Summary
Forest edges influence more than half the world’s forests and contribute to worldwide declines in biodiversity and ecosystem functions. However, predicting these declines is challenging in heterogeneous fragmented landscapes. We assembled an unmatched global dataset on species responses to fragmentation and developed a new statistical approach for quantifying edge impacts in heterogeneous landscapes to quantify edge-determined changes in abundance of 1673 vertebrate species. We show that 85% of species’ abundances are affected, either positively or negatively, by forest edges. Forest core species, which were more likely to be listed as threatened by the IUCN, only reached peak abundances at sites farther than 200-400 m from sharp high-contrast forest edges. Smaller-bodied amphibians, larger reptiles and medium-sized non-volant mammals experienced a larger reduction in suitable habitat than other forest core species. Our results highlight the pervasive ability of forest edges to restructure ecological communities on a global scale.
Introduction
Fragmentation of forest ecosystems has critical and on-going impacts that erode biodiversity and ecological processes1–6. Fragmentation is a ubiquitous phenomenon, with nearly 20% of the world’s remaining forest now found within 100 m of an edge, 50% within 500 m and 70% within 1 km1. Efforts to understand and manage the impacts of fragmentation have thus become critical for effective conservation action7. Ecological effects emanating from edges between forest and non-forest habitat change biophysical environments for species8 and can drive species that otherwise inhabit core forest to extinction over spatial scales of more than 1 km9. Moreover, edge effects alter the amount of ‘effective’ habitat area in a landscape4,10, suggesting they are at least as important as habitat amount11 in driving biodiversity responses to land use change. However, our capacity to predict which species and ecosystem functions are likely to disappear first from edge-dominated landscapes is still limited. In particular, we lack consistent approaches to quantify the impacts of edge effects in a rigorous manner12 across species13 and key functional groups14, leading to potentially distorted projections of overall changes in biodiversity in fragmented landscapes.
Species’ traits frameworks15,16 should form a reliable, heuristic tool to predict species’ sensitivities to edge effects in the way that they do for predicting species’ extinction risks17,18. A paucity of meta-analyses in the fragmentation literature12 has prevented such frameworks from being tested robustly, despite an abundance of hypotheses and data. We expect, for example, that species body size — a commonly measured vertebrate trait that correlates with many extinction-promoting traits18 — will be significantly associated with how species respond to habitat edge effects. Forest ectotherms (i.e. amphibians, reptiles) should have desiccation-driven relationships responding to decreased humidity and increased temperature at forest edges and in the matrix8. Edge sensitivity should decrease with body size for amphibians as their desiccation tolerance increases due to reduced surface-to-volume ratio in larger species19. The opposite should be true for reptiles (and in particular snakes) whose often elongated body shape does not lend themselves to a similar decrease in surface to volume ratio. By contrast, we expect mobility and metabolism to drive relationships between body size of forest endotherms (i.e. mammals, birds) and their sensitivity to edges. Larger or more vagile forest species should have lower edge sensitivities compared to smaller-bodied species, because the former are better equipped to traverse and forage in the matrix as well as to detect suitable habitat and resources in a fragmented landscape20,21.
Simplistic approaches to quantifying edge effects treat landscapes as binary entities (e.g. forest versus non-forest) and quantify biodiversity responses to the nearest forest edge10. These ignore the role of the habitat that surrounds forests22 in human-modified landscapes (referred to as the “matrix”3), overlooks the additive effects of multiple edges that arise in fragments with irregular shapes23, and makes no predictions about the identity of species that might go extinct24. These unsophisticated approaches stand in contrast to widespread recognition that habitat quality varies continuously in space and shapes the contrast between forest and matrix25,26, thus modulating edge impacts in the landscape. Matrix habitat can in some cases provide resources for some species27, and in combination with species-specific requirements may determine whether forest edges act as ‘hard’ or ‘soft’ boundaries to species populations28. How species respond to edges affects abundance and persistence in a landscape9, with declines in abundance reliably indicating that a species is at increased risk of local extinctions29.
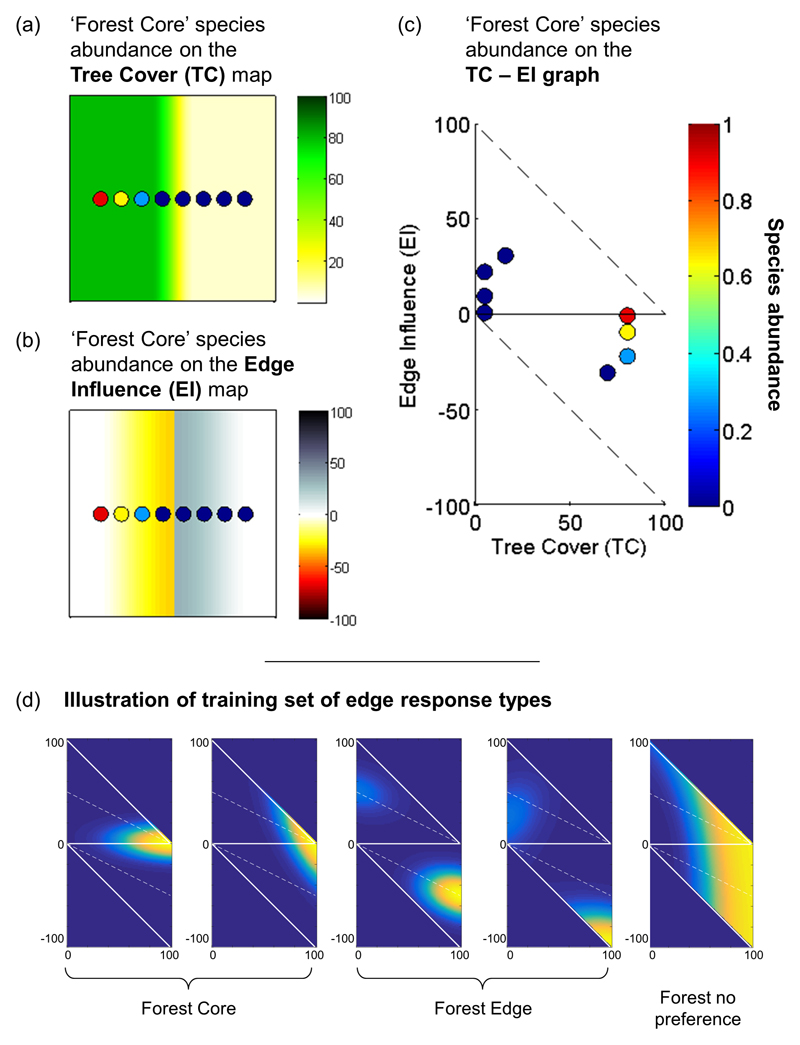
We use a novel approach to quantify the impacts of habitat edges on biodiversity. We map and quantify changes in the landscape-scale abundances30 of 1673 vertebrate species (103 amphibians, 146 reptiles, 1158 birds and 266 mammals) that can be attributed to edge effects in fragmented forest landscapes, using data collected in 22 landscapes distributed across seven major biogeographic realms (Fig. 1 and Extended Data Tables 1 and 2). Our approach defines two novel spatially explicit metrics, which together address two challenges that have so far prevented the detection of generalities in the edge responses of species. (1) Edge Influence (EI) assesses the configuration of landscapes and is calculated as a continuous, bounded spatial metric that quantifies local variations in percentage tree cover (Methods). We developed this metric to account specifically for the cumulative effects of multiple edges (including edge shape and patch size) that exacerbate the realised impact of habitat edges on species4,12,23 (Methods). Additionally, by computing EI from continuous gradients in percentage tree cover (measured at the levels of pixels and ranging from 0 to 100 %), as opposed to computing it from a binary classification of forest/non-forest habitat, we also account for variation in edge contrast and breadth (Methods) and thereby quantify the controlling influence of matrix habitat on the fragmented forest3. Absolute values of EI range from 0 (when there are no edges within a 1 km radius) to 100 (when a pixel is surrounded by different habitat for 1 km in all directions). EI does not correlate closely with any single traditional landscape fragmentation metric such as distance to the nearest edge, edge structure, fragment shape or fragment size, but rather aims to represent them all in one metric. (2) We measured the Edge Sensitivity (ES) of species as a biologically meaningful metric of changes in abundance12. ES is the proportion of the EI range that is avoided by the species (Methods), and is a bounded metric that ranges from 0.0 (inclusive) to 1.0 (exclusive). Species whose ES is equal to 0 have no change in local abundance due to edge effects, whereas species whose ES is close to 1 are restricted to a specific habitat because of edge effects (e.g. abundant in core habitat only or at edges only). Because ES is defined on a bounded landscape metric, it facilitates rigorous quantification and comparison of species’ edge responses between landscapes.
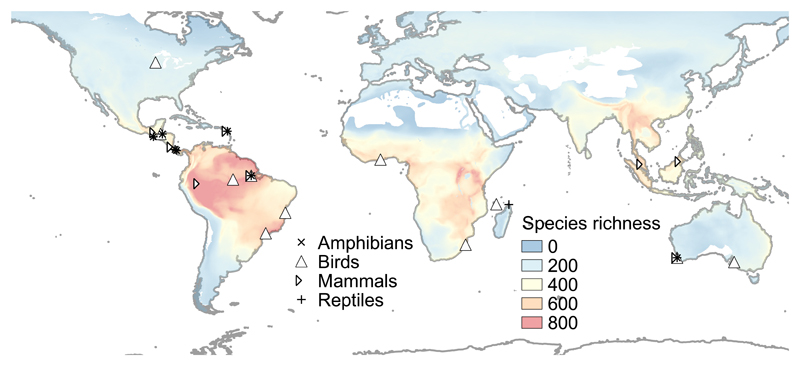
Fig. 1. Global distribution of the 22 study landscapes.
Some of these were sampled for more than one vertebrate group. We sampled abundance data from a total of 1673 vertebrate species (103 amphibians, 146 reptiles, 1158 birds and 266 mammals). Landscape centroids are shown on the background of vertebrate species richness maps showing the total number of bird, mammal, and amphibian species31 combined using data from Clinton Jenkins, BirdLife, and IUCN (Credits: Clinton Jenkins, Instituto de Pesquisas Ecológicas / SavingSpecies).
Pervasive impact of forest edges
For each species, we classified their observed abundance variations in the fragmented landscape with respect to EI and percentage tree cover as one of seven categorical edge response types9: forest core and matrix core (both edge-avoiding), forest edge and matrix edge (both edge-seeking), forest and matrix species with no preference regarding the edge, and generalist species (with no preference for either forest or matrix habitat). Edge responses of species that could not be classified into one of these types are referred to as unknown. We used a Naïve Bayes classifier to estimate the most likely edge response type for each species from a training set comprising simulated abundance patterns defining each edge response type (Methods).
We found that the abundance of 85% of all vertebrate species were affected by forest edges (46% positively and 39% negatively), excluding 369 species of unknown edge responses. The most common edge response type was forest core with 519 species, followed by forest edge (338 species), matrix edge (165 species), forest and matrix with no preference regarding the edge (112 and 34 species), matrix core (80 species), and generalist (56 species). The apparent ‘good news’ that marginally more species were positively rather than negatively impacted by edges should be interpreted with caution. Simple vote-counting the number of positive vs negative impacts, and assuming that one cancels out the other, ignores the more important fact that 85% of species are impacted and that the resultant community that now persists near edges bears little resemblance to that of forest interiors. Such large turnover in vertebrate community composition at edges likely reflects dramatic changes to the ecological functioning of these modified forest habitats31. Species negatively affected by edges include threatened forest core species of immediate conservation concern, such as the Sunda pangolin (Manis javanica, ES = 0.72), the Bahia Tapaculo (Eleoscytalopus psychopompus, ES = 0.88), the Long-billed Black Cockatoo (Zanda baudinii, ES = 0.77) and Baird’s tapir (Tapirus bairdii, ES = 0.73). Species positively affected by edges include invasives such as (Canis lupus, forest edge, ES = 0.6), the green iguana (Iguana iguana, matrix edge, ES = 0.56) and the common boa (Boa constrictor, forest edge, ES = 0.61).
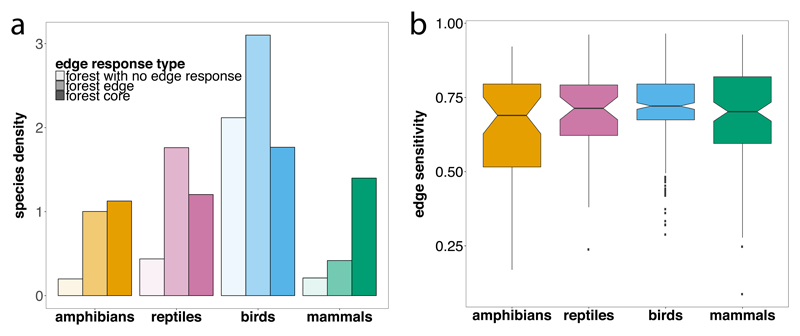
Taking into account sampling bias by computing species density (Methods) and excluding species whose edge response was unknown, we found that most species found in the forest and classified as species that preferred forest (i.e. forest core, forest edge, forest no preference) were sensitive to habitat edges, displaying either edge-seeking or edge-avoiding abundance distributions in the landscape (Fig. 2a). The abundances of 11%, 30%, 41% and 57% of bird, reptile, amphibian and mammal species, respectively, showed strong declines towards forest edges. We observed an analogous pattern for matrix-preferring species measured in the matrix (Extended Data Fig. 1a).
Fig. 2. Forest occupancy (a) and edge sensitivities for forest core species (b).
(a) Species density accounting for sampling bias in the datasets is shown for forest species, a subset of the seven edge response types (see Methods for details). (b) Edge sensitivity for forest core amphibian (n = 51) and reptile species (n = 49) (ectotherms) and forest core bird (n = 296) and mammals (n = 123) species (endotherms). Notched boxes show the median, 25th and 75th percentiles, error bars show 10th and 90th percentiles, and points show outliers. Notches display the 95% confidence interval around the median.
Edge sensitivities across species
As expected, species that were classified as having no preference for either edge or core habitat displayed the lowest edge sensitivities and were significantly less sensitive than species that were classified as preferring core habitats in either forest or matrix (Extended Data Fig. 2). The more edge sensitive a species is the less area it can use across fragmented landscapes. Although this is true for all edge response types, quantifying sensitivity is particularly critical for forest core species who are more likely to be threatened due to forest loss32 and whose suitable habitat area is decreasing due to fragmentation in addition to habitat loss resulting from deforestation5 (Methods). Thus, we particularly focus our analyses on the 519 forest core species (51 amphibians, 296 birds, 123 mammals, 49 reptiles; Extended Data Table 1).
Our data show that core forest habitat supported a larger number of amphibian, reptile and mammal species compared with forest edge, matrix core or matrix edge habitats (Extended Data Fig. 1b). Furthermore, forest core species were 3.7 times more likely to be listed as threatened on the IUCN Red List compared with species exhibiting other edge response types (two-sided 2-sample test for equality of proportions with continuity correction, P < 0.001) (see also Extended Data Table 3).
Edge sensitivities of forest core species varied more within than among all four vertebrate groups (Fig. 2b). However, on average, forest core species displayed edge sensitivities of ~ 0.7 across endotherms and ectotherms (Fig. 2b), which corresponds with a peak (or plateau) in species abundance from a minimum of 200-400 m away from sharp and high-contrast forest edges (Methods). This highlights how the amount of optimal forest habitat within fragmented forest patches can be much smaller than the total land area encompassed by the patch.
Of 277 high edge sensitivity species (ES ≥ 0.8) overall that have been assessed for the IUCN Red List (excluding ‘data deficient’ species), 8.6% were listed as threatened compared with just 3.3% of the 988 remaining species, demonstrating the conservation relevance of our edge sensitivity metric. Forest core species were more likely to have very high edge sensitivities (25.4% of forest core species) compared with forest species with other edge responses (20.6%) (two-sided 2-sample test for equality of proportions with continuity correction, P < 0.05). Very high edge sensitivities were particularly prevalent among forest core mammals (30.1% of species) and birds (24.0%), compared with forest core amphibian and reptile species (9.8% combined).
Size and edge sensitivity of ectotherms
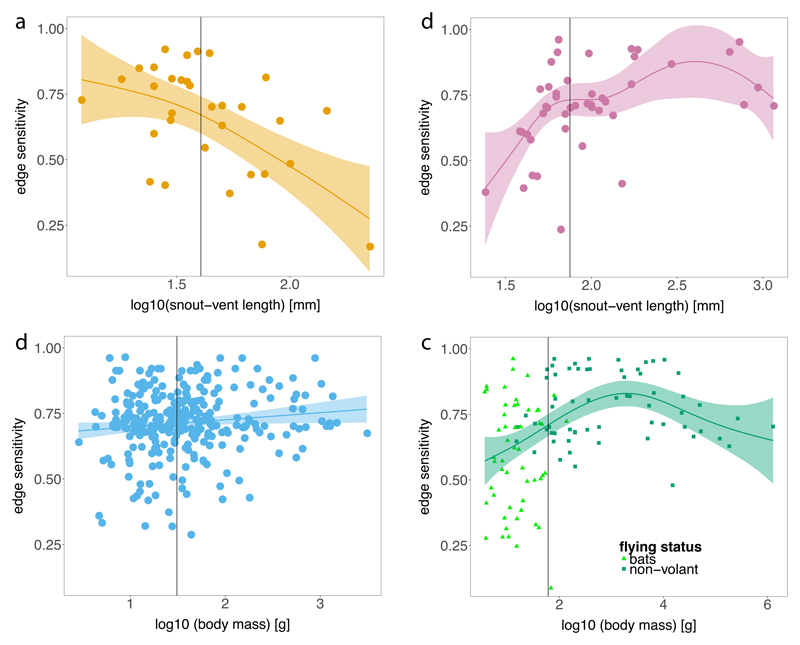
Edge sensitivity decreased with body size for forest core amphibians (generalized additive models, deviance explained = 39.6%, n = 32, P < 0.05) (Fig. 3a), but increased with body size for forest core reptile species (generalized additive models, deviance explained = 35.9%, n = 45, P < 0.01) (Fig. 3b). Avoiding overheating and severe water loss is likely to be an important driver of edge responses in forest core amphibians and reptiles, as most of the data were collected in tropical landscapes (Extended Data Tables 1 and 2), where year-round ambient temperatures are high but humidity can fluctuate considerably depending on microhabitat conditions33. Amphibians require moisture to maintain gas exchange, cultivate bacterial symbionts with immune-function and protect their eggs34. These physiological constraints make forest core amphibians, adapted to the high humidity interior of forests, prone to desiccation in dry environments such as habitats with lower tree cover, e.g. at the forest edge and in the matrix35. Small-bodied forest core amphibian species are particularly sensitive to forest edges (Fig. 3a) because their high surface area to volume ratios19 (except perhaps for salamander and newts) make them more susceptible to desiccation. By contrast, the body shape of forest core reptiles does not show a similar decrease in surface-to-volume ratio with increasing body size (Fig. 3b). Larger forest core reptiles are thus left more vulnerable to overheating in sun-exposed environments such as forest edges, particularly if they are too large to successfully exploit microhabitats such as shaded leaf litter (Fig. 3b).
Fig. 3. Edge sensitivity and body size in forest core vertebrates.
Relationships are shown for forest core amphibians, n = 32 (a), birds, n = 289 (b), mammals, n = 116 (c) and reptiles, n = 45 (d). Vertical lines in each panel indicate median body size of forest core species (amphibians, 40.5 mm; birds, 31.0 g; mammals, 61 g; reptiles, 75 mm). We excluded two amphibian species of the order Gymnophiona, who have an elongated body shape. Smoothed curves and 95% confidence bands were obtained from general additive models weighted by dataset reliability (Methods), which better explained the data than a null model for all taxa.
Size and edge sensitivity of endotherms
Edge sensitivity of forest core mammals displayed a significant hump-shaped relationship with body mass (generalized additive models, deviance explained = 23.3%, n = 116, P < 0.001), a pattern driven mainly by non-volant species (Fig. 3c). We attribute this relationship to the compound effects of species-specific means of locomotion (aerial or terrestrial) and energetic and other resource requirements. On average, forest core bats displayed significantly lower edge sensitivities (Mean ES ± SE = 0.59 ± 0.03, n = 53) compared with non-volant forest core mammals (0.77 ± 0.02, n = 63) (ANOVA with post-hoc Tukey HSD, P < 0.001). This suggests that the ability to fly may render mammals that prefer the forest interior less sensitive to changes in habitat. But forest core bats were also significantly smaller (P < 0.001) with only two species being slightly larger than the median body size of all studied forest core mammals (Fig. 3c).
Energy demands and home range size increase with body size in non-volant mammals36. Larger forest core mammals are less likely than smaller ones to meet their resource needs in highly fragmented landscapes comprising small forest patches with many edges but little core habitat to provide those resources37. Increasing energetic constraints are therefore hypothesized to account for the positive body size-edge sensitivity relationship for small to medium-sized forest core species (Fig. 3c). Yet, larger species are also predicted to roam more widely in search of resources in fragmented landscapes if habitat loss results in a loss of resource density38, decreasing their edge sensitivity in the landscape. This, together with other general features of large mammals, such as their lower vulnerability to predation39, may explain why the largest forest core mammals have lower edge sensitivities than do medium-sized species (which are also susceptible to hunting17).
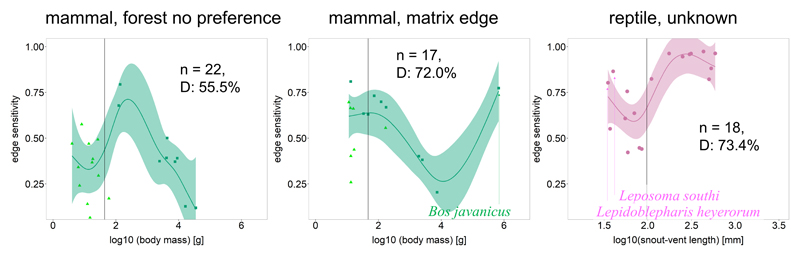
The combination of energetic constraints that are partly mitigated by dispersal capacity may also explain the similarly hump-shaped relationship of edge sensitivity with body mass in forest mammals that showed no edge preference (Extended Data Fig. 3). Conversely, dispersal capacity is likely to be the main driver explaining the decline in edge sensitivity with increasing body size in matrix edge mammals (Extended Data Fig. 3), with the exception of Bos javanicus, a large but threatened wild cattle species that displayed high edge sensitivity.
Edge sensitivity of forest core birds showed a weak increase with body size (generalized additive models, deviance explained = 1.5%, n = 289, P < 0.05). There was a tendency for small birds (< 31g, the median size of core forest birds analysed in this study) to have more variable responses (Fig. 3d), as also seen in bats (Fig. 3c). Some forest core bird species certainly are sensitive to forest edges (Fig. 2b), especially in tropical landscapes and during the non-breeding period40, yet there is little evidence in our data to support a body size link of edge sensitivity, probably because other traits such as trophic guild are more important41.
Other species traits & edge sensitivity
The ability of some endotherms to adapt to a diverse array of environments20 may enable them to respond better to habitat changes in a landscape20. By contrast, many amphibian species are habitat specialists with small home ranges42 and should be susceptible to changes in their environment. However, for both forest core endotherms and forest core ectotherms, our data do not support a habitat specialisation effect. Single predictor models of habitat trait-edge sensitivity models were not significant, and the direction of the coefficient for habitat traits retained in multiple predictor models could not be estimated with confidence except for forest core reptiles (Extended Data Tables 4 a-d). For forest core endotherms, our data instead emphasize the importance of species locomotion, which correlates with a species’ vulnerability to hunting or predation when traversing non-forest habitats: edge sensitivity was consistently higher in non-volant mammals compared to volant species with similar habitat breadths (Extended Data Table 4c).
Birds in particular may additionally be more susceptible to biophysical drivers such as disturbance history5 confounding the detection of patterns between life history traits and species responses to edges separating forest from non-forest habitat. This may explain why we found no evidence for direct effects of diet, range size, migratory status or clutch size on edge sensitivities of core forest birds in single predictor-models (Methods). Multiple-predictor models for edge sensitivities of core forest birds retained range size, body mass, migratory status, forest dependency and number of habitats (Extended Data Table 4d). Yet, none of the predictor coefficients were significant and the overall deviance explained by the model was negligible.
A ubiquitous phenomenon
Tracking changes in species’ abundances in response to edge effects allows us to predict biodiversity responses to forest loss and fragmentation at scales useful for land management. This is an important difference compared with previous global analyses and projections of biodiversity responses to global land use changes43, which do not account for the continuous variation in habitat quality of either matrix or forest habitat24 that are known to affect species and the ecosystem processes that they control44.
Considering edge effects (and hence landscape configuration and forest-matrix contrast) is at least as important as habitat amount when predicting species richness from habitat distribution in a landscape. Although forest core endotherms and ectotherms vary greatly in how their abundance changes in response to edge effects, on average they reach peak abundances in forest habitats farther than 200-400 m from sharp high-contrast forest edges. This seems to corroborate the traditional perception that edge effects operate within a relatively small spatial window of just a few hundred metres45–47. We cannot, however, exclude the possibility that the effect of edges on core species extend further within the forest, but rigorously testing this would require data from many more studies examining edge effects over scales of one kilometre or more9, which are currently rare. Regardless of whether larger-scale edge effects are as ubiquitous as small-scale effects, our data strongly indicate that small forest fragments with no forest located farther than 200-400 m from sharp high contrast edges (or alternatively, with no forest located farther than 100 m from low contrast edges) should probably be seen as extended forest edge habitat48. Such habitats may support lower abundances of forest core species and may act as a stepping stone or corridor for improving patch interconnectedness49, but maximum abundances for many species will only be achieved within much larger core forest fragments. Distances to edges given here are, however, only indicative. In practice, to account for multiple edges and forest-matrix contrast, it will be necessary to compute the EI map, using for example our software29, and delineate forest areas of EI < 30 as suitable for most forest core species.
Anthropogenic disturbances to tropical forests were recently shown to double biodiversity losses incurred directly from deforestation5. Our data demonstrate this pattern, observed in the Amazon, holds globally. Approximately half of the global forest area lies within 500 m of a forest edge1, likely of high contrast, the range over which the abundances of many core forest species can be diminished. The direct implication is that less than 50% of Earth’s remaining forests can be considered free from edge effects, yet even that proportion is under threat from the chaotic expansion of road networks, selective logging, wildfires, widespread hunting and other human encroachment into the last intact forest frontiers50.
Methods
Species abundance data and species traits data
We compiled primary biodiversity datasets containing abundance measurements at plot level acquired in 22 anthropogenically fragmented forest landscapes around the world (BIOFRAG database2). All landscapes encompassed anthropogenic forest edges and - except for one landscape which is dominated by forests with only a small amount of habitat conversion in the north-west corner - a mosaic of natural forests and other land uses (Extended Data Table 2). In seven of the landscapes, the natural forests were bordered at least in part by managed, plantation forest. Eighteen of the 22 landscapes were from continents with the remaining four from islands, and six of the 22 landscapes could reasonably be described as coastal (Extended Data Table 2). For our analysis, we only used datasets that measured abundance of vertebrates in at least nine plots per landscape. We only used datasets for which geographic coordinates of plots were provided at high spatial accuracy by the dataset authors, as the location of each plot in relation to forest edges was important. Datasets represented full gradients of distance to edge and edge influence. All datasets in our analysis were from community-level surveys of a focal taxonomic group (rather than sampling for a target list of species). The final datasets used in this analysis came from 22 landscapes, with some landscapes sampled for more than one taxonomic group in separate or combined studies (Fig. 1)51–71.
The majority of taxa represented in the datasets were true species (i.e. not morpho-species) (Extended Data Table 1). We matched taxonomic names given by the dataset author using steps outlined in Pfeifer et al.2 to obtain the full taxonomic classification for each species. We used lets.iucn and let.iucn.ha functions in the letsR72 package to extract, for each true species from the IUCN online database, the Red List conservation status (IUCN status), and habitat information (IUCN Tree: species present in forests + savannah or shrub habitats only, IUCN Forest: species present in forests only, IUCN Habitat: number of main IUCN habitat categories listed).
For each species, we extracted life history trait data from literature and database sources. For amphibians and reptiles, we extracted trait data (body size: maximum snout-vent length in mm and maximum total length in mm for snakes; mean clutch size; thermal niche: average temperature and temperature range; adult and larvae habitats; vertical stratification (i.e. arboreal, semi-arboreal, terrestrial) from academic literature73–113, region - specific guide books114–116, text books117–119, and websites (all last accessed 24/06/2016) including http://amphibiaweb.org/, http://frogs.org.au/, http://www.anolislizards.myspecies.info/, http://www.reptile-database.org/db-info/news.html, http://www.iucnredlist.org/, http://research.amnh.org/vz/herpetology/amphibia/index.php, http://eol.org/, and http://tolweb.org/tree/. For birds, we extracted information on body size (mean body mass in g), range size, migratory status (Not Migrating, Altitudinal Migrant, Full Migrant, Nomadic), generation length in years and mean clutch size from the trait database compiled by Bird International. We extracted information on bird diet from the Willman et al.120 global dataset, focussing on the Diet-5Cat attribute (i.e. assignment to the dominant category among five categories based on the summed scores of constituent individual diets: plant and seed-eating species; fruit and nectar-eating species; invertebrate eating species; vertebrate, fish-eating, and scavenging species; omnivores). For mammals, we extracted body size (mean body mass in g), trophic status, litter size and litter numbers per year, maximum longevity in months, migratory behaviour, range extent in km and age at first birth from the PanTHERIA database121 complemented by information from http://animaldiversity.org/accounts/Mammalia/ (last accessed 11/05/2016). We also recorded whether or not species can fly (volant: all from the order Chiroptera, non-volant)
Quantifying abundance responses to variations in tree cover
We analysed a species’ abundance distribution in the landscape with respect to two spatial variables, percentage of Tree Cover (TC) and Edge Influence (EI), to characterise both the species’ edge response and the species’ habitat preference. For each landscape we obtained 30m pixel resolution percentage TC maps122, which were generated from Landsat imagery using percent tree cover training data and decision trees classification algorithm implemented in the Google Earth Engine. These maps define tree cover in the year 2000 as canopy closure for all vegetation taller than 5m, encoded as a percentage per output grid cell and ranging between 0 and 100%.
Quantifying Edge Influence (EI) within and among landscapes
We computed the EI metric from the regional standard deviation of TC (a measure of regional heterogeneity), and the regional average TC subtracted to point TC (a measure of point heterogeneity and direction)30. EI is the maximum of regional and point heterogeneity for each pixel and has the sign of the point heterogeneity (Eq. 1).
| Eq.1 |
Regional average and standard deviation of TC were computed using a Gaussian filter of 1 km radius, the distance previously shown to impact animal abundance9, to ensure that all TC variations (i.e. edges) contained within a window of 1 km radius contribute to the value of EI. Absolute values of EI range from 0 (no edges within a 1 km radius) to 100 (one pixel surrounded by different habitat for 1 km in all directions). The sign of EI is determined by the point heterogeneity (regional average TC minus point TC): forest habitat near the matrix has a negative EI and matrix habitat near the forest has a positive EI (Extended Data Fig. 4).
The amplitude of EI depends on the landscape configuration (Extended Data Fig. 5a) and forest - matrix contrast (Extended Data Fig. 5b). EI measured at a focal point increases as the point approaches all nearby edges, and hence varies with the shape and with the size of the forest patch (Extended Data Fig. 5a). EI also varies with the contrast between forest and matrix habitats, i.e. the contrast in TC (Extended Data Fig. 5b). Hence, there is no general relationship between EI and the distance to a defined edge, and no direct relationship between the % forest cover in a buffer as EI is sensitive to contrast in TC whereas % forest cover is computed from a binary forest-non-forest map.
Categorising species into edge response types
Species abundance within each landscape was plotted in 2D space based on TC and EI values (TC - EI graph in Universal Transverse Mercator WGS 84 projection; Extended Data Fig. 6c). We defined seven edge response types9: “forest core”, “forest edge”, “forest no preference”, “matrix core”, “matrix edge”, “matrix no preference”, and “generalist” species.
We used a Naïve Bayes classifier to estimate the most likely edge response type for each species from a training set of simulated abundance patterns on the TC - EI graph (see Extended Data Fig. 4 for the TC - EI graph, Extended Data Fig. 6d for an illustration of a training set and Lefebvre et al.30, particularly pages 23 & 24 in the user manual for an illustration of classification). The training set contained, on average, 15 different abundance patterns for each edge response type to fully describe each type (span all possible patterns that may be classified as a specific type when measured on the TC - EI graph). We created the training sets using sigmoidal surfaces of varying means (location of maximum abundance) and standard deviations (spread) along the TC and EI axis, thereby defining areas of high and low abundance on the TC - EI graph. For “forest” and “matrix” types, the location of maximum abundance along the TC axis ranged from 60% to 100% and from 0% to 20%, respectively. We defined the training set by assuming that a species that is most abundant for TC > 60 has a high probability to be a forest species, whereas a species most abundant for TC around 50 is likely to be a forest species but retains a significant probability to be a matrix species (sigmoidal threshold). The classification of the preferred habitat depends on the full shape of the species abundance curve along the TC axis and how it compares to the training set patterns we defined. Similarly, we defined “core” and “edge” types in the training set with the location of maximum abundance ranging from |EI| = 0 to 10, and from |EI| = 30 to 100, respectively. By definition types of “no preference” have flat abundance along the EI axis, whereas “generalist” types have flat abundance along the TC axis. Location and spread parameters of sigmoid curves along the TC and EI axis were combined to create an ensemble of abundance surfaces describing each categorical edge response type in the TC - EI graph (see examples provided in Extended Data Fig. 6d). The collection of these simulated abundance patterns on the TC - EI graph forms the training set. The classifier compares the measured abundance distribution of each species to the ensemble of abundance patterns for each type in the training set and estimates the most likely match, depending on the area (or areas) in which the species was most abundant on the TC - EI graph and the shape of the abundance surface. For example, species whose abundance increases with TC are very likely to be classified as forest even if they are mostly abundant for TC below 60%.
Species that did not match any defined type were classified as “unknown” (e.g. species abundant in both the matrix core and forest edge but not on the matrix edge). Our approach of defining a training set to use a classifier is effective to categorize species with similar edge response pertaining to known types and is more flexible than fitting a parametric model to each species’ abundance distribution or using thresholds.
Quantifying edge sensitivity (ES) for each species
We developed the edge sensitivity (ES) metric to quantify and compare the edge responses of species that were measured in different landscapes but on the same scale, and to do so independently of landscape configuration123. ES is derived from comparing the species’ abundance surface on the TC - EI graph with the abundance surface it would have if it was insensitive to edge effects. A species’ ES hence corresponds to the proportion of the EI spectrum that is not occupied by this species.
We obtained each species’ abundance surface by linearly interpolating its abundance to the full graph (for TC ∈ [0,100] ∈ ℕ, and EI ∈ [0 – TC, 100 – TC] ∀ TC), assuming zero abundance for locations with no measurements. We estimated the abundance surface for each species assuming it was insensitive to edge effects by obtaining the maximum abundance at each TC value, and replicating maximum abundance along the EI axis of the graph, so that the abundance surface varies with TC only, and not with EI. We then computed ES from the ratio of the sum of the species abundance surface on the TC-EI graph and the sum of the abundance surface the species would have if it was insensitive to edge effects (“EI insensitive abundance surface”):
| Eq.2 |
Because the “EI insensitive abundance surface” is computed from the maximum for each TC of the species abundance surface, its sum is larger or equal to that of the species abundance surface, therefore ES is bounded between zero and one. Species with ES values equal to zero are species whose abundance is not influenced by the presence of habitat edges. Species with ES values larger than zero are species that either increase or decrease in abundance in response to edge effects. Species with values close to one are species that are only abundant for a specific edge influence value.
ES does not quantify the abundance variation of a species directly, as this depends on the configuration of the landscape. Also, ES does not quantify whether species abundance increases or decreases with the presence of edges as this depends on the EI values preferred by the species (i.e. low values for core species, high values for edge species). ES quantifies the length of the range of EI values for which a species is abundant: if the range is as wide as the EI spectrum (i.e. the species is abundant for large portions of the EI domain) then the species is not sensitive to edge effects and ES is low (and the species has a high tolerance to habitat change). If the range is small compared to the EI spectrum (i.e. the species is abundant at a small portion of the EI domain only) then the species is sensitive to EI, and ES is high (and the species has low tolerance to habitat change). Species whose ES value is close to 1 can only be abundant in narrow ranges of EI, .e.g. |EI| < 10 (core species) or 45<|EI|<55 (edge species).
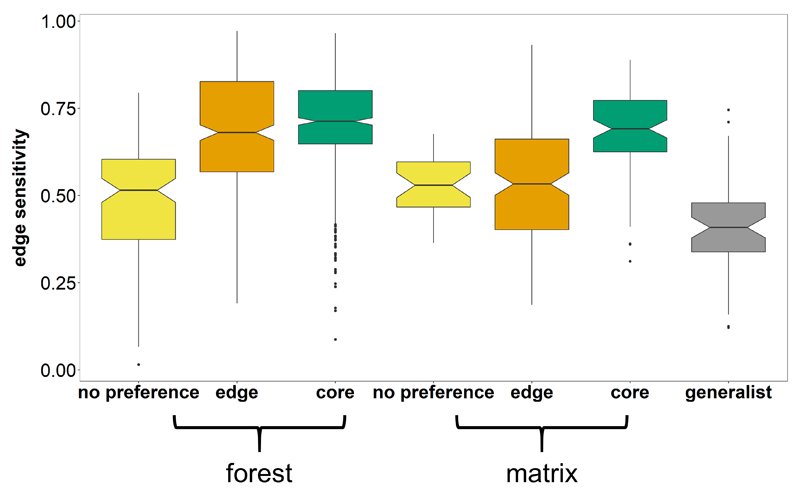
The ES metric is useful to compare species sensitivity for edges, and its computation is independent from the species categorisation described in the previous section. Two species with the same ES may have different predictions about the spatial distribution of their preferred habitat if they belong to different edge response types. Core forest species with ES > 0.7 will only be found within the forest interior far away from edges, whereas core forest species with ES of ~ 0.6 will be found near edges of large forest patches but not in peninsulas or small forest patches. Core forest species with ES < 0.6 will be found throughout the forest and in large forest patches but not in the smallest forest patches (size depending on the window size used to compute EI, which was 1 km in this study). We compared the distribution of ES for forest core species within taxonomic groups using notched boxplots (Fig. 2b), thereby notches display the 95% confidence interval around the median. If box notches do not overlap there is strong evidence that medians differ.
ES cannot generally be converted to a “distance to nearest edge” equivalent as it is based on Edge Influence (EI), which varies depending on landscape configuration (Extended Data Fig. 5a) and patch contrast (Extended Data Fig. 5b). However, in the special case that a species’ abundance was measured across a straight edge of constant and maximum contrast, core forest species with ES = 0.5 will be abundant up to this edge, and core forest species with ES = 0.7 will be abundant up to 400 m from this edge (for an EI computed with a 1 km window). A core forest species of low sensitivity would also be found near edges and even in small forest patches, albeit at lower abundance.
We provide these distance estimates as indication only, as there is no direct relationship between distance to the nearest edge and EI. In practice, instead of computing the distance to nearest edges using binary forest - non-forest maps, we urge decision-makers to utilise EI maps computed from bounded landscape measurements (e.g. percentage tree cover) using the provided software30. This would allow them to identify areas where EI is below 30 as suitable for most forest core species (whose ES is around 0.7) thereby taking into account edges varying in contrast, breadth and shape.
Rating datasets based on their capacity to assess species’ responses to edges
Each dataset was rated based on the accuracy of its TC map and the distribution of sampling points within the TC and EI spectra. To evaluate TC map accuracy we computed the proportion of sampling points whose TC value matches the description given by the dataset authors (e.g. the TC value of points identified as “forest” should be over 50%). We also rated the sampling design based on the distribution of plots on the TC - EI graph, because accurate classification of species responses requires data to be collected from each habitat type (forest core, forest edge, matrix edge and matrix core). We downgraded the dataset rating for each missing category. Datasets ratings were then used as weights when comparing ES of species across datasets.
Estimating the relative number of species belonging to edge response types
Due to sampling bias present in most datasets (for example, many datasets include more sample sites in core forest compared to forest edges), simple counts of the number of species belonging to each edge response type partly reflects the relative abundance of measurement locations within different habitat categories (Extended Data Table 1). For example, out of 103 amphibian species, 49 were categorised as core forest species. This could arise either because 49/103 = 48% of amphibian species show a preference for core forest habitats, or alternatively because 48% of sampling locations were in core forest habitats, or a mixture of both. Therefore, the number of sampling sites within different habitat categories must be considered when estimating the number of species belonging to each edge response type.
We addressed the ambiguity resulting from sampling bias across different habitat categories by computing the average number of species per site (termed “species density” or SD). Species density was computed separately for sites located within each of the four habitat categories (H: forest core, forest edge, matrix edge and matrix core) and for species classified in each of the seven edge response types. Thus, for each H and each species edge response type (T) we computed the average number of species of T recorded in sites located in H, formally termed “species density of species of type T in habitat H” and denoted :
| Eq.3 |
For example, the average number of core forest species (FC) recorded in sites located in forest core habitat was calculated as:
| Eq.4 |
the average number of core forest species recorded in sites located in the forest edge (FE) as:
| Eq.5 |
the average number of forest edge species recorded in sites located in the forest core as:
| Eq.6 |
and so on for each combination of T and H.
Species densities within the forest habitat, including the density of forest core species in the forest (F), were determined as the average of species densities for the forest core and forest edge habitats:
| Eq.7 |
Similarly, the average number of forest edge species in the forest was given by
| Eq.8 |
and the average number of forest no preference (NEP) species in the forest was given by
| Eq.9 |
This corresponds to the average number of species of edge response type T per forest site weighted by the number of sites in the forest core and the forest edge (Fig. 2a: forest occupancy per edge response type). If there were the same number of sites in the forest core and the forest edge then would simplify to the average number of species of type T per site in the forest. However, we weighted the average number of species per forest site (number of forest sites n = 4359: 203 for both amphibians and reptiles, 1805 for birds, 2148 for mammals) so that the contributions of core and edge habitats are equivalent. The weighted average allows us to compare for example the number of FC and FE species in the forest as if the same areas of edge and core forest habitats had been sampled (Fig. 2a).
We also quantified the average number of species (regardless of edge response type) per dataset in each habitat category to identify the habitat that can support the largest number of species.
| Eq.10 |
SDH was computed for all four habitat categories (Extended Data Fig 1b). To compute SD, sampling sites and species were pooled from all landscapes used in this study, i.e. SD was computed across rather than within landscapes.
Modelling edge sensitivity as a function of species life history traits
To test whether body size predicts species responses to edges, we used general additive models implemented in the mgcv package123 (using log10-transformed body size as predictor), with smoothers fitted separately for each taxonomic group. We used dataset ratings (see above) as a weighting factor for the smoothing. Data were visualized using the R package ggplot2124.
We also wanted to know whether we can use additional species’ traits, in particular their habitat specialisation, as a proxy for abundance when predicting sensitivities to habitat edge. Within each taxonomic group, we first tested for single-predictor relationships between edge sensitivity of core forest species and their life history traits (see above). We then fitted multiple predictor general linear models using automated model selection via information theoretic approaches and multi-model averaging using Maximum Likelihood. First, we constructed a global model for each taxonomic group, modelling edge sensitivity as a function of predictors. We excluded highly inter-correlated predictors (V > 0.5, R2 > 0.5, P > 0.6) from these models using Pearson's Chi-squared test with Yates' continuity correction and Cramer’s V measure of association to test for correlations among categorical predictors (lsr package), Pearson's product-moment correlation P for associations between numeric predictors and the coefficient of determination R2 of linear models for relationships between numeric and categorical predictors. For each global model, we used the dredge function in the R MuMIn package v1.10.5 (Barton 2014), which constructs models using all possible combinations of the explanatory variables supplied in each global model. These models were ranked, relative to the best model, based on the change in the Akaike Information Criterion (delta AIC). A multi-model average (final model) was calculated across all models with delta AIC < 2.
Global models were restricted to a subset of life history traits in mammals, amphibians and reptiles due to a large number of missing values. Predictors in the global models for ectotherms include IUCN Habitats, IUCN Forest, IUCN Tree (this variable correlated strongly with IUCN Forest and was excluded together with its two-way interaction from the mammal and the amphibian models), body size (decadic logarithmic; in mm), and two-way interactions of body size with each habitat trait. Predictors in the global models for endotherms include IUCN Habitats, IUCN Forest (this variable correlated strongly with IUCN Habitats and was excluded together with its two-way interaction from the reptile model), IUCN Tree, body mass (decadic logarithmic; in g), and two-way interactions of body mass with each habitat trait. For mammals, we also included body mass squared (given the hump-shaped relationship with edge sensitivity, Fig. 3c), flying status, and two – way interactions of flying status with body mass, and habitat traits. For birds, we also included: range size, mean clutch size, migratory status, diet and two-way interactions of migratory status with body mass and habitat traits, and of body mass with diet and extent of occurrence.
Code availability
We used R 3.2.1 statistical software for all statistical analyses. We used in house generated software for analyses central to the manuscript: computing edge influence, categorising species into edge response types, quantifying edge sensitivity, rating datasets and estimating the relative number of species belonging to edge response types. Details on these analyses are described in the Methods section of the manuscript. The software itself is accessible at https://github.com/VeroL/BioFrag (see reference 30in the manuscript).
Data availability
The *xls and *kml data that support the findings of this study are available in figshare with the identifier doi: 10.6084/m9.figshare.4573504. Original BIOFRAG data are available on request from the corresponding author but restrictions apply to the availability of these data, which are not publicly available. Data are however available from the authors upon reasonable request and with permission of dataset authors as specified in the BIOFRAG database2 (https://biofrag.wordpress.com/).
Extended Data
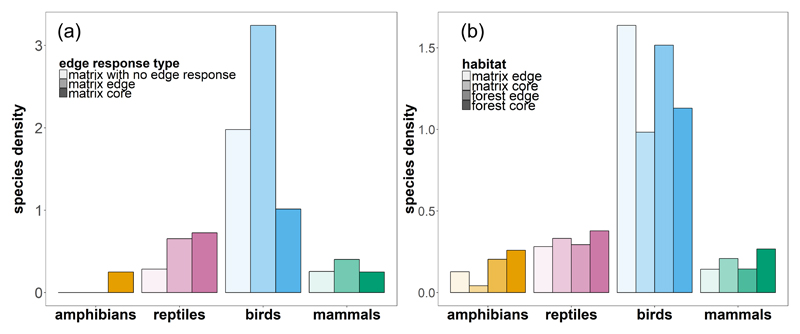
Extended Data, Fig. 1. Matrix occupancy by matrix species per edge response type and average number of species per habitat category.
(a) Average number of species per matrix site (number of matrix sites = 727, 7 for amphibians, 659 for birds, 51 for mammals and 10 for reptiles), weighted so that the contributions of core and edge habitats are equivalent (Methods, Eq. 7-9). Only species classified as preferring the matrix are shown (i.e. matrix core, matrix edge, matrix with no edge response). (b) Average number of species (regardless of edge response type) in each habitat category showing which habitat can support the largest number of species after addressing the ambiguity resulting from sampling bias across different landscape configurations (Methods, Eq.10). Plots were categorised by their locations into: forest core (n=2955), forest edge (n=1404), matrix core (n=388), and matrix edge plots (n=339). For each configuration we computed the average number of species present per habitat category plot, which identifies the habitat that can support larger numbers of species. For amphibians, reptiles and mammals, core forest habitat supported more species than did forest edge, core matrix or matrix edge habitats. In contrast, bird species were found in larger numbers in edge habitats (in forest and matrix) than in core habitats.
Extended Data, Fig. 2. Distribution of edge sensitivities for seven recognised edge response types.
Forest core species (n = 519) and matrix core species (n = 80) displayed significantly higher edge sensitivities compared to generalists (n = 56) and to forest (n = 112) and matrix species (n = 34) with no preference for either edge or core habitats (two-sided Pairwise Wilcoxon Signed-Rank Test with Bonferroni correction: P < 0.001). We excluded species that could not be classified (n = 113). Forest edge species (n = 338) had significantly higher edge sensitivities compared to forest no preference, matrix no preference, generalist and matrix edge species (P < 0.001). Matrix edge species (n = 165) also displayed significantly lower edge sensitivities compared to matrix core species and higher edge sensitivities compared to generalists (P < 0.001). Notched boxes show the median, 25th and 75th percentiles, error bars show 10th and 90th percentiles, and points show outliers. Notches display the 95% confidence interval around the median.
Extended Data, Fig. 3. Significant relationship between edge sensitivity and body size across edge response types.
(except forest core species that are shown in Figure 3 in main manuscript). Vertical lines in each panel indicate median body size of the species per taxonomic group and edge response type (mammals forest no preference, 43.8 g; mammals matrix edge, 47.0 g; reptiles, unknown 97.5 mm). Smoothed curves and 95% confidence bands were obtained from general additive models (GAMs), with the model weighted by a variable that reflects dataset reliability (Methods). GAMs better explained the data than a null model for taxa and edge response types shown. Edge sensitivity ranges from 0.0 (no declines in local abundance due to edge effects) to 1.0 (local extinction due to edge effects).
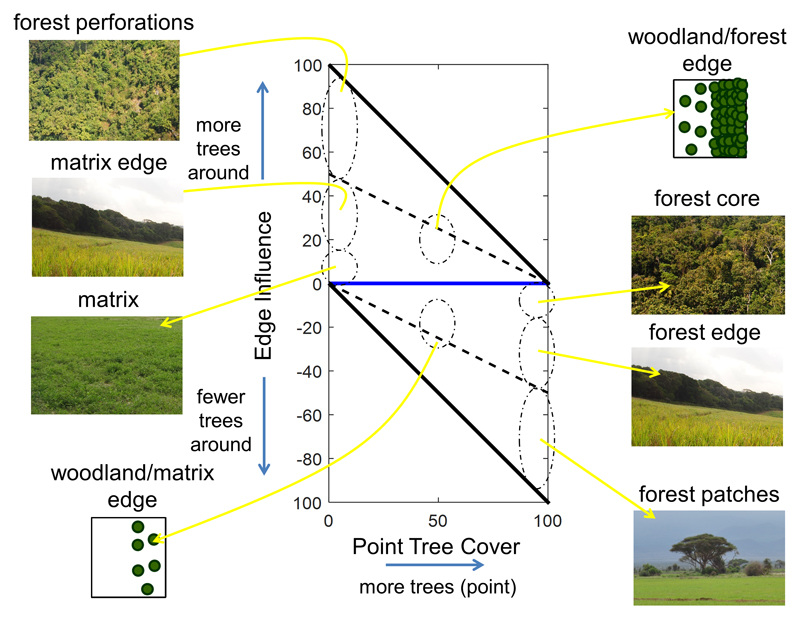
Extended Data, Fig. 4. Illustration of the TC – EI graph.
Combinations of point TC and EI characterize different landscape configurations, and some combinations are impossible by design (grey areas). The x - axis represents the percentage of tree cover at the scale of a pixel. The y - axis represents the EI metric, computed from the regional standard deviation of TC (a measure of regional heterogeneity), and the regional average TC subtracted to point TC (a measure of point heterogeneity and direction).
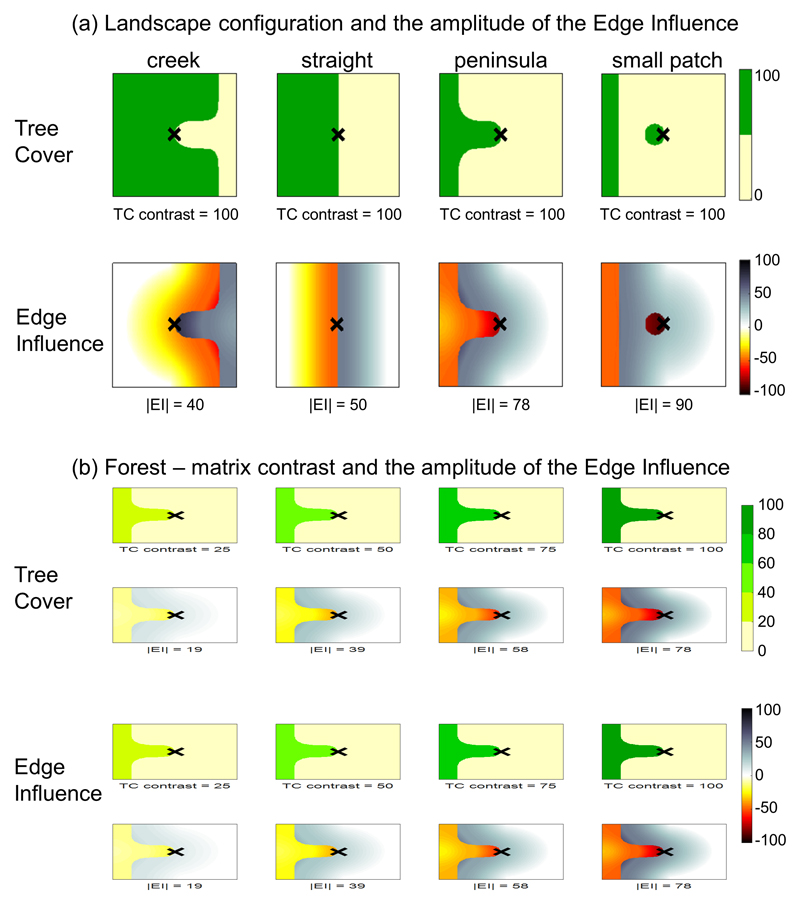
Extended Data, Fig. 5. Variations of Edge Influence (EI) with Tree Cover (TC) configuration (a) and contrast (b).
(a, top row) Four examples of landscape configurations comprising dense tree cover habitats (green) and matrix (cream). From left to right: creek edge, straight edge, peninsula edge and small forest patch. (a, bottom row) EI maps that correspond to above landscape configurations. The EI value at the central point (cross) is given for each configuration. The central point is always located on an edge and its distance to nearest edge is always zero. Nonetheless, EI increases in absolute value as the central point is increasingly surrounded by a different type of habitat. (b, top row) Four examples of peninsula edges between matrix (cream, TC=0%) and habitats of varying tree density (shades of green). From left to right: 25%, 50%, 75% and 100%. (b, bottom row) EI maps that correspond to above landscape contrasts. The EI value at the central point (cross) is given for each configuration. The central point is always located on an edge and its distance to nearest edge is always zero. EI increases as the edge contrast increases.
Extended Data, Fig. 6. Computing species abundance surfaces on the TC - EI graph and simulated edge response types on the TC – EI graph.
(a) Plots superimposed on an hypothetical TC map. Marker colours correspond to the abundance of a hypothetical species and follow the colour bar shown in (c). (b) EI map corresponding to (a). (c) TC - EI graph: species abundance (warm colour = higher abundance) is plotted as a function of TC and EI measured at the species’ plots. In this example, the species is predominantly found in sites characterised by high TC and low |EI|, and would be classified as a core forest species. (d) Illustration of the training set of edge response types used for classification. Each of the 7 response type has around 15 patterns associated with it in the training set; here we show 2 examples for the forest core type and forest edge type and one example for the forest no-preference type. Each graph is a TC – EI graph with TC on the x-axis and EI on the y-axis. Warmer colours means high abundance, dark blue is 0.
Extended Data, Table 1. Summary statistics of species and landscapes assessed in our study.
We include information of the number of species measured across datasets (n), the number of those species that were not morpho-species (n, true) and that were assessed by IUCN (n, IUCN), and the number of landscapes (LS) sampled overall and in the tropics only (in parentheses). The number of forest core (n, fc) species (all and true species only) after grouping species into edge response types based on their abundance distribution in the fragmented landscapes is also shown. Note that 299 birds (25.8%), 35 mammals (13.2%), 21 reptiles (14.4%) and 14 amphibians (13.6%) could not be categorised, as their abundance in the landscape was either too low or too variable to reliably classify them into any of the edge response types.
| Taxon | n | n, true | n, IUCN | LS (tropical) | n, fc (tropical) | n, fc + true (tropical) |
|---|---|---|---|---|---|---|
| Amphibians | 103 | 72 | 72 | 7 (6) | 51 (48) | 35 (32) |
| Birds | 1158 | 1139 | 1139 | 11 (7) | 296 (275) | 293 (273) |
| Mammals | 266 | 260 | 258 | 8 (7) | 123 (121) | 118 (117) |
| Reptiles | 146 | 124 | 49 | 8 (7) | 49 (41) | 45 (37) |
Extended Data, Table 2. Attributes describing the geographic context for each landscape.
PA - Protected Area, within - w, outside - o, within & outside - wo, primarily within - pw. Islands shown in bold in the column ‘Geographic context’. Landscape minimum convex polygons created to encompass the plots sampled in each landscape are available for display as *kml. All landscapes have anthropogenic forest edges present in them. The majority encompass a mosaic of natural forests and other land uses. Only one landscape (LS_30, Madagascar) is forest-dominated with few anthropogenic edges present at the northern edge.
| Landscape | Ocean present in landscape | Geographic context | Forest within & outside PAs | Plots within & outside PAs | Land use in the matrix |
|---|---|---|---|---|---|
| LS_01 | yes | Africa | pw | wo | Crops, Plantation forest |
| LS_02 | no | S America | wo | wo | Clear cuts |
| LS_03 | yes | Islanda | oe | o | Clear cuts, Crops, Cattle pasture, Settlements |
| LS_06 | no | S America | o | o | Clear cuts, Crops |
| LS_10 | yes | Australia | pw | wo | Clear cuts, Crops, Settlements |
| LS_15 | no | Islandb | oe | o | Clear cuts, Grassland, Settlements |
| LS_16 | no | SE Asia | wo | wo | Plantation forest (oil palm rubber) |
| LS_18 | no | S America | o | o | Clear cuts, Crops, Plantatic forest (Eucalyptus) |
| LS_25 | no | N America | o | o | Savannah, Grassland |
| LS_30 | no | Islandc | oe,f | o | Clear cuts, Orchards |
| LS_37 | no | C America | wo | wo | Grassland |
| LS_38 | no | C America | wo | wo | Crops, Plantation forest, Settlements |
| LS_39 | no | C America | wo | wo | Clear cut, Settlements |
| LS_40 | yes | C America | wo | wo | Clear cut, Crops, Settlements |
| LS_42 | yes | C America | pw | wo | Cattle pasture, Crops, Plantation forest |
| LS_44 | no | Australia | wo | wo | Plantation forest |
| LS_46 | no | C America | wo | wo | Crops, Grassland, Settlements |
| LS_47 | no | S America | wo | wo | Clear cuts, Settlements |
| LS_57 | no | C America | wo | wo | Crops, Pasture, Settlements |
| LS_59 | no | Islandd | wo | wo | Clear cuts, Plantation fore (oil palm) |
| LS_60 | no | S America | w | w | Pasture, Plantation forest (rubber, eucalyptus, cocoa) |
| LS_62 | yes | Africa | wo | wo | Crops, Plantation forest |
Extended Data, Table 3. Number of threatened and not threatened species for forest core and all other species in each taxonomic group.
We excluded species that were not assessed or that were listed as ‘data deficient’ by the IUCN Red Lists (IUCN status data were not accessible for the majority of reptile species). We used a two-sided 2-sample test for equality of proportions with continuity correction and confidence level = 0.95. P value is significant if forest core species were more threatened than species of other edge response types.
| Taxon | p | Forest core species | Not forest core species | ||
|---|---|---|---|---|---|
| Not threatened | Threatened | Not threatened | Threatened | ||
| Amphibians | 1.0 | 32 | 3 | 32 | 3 |
| Birds | < 0.01 | 280 | 13 | 835 | 10 |
| Mammals | < 0.05 | 92 | 21 | 120 | 11 |
| Reptiles | 1.0 | 9 | 0 | 37 | 1 |
Extended Data, Table 4. Importance of predictor variables in explaining Edge Sensitivities of forest core ectotherms and forest core endotherms.
I, Importance; Coeff, Coefficient; P, significance of coefficient estimate; 2.5% and 97.5%, lower and upper limits for coefficient estimates; outputs as conditional average. L - only one species identified as IUCN forest dependent. We fitted two-sided general linear models and selected models from a global model for edge sensitivity via information theoretic approaches and multi-model averaging. Predictors in global models are detailed in Methods. This yielded 1 model for reptiles (n = 9 species), 5 models for amphibians (n = 34 species), 7 models for mammals (n = 111 species) and 20 models for birds (n = 190). The deviance explained by the final model was 98% (reptiles), 31% (amphibians), 24% (mammals) and 3% (birds).
| 4a Predictors retained, Reptiles | I | Coeff | P | 2.5% | 97.5% |
|
| |||||
| Body size | - | 3.11 | < 0.01 | 2.33 | 3.89 |
| IUCN Tree | - | 2.94 | < 0.01 | 2.02 | 3.86 |
| IUCN Habitats | - | 2.53 | < 0.01 | 1.88 | 3.17 |
| Body size : IUCN Tree | - | -1.54 | < 0.01 | -2.04 | -1.04 |
| IUCN Habitats : Body size | - | -1.34 | < 0.01 | -1.69 | -1.00 |
|
| |||||
|
| |||||
| 4b Predictors retained, Amphibians | I | Coeff | P | 2.5% | 97.5% |
|
| |||||
| IUCN Habitats | 1.00 | 0.03 | 0.73 | -0.16 | 0.23 |
| Body size | 1.00 | -0.02 | 0.77 | -0.17 | 0.13 |
| IUCN Forest | 0.89 | -0.36 | 0.07 | -0.75 | 0.02 |
| Body size: IUCN Habitats | 0.56 | -0.03 | 0.18 | -0.07 | 0.01 |
| Body size: IUCN Forest | 0.45 | - | L | - | - |
|
| |||||
|
| |||||
| 4c Predictors retained, Mammals | I | Coeff | P | 2.5% | 97.5% |
|
| |||||
| Non-volant | 1.00 | 0.20 | < 0.001 | 0.10 | 0.30 |
| IUCN Habitats | 0.24 | 0.02 | 0.40 | -0.03 | 0.07 |
| IUCN Forest | 0.23 | -0.04 | 0.39 | -0.14 | 0.06 |
| (Body size)2 | 0.13 | -0.00 | 0.55 | -0.01 | 0.00 |
| IUCN Habitats : Non-volant | 0.12 | -0.04 | 0.16 | -0.10 | 0.01 |
| IUCN Forest : Non-volant | 0.11 | 0.09 | 0.21 | -0.05 | 0.23 |
| Body size | 0.11 | -0.01 | 0.78 | -0.04 | 0.03 |
|
| |||||
|
| |||||
| 4d Predictors retained, Birds | I | Coeff | P | 2.5% | 97.5% |
|
| |||||
| IUCN Forest | 0.51 | -0.04 | 0.27 | -0.10 | 0.03 |
| IUCN Tree | 0.29 | 0.00 | 0.97 | -0.16 | 0.17 |
| Body size | 0.26 | 0.01 | 0.36 | -0.02 | 0.04 |
| Migrant = Full Migrant | 0.16 | 0.13 | 0.10 | -0.03 | 0.29 |
| Migrant = Nomadic | - | 0.06 | 0.70 | -0.24 | 0.35 |
| Migrant = Not migrating | - | 0.13 | 0.08 | -0.02 | 0.28 |
| Range size | 0.09 | 0.00 | 0.50 | -0.00 | 0.00 |
| IUCN Habitats | 0.08 | 0.00 | 0.93 | -0.02 | 0.02 |
| Mean clutch | 0.08 | -0.01 | 0.55 | -0.02 | 0.01 |
| IUCN Forest : Full Migrant | 0.07 | 0.05 | 0.45 | -0.08 | 0.19 |
| IUCN Forest : Full Nomadic | - | 0.30 | 0.04 | 0.02 | 0.58 |
| IUCN Forest : Body size | 0.05 | 0.04 | 0.23 | -0.02 | 0.10 |
| IUCN Tree : Full Migrant | 0.05 | -0.12 | 0.45 | -0.42 | 0.18 |
| IUCN Tree : Nomadic | - | 0.12 | 0.56 | -0.27 | 0.51 |
| IUCN Tree : Not migrating | - | -0.18 | 0.21 | -0.46 | 0.10 |
Supplementary Material
Acknowledgements
We thank Ben Phalan, Philip Stouffer, Hugh Possingham and the Western Australian Department of Parks and Wildlife for supplying additional data from Ghana, Brazil, Australia, and Western Australia, respectively. We are grateful to Jason Tylianakis for providing comments on an earlier draft of the manuscript. M.P., V.L. and R.M.E. were supported by European Research Council Project number 281986. This paper represents a contribution to Imperial College’s Grand Challenges in Ecosystems and the Environment Initiative.
Footnotes
Author Contributions: M.P., V.L. and R.M.E. designed the study and wrote the first draft of the manuscript. M.P. conducted all analyses and V.L. developed the methodology. R.M.E and all other authors contributed data. All authors commented on manuscript drafts.
Author Information: Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Haddad NM, et al. Habitat fragmentation and its lasting impact on Earth’s ecosystems. Sci Adv. 2015;1:e1500052–e1500052. doi: 10.1126/sciadv.1500052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfeifer M, et al. BIOFRAG - a new database for analyzing BIOdiversity responses to forest FRAGmentation. Ecol Evol. 2014;4:1524–37. doi: 10.1002/ece3.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Debinski DM. Forest fragmentation and matrix effects: the matrix does matter. J Biogeogr. 2006;33:1791–1792. [Google Scholar]
- 4.Fletcher RJ. Multiple edge effects and their implications in fragmented landscapes. J Anim Ecol. 2005;74:342–352. [Google Scholar]
- 5.Barlow J, et al. Anthropogenic disturbance in tropical forests can double biodiversity loss from deforestation. Nature. 2016;53:144–147. doi: 10.1038/nature18326. [DOI] [PubMed] [Google Scholar]
- 6.Rosa IMD, Smith MJ, Wearn OR, Purves D, Ewers RM. The Environmental Legacy of Modern Tropical Deforestation. Current Biology. 2016;26:2161–2166. doi: 10.1016/j.cub.2016.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindenmayer DB, Fischer J. Habitat Fragmentation and Landscape Change: An Ecological and Conservation Synthesis. Island Press; Washington DC: 2013. [Google Scholar]
- 8.Didham RK, Lawton JH. Edge Structure Determines the Magnitude of Changes in Microclimate and Vegetation Structure in Tropical Forest Fragments1. Biotropica. 1999;31:17–30. [Google Scholar]
- 9.Ewers RM, Didham RK. Pervasive impact of large-scale edge effects on a beetle community. Proc Natl Acad Sci. 2008;105:5426–5429. doi: 10.1073/pnas.0800460105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ewers RM, Thorpe S, Didham RK. Synergistic Interactions between Edge and Area Effects in a Heavily Fragmented Landscape. Ecology. 2010;88:96–106. doi: 10.1890/0012-9658(2007)88[96:sibeaa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 11.Fahrig L. Rehtinking patch size and isolation effects: the habitat amount hyothesis. J Biogeo. 2013;40:1649–1663. [Google Scholar]
- 12.Ewers RM, Marsh CJ, Wearn OR. Making statistics biologically relevant in fragmented landscapes. Trends Ecol Evol. 2010;25:699–704. doi: 10.1016/j.tree.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Arroyo-Rodríguez V, Dias PAD. Effects of habitat fragmentation and disturbance on howler monkeys: A review. Am J Primatol. 2010;72:1–16. doi: 10.1002/ajp.20753. [DOI] [PubMed] [Google Scholar]
- 14.Vetter D, Hansbauer MM, Végvári Z, Storch I. Predictors of forest fragmentation sensitivity in Neotropical vertebrates: a quantitative review. Ecography. 2011;34:1–8. [Google Scholar]
- 15.Pearson RG, et al. Life history and spatial traits predict extinction risk due to climate change. Nat Clim Chang. 2014;4:217–221. [Google Scholar]
- 16.Sodhi NS, Liow LH, Bazzaz F. Avian Extinctions from Tropical and Subtropical Forests. Annu Rev Ecol Evol Syst. 2004;35:323–345. [Google Scholar]
- 17.Cardillo M, et al. Multiple causes of high extinction risk in large mammal species. Science. 2005;309:1239–41. doi: 10.1126/science.1116030. [DOI] [PubMed] [Google Scholar]
- 18.Purvis A, Gittleman JL, Cowlishaw G, Mace GM. Predicting extinction risk in declining species. Proc Biol Sci. 2000;267:1947–1952. doi: 10.1098/rspb.2000.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olalla-Tárraga MÁ, Rodríguez MÁ. Energy and interspecific body size patterns of amphibian faunas in Europe and North America: anurans follow Bergmann’s rule, urodeles its converse. Glob Ecol Biogeogr. 2007;16:606–617. [Google Scholar]
- 20.Gehring TM, Swihart RK. Body size, niche breadth, and ecologically scaled responses to habitat fragmentation: mammalian predators in an agricultural landscape. Biol Conserv. 2003;109:283–295. [Google Scholar]
- 21.Newbold T, et al. Ecological traits affect the response of tropical forest bird species to land-use intensity. Proc R Soc B Biol Sci. 2013;280 doi: 10.1098/rspb.2012.2131. 20122131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendenhall CD, et al. Countryside biogeography of Neotropical reptiles and amphibians. Ecology. 2014;95:856–870. doi: 10.1890/12-2017.1. [DOI] [PubMed] [Google Scholar]
- 23.Malcolm JR. Edge Effects in Central Amazonian Forest Fragments. Ecology. 1994;75:2438–2445. [Google Scholar]
- 24.Didham RK, Kapos V, Ewers RM. Rethinking the conceptual foundations of habitat fragmentation research. Oikos. 2012;121:161–170. [Google Scholar]
- 25.Kupfer JA, Malanson GP, Franklin SB. Not seeing the ocean for the islands: The mediating influence of matrix-based processes on forest fragmentation effects. Glob Ecol Biogeogr. 2006;15:8–20. [Google Scholar]
- 26.Laurance WF, et al. Habitat fragmentation, variable edge effects, and the landscape-divergence hypothesis. PLoS One. 2007;2:e1017. doi: 10.1371/journal.pone.0001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendenhall CD, Karp DS, Meyer CFJ, Hadly Ea, Daily GC. Predicting biodiversity change and averting collapse in agricultural landscapes. Nature. 2014;509:213–7. doi: 10.1038/nature13139. [DOI] [PubMed] [Google Scholar]
- 28.Tscharntke T, et al. Landscape moderation of biodiversity patterns and processes-eight hypotheses. Biol Rev. 2012;87:661–685. doi: 10.1111/j.1469-185X.2011.00216.x. [DOI] [PubMed] [Google Scholar]
- 29.Schneider-Maunoury L, et al. Abundance signals of amphibians and reptiles indicate strong edge effects in Neotropical fragmented forest landscapes. Biol Conserv. 2016;200:207–215. [Google Scholar]
- 30.Lefebvre V, Pfeifer M, Ewers RM. BioFrag | Edge response -The Biofrag software. 2016 Available at: https://github.com/VeroL/BioFrag.
- 31.De Coster GC, Banks-Leite C, Metzger JP. Atlantic forest bird communities provide different but not fewer functions after habitat loss. Proc R Soc B. 2015;282 doi: 10.1098/rspb.2014.2844. 20142844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenkins CN, Pimm SL, Joppa LN. Global patterns of terrestrial vertebrate diversity and conservation. Proc Natl Acad Sci. 2013;110:E2602–E2610. doi: 10.1073/pnas.1302251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardwick SRSR, et al. The relationship between leaf area index and microclimate in tropical forest and oil palm plantation: Forest disturbance drives changes in microclimate. Agric For Meteorol. 2015;201:187–195. doi: 10.1016/j.agrformet.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watling JI, Braga L. Desiccation resistance explains amphibian distributions in a fragmented tropical forest landscape. Landsc Ecol. 2015;30:1449–1459. [Google Scholar]
- 35.Tuff KT, Tuff T, Davies KF. A framework for integrating thermal biology into fragmentation research. Ecol Lett. 2016;19:361–374. doi: 10.1111/ele.12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindstedt SL, Miller BJ, Buskirk SW. Home range, time, and body size in mammals. Ecology. 1986;67:413–418. [Google Scholar]
- 37.Kosydar AJ. Can life histories predict the effects of habitat fragmentation? A meta-analysis with terrestrial mammals. Appl Ecol Environ Res. 2014;12:505–521. [Google Scholar]
- 38.Haskell JP, Ritchie ME, Olff H. Fractal geometry predicts varying body size scaling relationships for mammal and bird home ranges. Nature. 2002;418:527–530. doi: 10.1038/nature00840. [DOI] [PubMed] [Google Scholar]
- 39.Tucker MA, Rogers TL. Examining predator–prey body size, trophic level and body mass across marine and terrestrial mammals. Proc R Soc London B Biol Sci. 2014;281 doi: 10.1098/rspb.2014.2103. 20142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuckerberg B, Fink D, La Sorte FA, Hochachka WM, Kelling S. Novel seasonal land cover associations for eastern North American forest birds identified through dynamic species distribution modelling. Divers Distrib. 2016;22:717–730. [Google Scholar]
- 41.Watson JEM, Whittaker RJ, Dawson TP. Habitat structure and proximity to forest edge affect the abundance and distribution of forest-dependent birds in tropical coastal forests of southeastern Madagascar. Biol Conserv. 2004;120:311–327. [Google Scholar]
- 42.Pittman SE, Osbourn MS, Semlitsch RD. Movement ecology of amphibians: A missing component for understanding population declines. Biol Conserv. 2014;169:44–53. [Google Scholar]
- 43.Newbold T, et al. Global effects of land use on local terrestrial biodiversity. Nature. 2015;520:45–50. doi: 10.1038/nature14324. [DOI] [PubMed] [Google Scholar]
- 44.Ewers RMRM, et al. Logging cuts the functional importance of invertebrates in tropical rainforest. Nat Commun. 2015;6:6836. doi: 10.1038/ncomms7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ries L, et al. Ecological responses to habitat edges: Mechanisms, Models, and Variability Explained. Annu Rev Ecol Evol Syst. 2004;35:491–522. [Google Scholar]
- 46.Harper Ka, et al. Edge influence on forest structure and composition in fragmented landscapes. Conserv Biol. 2005;19:768–782. [Google Scholar]
- 47.Murcia C. Edge effects in fragmented forests: implications for conservation. Trends Ecol Evol. 1995;10:58–62. doi: 10.1016/S0169-5347(00)88977-6. [DOI] [PubMed] [Google Scholar]
- 48.Broadbent EN, et al. Forest fragmentation and edge effects from deforestation and selective logging in the Brazilian Amazon. Biol Conserv. 2008;141:1745–1757. [Google Scholar]
- 49.Ribeiro MC, Metzger JP, Martensen AC, Ponzoni FJ, Hirota MM. The Brazilian Atlantic Forest: How much is left, and how is the remaining forest distributed? Implications for conservation. Biol Conserv. 2009;142:1141–1153. [Google Scholar]
- 50.Laurance WF, et al. A global strategy for road building. Nature. 2014;513:229–232. doi: 10.1038/nature13717. [DOI] [PubMed] [Google Scholar]
- 51.McCaw WL, Robinson RM, Williams MR. Integrated biodiversity monitoring for the jarrah (Eucalyptus marginata) forest in south-west Western Australia: The Forestcheck project. Aust For. 2011;74:240–253. [Google Scholar]
- 52.Szabo JK, Vesk PA, Baxter PWJ, Possingham HP. Paying the extinction debt: Woodland birds in the Mount Lofty Ranges, South Australia. Emu. 2011;111:59–70. [Google Scholar]
- 53.Marsh CJ, Lewis OT, Said I, Ewers RM. Community-level diversity modelling of birds and butterflies on Anjouan, Comoro Islands. Biol Conserv. 2010;143:1364–1374. [Google Scholar]
- 54.Norris K, et al. Biodiversity in a forest-agriculture mosaic - The changing face of West African rainforests. Biol Conserv. 2010;143:2341–2350. [Google Scholar]
- 55.Banks-Leite C, Ewers RM, Metzger JP. Edge effects as the principal cause of area effects on birds in fragmented secondary forest. Oikos. 2010;119:918–926. [Google Scholar]
- 56.Bar-Massada A, Wood EM, Pidgeon AM, Radeloff VC. Complex effects of scale on the relationships of landscape pattern versus avian species richness and community structure in a woodland savanna mosaic. Ecography. 2012;35:393–411. [Google Scholar]
- 57.Cerezo A, Perelman S, Robbins CS. Landscape-level impact of tropical forest loss and fragmentation on bird occurrence in eastern Guatemala. Ecol Modell. 2010;221:512–526. [Google Scholar]
- 58.Gardner TA, et al. The value of primary, secondary, and plantation forests for a neotropical herpetofauna. Conserv Biol. 2007;21:775–787. doi: 10.1111/j.1523-1739.2007.00659.x. [DOI] [PubMed] [Google Scholar]
- 59.Barlow J, Mestre LAM, Gardner TA, Peres CA. The value of primary, secondary and plantation forests for Amazonian birds. Biol Conserv. 2007;136:212–231. doi: 10.1111/j.1523-1739.2007.00659.x. [DOI] [PubMed] [Google Scholar]
- 60.Morante-Filho JCJC, Faria D, Mariano-Neto E, Rhodes J. Birds in anthropogenic landscapes: The responses of ecological groups to forest loss in the Brazilian Atlantic forest. PLoS One. 2015;10:e0128923. doi: 10.1371/journal.pone.0128923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stouffer PC, Bierregaard RO, Strong C, Lovejoy TE. Long-term landscape change and bird abundance in Amazonian rainforest fragments. Conserv Biol. 2006;20:1212–1223. doi: 10.1111/j.1523-1739.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- 62.Barlow J, et al. Quantifying the biodiversity value of tropical primary, secondary, and plantation forests. Proc Natl Acad Sci U S A. 2007;104:18555–60. doi: 10.1073/pnas.0703333104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wells K, Kalko EKV, Lakim MB, Pfeiffer M. Effects of rain forest logging on species richness and assemblage composition of small mammals in Southeast Asia. J Biogeogr. 2007;34:1087–1099. [Google Scholar]
- 64.Young RP, editor. A biodiversity assessment of the Centre Hills, Montserrat. 2008 [Google Scholar]
- 65.Klingbeil BT, Willig MR. Seasonal differences in population-, ensemble- and community-level responses of bats to landscape structure in Amazonia. Oikos. 2010;119:1654–1664. [Google Scholar]
- 66.Cisneros LM, et al. Multiple dimensions of bat biodiversity along an extensive tropical elevational gradient. J Anim Ecol. 2014;83:1124–1136. doi: 10.1111/1365-2656.12201. [DOI] [PubMed] [Google Scholar]
- 67.Garmendia A, Arroyo-Rodríguez V, Estrada A, Naranjo EJ, Stoner KE. Landscape and patch attributes impacting medium- and large-sized terrestrial mammals in a fragmented rain forest. J Trop Ecol. 2013;29:331–344. [Google Scholar]
- 68.Struebig MJ, Kingston T, Zubaid A, Mohd-Adnan A, Rossiter SJ. Conservation value of forest fragments to Palaeotropical bats. Biol Conserv. 2008;141:2112–2126. [Google Scholar]
- 69.Wayne AF, Liddelow GL, Williams MR. Forestcheck: Terrestrial vertebrate associations with fox control and silviculture in jarrah (Eucalyptus marginata) forest. Aust For. 2011;74:336–349. [Google Scholar]
- 70.D’Cruze N, Kumar S. Effects of anthropogenic activities on lizard communities in northern Madagascar. Anim Conserv. 2011;14:542–552. [Google Scholar]
- 71.Olivier PI, van Aarde RJ, Lombard AT. The use of habitat suitability models and species-area relationships to predict extinction debts in coastal forests, South Africa. Divers Distrib. 2013;19:1353–1365. [Google Scholar]
- 72.Vilela B, Villalobos F. LetsR: A new R package for data handling and analysis in macroecology. Methods Ecol Evol. 2015;6:1229–1234. [Google Scholar]
- 73.Cooper N, Bielby J, Thomas GH, Purvis A. Macroecology and extinction risk correlates of frogs. Glob Ecol Biogeogr. 2008;17:211–221. [Google Scholar]
- 74.Bielby J, Cooper N, Cunningham AAa, Garner TWJWJ, Purvis A. Predicting susceptibility to future declines in the world’s frogs. Conserv Lett. 2008;1:82–90. [Google Scholar]
- 75.Vidal-García M, Byrne PG, Roberts JD, Keogh JS. The role of phylogeny and ecology in shaping morphology in 21 genera and 127 species of Australo-Papuan myobatrachid frogs. J Evol Biol. 2014;27:181–92. doi: 10.1111/jeb.12292. [DOI] [PubMed] [Google Scholar]
- 76.Feldman A, Sabath N, Pyron RA, Mayrose I, Meiri S. Body sizes and diversification rates of lizards, snakes, amphisbaenians and the tuatara. Glob Ecol Biogeogr. 2016;25:187–197. [Google Scholar]
- 77.Meiri S. Evolution and ecology of lizard body sizes. Glob Ecol Biogeogr. 2008;17:724–734. [Google Scholar]
- 78.Andrews RM, Sexton OJ. Water Relations of the Eggs of Anolis Auratus and Anolis Limifrons. Ecology. 1981;62:556–562. [Google Scholar]
- 79.Ballinger RE, Marion KR, Sexton OJ. Thermal Ecology of the Lizard, Anolis Limifrons with Comparative Notes on Three Additional Panamanian Anoles. Ecol Soc Am. 1970;51:246–254. [Google Scholar]
- 80.Avila-Pires TCST. Lizards of Brazilian Amazonia (Reptilia: Squamata) Zoologische verhandelingen. 1995;299 [Google Scholar]
- 81.Bickel R, Losos JB. Patterns of morphological variation and correlates of habitat use in Chameleons. Biol J Linn Soc. 2002;76:91–103. [Google Scholar]
- 82.Brattstrom BH. Amphibian Temperature Regulation Studies in the Field and Laboratory. Am Zool. 1979;19:345–356. [Google Scholar]
- 83.Brattstrom BH. Body Temperatures of Reptiles. Am Midl Nat. 1965;73:376–422. [Google Scholar]
- 84.Brattstrom B. A preliminary review of the thermal requirements of amphibians. Ecology. 1963;44:238–255. [Google Scholar]
- 85.Campbell HW. Ecological observations on Anolis lionotus and Anolis poecilopus (Reptilia, Sauria) in Panama. Am Museum Nat Hist. 1973;2516:1–29. [Google Scholar]
- 86.Carvalho EAR, Lima AP, Magnusson WE, Albernaz ALKM. Long-term effect of forest fragmentation on the Amazonian gekkonid lizards, Coleodactylus amazonicus and Gonatodes humeralis. Austral Ecol. 2008;33:723–729. [Google Scholar]
- 87.Censky EJ, Mccoy CJ. Cycles of Five Species of Snakes (Reptilia: Colubridae) Female Reproductive the Yucatan Peninsula, Mexico. Biotropica. 1988;20:326–333. [Google Scholar]
- 88.Christie K, Stokes VL, Craig MD, Hobbs RJ. Microhabitat preference of Egernia napoleonis in undisturbed jarrah forest, and availability and introduction of microhabitats to encourage colonization of restored forest. Restor Ecol. 2013;21:722–728. [Google Scholar]
- 89.Costa HC, Barros AB, Sueiro LR, Feio RN. The blunt-headed vine snake, Imantodes cenchoa (Linnaeus, 1758), in Minas Gerais, southeastern Brazil. Biotemas. 2010;23:173–176. [Google Scholar]
- 90.Duellman WE, Hedges SB. Three New Species of Pristimantis (Anura: Leptodactylidae) From the Cordillera De Huancabamba in Northern Peru. Herpetologica. 2007;6:119–135. [Google Scholar]
- 91.Harvey MB, Rivas GA, Manzanilla J. Redescription of Stenocercus erythrogaster (Hallowell) Copeia. 2004;2004:940–944. [Google Scholar]
- 92.Henderson RW. Lesser Antillean snake faunas: distribution, ecology, and conservation concerns. Oryx. 2004;38:311–320. [Google Scholar]
- 93.Hirth HF. The ecology of two lizards on a tropical beach. Ecol Monogr. 1963;33:83–112. [Google Scholar]
- 94.Hoogmoed MS, Ávila-Pires TCS. Studies on the species of the South American lizard genus Arthrosaura Boulenger (Reptilia: Sauria: Teiidae), with the resurrection of two species. Zool Meded Leiden. 1992;66:453–484. [Google Scholar]
- 95.Irschick DJ, Vitt LJ, Zani PA, Losos JB. A comparison of evolutionary radiations in mainland and Caribbean Anolis lizards. Ecology. 1997;78:2191–2203. [Google Scholar]
- 96.Kanowski JJ, Reis TM, Catterall CP, Piper SD. Factors affecting the use of reforested sites by reptiles in cleared rainforest landscapes in tropical and subtropical Australia. Restor Ecol. 2006;14:67–76. [Google Scholar]
- 97.Koehler G, Alt S, Gruenfelder C, Dehling M, Sunyer J. Morphological variation in Central American leaf-litter anoles: Norops humilis, N. quaggulus and N. uniformis. Salamandra. 2006;42:239–254. [Google Scholar]
- 98.Mesquita DO, Colli GR. Geographical Variation in the Ecology of Populations of Some Brazilian Species of Cnemidophorus (Squamata, Teiidae) Copeia. 2003;2003:285–298. [Google Scholar]
- 99.Montgomery CE, Reed RN, Shaw HJ, Boback SM, Walker JM. Distribution, Habitat, Size, and Color Pattern of Cnemidophorus lemniscatus (Sauria: Teiidae) on Cayo Cochino Pequeño, Honduras. Southwest Nat. 2007;52:38–45. [Google Scholar]
- 100.Moreno-Arias RA, Medina-Rangel GF, Castano-Mora OV. Lowland reptiles of Yacopi (Cundinamarca, Colombia) Rev la Acad Colomb Ciencias Exactas Fis y Nat. 2008;32:93–103. [Google Scholar]
- 101.Nichols OG, Bamford MJ. Reptile and frog utilisation of rehabilitated bauxite minesites and dieback-affected sites in Western Australia’s Jarrah Eucalyptus marginata forest. Biol Conserv. 1985;34:227–249. [Google Scholar]
- 102.Presch W. Evolutionary history of the South American microteiid lizards (Teiidae: Gymnophthalminae) Copeia. 1980:36–56. [Google Scholar]
- 103.Puente M, Raselimanana AP, Vences M. Rediscovery and redescription of the Malagasy dwarf gecko Lygodactylus klemmeri. Zootaxa. 2005:31–35. [Google Scholar]
- 104.Rossman DA. Morphological variation in the endemic Colombian water snake, Helicops danieli (Serpentes, Xenododontidae) Revista de la Academia Colombiana de Ciencias Exactas Fisicas y Naturales. 2003;26:589–594. [Google Scholar]
- 105.Sajdak RA, Henderson R. Status of West Indian racers in the Lesser Antilles. Oryx. 1991;25:33–38. [Google Scholar]
- 106.Sales RFD, Ribeiro LB, Jorge JS, Freire EMX. Habitat use, daily activity periods, and thermal ecology of Ameiva ameiva (Squamata: Teiidae) in a caatinga area of northeastern Brazil. Phyllomedusa. 2011;10:165–176. [Google Scholar]
- 107.Seebacher F, Franklin CE. Physiological mechanisms of thermoregulation in reptiles: A review. J Comp Physiol B. 2005;175:533–541. doi: 10.1007/s00360-005-0007-1. [DOI] [PubMed] [Google Scholar]
- 108.Seib R. Euryphagy in a Tropical Snake, Coniophanes fissidens. Biotropica. 1985;17:57–64. [Google Scholar]
- 109.Vitt LJ, Zani Pa, Lima aCM. Heliotherms in tropical rain forest: the ecology of Kentropyx calcarata (Teiidae) and Mabuya nigropunctata (Scincidae) in the Curuá-Una of Brazil. J Trop Ecol. 1997;13:199–220. [Google Scholar]
- 110.Vitt LJ, et al. History and the Global Ecology of Squamate reptiles. Am Nat. 2003;162:44–60. doi: 10.1086/375172. [DOI] [PubMed] [Google Scholar]
- 111.Van Devender RW. Comparative demography of the lizard Bailiscus basiliscus. Herpetologica. 1998;58:375–379. [Google Scholar]
- 112.Vitt LJ, Zani PA, Caldwell JP, de Araujo MC, Magnusson WE. Ecology of whiptail lizards (Cnemidophorus) in the Amazon region of Brazil. Copeia. 1997;4:745–757. [Google Scholar]
- 113.Wilson LD, Townsend JH. The herpetofauna of the rainforests of Honduras. Caribb J Sci. 2006;42:88–113. [Google Scholar]
- 114.Campbell J. Amphibians and Reptiles of Northern Guatemala, the Yucatan, and Belize (Animal Natural History Series) University of Oklahoma Press; 1999. [Google Scholar]
- 115.Cochran D. Frogs of southeastern Brazil, 1954, Smithsonian Institution, United States National Museum Bulletin. Smithsonian Institution Press; 1954. [Google Scholar]
- 116.Henkel F, Schmidt W. Amphibians and Reptiles of Madagascar, the Mascarene, the Seychelles, and the Comoro Islands. Krieger Pub, Co; 2000. [Google Scholar]
- 117.Vitt LJ, Caldwell JP. Herpetology: An Introductory Biology of Amphibians and Reptiles. Fourth Edition. 2013. Herpetology: An Introductory Biology of Amphibians and Reptiles. [Google Scholar]
- 118.Duellman WE, Trueb L. Biology of Amphibians. McGrawHill Inc; New York: 1994. [DOI] [Google Scholar]
- 119.Savage J. The amphibians and reptiles of Costa Rica: A herpetofauna between two continents, between two seas Uma ética para quantos? 2002;XXXIII [Google Scholar]
- 120.Wilman H, et al. EltonTraits 1.0: Species-level foraging attributes of the world’s birds and mammals. Ecology. 2014;95:2027–2027. [Google Scholar]
- 121.Jones KE, et al. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology. 2009;90:2648–2648. [Google Scholar]
- 122.Hansen MC, et al. High-resolution global maps of 21st-century forest cover change. Science. 2013;342:850–3. doi: 10.1126/science.1244693. [DOI] [PubMed] [Google Scholar]
- 123.Wood S. Package: mgcv 1.8-9 Mixed GAM Computation Vehicle with GCV/AIC/REML Smoothness Estimation. R 3.2.3. 2015 doi: 10.1111/j.1541-0420.2007.00905_3.x. [DOI] [Google Scholar]
- 124.Wickham H. ggplot2. Elegant Graphics for Data Analysis. 2009 doi: 10.1007/978-0-387-98141-3. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The *xls and *kml data that support the findings of this study are available in figshare with the identifier doi: 10.6084/m9.figshare.4573504. Original BIOFRAG data are available on request from the corresponding author but restrictions apply to the availability of these data, which are not publicly available. Data are however available from the authors upon reasonable request and with permission of dataset authors as specified in the BIOFRAG database2 (https://biofrag.wordpress.com/).