Abstract
To further comprehend the genome features of Cephalloscyllium umbratile (Carcharhiniformes), an endangered species, the complete mitochondrial DNA (mtDNA) was firstly sequenced and annotated. The full-length mtDNA of C. umbratile was 16,697 bp and contained ribosomal RNA (rRNA) genes, 13 protein-coding genes (PCGs), 23 transfer RNA (tRNA) genes, and a major non-coding control region. Each PCG was initiated by an authoritative ATN codon, except for COX1 initiated by a GTG codon. Seven of 13 PCGs had a typical TAA termination codon, while others terminated with a single T or TA. Moreover, the relative synonymous codon usage of the 13 PCGs was consistent with that of other published Carcharhiniformes. All tRNA genes had typical clover-leaf secondary structures, except for tRNA-Ser (GCT), which lacked the dihydrouridine ‘DHU’ arm. Furthermore, the analysis of the average Ka/Ks in the 13 PCGs of three Carcharhiniformes species indicated a strong purifying selection within this group. In addition, phylogenetic analysis revealed that C. umbratile was closely related to Glyphis glyphis and Glyphis garricki. Our data supply a useful resource for further studies on genetic diversity and population structure of C. umbratile.
Introduction
Cephalloscyllium umbratile (Cephaloscyllium, Scyliorhinidae, Chondrichthyes), belonging the Carcharhiniformes order, is one of the most important aquarium and reef fish, and mainly distribute in the coastwise of China, Vietnam and Japan. Due to small amount, it is regarded as endangered species, and absorbed in red list of International Union for Conservation of Nature (IUCN)1. Since the information about C. umbratile has been generally scarce, with the development of offshore fishery, increasing research interest has been developed in conservation as well as in scientific and economic topics regarding reef fish2,3.
In Chondrichthyes, the typical complete mitochondrial DNA (mtDNA) was circular and approximately 17 kb in length with correspondingly conserved gene content which encoded 37 genes, including 22 transfer RNA (tRNA), 13 protein-coding genes (PCGs), 2 ribosomal RNA (rRNA), a major non-coding control region (D-loop region), and an A + T-rich region4,5. Furthermore, genomic information is considered to be reliable for the efficient implementation strategies to study evolutionary relationships, phylogeography and phylogeny6,7. Due to its conserved gene content, maternal inheritance, a small genome size, relatively fast evolutionary rate, high copy number and lack of intermolecular genetic recombination8–10, mtDNA has been broadly adopted in species identification11,12, genome evolution13–16 and nonsynonymous (Ka) and synonymous (Ks) substitutions of many species17–23.
Moreover, Carcharhiniformes include about 49 genera and over 200 species, and many of them are important economic categories. Nevertheless, several evidences gathered with genome synteny analysis have revealed a number of shared unique mitochondrial gene features in Chondrichthyes, towards a better understanding of the functions and evolution of Chondrichthyes24–27. So far, there was still a notably lack of mtDNA information in Carcharhiniformes. In order to provide a theoretical foundation for the conservation strategy of C. umbratile within Scyliorhinidae and new sight for further studies of phylogenetically-informative sequence data, in the current study the complete mtDNA of C. umbratile was sequenced, assembled and annotated, and compared with other members of Carcharhiniformes.
Results and Discussion
Genome size and organization
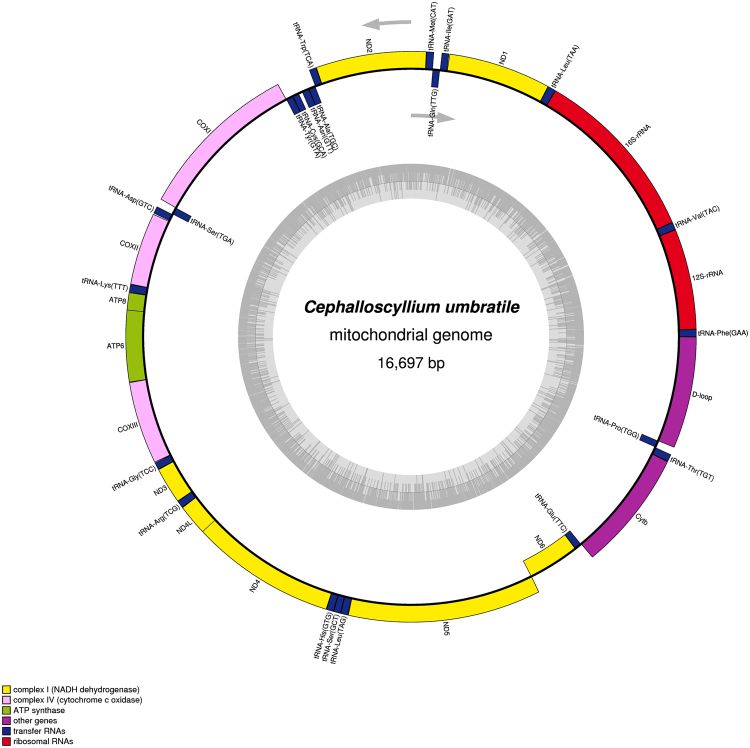
About 1.5 G raw data is generated with reads length 125 bp. Sequencing coverage and depth (X) of mtDNA data is 100% and approximately 394.23, respectively. Reads number is 52,660 and total bases (bp) is 6,582,500. The mtDNA of C. umbratile was a closed-circular DNA molecule of 16,697 bp in length (GenBank: KX354996; Fig. 1, Table 1), which was comparable to other Carcharhiniformes mtDNA ranging from 16,697 bp in Scyliorhinus canicula 25 to 16,719 bp in Carcharhinus acronotus 28. Nucleotide BLAST (blastn) of the whole C. umbratile mtDNA against other Carcharhiniformes revealed sequence identities with closely related species of 88% (S. canicula), 84% (Proscyllium habereri), and 84% (Pseudotriakis microdon) and with distantly related species of 82% (Scoliodon laticaudus), 82% (Hemigaleus microstoma), 82% (Hemipristis elongata) (Supplementary Table 1). The mtDNA of C. umbratile contained 2 rRNA genes, 13 PCGs, 22 tRNA genes and D-loop region. The arrangement of the genes was identical to that of other Scyliorhinidae mtDNA (Table 1)29,30. Among these genes, 29 genes (12 PCGs, 2 rRNA genes and 15 tRNA genes) are located on the heavy strand (H-strand) and the others (1 PCGs and 8 tRNA genes) are located on the light strand (L-strand) (Table 1). These obvious features have also been reported in other Carcharhiniformes species31,32 and could be regarded as effective markers for authentication at genus and species level.
Figure 1.
Map of the Cephalloscyllium umbratile mitochondrial genome. The genes outside the circle are transcribed clockwise, while the genes inside are transcribed counterclockwise. Gene blocks are filled with different colors as the cutline shows. The inner ring shadow indicates the GC content of the genome.
Table 1.
Sequence characteristics of Cephalloscyllium umbratile mitochondrial genome.
| Locus name | One Letter code | From | to | Size | Strand | Nr.of Aminao Acids | Anti-Coden | Inferred Initiation Coden | Inferred Termination Coden | GC_Percent | Intergenic nucleotides* |
|---|---|---|---|---|---|---|---|---|---|---|---|
| tRNA-Phe | F | 1 | 69 | 69 | H | GAA | 37.68% | 0 | |||
| 12S-rRNA | 70 | 1023 | 954 | H | 42.98% | 0 | |||||
| tRNA-Val | V | 1024 | 1095 | 72 | H | TAC | 40.28% | 0 | |||
| 16S-rRNA | 1096 | 2764 | 1669 | H | 36.01% | 0 | |||||
| tRNA-Leu | L | 2765 | 2839 | 75 | H | TAA | 44.00% | 0 | |||
| ND1 | 2840 | 3814 | 975 | H | 324 | ATG | TAA | 38.97% | 3 | ||
| tRNA-Ile | I | 3818 | 3886 | 69 | H | GAT | 39.13% | 1 | |||
| tRNA-Gln | Q | 3888 | 3959 | 72 | L | TTG | 29.17% | 0 | |||
| tRNA-Met | M | 3960 | 4029 | 70 | H | CAT | 40.00% | 0 | |||
| ND2 | 4030 | 5075 | 1046 | H | 348 | ATG | TA | 37.86% | 0 | ||
| tRNA-Trp | W | 5076 | 5144 | 69 | H | TCA | 33.33% | 1 | |||
| tRNA-Ala | A | 5146 | 5214 | 69 | L | TGC | 31.88% | 0 | |||
| tRNA-Asn | N | 5215 | 5287 | 73 | L | GTT | 34.25% | 36 | |||
| tRNA-Cys | C | 5324 | 5389 | 66 | L | GCA | 51.52% | 1 | |||
| tRNA-Tyr | Y | 5391 | 5460 | 70 | L | GTA | 47.14% | 1 | |||
| COXI | 5462 | 7015 | 1554 | H | 517 | GTG | TAA | 38.61% | 0 | ||
| tRNA-Ser | S | 7016 | 7086 | 71 | L | TGA | 45.07% | 3 | |||
| tRNA-Asp | D | 7090 | 7159 | 70 | H | GTC | 32.86% | 7 | |||
| COXII | 7167 | 7857 | 691 | H | 230 | ATG | T | 38.35% | 0 | ||
| tRNA-Lys | K | 7858 | 7932 | 75 | H | TTT | 44.00% | 1 | |||
| ATP8 | 7934 | 8101 | 168 | H | 55 | ATG | TAA | 30.95% | −22 | ||
| ATP6 | 8080 | 8774 | 695 | H | 231 | ATG | TA | 37.55% | 0 | ||
| COXIII | 8775 | 9560 | 786 | H | 261 | ATG | TAA | 42.88% | 2 | ||
| tRNA-Gly | G | 9563 | 9632 | 70 | H | TCC | 27.14% | 0 | |||
| ND3 | 9633 | 9981 | 349 | H | 116 | ATG | T | 40.97% | 0 | ||
| tRNA-Arg | R | 9982 | 10051 | 70 | H | TCG | 32.86% | 0 | |||
| ND4L | 10052 | 10348 | 297 | H | 98 | ATG | TAA | 38.72% | −7 | ||
| ND4 | 10342 | 11722 | 1381 | H | 460 | ATG | T | 37.73% | 0 | ||
| tRNA-His | H | 11723 | 11791 | 69 | H | GTG | 18.84% | 0 | |||
| tRNA-Ser | S | 11792 | 11858 | 67 | H | GCT | 37.31% | 0 | |||
| tRNA-Leu | L | 11859 | 11930 | 72 | H | TAG | 48.61% | 0 | |||
| ND5 | 11931 | 13760 | 1830 | H | 609 | ATG | TAA | 35.85% | −4 | ||
| ND6 | 13757 | 14278 | 522 | L | 173 | ATG | TAA | 36.97% | 0 | ||
| tRNA-Glu | E | 14279 | 14348 | 70 | L | TTC | 32.86% | 2 | |||
| Cytb | 14351 | 15495 | 1145 | H | 381 | ATG | TA | 39.91% | 0 | ||
| tRNA-Thr | T | 15496 | 15567 | 72 | H | TGT | 51.39% | 2 | |||
| tRNA-Pro | P | 15570 | 15638 | 69 | L | TGG | 49.28% | 0 | |||
| D-loop | 15639 | 16697 | 1059 | H | 31.35% | 0 |
+ and − correspond to the H and L strands, respectively.
The nucleotide composition of the mtDNA is biased toward A + T nucleotides (52.9%), which made up of 61.8%, 61.4%, 61.5% and 68.7% in the PCGs, tRNA, rRNA and D-loop region, respectively (Table 2). However, the A + T nucleotide composition in C. umbratile was the lowest among Carcharhiniformes. The positive AT skew (0.025) observed here with the presence of more As than Ts, was similar to that only in Sphyrna tiburo (0.031), nevertheless, mtDNA in majority of Carcharhiniformes showed negative AT skew (Table 2). The GC skew ranged from −0.324 in S. tiburo to 0.040 in C. macloti (Table 2). The C. umbratile mtDNA was negative (−0.245), indicating the presence of more Cs than Gs.
Table 2.
Nucleotide composition of the mitochondrial genome in different Carcharhiniformes mtDNA.
| Species | Size (bp) | A% | T% | G% | C% | A + T % | AT skewness | GC skewness |
|---|---|---|---|---|---|---|---|---|
| Whole mitogenome | ||||||||
| C.umbratile | 16896 | 27.08 | 25.78 | 17.81 | 29.34 | 52.86 | 0.025 | −0.245 |
| S. canicula | 16697 | 30.80 | 31.20 | 14.12 | 23.87 | 62.00 | −0.006 | −0.257 |
| S. tiburo | 16723 | 31.26 | 29.38 | 13.24 | 25.94 | 60.64 | 0.031 | −0.324 |
| P. habereri | 16708 | 30.88 | 31.19 | 14.18 | 23.75 | 62.07 | −0.005 | −0.252 |
| C. acronotus | 16719 | 31.48 | 30.22 | 13.18 | 25.20 | 61.65 | −0.311 | 0.017 |
| C.amblyrhynchoides | 16705 | 31.40 | 30.34 | 13.15 | 25.03 | 61.79 | −0.313 | 0.020 |
| C. amboinensis | 16704 | 31.57 | 30.42 | 13.06 | 24.95 | 62.00 | −0.313 | 0.019 |
| C. brevipinna | 16706 | 31.35 | 30.13 | 13.24 | 25.28 | 61.47 | −0.313 | 0.020 |
| C. leucas | 16704 | 31.47 | 31.10 | 13.11 | 24.32 | 62.57 | −0.300 | 0.006 |
| C.longimanus | 16706 | 31.49 | 30.01 | 13.12 | 25.38 | 61.50 | −0.318 | 0.024 |
| C.macloti | 16701 | 31.61 | 29.19 | 13.02 | 26.18 | 60.80 | −0.336 | 0.040 |
| C.melanopterus | 16706 | 31.28 | 30.06 | 13.32 | 25.33 | 61.35 | −0.311 | 0.020 |
| C. plumbeus | 16706 | 31.25 | 29.89 | 13.32 | 25.54 | 61.14 | −0.314 | 0.022 |
| C. sorrah | 16707 | 31.45 | 29.60 | 13.17 | 25.77 | 61.05 | −0.323 | 0.030 |
| L.tephrodes | 16705 | 31.43 | 29.77 | 13.02 | 25.70 | 61.25 | −0.328 | 0.027 |
| L.macrorhinus | 16702 | 31.71 | 29.36 | 13.14 | 25.80 | 61.06 | −0.325 | 0.039 |
| P. microdon | 16700 | 31.30 | 32.32 | 13.63 | 22.75 | 63.62 | −0.251 | −0.016 |
| T. obesus | 16700 | 31.38 | 29.65 | 13.19 | 25.78 | 61.03 | −0.323 | 0.028 |
| Protein-coding genes | ||||||||
| C.umbratile | 11440 | 28.73 | 33.02 | 13.74 | 24.51 | 61.75 | −0.282 | −0.069 |
| S. canicula | 11430 | 28.71 | 33.15 | 13.85 | 24.30 | 61.85 | −0.274 | −0.072 |
| S. tiburo | 11430 | 28.85 | 31.09 | 13.06 | 26.99 | 59.95 | −0.348 | −0.037 |
| P. habereri | 11430 | 28.83 | 33.25 | 13.74 | 24.18 | 62.08 | −0.275 | −0.071 |
| C. acronotus | 11429 | 29.44 | 31.95 | 12.58 | 26.02 | 61.40 | −0.348 | −0.041 |
| C.amblyrhynchoides | 11430 | 29.45 | 32.30 | 12.59 | 25.66 | 61.75 | −0.342 | −0.046 |
| C. amboinensis | 11430 | 29.58 | 32.32 | 12.49 | 25.61 | 61.90 | −0.344 | −0.044 |
| C. brevipinna | 11430 | 29.36 | 31.92 | 12.65 | 26.06 | 61.29 | −0.346 | −0.042 |
| C. leucas | 11430 | 29.43 | 33.08 | 12.55 | 24.94 | 62.51 | −0.331 | −0.058 |
| C.longimanus | 11430 | 29.55 | 31.85 | 12.53 | 26.07 | 61.40 | −0.351 | −0.038 |
| C.macloti | 11430 | 29.42 | 30.83 | 12.62 | 27.13 | 60.25 | −0.365 | −0.023 |
| C.melanopterus | 11430 | 29.32 | 31.96 | 12.77 | 25.93 | 61.29 | −0.340 | −0.043 |
| C. plumbeus | 11430 | 29.22 | 31.72 | 12.81 | 26.25 | 60.94 | −0.344 | −0.041 |
| C. sorrah | 11430 | 29.34 | 31.41 | 12.74 | 26.52 | 60.74 | −0.351 | −0.034 |
| L.tephrodes | 11247 | 29.23 | 31.52 | 9.29 | 26.69 | 62.80 | −0.484 | −0.038 |
| L.macrorhinus | 11430 | 29.51 | 30.84 | 12.69 | 26.96 | 60.35 | −0.360 | −0.022 |
| P. microdon | 11496 | 29.51 | 34.71 | 13.21 | 22.57 | 64.21 | −0.262 | −0.081 |
| T. obesus | 11430 | 29.22 | 31.35 | 12.78 | 26.65 | 60.57 | −0.352 | −0.035 |
| tRNA | ||||||||
| C.umbratile | 1538 | 32.51 | 28.87 | 17.43 | 21.20 | 61.38 | −0.098 | 0.059 |
| S. canicula | 1551 | 31.53 | 30.82 | 20.12 | 17.54 | 62.35 | 0.068 | 0.011 |
| S. tiburo | 1551 | 32.62 | 27.98 | 17.21 | 22.18 | 60.61 | −0.126 | 0.077 |
| P. habereri | 1553 | 30.71 | 29.75 | 21.31 | 18.22 | 60.46 | 0.078 | 0.016 |
| C. acronotus | 1552 | 30.86 | 29.70 | 21.20 | 30.86 | 60.57 | 0.075 | 0.019 |
| C.amblyrhynchoides | 1551 | 32.62 | 27.92 | 17.28 | 32.62 | 60.54 | −0.124 | 0.078 |
| C. amboinensis | 1548 | 32.62 | 27.78 | 17.31 | 32.62 | 60.40 | −0.126 | 0.080 |
| C. brevipinna | 1550 | 30.77 | 29.55 | 21.35 | 30.77 | 60.32 | 0.076 | 0.020 |
| C. leucas | 1552 | 0.069 | 32.73 | 28.48 | 17.14 | 61.21 | −0.116 | 0.069 |
| C.longimanus | 1553 | 0.077 | 32.39 | 27.75 | 17.51 | 60.14 | −0.121 | 0.077 |
| C.macloti | 1542 | 0.074 | 32.49 | 28.02 | 17.32 | 60.51 | −0.123 | 0.074 |
| C.melanopterus | 1551 | 0.076 | 32.43 | 27.85 | 17.54 | 60.28 | −0.117 | 0.076 |
| C. plumbeus | 1551 | 0.071 | 32.17 | 27.92 | 17.73 | 60.09 | −0.111 | 0.071 |
| C. sorrah | 1552 | 0.003 | 27.90 | 27.71 | 17.53 | 58.23 | −0.121 | 0.003 |
| L.tephrodes | 1551 | 0.080 | 32.75 | 27.92 | 17.21 | 60.67 | −0.125 | 0.080 |
| L.macrorhinus | 1552 | 31.25 | 30.15 | 20.75 | 17.85 | 61.4 | 0.075 | 0.018 |
| P. microdon | 1551 | 31.85 | 28.76 | 17.73 | 21.66 | 60.61 | −0.100 | 0.051 |
| T. obesus | 1552 | 32.73 | 27.90 | 17.27 | 22.10 | 60.63 | −0.123 | 0.080 |
| rRNA | ||||||||
| C.umbratile | 2623 | 34.77 | 26.69 | 17.69 | 20.85 | 61.46 | −0.082 | 0.132 |
| S. canicula | 2630 | 34.26 | 26.50 | 18.02 | 21.22 | 60.76 | −0.081 | 0.128 |
| S. tiburo | 2623 | 35.46 | 26.12 | 17.35 | 21.08 | 61.57 | −0.097 | 0.152 |
| P. habereri | 2619 | 35.01 | 26.42 | 17.83 | 20.73 | 61.44 | −0.075 | 0.140 |
| C. acronotus | 2629 | 35.34 | 26.21 | 17.15 | 21.30 | 61.54 | −0.108 | 0.148 |
| C.amblyrhynchoides | 2624 | 35.21 | 25.88 | 17.34 | 21.57 | 61.09 | −0.109 | 0.153 |
| C. amboinensis | 2627 | 35.40 | 26.19 | 17.17 | 21.24 | 61.59 | −0.106 | 0.150 |
| C. brevipinna | 2626 | 35.15 | 25.89 | 17.40 | 21.55 | 61.04 | −0.107 | 0.152 |
| C. leucas | 2624 | 35.18 | 26.68 | 17.38 | 20.77 | 61.85 | −0.089 | 0.137 |
| C.longimanus | 2625 | 35.20 | 25.71 | 17.33 | 21.75 | 60.91 | −0.113 | 0.156 |
| C.macloti | 2622 | 35.28 | 25.36 | 17.28 | 22.08 | 60.64 | −0.122 | 0.164 |
| C.melanopterus | 2626 | 35.03 | 25.55 | 17.48 | 21.93 | 60.59 | −0.113 | 0.157 |
| C. plumbeus | 2629 | 35.26 | 25.45 | 17.27 | 22.02 | 60.71 | −0.121 | 0.162 |
| C. sorrah | 2627 | 35.25 | 25.58 | 17.24 | 21.93 | 60.83 | −0.120 | 0.159 |
| L.tephrodes | 2624 | 35.37 | 25.69 | 17.15 | 21.72 | 61.10 | −0.118 | 0.159 |
| L.macrorhinus | 2625 | 35.73 | 26.10 | 17.10 | 21.07 | 61.83 | −0.104 | 0.156 |
| P. microdon | 2624 | 35.02 | 26.64 | 17.72 | 20.62 | 61.66 | −0.076 | 0.136 |
| T. obesus | 2622 | 35.51 | 25.55 | 17.09 | 21.85 | 61.06 | −0.122 | 0.163 |
| Control region | ||||||||
| C.umbratile | 1059 | 34.09 | 34.56 | 12.94 | 18.41 | 68.65 | −0.175 | −0.007 |
| S. canicula | 1051 | 33.21 | 33.59 | 13.23 | 19.89 | 66.86 | −0.201 | −0.006 |
| S. tiburo | 1087 | 31.83 | 32.84 | 12.60 | 21.07 | 65.76 | −0.251 | −0.016 |
| P. habereri | 1067 | 32.61 | 33.55 | 13.96 | 19.87 | 66.17 | −0.175 | −0.014 |
| C. acronotus | 1076 | 31.69 | 35.13 | 13.57 | 19.61 | 66.82 | −0.182 | −0.051 |
| C.amblyrhynchoides | 1067 | 31.40 | 35.05 | 13.59 | 19.96 | 66.45 | −0.190 | −0.055 |
| C. amboinensis | 1067 | 31.68 | 35.43 | 13.40 | 19.49 | 67.10 | −0.185 | −0.056 |
| C. brevipinna | 1068 | 31.74 | 35.11 | 13.67 | 19.48 | 66.85 | −0.175 | −0.050 |
| C. leucas | 1066 | 32.27 | 35.08 | 13.32 | 19.32 | 67.35 | −0.184 | −0.042 |
| C.longimanus | 1066 | 31.24 | 35.27 | 13.51 | 19.98 | 66.51 | −0.193 | −0.061 |
| C.macloti | 1066 | 33.40 | 34.80 | 12.38 | 19.42 | 68.20 | −0.221 | −0.021 |
| C.melanopterus | 1067 | 31.58 | 34.58 | 13.40 | 20.43 | 66.17 | −0.208 | −0.045 |
| C. plumbeus | 1063 | 31.14 | 35.47 | 13.55 | 19.85 | 66.60 | −0.189 | −0.065 |
| C. sorrah | 1066 | 31.99 | 34.80 | 13.23 | 19.98 | 66.79 | −0.203 | −0.042 |
| L.tephrodes | 1069 | 32.18 | 34.89 | 13.38 | 19.27 | 67.26 | −0.181 | −0.040 |
| L.macrorhinus | 1063 | 32.64 | 34.24 | 13.26 | 19.85 | 66.89 | −0.199 | −0.024 |
| P. microdon | 1058 | 33.74 | 34.03 | 11.81 | 20.42 | 67.77 | −0.267 | −0.004 |
| T. obesus | 1064 | 31.48 | 35.53 | 13.72 | 19.27 | 67.01 | −0.168 | −0.060 |
Note: The A + T biases of whole mitogenome, protein-coding genes, tRNA, rRNA and control regions were calculated by AT-skew = (A − T)/(A + T) and GC-skew = (G − C)/(G + C), respectively.
Protein-coding gene features
The PCG region formed 68.5% of the C. umbratile mitogenome, and was 11,440 bp long. Furthermore, a contrast of nucleotide composition, AT-skew, and GC-skew of Carcharhiniformes PCGs were exhibited in Table 2. A + T content of the rRNA genes was 61.75%. The AT skew value (−0.282) of the PCG region in the C. umbratile mtDNA was higher than that of several reported mtDNA, nevertheless the negative GC skew (−0.069) was similar to that observed in other fish33,34.
Each PCG was initiated by a canonical ATN codon, except for COXI, which was initiated by a GTG codon (Table 1). Similar results have been documented in other Carcharhiniformes35,36. Seven of 13 PCGs (ND1, COXI, ATP8, COXIII, ND4L, ND5, ND6) used a typical TAA termination codon, which was typical for Carcharhiniformes mtDNA35,36; whereas COXII, ND3 and ND4 terminated with a single T and ATP6, ND2 and Cytb terminated with TA (Table 1). It was akin to sequenced mtDNA of Carcharhiniformes, including Triaenodon obesus 37, Carcharhinus macloti 38, Mustelus griseus 39, S. canicula 25 and C. acronotus 28.
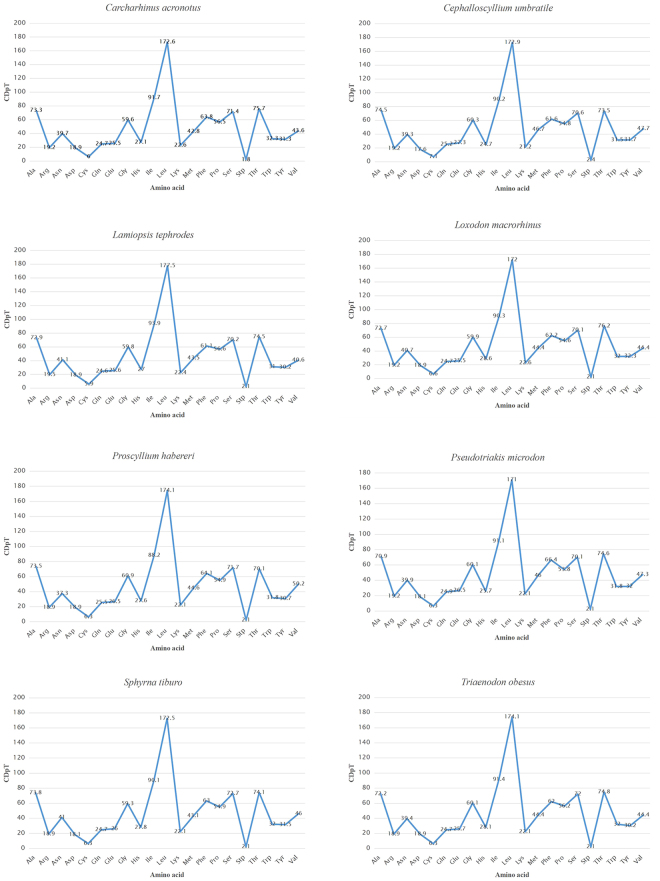
A total of 3,803 amino acids of PCGs are encoded in C. umbratile. In addition, the codon usage is shown in Table 3. The most frequent amino acids in the PCGs of C. umbratile were Leucine (17.3%), Isoleucine (9.02%) and Alanine (7.45%) (Table 3). Relative synonymous codon usage (RSCU) analysis of PCGs in C. umbratile revealed that the codons encoding Leu, Thr, Ala, Arg, Gln, Gly, Pro and Ser were the most frequently present, nevertheless those encoding Asn, Asp, Cys and Lys were rare (Fig. 2). In the PCGs of the eight species examined, codon distributions and amino acid content were corresponding among species (Fig. 3). It was declared that conserved amino acid sequences were present among those fish28,32,40. Moreover, codons with A or T in the third position were overused in comparison to other synonymous codons, for example, the codons for glutamine CAG and GAG were rare, while the synonymous codons CAA and GAA were prevalent (Fig. 4), which is consistent with previous observations of Carcharhiniformes36.
Table 3.
Codon usage of Cephalloscyllium umbratile mitochondrial protein-coding genes.
| Amino acid | Codon | Number | Frequency (%) | RSCU | Amino acid | Codon | Number | Frequency (%) | RSCU |
|---|---|---|---|---|---|---|---|---|---|
| Ala | GCC | 117 | 3.07 | 1.65 | CAC | 56 | 1.47 | 1.19 | |
| GCA | 87 | 2.28 | 1.23 | CAT | 38 | 0.99 | 0.81 | ||
| GCT | 76 | 1.99 | 1.07 | Ile | ATT | 246 | 6.45 | 1.43 | |
| GCG | 4 | 0.11 | 0.06 | ATC | 98 | 2.57 | 0.57 | ||
| Arg | CGA | 40 | 1.05 | 2.19 | Leu | TTA | 229 | 6.01 | 2.08 |
| CGT | 16 | 0.42 | 0.88 | CTA | 153 | 4.02 | 1.39 | ||
| CGC | 13 | 0.34 | 0.71 | CTT | 145 | 3.81 | 1.32 | ||
| CGG | 4 | 0.10 | 0.22 | CTC | 90 | 2.36 | 0.82 | ||
| Asn | AAT | 96 | 2.52 | 1.28 | TTG | 24 | 0.63 | 0.22 | |
| AAC | 54 | 1.42 | 0.72 | CTG | 18 | 0.47 | 0.16 | ||
| Asp | GAT | 44 | 1.15 | 1.31 | Lys | AAA | 77 | 2.02 | 1.90 |
| GAC | 23 | 0.60 | 0.69 | AAG | 4 | 0.10 | 0.10 | ||
| Cys | TGT | 16 | 0.42 | 1.19 | Met | ATA | 136 | 3.57 | 1.53 |
| TGC | 11 | 0.29 | 0.81 | ATG | 42 | 1.10 | 0.47 | ||
| Gln | CAA | 89 | 2.34 | 1.85 | Phe | TTT | 146 | 3.83 | 1.24 |
| CAG | 7 | 0.18 | 0.15 | TTC | 89 | 2.34 | 0.76 | ||
| GAA | 89 | 2.34 | 1.71 | Pro | CCA | 87 | 2.28 | 1.67 | |
| GAG | 15 | 0.39 | 0.29 | CCC | 76 | 1.99 | 1.45 | ||
| Gly | GGA | 88 | 2.31 | 1.53 | CCT | 42 | 1.10 | 0.80 | |
| GGC | 57 | 1.50 | 0.99 | CCG | 4 | 0.10 | 0.08 | ||
| GGT | 51 | 1.34 | 0.89 | Ser | TCA | 89 | 2.34 | 1.99 | |
| GGG | 34 | 0.89 | 0.59 | TCT | 62 | 1.63 | 1.38 | ||
| His | CAC | 56 | 1.47 | 1.19 | TCC | 58 | 1.52 | 1.29 | |
| Amino acid | Codon | Number | Frequency (%) | RSCU | Amino acid | Codon | Number | Frequency (%) | RSCU |
| AGC | 34 | 0.89 | 0.76 | ACG | 7 | 0.18 | 0.1 | ||
| AGT | 21 | 0.55 | 0.47 | Trp | TGA | 107 | 2.81 | 1.78 | |
| TCG | 5 | 0.13 | 0.11 | TGG | 13 | 0.34 | 0.22 | ||
| Stp* | TAA | 7 | 0.18 | 4 | Tyr | TAT | 88 | 2.31 | 1.45 |
| AGA | 0 | 0 | 0 | TAC | 33 | 0.87 | 0.55 | ||
| AGG | 0 | 0 | 0 | Val | GTA | 80 | 2.10 | 1.76 | |
| TAG | 0 | 0 | 0 | GTT | 52 | 1.36 | 1.14 | ||
| Thr | ACA | 117 | 3.07 | 1.67 | GTC | 31 | 0.81 | 0.68 | |
| ACC | 99 | 2.60 | 1.41 | GTG | 19 | 0.50 | 0.42 | ||
| ACT | 57 | 1.50 | 0.81 |
Figure 2.
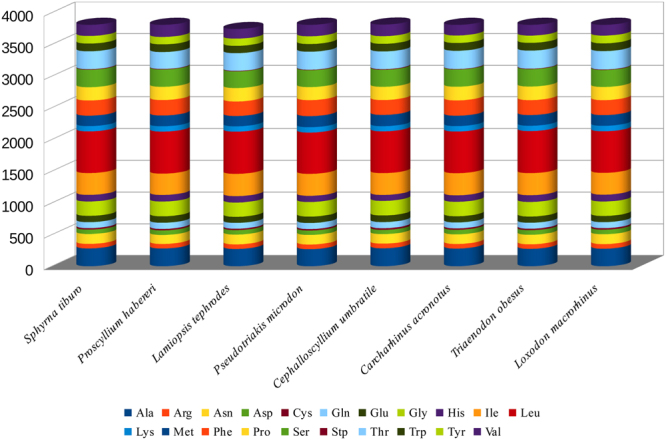
Comparison of codon usage within the mitochondrial genome of members of the Carcharhiniformes. Species (Sphyrna tiburo, Proscyllium habereri, Lamiopsis tephrodes, Pseudotriakis microdon, Cephalloscyllium umbratile, Carcharhinus acronotus, Triaenodon obesus, Loxodon macrorhinus) represent the superfamily to which the species belongs (Sphyrna, Proscyllium, Lamiopsis, Pseudotriakis, Cephaloscyllium, Carcharhinus, Triaenodon, Loxodon).
Figure 3.
Codon distribution in members of eight superfamilies in the Carcharhiniformes. CDspT = codons per thousand codons.
Figure 4.
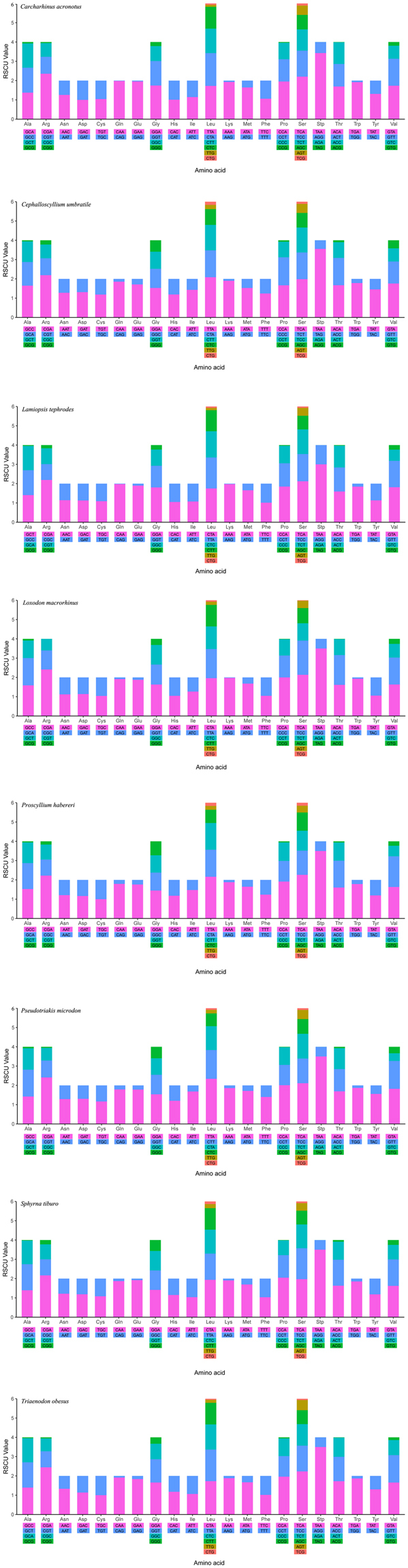
Relative Synonymous Codon Usage (RSCU) of the mitochondrial genome of eight superfamilies in the Carcharhiniformes. Codon families are plotted on the x-axis. Codons indicated above the bar are not present in the mitogenome.
Transfer RNAs and ribosomal RNAs
The representative complement structures of 22 tRNAs were identified in the C. umbratile mtDNA, ranging from 62 bp (tRNAThr) to 76 bp (tRNALys)35,36 for 1,538 bp in total (Table 1). Of those, the highest A + T content of tRNAs was S. canicula and the lowest was C. sorrah. Fifteen tRNA genes were encoded on the H strand while the remains were located in the L strand (Table 1). The overall A+T content of tRNAs was 61.38% which was approximate to that observed in Loxodon macrorhinus (61.4%). The negative AT skew (−0.098) and positive GC skew (0.059) showed in the C. umbratile mtDNA were also analogous with several sequenced Carcharhiniformes (Table 2).
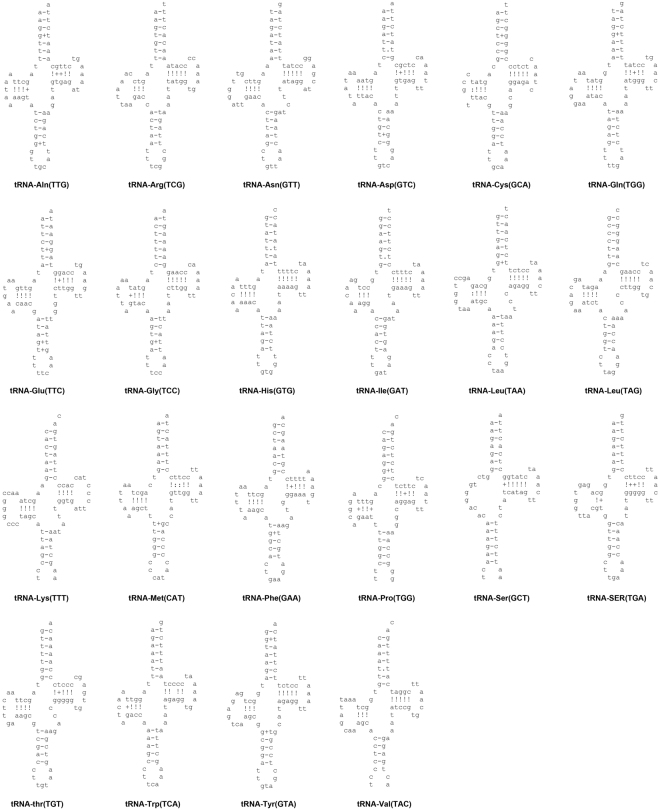
The forecasted tRNAs were shown in Fig. 5. All of the tRNAs could be folded into classic clover-leaf secondary structures in C. umbratile, except for tRNA-Ser (GCT), which lacked the dihydrouridine ‘DHU’ arm (Fig. 5). The ‘DHU’ arm of this tRNA was a large loop instead of the conserved stem-and-loop structure. Due to a representative characteristics41, it was also observed in other Chondrichthyes mtDNA, including Chiloscyllium griseum 42 T. obesus 37 and so on. Fifteen of the tRNA genes were each observed to have at least one G-T mismatches in their respective secondary structures, which forming a weak bond. Five T-T mismatches were present in the respective amino acid acceptor stems of tRNA Asp(GTC), tRNA Cys(GCA), tRNA His(GTG), tRNA Ile(GAT) and tRNAMet(CAT) (Fig. 5). Interestingly, A-G mismatch was also present in tRNA-Leu (TAA). Unmatched base pairs perceived in tRNA sequences can be amended by RNA-editing mechanisms that were well known for vertebrate mtDNA43.
Figure 5.
Putative secondary structures for 22 tRNA genes in mitochondrial genome of Cephalloscyllium umbratile. Watson-Crick and GT bonds are illustrated by “−” and “+”, respectively.
The A + T content of the rRNA genes was 61.46%, indicating an A+C-rich trend as in other Scyliorhinidae fish25. AT and GC skews were negative (−0.082) and positive (0.132), respectively (Table 2). The 12S rRNA and 16S rRNA subunit gene of C. umbratile was 954 bp and 1,668 bp in length, respectively. As in other vertebrates44, both two genes are separated by the tRNA Val gene, and located between tRNA Phe and tRNA Leu(UUR) (Fig. 1, Table 1). The overall content of the rRNA was analogous to that observed for other Carcharhiniformes.
The control region
The length of D-loop region of C. umbratile was 1,059 bp, which was less long than majority of Carcharhiniformes. The A + T content was 68.65%, and equal with other Carcharhiniformes (Table 2), which was consistent with the findings of previous reports on other teleosts33,45,46. Moreover, both of the AT-skew and GC-skew were strongly negative (Table 2).
Overlapping and intergenic spacer regions
There were three gene boundaries where bases overlapped between adjacent genes, ranging from 4–22 bp in size. The longest overlapping region was 22 bp between ATP8 and ATP6 (Table 1) which has been documented in several other Chondrichthyes mtDNA4,25,32. Moreover, intergenic spacers of C. umbratile were spread over 12 locations and ranged from 1–36 bp, making up 60 bp in total, and the longest intergenic spacer region (36 bp) was between tRNA Asn and tRNA Cys (Table 1).
Synonymous and nonsynonymous substitutions
The ratio of Ka/Ks is generally regarded as a pointer of selective pressure and evolutionary relations at the molecular level among homogenous or heterogeneous species47,48. It is reported that Ka/Ks > 1, Ka/Ks = 1, and Ka/Ks < 1 popularly declared positive selection, neutral mutation and negative selection, respectively49. To investigate the evolutionary rate differences in three Carcharhiniformes mtDNA (C. umbratile, S. canicula and P. habereri), sequence divergences by counting Ka and Ks substitution rates were next calculated. The Ka/Ks values of 13 PCGs varied from 0.0198 (COXI) to 0.5322 (ATP8) and were less than 0.6 (Ka was lower than Ks) for all other genes which indicated a strong purifying and negative selection in those fishes (Fig. 6). Our result of the Ka/Ks ratio illustrated that the multitudinous genes evolved under strong negative selection which meant natural selection against profitless mutations with negative selective coefficients50. The percentages of variable sites of SC/PH were the highest in COXIII and ND1 among the groups, while the percentages was the least in COXI gene, which indicated that COXIII and ND1 were under the least selective pressure, and COXI was under the most selective pressure among all mitochondrial proteins. In C. umbratile and S. canicula, the ratio of Ka/Ks was the least in all 13 protein-coding genes compared to P. habereri, implying that these two Scyliorhinidae fish had the closer phylogenetic relationship than P. habereri, which was consistent with their rozmieszczenie naturalne and ecological habit25.
Figure 6.
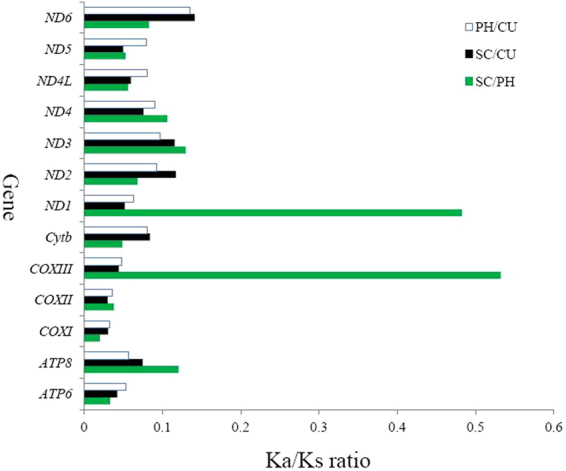
Ka/Ks ratios for the 13 mitochondrial protein-coding genes among the reference Cephalloscyllium umbratile (CU), Scyliorhinus canicula (SC), Proscyllium habereri (PH).
Phylogeny
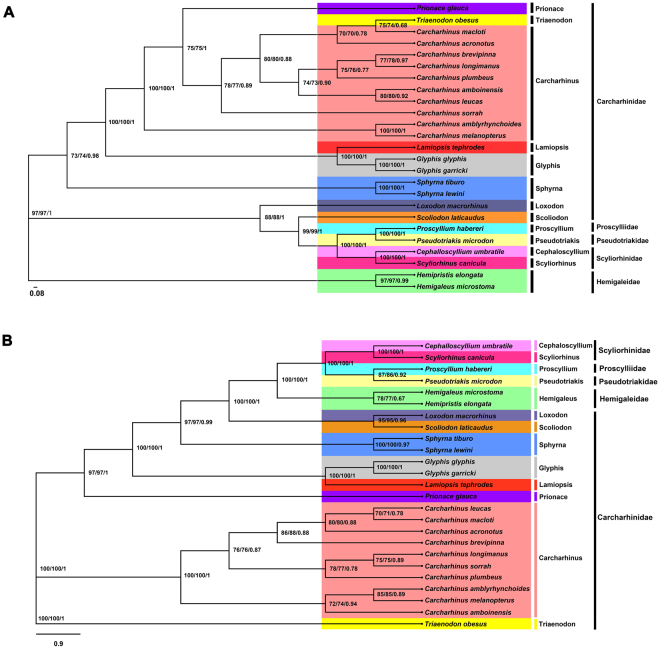
To understand the phylogenetic relationships among Carcharhiniformes, base on Maximum Likelihood (ML), Neighbor Joining (NJ) and Bayesian Inference (BI) methods, a dataset of 25 species containing the concatenated nucleic acid and amino acid sequences of 13 PCGs was used to generate phylogenetic relationships (Fig. 7). The topologies of the 6 phylogenetic trees were analogical in our study. The results implied that strong statistics supported for the following relationship among the 5 Superfamily (Scyliorhinidae, Carcharhinidae, Hemigaleidae, Proscylliidae, Pseudotriakidae) (Fig. 7A,B). This clustered pattern of 5 Superfamily was broadly consistent with previous studies32,42,51–53. Furthermore, based on all of ML, NJ and BI methods, 5 superfamily divided into 13 closely genera, and C. umbratile (Cephaloscyllium) was most closely related to S. canicula (Scyliorhinus) in Scyliorhinidae, which was accord with the tendency of nucleotide sequence identity and a recent study51,54–57. Scyliorhinidae was most closely related to Proscylliidae. Additionally, further taxon sampling within Scyliorhinidae and related superfamilies is required to resolve the location of Scyliorhinidae in Carcharhiniformes.
Figure 7.
Phylogenetic trees of Cephalloscyllium umbratile relationships from the nucleotide (A) and amino acid datasets (B). Sequences alignment of mtDNA were analyzed using the MEGA 6.0 and Phylobayes 3.3 f software with Maximum likelihood (ML), Maximum parsimony (MP) and Bayesian inference (BI) method, respectively. The accession numbers of the sequences used in the phylogenetic analysis are listed in Supplementary Table 1.
Materials and Methods
Sample collection and mitochondrial DNA extraction
C. umbratile juveniles were collected from South China Sea (Longitude 5°20.267′ N and latitude 109°48.435′ E) in September 2014 and directly frozen. Muscle tissues were used for DNA extraction according to the Genomic DNA Extraction Kit’s instructions (TaKaRa MiniBEST Universal Genomic DNA Extraction Kit Ver.5.0, Japan). The quantity (concentration) of isolated total DNA was determined by NANODROP 2000 spectrophotometer (Thermo Scientific, USA). Furthermore, quality of extracted DNA was assessed by electrophoresis on a 1% agarose gel stained with Gel Red™ (Biotium).
Genome sequencing
According to NEBNext DNA sample libraries kit (NEB, New England)‘s instructions, the normalized DNA (4 μg) was used to structure the paired-end library. Size and quantification estimation of the library were implemented by a Bioanalyzer 2100 High Sensitivity DNA chip (Agilent, USA). Illumina HiSeq. 2500 (2 × 101 bp paired-end reads) (Illumina, USA) was used to sequence the normalized library (2 nM).
Genome assembly and annotation
A de novo assembly of the paired-end HiSeq reads was performed using SeqMan NGen (http://www.dnastar.com/t-tutorials-seqman-ngen.aspx) (DNASTAR Inc., Madison, WI, USA)58. Assembly parameters minimum match percentage, match spacing, match size, gap penalty, mismatch penalty, maximum gap length and expected genome length were set to 93, 10, 50, 30, 20, 6% and 16,000, respectively. Accordance sequence was exported and ends were manually edited to remove duplicated nucleotides. Subsequently, the assembled sequences were aligned to NCBI nt database with blastn method (https://blast.ncbi.nlm.nih.gov/). Sequences that mapping to Carcharhiniformes mtDNA were considered as C. umbratile mtDNA. To verify the accuracy of the assembled mtDNA sequence, the primers (Supplementary Table 2) were used to amplify the genome sequence. The procedure of PCR amplification was referred from Sun et al.59. To determine whether this method was accurate, the sequence segments of same genomic region obtained from Sanger sequencing and shotgun assembly were compared. If they were identical, that meaning this method was precise. Moreover, the PCGs, rRNA genes, tRNA genes and D-loop region of mtDNA were annotated by MitoAnnotator (http://mitofish.aori.u-tokyo.ac.jp/annotation/input.html)60 with parameters of complete circular genome. The mtDNA sequence of C. umbratile has been deposited in the GenBank database under accession numbers KX354996.
Genome sequence analysis
tRNAscan-SE Search Server 1.21 program was used to primordially determine Transfer RNAs61,62. The gene map of C. umbratile mtDNA was built by OGDRAW1.2 and embellished manually63. The strand skew values were reckoned in terms of the formulae by Perna and Kocher (1995)64. The mode of “models- > Compute Codon Usage Bias” was chose to obtain RSCU in MEGA 6.065. To determine the evolutionary branching of the Carcharhiniformes lineage, codon usage in the 13 PCGs and the rates of Ka/Ks substitutions in the mtDNA of Carcharhiniformes were calculated by DnaSP 5.10.0166. To describe base composition, we analyzed skew as described as below: AT-skew = (A − T)/(A + T) and GC-skew = (G − C)/(G+C)67.
Phylogenetic analysis
To discuss the phylogenetic position of Carcharhiniformes, a total of 25 species of 13 PCG sequences were used to perform phylogenetic analysis, including those of C. umbratile. Alignments of the 13 concatenated PCGs nucleotide and amino acid sequences were conducted using ClustalX version 2.0 with default parameters68. Phylogenetic analyses for each concatenated dataset was performed using ML, MP and BI methods with MEGA 6.0 and Phylobayes 3.3 f, respectively65,69. The methods of ML and MP analysis were performed with GTR+I+G model and Subtree-Purning-Regrafting (SPR) model using MEGA 6.0, respectively. The evaluation of node accuracy was done by using 1,000 bootstrap replicates in MEGA 6.0 with default parameters. Furthermore, BI analysis was selecting the CAT-GTR model, two independent Markov chain Monte Carlo (MCMC) chains were run for 10,000 cycles. The phylogenetic tree was embellished using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Electronic supplementary material
Acknowledgements
This work was supported by the Special Scientific Research Funds for Central Non-profit Institute, Chinese Academy of Fishery Sciences (2016HY-JC0304), the Science and Technology Infrastructure Construction Project of Guangdong Province (2014A030305005, 2015A030303008), National Infrastructure of Fishery Germplasm Resource Project (2017DKA30470).
Author Contributions
K.C.Z. and D.C.Z. designed the research and wrote the paper. N.W. and H.Y.G. performed the research. Y.Y.L. and N.Z. analyzed the data, S.G.J. contributed reagents/materials/analysis tools.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-15702-0.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Iglésias, S., Tanaka, S. & Nakaya, K. Cephaloscyllium umbratile. The IUCN Red List of Threatened Species: e.T161724A5488954. 10.2305/IUCN.UK.2009-2.RLTS.T161724A5488954.en (2009).
- 2.Momigliano P, Harcourt R, Robbins WD, Stow A. Connectivity in grey reef sharks (Carcharhinus amblyrhynchos) determined using empirical and simulated genetic data. Sci. Rep. 2015;5:13229. doi: 10.1038/srep13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delser PM, et al. Population genomics of C. melanopterus using target gene capture data: demographic inferences and conservation perspectives. Sci. Rep. 2016;6:33753. doi: 10.1038/srep33753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chai AH, Yamaguchi A, Furumitsu K, Zhang J. Mitochondrial genome of Japanese angel shark Squatina japonica (Chondrichthyes: Squatinidae) Mitochondrial DNA Part A. 2016;27(2):832–833. doi: 10.3109/19401736.2014.919463. [DOI] [PubMed] [Google Scholar]
- 5.Chen X, Shen XJ, Arunrugstichai S, Ai WM, Xiang D. Complete mitochondrial genome of the blacktip reef shark Carcharhinus melanopterus (Carcharhiniformes: Cfarcharhinidae) Mitochondrial DNA Part A. 2016;27(2):873–874. doi: 10.3109/19401736.2014.919483. [DOI] [PubMed] [Google Scholar]
- 6.Avise, J. C. Phylogeography: the history and formation of species. Massachusetts: Harvard university press (2000).
- 7.Moritz C. Uses of Molecular Phylogenies for Conservation. Philos T R Soc B. 1995;349:113–118. doi: 10.1098/rstb.1995.0097. [DOI] [Google Scholar]
- 8.Hao W, Richardson AO, Zheng Y, Palmer JD. Gorgeous mosaic of mitochondrial genes created by horizontal transfer and gene conversion. P Natl A Sci India A. 2010;107:21576–21581. doi: 10.1073/pnas.1016295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lei RH, Shore GD, Brenneman RA, et al. Complete sequence and gene organization of the mitochondrial genome for Hubbard’s sportive lemur (Lepilemur hubbardorum) Gene. 2010;464(1-2):44–49. doi: 10.1016/j.gene.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Zhang DC, et al. Shotgun assembly of the mitochondrial genome from Fenneropenaeus penicillatus with phylogenetic consideration. Marine. Genomics. 2015;24:379–386. doi: 10.1016/j.margen.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Ye J, Fang R, Yi JP, Zhou GL, Zheng JZ. The complete sequence determination and analysis of four species of Bactrocera mitochondrial genome. Plant Quar. 2010;24(3):11–14. [Google Scholar]
- 12.Krzywinski J, et al. Analysis of the evolutionary forces shaping mitochondrial genomes of a Neotropical malaria vector complex. Mol. Phylogenet. Evol. 2011;58(3):469–477. doi: 10.1016/j.ympev.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma H, et al. The complete mitochondrial genome sequence and gene organization of the mud crab (Scylla paramamosain) with phylogenetic consideration. Gene. 2013;519(1):120–127. doi: 10.1016/j.gene.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 14.Cameron SL. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu. Rev. Entomol. 2014;59:95–117. doi: 10.1146/annurev-ento-011613-162007. [DOI] [PubMed] [Google Scholar]
- 15.Williams ST, Foster PG, Littlewood DT. The complete mitochondrial genome of a turbinid vetigastropod from MiSeq Illumina sequencing of genomic DNA and steps towards a resolved gastropod phylogeny. Gene. 2014;533:38–47. doi: 10.1016/j.gene.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Chen H, Lin LL, Chen X, Ai WM, Chen SB. Complete mitochondrial genome and the phylogenetic position of the Blotchy swell shark Cephaloscyllium umbratile. Mitochondrial DNA Part A. 2016;27(4):3045–3047. doi: 10.3109/19401736.2015.1063127. [DOI] [PubMed] [Google Scholar]
- 17.Simon C, et al. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994;87:651–701. doi: 10.1093/aesa/87.6.651. [DOI] [Google Scholar]
- 18.Dowton M, Castro L, Austin A. Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: the examination of genome ‘morphology’. Invertebr. Syst. 2002;16:345–356. doi: 10.1071/IS02003. [DOI] [Google Scholar]
- 19.Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach AT. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev. Ecol. Evol. Syst. 2006;37:545–579. doi: 10.1146/annurev.ecolsys.37.091305.110018. [DOI] [Google Scholar]
- 20.Oliveira MT, et al. Structure and evolution of the mitochondrial genomes of Haematobia irritans and Stomoxis calcitrans: the Muscidae (Diptera: Calyptratae) perspective. Mol. Phylogenet. Evol. 2008;48:850–857. doi: 10.1016/j.ympev.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 21.Moreno M, et al. Complete mtDNA genomes of Anopheles darlingi and an approach to anopheline divergence time. Malar. J. 2010;9:127. doi: 10.1186/1475-2875-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behura SK, et al. Complete sequences of mitochondria genomes of Aedes aegypti and Culex quinquefasciatus and comparative analysis of mitochondrial DNA fragments inserted in the nuclear genomes. Insect Biochem. Mol. Biol. 2011;41:770–777. doi: 10.1016/j.ibmb.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cameron SL. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst. Entomol. 2014;39(3):400–411. doi: 10.1111/syen.12071. [DOI] [Google Scholar]
- 24.Delarbre C, Barriel V, Tillier S, Janvier P, Gachelin G. The main features of the craniate mitochondrial DNA between the ND1 and the COI genes were established in the common ancestor with the lancelet. Molecular Biol. and Evol. 1997;14(8):807–813. doi: 10.1093/oxfordjournals.molbev.a025821. [DOI] [PubMed] [Google Scholar]
- 25.Delarbre C, et al. The complete nucleotide sequence of the mitochondrial DNA of the dogfish. Scyliorhinus canicula. Genetics. 1998;150(1):331–344. doi: 10.1093/genetics/150.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasmussen AS, Arnason U. Molecular studies suggest that cartilaginous fishes have a terminal position in the piscine tree. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(5):2177–2182. doi: 10.1073/pnas.96.5.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H, Ding W, Shan L, Chen X, Ai W. Complete mitochondrial genome and the phylogenetic position of the blackspotted catshark Halaelurus burgeri (Carcharhiniformes: Scyliorhinidae) Mitochondrial DNA B Resour. 2016;1(1):369–370. doi: 10.1080/23802359.2016.1168722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang L, Matthes-Rosana KA, Naylor GJP. Complete mitochondrial genome of the blacknose shark Carcharhinus acronotus (Elasmobranchii: Carcharhinidae) Mitochondrial DNA Part A. 2016;27(1):169–170. doi: 10.3109/19401736.2013.878928. [DOI] [PubMed] [Google Scholar]
- 29.Oh DJ, Kim JY, Lee JA, Jung YH. Complete mitochondrial genome of the multicolorfin rainbowfish and Halichoeres poecilopterus (Perciformes, Labridae) Korean J. Genet. 2007;29(1):65–72. [Google Scholar]
- 30.Han, Y. et al. The complete mitochondrial genome of Cheilinus undulates based on high-throughput sequencing technique. MitochondrialDNA 1–3; 10.3109/19401736.2014.971276 (2014). [DOI] [PubMed]
- 31.Chen X, Xiang D, Xu YZW, Shi XF. Complete mitochondrial genome of the scalloped hammerhead Sphyrna lewini (Carcharhiniformes: Sphyrnidae). Mitochondrial. DNA. 2015;26(4):621–622. doi: 10.3109/19401736.2013.834432. [DOI] [PubMed] [Google Scholar]
- 32.Pindaro DJ, Natalia BV, Douglas HA, Manuel UA. Complete mitochondrial DNA genome of bonnethead shark, Sphyrna tiburo, and phylogenetic relationships among main superorders of modern elasmobranchs. Meta gene. 2016;7:48–55. doi: 10.1016/j.mgene.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi, X. F., Tian, P., Lin R. C., Huang D. Y. & Wang J. J. Characterization of the Complete Mitochondrial Genome Sequence of the Globose Head Whiptail Cetonurus globiceps (Gadiformes: Macrouridae) and Its Phylogenetic Analysis. Plos One, 10.1371/journal.pone.0153666 (2016). [DOI] [PMC free article] [PubMed]
- 34.Zhu KC, et al. Characterization of complete mitochondrial genome of fives tripe wrasse (Thalassoma quinquevittatum, Lay & Bennett, 1839) and phylogenetic analysis. Gene. 2017;98:71–78. doi: 10.1016/j.gene.2016.10.042. [DOI] [PubMed] [Google Scholar]
- 35.Alam MT, Petit RA, Read TD, Dove ADM. The complete mitochondrial genome sequence of the world’s largest fish, the whale shark (Rhincodon typus), and its comparison with those of related shark species. Gene. 2016;539(1):44–49. doi: 10.1016/j.gene.2014.01.064. [DOI] [PubMed] [Google Scholar]
- 36.Periasamy R, Chen X, Ingole B, Liu WA. Complete mitochondrial genome of the Spadenose shark Scoliodon laticaudus (Carcharhiniformes: Carcharhinidae) Mitochondrial DNA Part A. 2016;27(5):3248–3249. doi: 10.3109/19401736.2015.1007368. [DOI] [PubMed] [Google Scholar]
- 37.Chen X, Sonchaeng P, Yuvanatemiya V, Nuangsaeng B, Ai WM. Complete mitochondrial genome of the whitetip reef shark Triaenodon obesus (Carcharhiniformes: Carcharhinidae) Mitochondrial DNA Part A. 2016;27(2):947–948. doi: 10.3109/19401736.2014.926499. [DOI] [PubMed] [Google Scholar]
- 38.Chen X, Liu M, Xiao JM, Yang WD, Peng ZQ. Complete mitochondrial genome of the hardnose shark Carcharhinus macloti (Carcharhiniformes: Carcharhinidae) Mitochondrial DNA Part A. 2016;27(2):1090–1091. doi: 10.3109/19401736.2014.930836. [DOI] [PubMed] [Google Scholar]
- 39.Chen X, Peng ZQ, Pan LH, Shi XF, Cai L. Mitochondrial genome of the spotless smooth-hound Mustelus griseus (Carcharhiniformes: Triakidae) Mitochondrial DNA Part A. 2016;27(1):78–79. doi: 10.3109/19401736.2013.873908. [DOI] [PubMed] [Google Scholar]
- 40.Wang JJ, Chen H, Lin LL, Ai WM, Chen X. Mitochondrial genome and phylogenetic position of the sliteye shark Loxodon macrorhinus. Mitochondrial DNA Part A. 2016;27(6):4288–4289. doi: 10.3109/19401736.2015.1082099. [DOI] [PubMed] [Google Scholar]
- 41.Wolstenholme DR. Animal mitochondrial DNA: structure and evolution. Int. Rev. Cytol. 1992;141:173–216. doi: 10.1016/S0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]
- 42.Chen X, et al. The complete mitochondrial genome of the grey bamboo shark (Chiloscyllium griseum) (Orectolobiformes: Hemiscylliidae): genomic characterization and phylogenetic application. ACTA Oceanologica Sinica. 2013;32(4):59–65. doi: 10.1007/s13131-013-0298-0. [DOI] [Google Scholar]
- 43.Janke A, Pääbo S. Editing of a tRNA anticodon in marsupial mitochondria changes its codon recognition. Nucleic. Acids. Res. 1993;21(7):1523–1525. doi: 10.1093/nar/21.7.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Satoh TP, Miya M, Endo H, Nishida M. Round and pointed-head grenadier fishes (Actinopterygii: Gadiformes) represent a single sister group: Evidence from the complete mitochondrial genome sequences. Mol Phylogenet Evol. 2006;40:129–138. doi: 10.1016/j.ympev.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 45.Breines R, Ursvik A, Nymark M, Johansen SD, Coucheron DH. Complete mitochondrial genome sequences of the Arctic Ocean codfishes Arctogadus glacialis and Boreogadus saida reveal oriL and tRNA gene duplications. Polar Biol. 2008;31:1245–1252. doi: 10.1007/s00300-008-0463-7. [DOI] [Google Scholar]
- 46.Wu QL, et al. The complete mitochondrial genome of Taeniogonalos taihorina (Bischoff) (Hymenoptera: Trigonalyidae) reveals a novel gene rearrangement pattern in the Hymenoptera. Gene. 2014;543:76–84. doi: 10.1016/j.gene.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Shen X, et al. The complete mitochondrial genomes of two common shrimps (Litopenaeus vannamei and Fenneropenaeus chinensis) and their phylogenomic considerations. Gene. 2007;403(1-2):98–109. doi: 10.1016/j.gene.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 48.Li, X. J., Huang, Y. & Lei, F. M. Comparative mitochondrial genomics and phylogenetic relationships of the Crossoptilon species (Phasianidae, Galliformes). BMC Genomics, 16(42), 10.1186/s12864-015-1234-9 (2015). [DOI] [PMC free article] [PubMed]
- 49.Nei, M. & Kumar, S. Molecular Evolution and Phylogenetics. New York: Oxford University Press (2000).
- 50.Yang Z, Bielawski JP. Statistical methods for detecting molecular adaptation. Trends Ecol Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Velez-Zuazo X, Agnarsson I. Shark tales: A molecular species-level phylogeny of sharks (Selachimorpha, Chondrichthyes) Mole. Phyl and Evol. 2011;58(2):207–217. doi: 10.1016/j.ympev.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 52.Swift, D. G. et al. Evidence of positive selection associated with placental loss in tiger sharks. BMC Evol. Biol. 16, 10.1186/s12862-016-0696-y (2016). [DOI] [PMC free article] [PubMed]
- 53.Liu, S. Y. V., Chan, C. L. C., Lin, O., Hu, C. S. & Chen, C. A. DNA Barcoding of Shark Meats Identify Species Composition and CITES-Listed Species from the Markets in Taiwan. PLOS One8(11), 10.1371/journal.pone.0079373 (2013). [DOI] [PMC free article] [PubMed]
- 54.Stehmann M, Herman J, Hovestadt-Euler M, Hovestadt DC. Contributions to the study of the comparative morphology of teeth and other relevant ichthyodorulites in living supraspecific taxa of chondrichthyan fishes - Part A: Selachii. Addendum to 1: Order Hexanchiformes - Family Hexanchidae, 2: Order Carcharhiniformes, 2a: Family Triakidae, 2b: Family Scyliorhinidae, 2c: Family Carcharhinidae, Hemigaleidae, Leptochairiidae, Sphyrnidae, Proscylliidae and Pseudotriakidae, 3: Order Qqualiformes: Family Echinorhinidae, Oxynotidae and Squalidae. Tooth vascularization and phylogenetic interpretation. Bulletin de l’Institut Royal des Sciences Naturelles de Belgique Biologie. 2003;75:5–26. [Google Scholar]
- 55.Moftah M, Aziz SHA, Elramah S, Favereaux A. Classification of Sharks in the Egyptian Mediterranean Waters Using Morphological and DNA Barcoding Approaches. PLoS ONE. 2011;6(4):1–7. doi: 10.1371/journal.pone.0027001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gruber DF, et al. Biofluorescence in Catsharks (Scyliorhinidae): Fundamental Description and Relevance for Elasmobranch Visual Ecology. Sci. Rep. 2016;6:24751. doi: 10.1038/srep24751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Naylor, G. J. P. et al. Elasmobranch phylogeny: a mitochondrial estimate based on 595 species. In: Carrier, J. C., Musick, J. A. & Heithaus, M. R. (Eds.), Biology of sharks and their relatives. Second Edition. CRC Press, Boca Raton, pp. 31–56 (2012).
- 58.SeqMan NGen, version 3. DNASTAR Inc. Madison, WI, USA (2012).
- 59.Sun YX, et al. Characterization of the Complete Mitochondrial Genome of Leucoma salicis (Lepidoptera: Lymantriidae) and Comparison with Other Lepidopteran Insects. Sci. Rep. 2016;6:39153. doi: 10.1038/srep39153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iwasaki Fukunaga T, Isagozawa R, Nishida MW. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol. Biol. Evol. 2013;30:2531–2540. doi: 10.1093/molbev/mst141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2007;33:686–689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lohse, M., Drechsel, O., Kahlau, S. & Bock, R. Organellar Genome DRAW a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 575–581; 10.1093/nar/gkt289 (2013). [DOI] [PMC free article] [PubMed]
- 64.Perna NT, Kocher TD. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995;41(3):353–358. doi: 10.1007/BF01215182. [DOI] [PubMed] [Google Scholar]
- 65.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rozas, J. DNA sequence polymorphism analysis using DnaSP. In: Posada, D. (Ed.), Bioinformatics for DNA Sequence Analysis Methods in Molecular Biology Series vol. 537. Humana Press, NJ, USA, pp. 337–350 (2009). [DOI] [PubMed]
- 67.Junqueira ACM, et al. The mitochondrial genome of the blowfly Chrysomya chloropyga (Diptera: Calliphoridae) Gene. 2004;339:7–15. doi: 10.1016/j.gene.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 68.Larkin MA, et al. ClustalW and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 69.Nicolas L, Thomas L, Samuel B. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25(17):2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.