Abstract
DYRK1A, located on chromosome 21, is a major candidate gene of Down syndrome (DS, trisomy21), and its overexpression is associated with abnormal phenotype of Down syndrome patients. The defects of gonads and germ cells in Down Syndrome suggest that overexpression of DYRK1A has potential effect on primordial germ cells (PGCs) development. Human and zebrafish DYRK1A protein sequence possess 75.6% similarity and same function domains, suggesting the evolutional conservation. Here, we used zebrafish model to detect the definite role of excessive expression of DYRK1A in PGCs development during embryogenesis. We injected DYRK1A mRNA into embryos and detected the PGCs marker gene vasa and nanos1. Results showed depletion in numbers and disordering migration of PGCs in human or zebrafish DYRK1A overexpressed zebrafish embryos. Quantitative proteome analysis indicated that embryonic proteins were significantly altered in DYRK1A overexpressed embryos. Of note, ca15b and piwil1, two identified critical factors for PGCs development, showed ectopic expression induced by overexpressed DYRK1A. In brief, we demonstrate that overexpression of DYRK1A, a candidate gene of Down’s syndrome, impairs PGCs development during early embryogenesis by altering key factors in embryos. Importantly, our work may provide a conceivable mechanism for the gonads and germ cells defects of Down syndrome patients.
Introduction
In 1966 of last century, Finch et al. reported that the chromosomal aberration leads to sub- or infertility in carriers of trisomy 21, that is, Down syndrome1. Hojager et al.2 showed a reduced follicle number and a retarded follicle growth in prepubertal female patients gonads2. The tubuli seminiferi contain a reduced number of germ cells or are completely devoid of germ cells in the testes of adult individuals3,4, which caused by aneuploid conditions meiotic defects5,6. Leffler et al. in year 1999 showed migration delay and reduction of PGCs in trisomy 16 mouse7, an animal model for Down’s syndrome.
DYRK1A gene in human maps to the Down syndrome critical region q22.2 of chromosome 218–12. DYRK1A plays pivotal regulatory roles in the signaling of cell proliferation and development, which has dual substrate specificities. Autophosphorylation for self-activation takes place on the tyrosine-321 residue in the active loop of the catalytic domain13,14, and target protein phosphorylation occurs on serine/threonine residues15. DYRK1A has been reported to phosphorylate or interact with several proteins, including STAT3, FHKR, Gli-1, eIF2Be, Tau, dynamin, glycogen synthase, 14-3-3, CREB, cyclin L2, Arip4, Hip-1, PAHX-AP1, and SF3b1, suggesting that DYRK1A participates in multiple biological pathways by diverse array of interactions15–17.
Compared with the healthy person, the expression of DYRK1A in Down Syndrome patients shows increased pattern18. The many features of Down’s syndrome include neurological, skeletal, cardiovascular and immunological defects, and are generally thought to originate from a 1.5-fold increase in the dosage of genes, including DYRK1A, within a critical region of chromosome 21, which is present in triplicate in all cases of Down’s syndrome19. The defects of gonads and germ cells in Down Syndrome patients suggest that overexpression of DYRK1A has potential function on primordial germ cells (PGCs) development. Importantly, the definite role of increased dosage of DYRK1A in PGCs development remains indistinct.
Model organism zebrafish is relevant to higher vertebrates significantly, with highly manageable genetic manipulation, which making it an exceptional animal model for exploring molecular mechanism regulating key developmental processes. At very early embryogenesis, zebrafish PGCs are segregated from the somatic lineage, and start a characteristic performance of migration toward the genital ridges shortly thereafter20,21. This migration process is completed within the first developmental day, and can be detected at high resolution using simple microscopy, due to the optical transparency of zebrafish embryos. Hence, zebrafish embryo has been considered to be excellent in vivo model for investigation of PGC migration22.
Zebrafish PGCs are specified in different locations in the embryo and migrate toward the location where the gonad develops, the site where they eventually differentiate into gametes, sperm and egg. Following their specification during early embryonic stages, PGCs polarize and acquire motility. As they migrate, PGCs are presented with attractive and repulsive guidance cues provided by somatic cells along the migration path23, which exist complicated developmental and cellular mechanisms. Some key factors play critical role for this migration process, such as Piwil1 and Ca15b23–25.
Here, we used zebrafish model and two PGCs markers vasa and nanos1 to investigate the function of overexpression DYRK1A on PGCs during embryo development. Results showed that PGCs in DYRK1A overexpressed embryos were decreased and disordered. Alone with the PGCs defect, critical factors for PGCs development were altered in aberrant DYRK1A injected embryos, suggesting the important and definite role of DYRK1A for PGCs development in early embryogenesis.
Results
DYRK1A protein is evolutionally conserved between zebrafish and human
Zebrafish DYRK1A protein, similar to human DYRK1A, shows 75.6% similarity to its human ortholog (Fig. 1a ). Particularly, they have same PKc_DYRK1 and S_TKc domains which are important for DYRK1A function (Fig. 1b and c ). The evolutional conservation suggests the critical role of DYRK1A for vertebrate and qualification of zebrafish as model organism to study DYRK1A molecular function.
Figure 1.
DYRK1A is evolutionally conserved between human and zebrafish. (a) Complete sequence alignment of human DYRK1A and zebrafish DYRK1A protein sequence. Conserved domain of human DYRK1A (b) and zebrafish DYRK1A (c) protein analyzed by NCBI conserved domains website.
Expression pattern of dyrk1a during zebrafish early embryogenesis
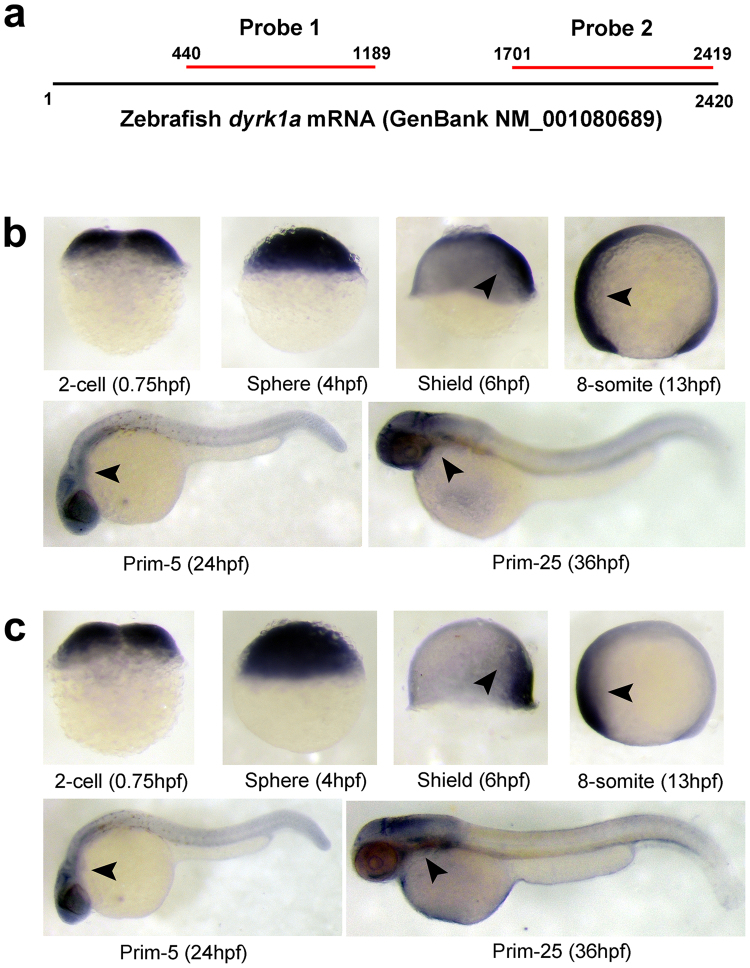
Expression of DYRK1A mRNA during zebrafish embryogenesis was examined by whole mount in situ hybridization (WISH), and two nonoverlapping probes for DYRK1A, localizing at positions 440–1189 bp (probe 1) and 1701–2419 bp (probe 2) (Fig. 2a ), were used to demonstrate specificity and obtain identical spatially restricted expression patterns26.
Figure 2.
Spatiotemporal expression pattern of DYRK1A transcript in zebrafish embryos. (a) Location of anti-sense RNA probe for detection of zebrafish DYRK1A using WISH. (b) Detection by Probe 1 at indicated stages. (c) Detection by Probe 2 at indicated stages. Arrows show the stronger expression region of DYRK1A. The same expression pattern of DYRK1A shown by two non-overlapping probes, demonstrating the specificity and identical spatially restricted expression patterns of DYRK1A in zebrafish early embryogenesis. Embryo orientations: 2-cell and Sphere stage, lateral views with the animal pole oriented at the top; Shield stage, lateral view with the dorsal side oriented at the right; 8-somite, Prim-5 and Prim-25 stage, lateral views with anterior oriented toward the left.
Results show that zebrafish DYRK1A mRNA appears in the 2-cell stage and in all blastodermal cells until the dome stage (Fig. 2b and c). At the onset of gastrulation, DYRK1A expression starts to concentrate on the dorsal side (Fig. 2b and c, Shield stage). When segmentation starts, DYRK1A is expressed in axis ubiquitously and shows stronger expression at anterior part of embryo (Fig. 2b and c, 8-somite stage), and then branchial and pharyngeal arches gradually gain stronger expression (Fig. 2b and c, Prim-5 and Prim-25 stage).
Next, expression of DYRK1A protein during zebrafish embryogenesis was examined by using whole-mount immunohistochemistry (WIHC). Results of WIHC show that spatiotemporal expression pattern of DYRK1A protein is similar to DYRK1A mRNA (Supplemental Figure S1). These results of expression pattern suggest potential and important role of DYRK1A in early embryogenesis, especially at blastula period of beginning of PGCs development.
Overexpression of DYRK1A impairs PGCs Maintenance and Migration
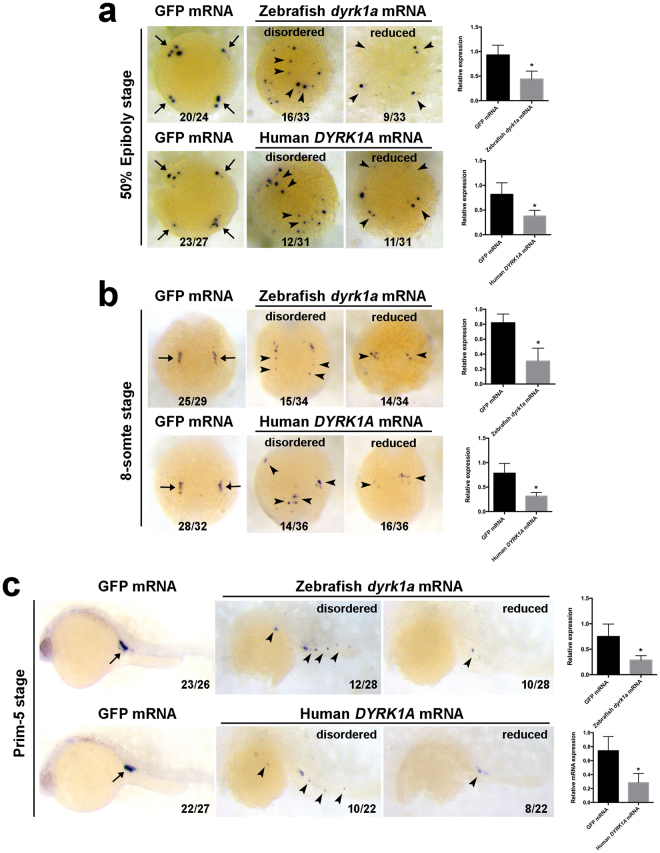
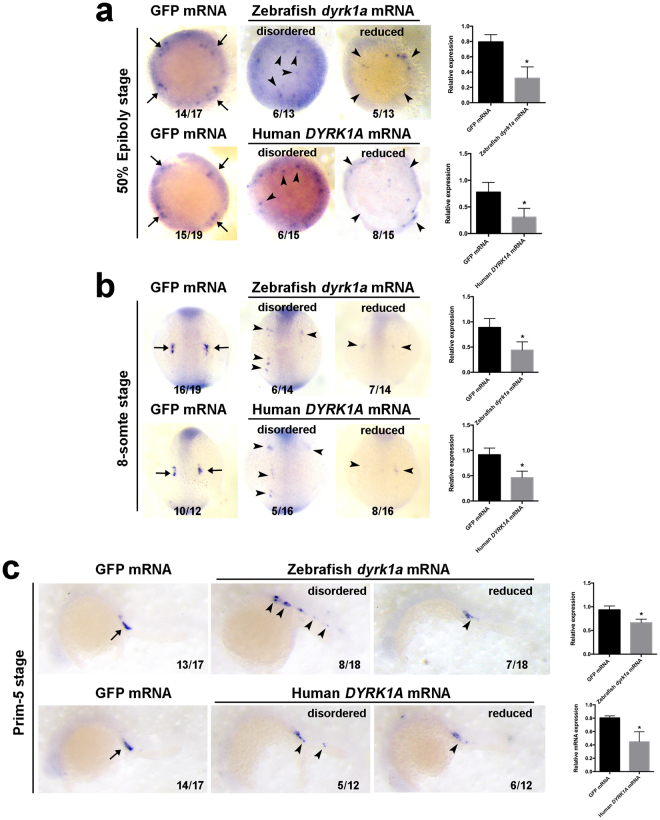
To determine whether DYRK1A overexpression effects on maintenance and migration of the PGCs, we followed the overexpression process in embryos in which the DYRK1A mRNA was injected. Since human and zebrafish DYRK1A protein sequence and domain are evolutionally conserved, we injected human and zebrafish DYRK1A mRNA individually to zebrafish embryos to investigate whether DYRK1A has conserved effect to PGCs development. PGCs were marked by gene nanos1 and vasa at 50% epiboly, 8-somite and Prim-5 stage, and we found that overexpression of human and zebrafish DYRK1A individually led to same abnormity of PGCs, showing ectopic locations and reduction (Figs 3 and 4), suggesting that human and zebrafish DYRK1A functions conservatively to PGCs development. To confirm the importance of DYRK1A conserved domain PKc in PGCs development, we constructed deletion mutants of DYRK1A lacking PKc domain. Embryos injected with mRNA of DYRK1A PKc domain deletion mutant showed that the mutation abrogated the abnormality of PGCs development (Supplemental Figure S2), suggesting the requirement of PKc domain for DYRK1A function.
Figure 3.
Overexpression of DYRK1A induces nanos1-marked PGCs deficiency in early zebrafish embryo. Analysis of localization and strength of nanos1 positive cells in DYRK1A overexpressed embryos by WISH at 50% Epiboly stage (a), 8-somite stage (b) and Prim-5 stage (c). Histogram representing the relative expression detected by qPCR in GFP injected and DYRK1A overexpressed embryos at corresponding assay. Embryo orientations: 50% Epiboly stage, top view with the dorsal oriented at the right; 8-somite, dorsal view with anterior oriented at the top; Prim-5 stage, lateral views with anterior oriented toward the left. Arrows show the normal location of PGCs, arrowheads demonstrate the aberrant position of PGCs induced by overexpressed DYRK1A. The numbers indicated in each picture are the number (left) of affected embryos with phenotype similar to what is shown in the picture and the total number (right) of observed embryos. The same number labeling was used thereafter.
Figure 4.
Overexpression of DYRK1A induces vasa-marked PGCs deficiency in early zebrafish embryo. Analysis of localization and strength of vasa positive cells in DYRK1A overexpressed embryos by WISH at 50% Epiboly stage (a), 8-somite stage (b) and Prim-5 stage (c). Histogram representing the relative expression detected by qPCR in GFP injected and DYRK1A overexpressed embryos at corresponding assay. Embryo orientations: 50% Epiboly stage, top view with the dorsal oriented at the right; 8-somite, dorsal view with anterior oriented at the top; Prim-5 stage, lateral views with anterior oriented toward the left. Arrows show the normal location of PGCs, arrowheads demonstrate the aberrant position of PGCs induced by overexpressed DYRK1A.
Furthermore, to investigate role of dyrk1a deficiency for PGCs development, we used DYRK1A popular inhibitor Epigallocatechin Gallate (EGCG) to treat WT embryos. Results showed that EGCG could not lead to PGCs abnormality, even though we tried wide range of concentration from 10 µm to 200 µm (Supplemental Figure S3). Taken together, similar to the situation of Down syndrome, DYRK1A with overexpressed state, but not deficiency, leads to abnormality of PGCs, depending on its PKc conserved domain.
Proteomics analysis shows aberrant expression of key factors essential for PGCs development in DYRK1A overexpressed embryos
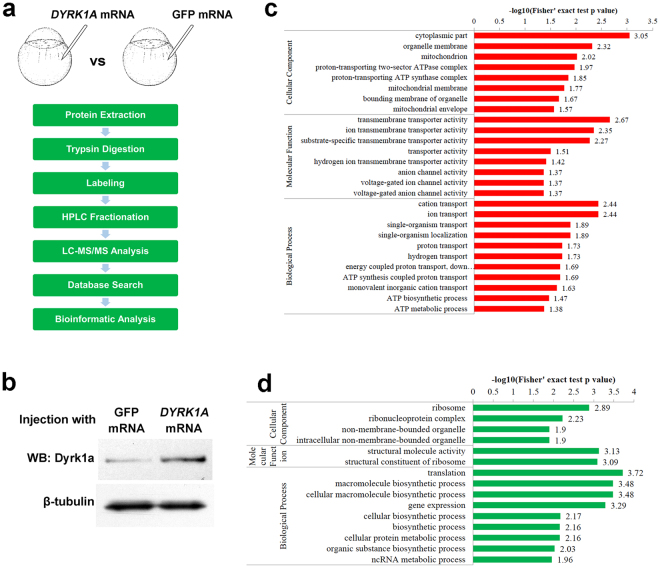
To discover the molecular mechanism of DYRK1A overexpression regulating PGCs development, we used an integrated approach involving TMT labeling and LC-MS/MS to quantify the dynamic changes of the whole proteome of zebrafish embryos at 50% epiboly stage (injection of human DYRK1A mRNA vs GFP mRNA) (Fig. 5a ). Western blot assay shows the increased protein level of DYRK1A in the embryos injected with DYRK1A mRNA (Fig. 5b and Supplemental Figure S4). In total, 1,573 proteins from embryos were identified in response to DYRK1A mRNA and GFP mRNA injection, among which 1,324 proteins were quantified. All the annotation and quantification information were presented in the Supplemental Table S1. Relative quantitation of proteins was divided into two categories. Quantitative ratio over 1.2 was considered up-regulation while quantitative ratio less than 1/1.2 was considered as down-regulation. Results showed that DYRK1A mRNA injection induced 265 differentially expressed proteins (141 up-regulated and 124 down-regulated).
Figure 5.
Proteomics analysis shows the altered proteins pattern essential for PGCs development in DYRK1A overexpressed embryos. (a) General work flow of Proteomics analysis in this work. (b) Embryos injected with DYRK1A mRNA show increased protein level of DYRK1A. The blots shown are cropped; the uncropped full-length gels are presented in the Supplementary Figure 1. Gene Ontology (GO)-based enrichment analysis of up-regulated (c) and down-regulated (d) proteins (DYRK1A mRNA vs GFP mRNA).
To characterize the function of these altered proteins, Gene Ontology (GO)-based classification analysis on the ontology of biological process, cellular component and molecular function was performed and reveals widely different distribution between DYRK1A mRNA and GFP mRNA injection (Supplemental Table S2).
To reveal the nature of the differentially expressed proteins upon DYRK1A mRNA and GFP mRNA injection, the GO functional enrichment of differentially quantified proteins was carried out. The biological process was firstly investigated (Fig. 5c), it is found that the up-regulated proteins in response to DYRK1A mRNA injection show enrichment of ion, hydrogen, ATP transport. In the down-regulated proteins, the enrichment includes macromolecule biosynthetic process, cellular protein metabolic process and ncRNA metabolic process (Fig. 5d).
Molecular function-based enrichment results were shown in Fig. 5c. It is found that the ion/substrate-specific/hydrogen ion transmembrane transporter activity and anion/voltage-gated ion channel activity in up-regulated proteins; and the structural molecule activity and structural constituent of ribosome in down-regulated proteins were enriched in DYRK1A mRNA injected embryos (Fig. 5d).
In the cellular component category, cytoplasmic part, organelle membrane, mitochondrion, proton-transporting ATPase complex and bounding membrane of organelle were enriched in the up-regulated proteins. In the down-regulated proteins, enrichment includes ribosome, ribonucleoprotein complex and non-membrane-bounded organelle (Fig. 5c and d).
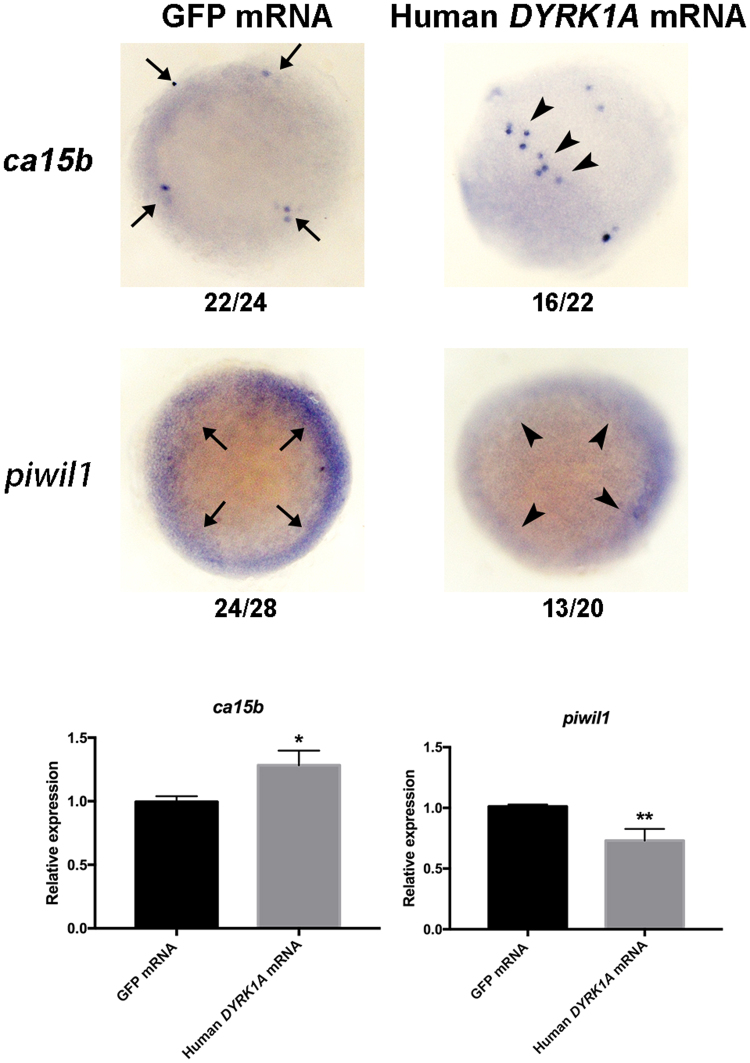
Of note, we found that expression of ca15b (ratio 1.429, P value 0.014260929) and piwil1 (ratio 0.755, P value 0.014935562) was significantly changed in DYRK1A mRNA injected embryos (Supplemental Table S3). Piwil1 is identified as a critical factor essential for PGCs maintenance and migration in zebrafish and medaka24,27. Ca15b, an enzyme expressed specifically in the PGCs, plays an important role in establishment of polar pH distribution for guided PGCs migration25. Subsequently, we used in situ hybridization and qPCR assays to verify the expression of ca15b and piwil1 in DYRK1A overexpressed embryos. Results showed that overexpressed DYRK1A led to upregulation of ca15b and downregulation of piwil1 indeed, which were consistent with the proteomics data (Fig. 6).
Figure 6.
Verification of aberrant expression of PGCs key factors in DYRK1A overexpressed embryos. Analysis of localization and strength of ca15b and piwil1 in DYRK1A overexpressed embryos by WISH. All of embryos are 50%-epiboly stage with top view. Arrows show the normal location of detected gene, arrowheads demonstrate the aberrant position induced by overexpressed DYRK1A. Histogram representing the relative expression of ca15b and piwil1 in GFP injected and DYRK1A overexpressed embryos detected by qPCR.
In conclusion, quantitative analysis of global proteome between DYRK1A mRNA and GFP mRNA injection indicates that overexpression of DYRK1A induces significant impacts in embryos, resulting the remarkably altered expression of many critical proteins for PGCs development, such as Ca15b and Piwil1.
Discussion
DYRK1A gene in human localizes to the Down syndrome critical region q22.2 of chromosome 21, and its overexpression is associated with abnormal phenotype of Down syndrome patients. Down syndrome shows severe defect of gonads and germ cells, however, the distinct function of increased DYRK1A dosage in PGCs remains obscured.
Both zebrafish and human DYRK1A protein have same PKc_DYRK1 and S_TKc domains, suggesting the evolutional conservation. We injected DYRK1A mRNA into embryos and detected the PGCs marker gene vasa and nanos1. Results demonstrated that overexpressed either zebrafish or human DYRK1A induced same abnormity of PGCs, showing reduction and disorder. These results are similar to that observed from trisomy 16 mouse7, indicating that overexpression of DYRK1A impairs PGCs maintenance and migration, and suggesting the evolutional functional conservation of DYRK1A in human and zebrafish.
Subsequently, we performed quantitative proteome analysis to uncover the molecular mechanism of overexpressed DYRK1A to PGCs development and early embryogenesis. It is a pity that the identified quantitative proteins are less than expectation, because of the obstacle from the high proportion of yolk proteins in early embryos. Even so, we still identified 1573 proteins and quantified 1324 proteins, and found 265 differentially expressed proteins. GO function classification analysis reveal that wide range of proteins are regulated by overexpressed DYRK1A, including transport of ion/hydrogen/ATP, process of macromolecule/protein metabolism/ncRNA metabolism, etc. Most importantly, we identify that piwil1 and ca15b gene expression are significantly changed in DYRK1A overexpressed embryos, the two genes are key factors which play critical role in PGCs development process.
Loss of zebrafish piwil1(ziwi) function results in a progressive loss of germ cells due to apoptosis during larval development24. Houwing et al. found that wild-type and piwil1 mutant gonads had similar germ cell numbers at two weeks of age24. However, Houwing et al. did not show PGCs status of piwil1 mutant and piwil1 expression pattern at blastula period of early embryogenesis in zebrafish. Li et al. showed that medaka piwil1 knockdown significantly reduces the number of PGCs in vivo and in vitro and affects the distribution of PGCs in early developing embryos. Surprisingly, depletion of zygotic medaka piwil1 severely and specifically affected PGC migration at early embryogenesis27. Interestingly, both zebrafish24 and medaka27 piwil1 is maternally provided and localizes to granules at the cleavage planes in four cell embryos similar to vasa mRNA. However, piwil1 RNA was widely distributed to many cells, but not concentrated in PGCs, at the blastula stage in medaka27, and we also found the similar expression pattern in zebrafish (Fig. 6), suggesting that piwil1 may play diverse role in embryogenesis and PGCs development at different stages.
Studies show that the gradient interpretation produces intracellular response, that is, elevation of pH at the front of PGCs drive their migration. Carbonic anhydrase 15b (Ca15b), an enzyme that is expressed specifically in the PGCs, plays an important role in establishment of polar pH distribution for guided PGCs migration. In ca15b knockdown embryos, a uniform low pH level is observed within PGCs along with an increased proportion of PGCs that failed to reach their target. At the meantime, lack of chemokine Cxcl12a gradient, reduction of local Rac1 activity and non-polarization of actin occur at the leading edge of the migrating cells in Ca15b knockdown embryos23,25. Then, the actin loses to polymerization and leads the loss of myosin activity and normal formation of cellular structures, finally, PGCs migration fail to track correctly28.
Methods
Ethics Statement
All experiments in this study were in accordance with the “Guide for the Care and Use of Laboratory Animals” (Eighth Edition, 2011. ILARCLS, National Research Council, Washington, D.C.) and were approved by the Animal Care and Use Committee of West China Second University Hospital, Sichuan University (Approval ID: HXDEYY20101105).
Zebrafish and embryos
Zebrafish WT embryos from AB strain were used. Embryos were obtained by natural mating and cultured in embryo medium29. Staging of the embryos was carried out according to Kimmel et al.30.
Constructs
Zebrafish DYRK1A full length mRNA sequence was from openbiosystems (Catalog Number MDR1734-202804618). The coding region with poly A sequence was amplified by primer pair zDYRK1A-cds (Supplemental Table S4) for capped mRNA synthesis; Human DYRK1A full length mRNA sequence was amplified by primer pair hDYRK1A-cds (Supplemental Table S4) from K562 cells and cloned into pcDNA3.1+ vector (Invitrogen) for capped mRNA synthesis. Fragments of PGCs marker genes vasa (GenBank# NM_131057) and nonos1 (GenBank# NM_131878) (primers are shown in Supplemental Table S4) were cloned into pEASY-T3 (Transgen) for antisense RNA probe synthesis. The ∆PKc domain expression plasmid of dyrk1a was constructed using KOD-Plus-Mutagenesis Kit (TOYOBO) using primer pair hDYRK1A-∆PKc and zDYRK1A-∆PKc (Supplemental Table S4).
RNA microinjection and reagent treatment
Capped mRNAs were synthesized using mMESSAGE mMACHINE® Kit (Ambion); Synthetic capped mRNAs were injected into single-cell embryos. Injection dose was an estimated amount received by a single embryo, ~30 pg mRNA of DYRK1A and GFP was injected into embryos. Epigallocatechin Gallate (EGCG, S2250) was from Selleck. At the beginning of blastula period (2.5 hpf), embryos (30 embryos in a well of 6-well plate with 3 ml culture water) were treated with EGCG for 2.5 h and then subject to whole-mount in situ hybridization and quantitative real time RT–PCR.
Zebrafish embryo in situ hybridization and immunohistochemistry
Whole-mount in situ hybridization (WISH) was carried out as previously described in Thisse et al.26 and Sun et al.31. After lineage by appropriate restriction enzymes, antisense RNAs for in situ hybridization were synthesized using DIG RNA Labeling Kit (SP6/T7) (Roche) and purified by MEGAclear (Ambion).
Whole-mount immunohistochemistry (WIHC) in zebrafish embryos was performed as previously described (Jia et al.32 and Brend et al.33) with modifications. Embryos were fixed in fresh 2% paraformaldehyde for overnight, permeabilized in 100% methanol at −20 °C for at least 1 hour. The embryos were bathed in PBS solution, then incubated in block solution (PBS plus 0.5% Triton X-100 and 1% BSA) for 1 hr at room temperature. Embryos were then incubated with primary antibody at 4 °C overnight followed incubation with secondary antibodies (HRP conjugate from Thermo Fisher Corporation) at room temperature for 1 hour. DAB kit (ZSGB-BIO) was used to develop color.
Quantitative real time RT–PCR (qPCR) analysis
Total RNA was prepared with TRIzol (Invitrogen, 15596-018) and cDNA was synthesized from 1 μg of RNA with PrimeScript RT reagent Kit (Takara, DRR037A). qPCR was performed with the SYBR Green detection method with 7500 real-time PCR system (Applied Biosystems). The primers used were shown in Supplemental Table S4.
Grayscale measurement and statistical analysis
ImageJ software was used to measure signal strength grayscale of whole-mount in situ hybridization assay. Statistical analyses were performed with a Student’s t test. Quantitative data show the mean + SD. Statistical significance is defined as *P < 0.05, **P < 0.01, ***P < 0.001.
Proteomics analysis of embryos
Antibodies for western blot were used: anti-DYRK1A (Santa cruz, sc-12568), anti-β-tubulin (Zen Bioscience, 200608). Quantitative proteome analysis was performed by PTM-Biolabs (HangZhou) Co., Ltd., detailed materials and methods are shown in Supplementary Information Materials and Methods.
Electronic supplementary material
Acknowledgements
The authors wish to thank Prof Hsiao Chang Chan for funding the quantitative analysis of global embryos proteome. This research was supported by grants from National Natural Science Foundation of China (81200339), China Postdoctoral Science Foundation (20110491723), Young Teacher foundation of Sichuan University (2011SCU11040), National Basic Research Program of China (2012CB944903) and National 973 project (2013CB967404).
Author Contributions
Y.L. and H.S. conceived and designed the experiments; Z.L. and M.L. performed the experiments; Y.L. and H.S. analyzed the data; H.W. coordinated the project. Y.L. and H.S. wrote the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-15730-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Finch RBJA, Finley WH, Finley SC, Tucker CC. Meiosis in trisomic Down’s syndrome. Ala J Med Sci. 1966;3:117–120. [PubMed] [Google Scholar]
- 2.Hojager B, Peters H, Byskov AG, Faber M. Follicular development in ovaries of children with Down’s syndrome. Acta Paediatr Scand. 1978;67:637–643. doi: 10.1111/j.1651-2227.1978.tb17815.x. [DOI] [PubMed] [Google Scholar]
- 3.Kjessler B, De la Chapelle A. Meiosis and spermatogenesis in two postpubertal males with Down’s syndrome: 47, XY, G+ Clinical genetics. 1971;2:50–57. doi: 10.1111/j.1399-0004.1971.tb00255.x. [DOI] [PubMed] [Google Scholar]
- 4.Johannisson R, et al. Down’s syndrome in the male. Reproductive pathology and meiotic studies. Hum Genet. 1983;63:132–138. doi: 10.1007/BF00291532. [DOI] [PubMed] [Google Scholar]
- 5.Speed RM. Meiotic configurations in female trisomy 21 foetuses. Hum Genet. 1984;66:176–180. doi: 10.1007/BF00286596. [DOI] [PubMed] [Google Scholar]
- 6.Mittwoch U, Mahadevaiah S, Setterfield LA. Chromosomal anomalies that cause male sterility in the mouse also reduce ovary size. Genet Res. 1984;44:219–224. doi: 10.1017/S0016672300026409. [DOI] [PubMed] [Google Scholar]
- 7.Leffler A, Ludwig M, Schmitt O, Busch LC. Germ cell migration and early development of the gonads in the trisomy 16 mouse–an animal model for Down’s syndrome. Ann Anat. 1999;181:247–252. doi: 10.1016/S0940-9602(99)80039-9. [DOI] [PubMed] [Google Scholar]
- 8.Rahmani Z, et al. Critical role of the D21S55 region on chromosome 21 in the pathogenesis of Down syndrome. Proc Natl Acad Sci USA. 1989;86:5958–5962. doi: 10.1073/pnas.86.15.5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahmani Z, et al. Down syndrome critical region around D21S55 on proximal 21q22.3. Am J Med Genet Suppl. 1990;7:98–103. doi: 10.1002/ajmg.1320370720. [DOI] [PubMed] [Google Scholar]
- 10.Delabar JM, et al. Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur J Hum Genet. 1993;1:114–124. doi: 10.1159/000472398. [DOI] [PubMed] [Google Scholar]
- 11.Shindoh N, et al. Cloning of a human homolog of the Drosophila minibrain/rat Dyrk gene from “the Down syndrome critical region” of chromosome 21. Biochem Biophys Res Commun. 1996;225:92–99. doi: 10.1006/bbrc.1996.1135. [DOI] [PubMed] [Google Scholar]
- 12.Song WJ, et al. Isolation of human and murine homologues of the Drosophila minibrain gene: human homologue maps to 21q22.2 in the Down syndrome “critical region”. Genomics. 1996;38:331–339. doi: 10.1006/geno.1996.0636. [DOI] [PubMed] [Google Scholar]
- 13.Lochhead PA, Sibbet G, Morrice N, Cleghon V. Activation-loop autophosphorylation is mediated by a novel transitional intermediate form of DYRKs. Cell. 2005;121:925–936. doi: 10.1016/j.cell.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 14.Kentrup H, et al. Dyrk, a dual specificity protein kinase with unique structural features whose activity is dependent on tyrosine residues between subdomains VII and VIII. The Journal of biological chemistry. 1996;271:3488–3495. doi: 10.1074/jbc.271.7.3488. [DOI] [PubMed] [Google Scholar]
- 15.Dierssen M, de Lagran MM. DYRK1A (dual-specificity tyrosine-phosphorylated and -regulated kinase 1A): a gene with dosage effect during development and neurogenesis. TheScientificWorldJournal. 2006;6:1911–1922. doi: 10.1100/tsw.2006.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galceran, J., de Graaf, K., Tejedor, F. J. & Becker, W. The MNB/DYRK1A protein kinase: genetic and biochemical properties. Journal of neural transmission. Supplementum, 139–148 (2003). [DOI] [PubMed]
- 17.Hammerle, B., Elizalde, C., Galceran, J., Becker, W. & Tejedor, F. J. The MNB/DYRK1A protein kinase: neurobiological functions and Down syndrome implications. Journal of neural transmission. Supplementum, 129–137 (2003). [DOI] [PubMed]
- 18.Guimera J, Casas C, Estivill X, Pritchard M. Human minibrain homologue (MNBH/DYRK1): characterization, alternative splicing, differential tissue expression, and overexpression in Down syndrome. Genomics. 1999;57:407–418. doi: 10.1006/geno.1999.5775. [DOI] [PubMed] [Google Scholar]
- 19.Arron JR, et al. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441:595–600. doi: 10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- 20.Weidinger G, Wolke U, Koprunner M, Klinger M, Raz E. Identification of tissues and patterning events required for distinct steps in early migration of zebrafish primordial germ cells. Development. 1999;126:5295–5307. doi: 10.1242/dev.126.23.5295. [DOI] [PubMed] [Google Scholar]
- 21.Weidinger G, et al. Regulation of zebrafish primordial germ cell migration by attraction towards an intermediate target. Development. 2002;129:25–36. doi: 10.1242/dev.129.1.25. [DOI] [PubMed] [Google Scholar]
- 22.Sang X, Curran MS, Wood AW. Paracrine insulin-like growth factor signaling influences primordial germ cell migration: in vivo evidence from the zebrafish model. Endocrinology. 2008;149:5035–5042. doi: 10.1210/en.2008-0534. [DOI] [PubMed] [Google Scholar]
- 23.Paksa A, Raz E. Zebrafish germ cells: motility and guided migration. Curr Opin Cell Biol. 2015;36:80–85. doi: 10.1016/j.ceb.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Houwing S, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 25.Tarbashevich K, Reichman-Fried M, Grimaldi C, Raz E. Chemokine-Dependent pH Elevation at the Cell Front Sustains Polarity in Directionally Migrating Zebrafish Germ Cells. Curr Biol. 2015;25:1096–1103. doi: 10.1016/j.cub.2015.02.071. [DOI] [PubMed] [Google Scholar]
- 26.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nature protocols. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 27.Li M, Hong N, Gui J, Hong Y. Medaka piwi is essential for primordial germ cell migration. Curr Mol Med. 2012;12:1040–1049. doi: 10.2174/156652412802480853. [DOI] [PubMed] [Google Scholar]
- 28.Jaglarz MK, Howard KR. The active migration of Drosophila primordial germ cells. Development. 1995;121:3495–3503. doi: 10.1242/dev.121.11.3495. [DOI] [PubMed] [Google Scholar]
- 29.Westerfield, M. The Zebrafish Book. (University of Oregon Press, 1993).
- 30.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 31.Sun H, et al. Zili inhibits transforming growth factor-beta signaling by interacting with Smad4. J Biol Chem. 2010;285:4243–4250. doi: 10.1074/jbc.M109.079533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia S, et al. Protein phosphatase 4 cooperates with Smads to promote BMP signaling in dorsoventral patterning of zebrafish embryos. Developmental cell. 2012;22:1065–1078. doi: 10.1016/j.devcel.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Brend, T. & Holley, S. A. Zebrafish whole mount high-resolution double fluorescent in situ hybridization. Journal of visualized experiments: JoVE, 10.3791/1229 (2009). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.