation period of 24 months. While there was no difference between the two strains at younger ages, starting with 12 months C57BL/6J-mt FVB/NJ females were significantly lighter than C57BL/6J females (two-way ANOVA, P = 0.0279, P = 0.0284, and P < 0.0001, respectively, Supplementary Fig. S1,B).
By indirect calorimetric cage system, we assessed the basal metabolism of C57BL/6J-mt FVB/NJ and C57BL/6J mice at the age of 3 months. While the two strains displayed an equal respiratory exchange ratio (RER), energy expenditure and food and water intake, C57BL/6J-mt FVB/NJ mice were significantly less active than C57BL/6J mice, indicating an enhanced metabolic base level rate in the C57BL/6J-mt FVB/NJ strain (P = 0.0331, t test, Supplementary Fig. S1,C–G).
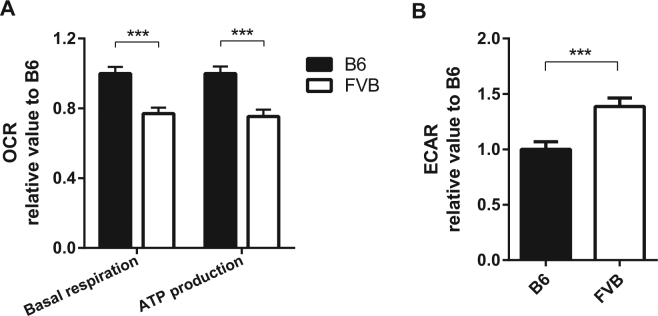
To evaluate the mitochondrially-related metabolic status, we measured the oxygen consumption and glycolysis levels in primary lymphocytes of C57BL/6J-mt FVB/NJ and C57BL/6J mice in a cell metabolism analyzer (Seahorse XF analyzer). C57BL/6J-mt FVB/NJ lymphocytes consumed significantly less oxygen than their C57BL/6J counterparts (Fig. 3A), but instead displayed higher levels of glycolysis (P = 0.0016, Fig. 3B). These data indicate that the mutation in mt-Atp8 impairs mitochondrial oxidative phosphorylation and consequently reduces ATP production, which is in turn compensated by an enhanced level of glycolysis.
Figure 3.
Reduced oxygen consumption levels and Increased levels of glycolysis in primary lymphocytes isolated from C57BL/6J-mt FVB/NJ compared with cells from C57BL/6J. (A) The oxygen consumption ratio (OCR) was measured in the primary lymphocytes from peripheral lymph nodes. Basal respiration and ATP production levels were significantly less in C57BL/6J-mt FVB/NJ compared to C57BL/6J. (B) The extracellular acidification rate (ECAR), which indicates glycolysis levels, in the primary lymphocytes was measured using a Seahorse XF analyzer. Data presented in the graph was normalized with the value of C57BL/6J. B6: C57BL/6J, FVB: C57BL/6J-mt FVB/NJ. ***P < 0.001, t test.
Notably, there was no difference in the overall lifespan of C57BL/6J-mt FVB/NJ and C57BL/6J mice under normal housing condition (Supplementary Fig. S2,A–C, and Supplementary Table S3).
Discussion
In this study, we provide the first direct evidence that mutations in the mitochondrial genes significantly impact the composition of the gut microbial community in mice. Among several mitochondrial genes participating in this regulation, the mt-Atp8 appears to be of particular importance, as the mutation (a single mutation, m.7778 G > T) in this gene alone leads to notable differences in gut microbial composition compared to C57BL/6J mice.
The link between mitochondria and commensal bacterial composition, particularly in the gut, is increasingly receiving attention31,32. Mitochondria are the cellular organelles that function as metabolic centers, controlling energy production by OXPHOS respiration, glycolysis and lipid metabolism. In the host cell metabolism, mitochondria utilize metabolites produced by the intestinal commensal bacteria, such as acetyl coenzyme A (acetyl-CoA) and fuel mitochondria to produce cellular energy in the form of ATP9. In addition to cellular bioenergetic functioning, mitochondria generate ROS during OXPHOS respiration, which plays a role in oxidative stress to damage cells, apoptosis and inflammatory signal transduction, in addition to being bactericidal33,34. Thus, mitochondria appear to be capable of influencing gut microbiota composition in multiple ways.
The communication of gut microbiota to mitochondria can be outlined as follows: Anaerobic bacteria ferment undigested dietary components such as dietary fibers and produce gases and organic acids, especially three short chain fatty acids (SCFAs): acetate, propionate and butyrate35. In addition, secondary bile acids such as lithocholic acids and deoxycholic acids are also produced by bacteria residing in the gut36. Regarding SCFAs, butyrate and propionate are known to inhibit the activity of histone deacetylases (HDACs) in colon cells and immune cells, particularly regulatory T cells37,38, accordingly increasing the hyperacetylation of histones, followed by controlling gene transcription involved in signal transduction such as FOXP3, NFκB, SIRT1 and PPARγ 35, which influence the inflammatory response and mitochondrial functioning. Acetate as well as butyrate are converted into acetyl-CoA and tricarboxylic acid (TCA) cycle intermediates in mitochondria, which are used in various bioenergetics processes39. Secondary bile acids interact with mitochondria by modulating transcription factors that relate to lipid and carbohydrate metabolism40, thus, these microbial products act as signaling molecules that modulate mitochondrial bioenergetic functioning. Recently, it was found that a shift in intestinal microbiota correlated with expression of a panel of mitochondrial proteins in Crohn´s disease patients, indicating that cross-talk between the microbiota and host mitochondria may be an important factor in disease pathology41.
Bacterial taxa whose abundance varies according to the conplastic strains investigated represent candidates for involvement in mitochondrial bioenergetic functions. At the phylum level, the most prominent differences in taxon abundance relative to C57BL/6J were observed in C57BL/6J-mt FVB/NJ, whereby a total of four phyla including the two major phyla Bacteroidetes and Firmicutes displayed significant differences. Moreover, the ratio of Firmicutes to Bacteroidetes, which reportedly correlates with metabolic phenotypes such as obesity17,29, was significantly increased in C57BL/6J-mt FVB/NJ. Thus, below we discuss taxon abundance differences with respect to C57BL/6J-mt FVB/NJ in particular.
Bacteroidetes and Firmicutes are the two most dominant bacterial phyla of the gut microbiota35. In line with analysis at the phylum level, the abundance of Bacteroidales S27-4 subgroup (belonging to Bacteroidetes) is significantly reduced in C57BL/6J-mt FVB/NJ. This family was previously shown to fluctuate in abundance in several mouse studies, including increased abundance in mice fed with a high-fat diet42, and mice in remission of colitis after treatment43. Thus, alterations in abundance of Bacteroidales S27-4 may reflect the metabolic status of the host. Further, OTUs belonging to Alistipes are among the indicator species of C57BL6-mt FVB/NJ, and reported to be increased in high-fat diet fed mice42. Alistipes abundance negatively correlates with D-pinitol, which is converted to myo-inositol and further processed to acetyl-CoA44, a key metabolite to fuel mitochondrial metabolism.
Other taxa associated to C57BL6-mt FVB/NJ belong to the Firmicutes, such as Lachnospiraceae spp. Members of this family are generally thought to exert anti-inflammatory effects through the biosynthesis of butyrate, whereas others have also been associated with increased disease susceptibility, including to lupus45 and diabetes46. These contrasting aspects may be due to differences in SCFA production among individual members of Lachnospiraceae 47. For example, some species convert lactate to propionate via the acrylate pathway48. Propionate is thought to regulate lipid synthesis in the liver49. Given that C57BL/6J- mtFVB/NJ is susceptible to many metabolic related disorders, it is conceivable that these bacteria may impact metabolic phenotypes in this strain.
Interestingly, Desulfovibrionaceae (Proteobacteria) are among the taxa exhibiting increased abundance in C57BL/6J-mt FVB/NJ. This family is of particular interest due to their capacity to produce hydrogen sulfide (H2S)50. Sulfate-reducing bacteria-derived H2S reportedly contribute to T cell activation and proliferation51, suggesting a contribution of Desulfobibrionaeceae to inflammatory phenotypes. Nevertheless, both pro-inflammatory and anti-inflammatory effects of sulfate-reducing bacteria have been reported52. Importantly, however, it was also reported that H2S regulates mitochondrial bioenergetic functioning, particularly on mitochondrial complex IV53.
Our phenotyping of C57BL/6J-mt FVB/NJ mice demonstrates a skewed cellular metabolism, i.e. reduction in OXPHOS respiration and increased glycolysis, which could alter cellular energy levels in the gut as well as in immune cells. In addition, it could lead to differences in metabolite profiles, including SCFA, which we have previously reported54. Changes in the host metabolic profile could accordingly impact other aspects of bacterial community structure in the gut, leading to further imbalances and potential disease susceptibility. On the other hand, C57BL/6J-mt FVB/NJ mice do not show obvious disease phenotypes in the absence of metabolic stress, and the lifespan of this strain was similar to that of C57BL/6J. The ratio of Firmicutes to Bacteroidetes was higher in C57BL/6J-mt FVB/NJ compared with C57BL/6J, indicating a potential association to obesity in C57BL/6J-mt FVB/NJ. Body weight of this strain shows no trends towards obesity under normal chow, which suggests a predisposition only under metabolic stress. Our findings indicate that the phenotypic differences observed in this strain in the absence of metabolic stress (i.e. increased glycolysis, less activity, dysbiosis in the gut) are tolerated without major health consequences until death. In contrast, metabolic stimulus such as dietary modification could exceed certain thresholds, resulting in more severe disease phenotypes. In line with this hypothesis, we previously reported that C57BL/6J-mt FVB/NJ mice are more susceptible to experimental metabolic diseases, such as insulin resistance55, diet-induced obesity and non-alcoholic steatohepatitis (NASH)54. Furthermore, another conplastic strain carrying the same mt-Atp8 mutation displays increased susceptibility to chronic inflammatory diseases, e.g. collagen-induced arthritis and lupus nephritis56 (Supplementary Table S4). Our findings offer a potential mechanistic explanation for these previously reported phenotypes: we hypothesize that the mt-Apt8 mutation primarily causes a shift in metabolism in the host, which results into the shift in the microbiota and thereby the metabolites in the gut. Alternatively, the shift in the microbiota could be an independent phenomenon of the mutation, which may contribute to the host’s metabolic disequilibrium.
In summary, our data highlight evidence that mutations in mtDNA influence the composition of gut microbiota, primarily by modulating mitochondrial functioning. The mt-Atp8 mutation in C57BL/6J-mt FVB/NJ mice significantly modulates key cellular metabolic processes, including the level of OXPHOS respiration and glycolysis, consequently changing the metabolite composition of host cells, which appears to lead to a shift of the microbial composition in the gut. The gut microbiota in turn has the potential to regulate gene expression involved in signaling, e.g. immune responses, which may further result in increased susceptibility to common diseases in the context of additional triggers (e.g. immunological or metabolical stimulus) in C57BL6J-mt FVB/NJ.
As shown in this study, the crosstalk between mtDNA/mitochondria and microbiota should be considered as a critical factor to elucidate the pathology and potential novel therapeutic options for common diseases.
Methods
Mice
C57BL/6J mice were obtained from Jackson Laboratory (Bar Harbor, USA) and bred in the animal facility of the University of Lübeck. The conplastic strains C57BL/6J-mt FVB/NJ and C57BL/6J-mt NZB/BlnJ were generated as described previously27. Conplastic strains were maintained by repeated backcrossing female conplastic offspring with male C57BL/6J mice, which were randomly selected from the C57BL/6J colony maintained in the same breeding facility room. With this breeding strategy there was ample opportunity for microbiota to be homogenized between the strains, i.e. genetic factors are not confounded by separate housing.
The mutations in the mtDNA of each conplastic strains are listed in Supplementary Table S5. Fresh fecal samples were collected from mice at three months of age from nine females and seven males per strain.
Animal use and all methods were approved by the Animal Care and Use Committee (V242-7224. 122-5, Kiel, Germany), and were performed in accordance with the relevant guidelines and regulations by certified personnel.
DNA extraction and 16S rRNA gene sequencing
DNA was extracted from fecal samples with the PowerSoil Kit (MoBio, Carlsbad, CA) following the manufacturer’s protocol. The 16S rRNA gene was amplified using uniquely barcoded primers flanking the V1 and V2 hypervariable regions (27F-338R) with fused MiSeq adapters in a 25 µl PCR as previously described28. The reaction mix consist of one µL of the template DNA, four µL of each forward and reverse primer (0.28 µM), 0.25 µL of Phusion Hot Start II DNA polymerase (0.5 units), 0.5 µL dNTPs (200µM each) and five µL of HF buffer. PCR amplification was performed with the condition described in Supplementary Table S6. The concentration of the PCR products was estimated on 1.5% agarose gels using the image software Quantum Capt v16.04 (Vilber Lourmat Deutschland GmbH, Eberhardzell, Germany) with a known concentration of DNA marker as the internal standard for band intensity measurement. The samples of individual gels were pooled into approximately equimolar subpools, as indicated by band intensity. The concentration of subpools was measured with the Quibit dsDNA BR Assay Kit (Life Technologies GmbH, Darmstadt, Germany), followed by purification with Agencourt AMPure Beads (Beckman Coulter GmbH, Krefeld, Germany) and quantification of library size and concentration using a Bioanalyzer (Agilent Technologies, Waldbronn, Germany). Subpools were mixed in equimolar amounts and stored at −20 °C until sequencing. Sequencing was performed on the Illumina MiSeq platform with v3 chemistry.
MiSeq data analysis
CASAVA version 1.8.2 was used to demultipex raw sequencing reads. Afterwards, reads were merged using fastq_mergepairs from VSEARCH version 1.9.957 with a maximum number of mismatches of 12 and the merged reads (contigs) between 270 and 330 bp long. Next, a quality filter step was applied as implemented in the fasta_filter command from the USEARCH package version 8.1.186158 with the expected numbers of errors set to 0.5. Chimeras were identified by uchime_ref 59 (USEARCH) with the RDP Gold database version 9 as a reference database and were removed from the contig set.
Taxonomic classification
Taxonomy was assigned to the genus level for all non-chimeric sequences using Mothur version 1.36.160 and the SILVA database version 12361 with 80% bootstrap support (1,000 iterations). Sequences of non-bacterial origin were removed from the data set and remaining sequences were aligned to the 16S rRNA V1-V2 region against the SILVA database. Next, 12,500 sequences per individual were chosen randomly to normalize for differences in read number between individuals. Operational taxonomic units (OTUs) were assigned at a 97% similarity threshold and the consensus sequence for each OTU was chosen using a distance-based method. Finally, we used FastTree version 2.1.462 to construct a phylogenetic tree with a generalized time-reversible (GTR) substitution model and the gamma option to rescale branch length. The resulting tree was rooted using the midpoint method for rooting. OTUs present in only one individual with one contig (singletons) were removed from the data prior to further analyses.
Alpha and beta diversity analysis
Three alpha diversity indices (Chao1, Shannon, Simpson) were estimated using the vegan package version 2.3–463. Unweighted UniFrac distance (beta diversity) was estimated using the phyloseq package version 1.14.064. To investigate differences in beta diversity, non-parametric analysis of variance was used as implemented in the adonis command (vegan package). To compute significance values for each factor, 999 permutations were run. Principle coordinate analysis was performed using the ordinate command as implemented in the phyloseq package. Constrained analysis of principle coordinates was carried out using capscale (vegan package) and significance of constraints was assessed with 999 permutations for each factor and for each cap-axis using an anova-like permutation test65.
Indicator species analysis
Indicator species analysis was performed as previously described66 using indicspecies version 1.7.5 with the specific site group function IndVal.g and 10,000 permutations to assess significance values. OTUs with an adjusted p-value (Benjamini-Hochberg correction) below 0.05 were considered as significant indicator species.
Statistical analyses
Unless stated otherwise, all statistical analyses were performed using R Version 3.2.467. When necessary, correction for multiple testing was applied using the Benjamini-Hochberg method68 as implemented in the p.adjust command (stats package for R). Throughout the manuscript Padj denotes corrected p-values, whereas P denotes uncorrected p-values. To test for differences between all three strains, Kruskal-Wallis tests were performed and Dunn tests were applied as post-hoc tests with Benjamini-Hochberg correction for multiple testing.
Phenotyping of C57BL/6J and C57BL/6J-mtFVB/NJ mice
See supplementary materials and methods.
Data availability
Sequencing data used for this study was submitted to the European Nucleotide Archive (ENA) and is available under accession number PRJEB14844.
Electronic supplementary material
Supplementary Table S1, Supplementary Table S2
Acknowledgements
The authors thank Katja Cloppenborg-Schmidt, Miriam Freitag, Miriam Daumann and Ines Stölting for excellent technical support. This work was supported by grants from the Bundesministerium für Bildung und Forschung (BMBF, 0315892B), the Deutsche Forschungsgemeinschaft (DFG EXC 306, GRK 1743, KFO 303), and the University of Lübeck (P01-2012).
Author Contributions
M.H. and S.M.I. designed the study. M.H., P.S., and A.S. performed experiments and analyzed the data. A.K. conducted the statistical analysis. J.F.B. contributed to the microbiota sequencing and interpretation of the gut microbiota data. O.J., M.S., J.R., and C.S. contributed to interpretation of the phenotype data. M.H., A.K., J.F.B., and S.M.I. wrote the manuscript with contributions from all other authors. S.M.I. directed the study.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Misa Hirose and Axel Künstner contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-15377-7.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiol. Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 2.O’Rahilly S. Human genetics illuminates the paths to metabolic disease. Nature. 2009;462:307–314. doi: 10.1038/nature08532. [DOI] [PubMed] [Google Scholar]
- 3.Zimmet P, Alberti KG, Shaw J. Global and societal implications of the diabetes epidemic. Nature. 2001;414:782–787. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
- 4.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 5.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallace DC, Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb Perspect Biol. 2013;5:a021220. doi: 10.1101/cshperspect.a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larsson N-G. Somatic mitochondrial DNA mutations in mammalian aging. Annu. Rev. Biochem. 2010;79:683–706. doi: 10.1146/annurev-biochem-060408-093701. [DOI] [PubMed] [Google Scholar]
- 8.Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 9.Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van den Ouweland JM, et al. Mutation in mitochondrial tRNA(Leu)(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat. Genet. 1992;1:368–371. doi: 10.1038/ng0892-368. [DOI] [PubMed] [Google Scholar]
- 11.Kokaze A, et al. Association of the mitochondrial DNA 5178 A/C polymorphism with serum lipid levels in the Japanese population. Hum. Genet. 2001;109:521–525. doi: 10.1007/s004390100602. [DOI] [PubMed] [Google Scholar]
- 12.Liguori R, et al. The mtDNA 15497 G/A polymorphism in cytochrome b in severe obese subjects from Southern Italy. Nutr Metab Cardiovasc Dis. 2006;16:466–470. doi: 10.1016/j.numecd.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Sawabe M, et al. Mitochondrial haplogroups A and M7a confer a genetic risk for coronary atherosclerosis in the Japanese elderly: an autopsy study of 1,536 patients. J. Atheroscler. Thromb. 2011;18:166–175. doi: 10.5551/jat.6742. [DOI] [PubMed] [Google Scholar]
- 14.Takasaki S. Mitochondrial SNPs associated with Japanese centenarians, Alzheimer’s patients, and Parkinson’s patients. Comput Biol Chem. 2008;32:332–337. doi: 10.1016/j.compbiolchem.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 15.Yu X, et al. mtDNA nt13708A variant increases the risk of multiple sclerosis. PLoS ONE. 2008;3:e1530. doi: 10.1371/journal.pone.0001530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jönsen A, et al. Mitochondrial DNA polymorphisms are associated with susceptibility and phenotype of systemic lupus erythematosus. Lupus. 2009;18:309–312. doi: 10.1177/0961203308097477. [DOI] [PubMed] [Google Scholar]
- 17.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 18.Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 19.Karlsson FH, et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. 2012;3:1245. doi: 10.1038/ncomms2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheperjans F, et al. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov. Disord. 2015;30:350–358. doi: 10.1002/mds.26069. [DOI] [PubMed] [Google Scholar]
- 21.Vaahtovuo J, Munukka E, Korkeamäki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. J. Rheumatol. 2008;35:1500–1505. [PubMed] [Google Scholar]
- 22.Hevia A, et al. Intestinal dysbiosis associated with systemic lupus erythematosus. MBio. 2014;5:e01548–1514. doi: 10.1128/mBio.01548-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma J, et al. mtDNA haplogroup and single nucleotide polymorphisms structure human microbiome communities. BMC Genomics. 2014;15:257. doi: 10.1186/1471-2164-15-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodrich JK, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferris SD, Sage RD, Prager EM, Ritte U, Wilson AC. Mitochondrial DNA evolution in mice. Genetics. 1983;105:681–721. doi: 10.1093/genetics/105.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goios A, Pereira L, Bogue M, Macaulay V, Amorim A. mtDNA phylogeny and evolution of laboratory mouse strains. Genome Res. 2007;17:293–298. doi: 10.1101/gr.5941007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu X, et al. Dissecting the effects of mtDNA variations on complex traits using mouse conplastic strains. Genome Res. 2009;19:159–165. doi: 10.1101/gr.078865.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rausch, P. et al. Analysis of factors contributing to variation in the C57BL/6J fecal microbiota across German animal facilities. Int. J. Med. Microbiol., 10.1016/j.ijmm.2016.03.004 (2016). [DOI] [PubMed]
- 29.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 30.Dufrene M, Legendre P. Species Assemblages and Indicator Species: The Need for a Flexible Asymmetrical Approach. Ecological Monographs. 1997;67:345–366. [Google Scholar]
- 31.Saint-Georges-Chaumet Y, Edeas M. Microbiota-mitochondria inter-talk: consequence for microbiota-host interaction. Pathog Dis. 2016;74:ftv096. doi: 10.1093/femspd/ftv096. [DOI] [PubMed] [Google Scholar]
- 32.Clark A, Mach N. The Crosstalk between the Gut Microbiota and Mitochondria during Exercise. Front Physiol. 2017;8:319. doi: 10.3389/fphys.2017.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Green DR, Galluzzi L, Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333:1109–1112. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sena LA, et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 36.Ajouz H, Mukherji D, Shamseddine A. Secondary bile acids: an underrecognized cause of colon cancer. World J Surg Oncol. 2014;12:164. doi: 10.1186/1477-7819-12-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamer HM, et al. Review article: the role of butyrate on colonic function. Aliment. Pharmacol. Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 38.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schug ZT, Vande Voorde J, Gottlieb E. The metabolic fate of acetate in cancer. Nat. Rev. Cancer. 2016;16:708–717. doi: 10.1038/nrc.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wahlström A, Sayin SI, Marschall H-U, Bäckhed F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 41.Mottawea W, et al. Altered intestinal microbiota–host mitochondria crosstalk in new onset Crohn’s disease. Nature Communications. 2016;7:ncomms13419. doi: 10.1038/ncomms13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Serino M, et al. Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut. 2012;61:543–553. doi: 10.1136/gutjnl-2011-301012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rooks MG, et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 2014;8:1403–1417. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Y, et al. Gut microbiota composition modifies fecal metabolic profiles in mice. J. Proteome Res. 2013;12:2987–2999. doi: 10.1021/pr400263n. [DOI] [PubMed] [Google Scholar]
- 45.Zhang H, Liao X, Sparks JB, Luo XM. Dynamics of gut microbiota in autoimmune lupus. Appl. Environ. Microbiol. 2014;80:7551–7560. doi: 10.1128/AEM.02676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kameyama K, Itoh K. Intestinal colonization by a Lachnospiraceae bacterium contributes to the development of diabetes in obese mice. Microbes Environ. 2014;29:427–430. doi: 10.1264/jsme2.ME14054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meehan CJ, Beiko RG. A Phylogenomic View of Ecological Specialization in the Lachnospiraceae, a Family of Digestive Tract-Associated Bacteria. Genome Biol Evol. 2014;6:703–713. doi: 10.1093/gbe/evu050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flint HJ, Duncan SH, Scott KP, Louis P. Links between diet, gut microbiota composition and gut metabolism. Proc Nutr Soc. 2015;74:13–22. doi: 10.1017/S0029665114001463. [DOI] [PubMed] [Google Scholar]
- 49.Hosseini E, Grootaert C, Verstraete W, Van de Wiele T. Propionate as a health-promoting microbial metabolite in the human gut. Nutr. Rev. 2011;69:245–258. doi: 10.1111/j.1753-4887.2011.00388.x. [DOI] [PubMed] [Google Scholar]
- 50.Scanlan PD, Shanahan F, Marchesi JR. Culture-independent analysis of desulfovibrios in the human distal colon of healthy, colorectal cancer and polypectomized individuals. FEMS Microbiol. Ecol. 2009;69:213–221. doi: 10.1111/j.1574-6941.2009.00709.x. [DOI] [PubMed] [Google Scholar]
- 51.Miller TW, et al. Hydrogen sulfide is an endogenous potentiator of T cell activation. J. Biol. Chem. 2012;287:4211–4221. doi: 10.1074/jbc.M111.307819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rey FE, et al. Metabolic niche of a prominent sulfate-reducing human gut bacterium. Proc. Natl. Acad. Sci. USA. 2013;110:13582–13587. doi: 10.1073/pnas.1312524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szabo C, et al. Regulation of mitochondrial bioenergetic function by hydrogen sulfide. Part I. Biochemical and physiological mechanisms. Br. J. Pharmacol. 2014;171:2099–2122. doi: 10.1111/bph.12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schröder T, et al. Mitochondrial gene polymorphisms alter hepatic cellular energy metabolism and aggravate diet-induced non-alcoholic steatohepatitis. Mol Metab. 2016;5:283–295. doi: 10.1016/j.molmet.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weiss H, et al. The mitochondrial Atp8 mutation induces mitochondrial ROS generation, secretory dysfunction, and β-cell mass adaptation in conplastic B6-mtFVB mice. Endocrinology. 2012;153:4666–4676. doi: 10.1210/en.2012-1296. [DOI] [PubMed] [Google Scholar]
- 56.Yu X, et al. The mtDNA nt7778 G/T polymorphism affects autoimmune diseases and reproductive performance in the mouse. Hum. Mol. Genet. 2009;18:4689–4698. doi: 10.1093/hmg/ddp432. [DOI] [PubMed] [Google Scholar]
- 57.GitHub - torognes/vsearch: Versatile open-source tool for metagenomics. Available at: https://github.com/torognes/vsearch (Accessed: 26th April 2016). [DOI] [PMC free article] [PubMed]
- 58.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 59.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pruesse E, Peplies J, Glöckner FO. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oksanen, J. et al. Vegan: Community Ecology Package. R package Version 2.0-2 (2011).
- 64.McMurdie PJ, Holmes S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLOS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anderson M, Willis TJ. Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology. 2003;84:511–525. doi: 10.1890/0012-9658(2003)084[0511:CAOPCA]2.0.CO;2. [DOI] [Google Scholar]
- 66.De Cáceres M, Legendre P, Moretti M. Improving indicator species analysis by combining groups of sites. Oikos. 2010;119:1674–1684. doi: 10.1111/j.1600-0706.2010.18334.x. [DOI] [Google Scholar]
- 67.R Core Team. T: A Language and Environment for Statistical Computing. (2015).
- 68.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1, Supplementary Table S2
Data Availability Statement
Sequencing data used for this study was submitted to the European Nucleotide Archive (ENA) and is available under accession number PRJEB14844.