Abstract
Noncoding RNAs (ncRNAs), including micro (mi)RNAs, long noncoding (lnc)RNAs, and circular (circ)RNAs, control specific gene expression programs by regulating transcriptional, post-transcriptional, and post-translational processes. Through their broad influence on protein expression and function, ncRNAs have been implicated in virtually all cellular processes such as proliferation, senescence, quiescence, differentiation, apoptosis, and the stress and immune responses. Senescence is a cellular phenotype associated with the physiologic decline of aging and with age-related pathologies. Besides their characteristic terminal growth arrest and differential gene expression programs, senescent cells are known to secrete potent pro-inflammatory, angiogenic, and tissue-remodeling factors. This important trait, known as the senescence-associated secretory phenotype (SASP), influences many biological processes such as tissue repair and regeneration, tumorigenesis, and the aging-associated pro-inflammatory state. Here, we review the microRNAs, lncRNAs, and circRNAs that influence the production of SASP factors and discuss the rising interest in SASP-regulatory ncRNAs as diagnostic and therapeutic targets.
1. Introduction
Cellular senescence is a state of terminal growth arrest in which cells are unresponsive to growth factor stimulation. This phenotype was initially described by Hayflick as the end of the lifespan of primary fibroblasts maintained in culture [1]. The senescence program is triggered when cells encounter stress conditions such as critically short telomeres, DNA damage, oncogenic activation, hypoxia, and oxidative stress [2]. Although senescent cells do not divide, they are metabolically active and exibit a distinct metabolic profile. They display a flattened and enlarged morphology, altered gene expression patterns, increased activity of a neutral β-galactosidase, and senescence-associated heterochromatic foci [3–7]. Another major feature of senescent cells is the senescence-associated secretory phenotype (SASP) [8], characterized by the production and secretion of regulatory factors including interleukins, cytokines, growth factors, angiogenic factors, and matrix metalloproteases [8, 9]. SASP affects the function of tissues and organs by attracting immune cells, enhancing angiogenesis, and remodeling the extracellular matrix [10, 11].
Cell senescence has numerous and complex effects on tissue homeostasis and health. It is necessary for tissue sculpting during development, enhances wound healing, and suppresses detrimental tissue fibrosis in response to damage to the liver, the pancreas, and the skin [12–17]. In addition, in young individuals, senescence has been shown to suppress tumor progression [18, 19]. However, senescence can also have detrimental effects. In older individuals, the accumulation of senescent cells during aging alters the physiologic function of tissues and organs leading to age-related diseases like cancer, cataracts, and atherosclerosis [20, 21]. The accumulation of senescent cells during aging has also been linked to the excessive production of SASP factors, which facilitate chronic inflammation and age-related diseases such as arthritis [15, 22, 23]. In addition, the inflammatory cytokines and growth factors secreted from senescent cells may promote tumor growth in old age by inducing angiogenesis and tumor cell proliferation [24–26].
Given that SASP is such a critical trait of senescent cells, understanding the regulation of SASP factor production provides direct insight into the mechanisms of aging. In this review, we discuss the regulatory RNAs that impact upon the SASP phenotype at all levels – transcriptional, post-transcriptional, and post-translational (Figure 1). We focus on specific noncoding (nc)RNAs that affect subsets of SASP factors, including microRNAs (miRNAs), long ncRNAs (lncRNAs), and circular RNAs (circRNAs).
Figure 1. Levels of SASP gene regulation by different ncRNAs.
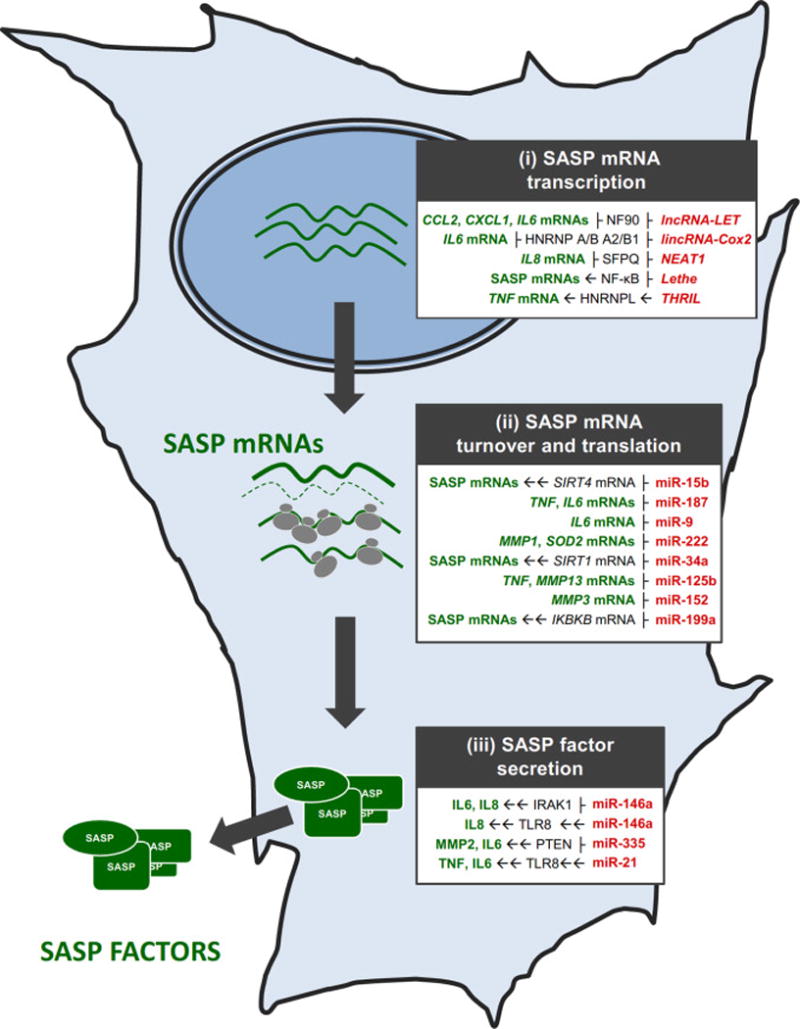
ncRNAs affecting SASP factor production and secretion in a senescent cell by influencing the transcription of SASP genes (i), the post-transcriptional fate of SASP mRNAs (ii), and the secretion of SASP factors (iii), indicated in gray boxes. Red, ncRNAs; green, SASP factors; black, mediators through which some ncRNAs affect SASP factor expression. ├, inhibition/repression, → direct induction/activation, →→ indirect induction/activation.
2. MicroRNAs
MicroRNAs (miRNAs) are small ncRNAs spanning ∼22 nucleotides. They are generated from primary transcripts (pri-miRNAs) transcribed from the genome by RNA polymerase II. Pri-miRNAs are processed in the nucleus by a complex comprising the ribonuclease DROSHA (HGNC) and DiGeorge critical region 8 (DGCR8) to generate microRNA precursors (pre-miRNAs) [27–31]. Pre-miRNAs are exported to the cytoplasm by Exportin 5 for further processing [32] and are cleaved in the cytoplasm by ribonuclease DICER1 to produce mature microRNAs [33], which are loaded into the RNA-induced silencing complex (RISC) [34]. Argonaute (AGO) proteins in the RISC direct microRNAs to specific mRNAs, typically forming partial complementarity between the microRNAs and segments of the mRNA 3′-untranslated region (UTR), which in turn lower the stability and/or translation of the target mRNA [35–38]. A few reports suggest that some microRNAs may instead upregulate the expression of target genes by competing with a translation suppressor or by other mechanisms [39, 40].
A single microRNA can target multiple mRNAs most human mRNAs are believed to be regulated by microRNAs [37]. Through this broad impact on gene expression, microRNAs can regulate diverse physiologic processes such as embryonic development and cell proliferation, differentiation, and apoptosis [41–45]. Many microRNAs have also been implicated in pathological conditions including cancer, diabetes, cardiovascular disease, neurodegeneration, and immune diseases [46–48]. Since identification of the first microRNA, lin-4, reported to regulate the lifespan of C. elegans [49], many studies have established that microRNAs are among the key regulators of cell senescence and aging [50, 51] in many species including humans.
2.1. MicroRNAs and SASP
Senescent cells are characterized by profound changes in metabolic and protein expression profiles, including SASP. As described above, SASP is characterized by increased production and secretion of various cytokines, growth factors and matrix metalloproteases [24]. Recently, several microRNAs have been shown to regulate the production and secretion of these factors in senescent cells, as discussed in this section (Table 1).
Table 1. ncRNAs affecting SASP factor production.
MicroRNAs, lncRNAs, and circRNAs implicated in regulating SASP factors. The specific ‘Target SASP factors’ affected by the regulatory ncRNAs are listed; ‘[SASP mRNAs]’ denotes general regulation of the SASP phenotype by a given ncRNA. ‘Effector molecules’ are proteins and microRNAs through which some ncRNAs affect SASP gene expression programs.
| ncRNA | Target SASP factor | Effector molecules | References | |
|---|---|---|---|---|
| microRNAs | miR-146a/b | IL6, IL8 | IRAK1, TLR8 | [52–54] |
| miR-335 | MMP2, IL6 | PTEN | [55] | |
| miR-15b | [SASP mRNAs] | SIRT4 | [56] | |
| miR-187 | IL6, TNF mRNAs | [57] | ||
| miR-9 | IL6 mRNA | [58] | ||
| miR-21 | IL6, TNF, IL10 | TLR8, PDCD4 | [59, 60] | |
| miR-222 | MMP1, SOD1 mRNAs | [61] | ||
| miR-34a | [SASP mRNAs] | SIRT1 | [62] | |
| miR-125b | TNF, MMP13 mRNAs | [63, 64] | ||
| miR-152 | MMP3 mRNA | [65] | ||
| miR-147 | TNF, IL6 | (negative feedback?) | [66] | |
| miR-199a | [SASP mRNAs] | IKBKB | [67] | |
| lncRNAs | lncRNA-LET | CCL2, CXCL1, IL6 mRNAs | NF90 | [81, 82] |
| LincRNA-Cox2 | IL6 mRNA | HNRNPA/B, HNRNPA2/B1 | [83] | |
| NEAT1 | IL8 mRNA | SFPQ | [85] | |
| lnc-IL7R | SELE, VCAM1, IL6, IL8 | (negative feedback?) | [86, 87] | |
| Lethe | [SASP mRNAs] | NF-κB | [88, 89] | |
| THRIL | TNF mRNA | HNRNPL | [90] | |
| circRNA | Circ-Foxo3 | [SASP mRNAs] | FAK, HIF1A, ID1, E2F1 | [97] |
| CircPVT1 | [SASP mRNAs] | Let-7 sequestration | [98] |
miR-146a/b
miR-146a and miR-146b were found to be more highly abundant in senescent than in quiescent fibroblasts. In primary human fibroblasts, miR-146a/b inhibit the secretion of SASP factors interleukin (IL)6 and IL8 by acting upon IRAK1 mRNA and lowering the production of IRAK1, a key factor of the IL-1α (IL1A) receptor signaling pathway [52]. Increased miR-146a/b expression in response to high levels of IL1A was proposed to function as a negative feedback loop to prevent excessive SASP activity [52]. However, miR-146a was induced in human diploid BJ fibroblasts immortalized by overexpression of telomerase, suggesting that in some cases, miR-146a might regulate SASP independently of senescence [53]. In trophoblasts, miR-146a-3p expression was elevated by antiphospholipid antibody (aPL) treatment through the activation of Toll-like receptor 4 (TLR4). This upregulation of miR-146a induced IL8 secretion by activating TLR8 [54].
miR-335
A recent report indicated that miR-335 was upregulated in normal senescent cells and cancer-associated senescent fibroblasts (CAF). Higher levels of miR-335 lowered the abundance of phosphatase and tensin homologue (PTEN), in turn causing a rise in SASP factors like MMP2 and IL6 [55]. Senescent cells showed increased secretion of the protein cyclooxygenase 2 (PTGS2/COX2) and the signaling lipid prostaglandin E2 (PGE2). The levels of miR-335 were downregulated by inhibition of PTSG2/COX2 using celecoxib, which restored PTEN expression and decreased SASP [55].
miR-15b
Sirtuin 4 (SIRT4) is implicated in senescence-associated mitochondrial dysfunction and SASP. Its high expression in human dermal fibroblasts undergoing replicative or stress-induced senescence was associated to decreased levels of miR-15b [56].
miR-187
IL10 is a potent anti-inflammatory molecule that suppresses cytokine expression both transcriptionally and post-transcriptionally. Activation of primary human monocytes with IL10 induced the expression of miR-187, which in turn suppressed production of two major SASP factors, TNF and IL6 [57].
miR-9
The tumor suppressor miR-9, downregulated in cervical adenocarcinoma, was found to inhibit the expression of several targets, including the SASP factor IL6 [58].
miR-21
miR-21 is secreted by cancer cells and acts as a ligand for TLR8 in immune cells, triggering the secretion of pro-inflammatory cytokines TNF and IL6 [59]. While programmed cell death protein 4 (PDCD4), a direct target of miR-21, activates NF-κB and suppresses IL10 production, upregulation of miR-21 in human peripheral blood mononuclear cells in response to lipopolysaccharide (LPS) suppressed PDCD4 production leading to lower NF-κB activity and greater IL10 production [60].
miR-222
miR-222 is upregulated in less aggressive metastatic cells of oral tongue squamous cell carcinoma (OTSCC). Functional analyses suggested that overexpression of miR-222 inhibited OTSCC cell invasion by inhibiting the SASP factor matrix metalloproteinase 1 (MMP1) [61]. Further analysis revealed that miR-222 inhibited the expression of MMP1 by directly targeting the 3′UTR of MMP1 mRNA as well as by inhibiting expression of manganese superoxide dismutase 2 (SOD2), an inducer of MMP1 production.
miR-34
Expression of the longevity-associated protein SIRT1 is repressed by miR-34a. An elevation in miR-34a abundance in human aortic smooth muscle cells led to significant downregulation of SIRT1 and caused higher production of pro-inflammatory SASP molecules, although the secretion of SASP factors did not appear to be influenced directly by SIRT1 [62].
miR-125b
Treatment of RAW 264.7 macrophages with LPS reduced the levels of miR-125b, a microRNA that targets TNF mRNA and reduces TNF levels [63]. These data suggest that the LPS-dependent decrease in miR-125b may contribute to the LPS-triggered increase in TNF and SASP. The tumor suppressor miR-125b was less abundant in cutaneous squamous cell carcinoma (cSCC) than in healthy skin, leading to higher production of the SASP factor MMP13 in cSCCs [64].
miR-152
miR-152 inhibits the expression of the SASP factor MMP3 by binding to the 3′UTR of MMP3 mRNA. In vitro invasion assays suggested that miR-152 significantly reduced the invasiveness of glioma cells, possibly by inhibiting MMP3 production [65].
miR-147
LPS also induced the expression of miR-147 in mouse macrophages via the TLR4–NF-κB axis. Overexpression of miR-147 was found to suppress the expression of pro-inflammatory cytokines TNF and IL6 in macrophages stimulated with TLR2/TLR3/TLR4 ligands. These findings reveal the existence of a negative feedback loop through which TLR stimulation increases miR-147 levels, in turn suppressing the excessive production of inflammatory cytokines [66].
miR-199a
Activation NF-κB requires a kinase IκB kinase-β (IKBKB), a crucial factor for the activation of TLR–MyD88–NF-κB pathway in ovarian cancer. Recent reports suggest that miR-199a was capable of suppressing the production of IKBKB, which led to the reduced secretion of SASP factors IL6, IL8, and MCP1, and suppressed tumor progression [67].
3. Long-noncoding RNAs (lncRNAs)
Recent advances in high-throughput transcriptome sequencing revealed that the human genome encodes thousands of long noncoding RNAs (lncRNAs) [68]. LncRNAs are defined as transcripts longer than 200 nucleotides that generally lack protein-coding capacity but are transcribed and processed like mRNAs [69]. LncRNAs can be classified based on their genomic origin into pseudogene-encoded lncRNAs, long intergenic RNAs, antisense lncRNAs, long intronic ncRNAs, etc [69]. LncRNAs have been implicated in the regulation of gene expression by controlling several key processes such as chromatin remodeling, transcription, mRNA stability, translation, and protein stability [70–75]. Recent reports indicate that lncRNAs are critical regulators of physiological processes such as cell division and differentiation, and numerous human diseases including cancer and neurodegeneration [76–78]. Recently, we screened for senescence-associated lncRNAs in human diploid fibroblasts and found that the expression of several lncRNAs was altered in senescent cells [79, 80]. In this section, we will discuss the role of lncRNAs in SASP.
3.1. LncRNAs and SASP
LncRNA-LET
lncRNA-LET (low expression in tumor) is less abundant in cancers including hepatocellular carcinoma, colorectal cancer, and squamous-cell lung carcinoma. Silencing of lncRNA-LET stabilizes and thereby allows the accumulation of nuclear factor 90 (NF90) [81]. Senescent cells express reduced levels of NF90, an RNA-binding protein (RBP) that suppresses the translation of several SASP factors including MCP1 (CCL2), GROa (CXCL1), and IL6 [82]. Thus, the reduction of NF90 levels in senescent cells is linked to the enhanced production of several SASP factors. Accordingly, NF90 appears to function as an effector of lncRNA-LET actions to maintain low levels of SASP factors.
LincRNA-Cox2
LincRNA-Cox2 expression is induced in mouse upon activation of TLRs through the TLR–MyD88–NF-κB pathway. LincRNA-Cox2 suppresses the transcription of different sets of proinflammatory genes by interacting with heterogeneous nuclear ribonucleoprotein (HNRNP) A/B and A2/B1. Silencing LincRNA-Cox2 upregulated chemokines CCL5 and CX3CL1 as well as chemokine receptor 1 (CCRl), while it downregulated other proteins including IL6 following treatment with the TLR activator Pam3CSK4 [83].
NEAT1
The lncRNA nuclear-enriched abundant transcript 1 (NEAT1) is required for the formation nuclear paraspeckles [84]. Induction of NEAT1 expression by viral infection or treatment with poly(I:C), a TLR3 agonist, enhanced the formation of paraspeckles. The splicing factor proline/glutamine-rich (SFPQ) inhibits IL8 transcription. Interestingly, NEAT1 facilitates the transcription of IL8 mRNA by relocating SFPQ to nuclear paraspeckles [85].
Lnc-IL7R
The levels of lnc-IL7R were elevated upregulated in response to treatment with LPS. Silencing of lnc-IL7R reduced trimethylation of histone H3 at lysine 27 (H3K27me3) leading to a decrease in the levels of the inflammatory mediators E-selectin (SELE), VCAM1, IL6, and IL8 [86]. These findings suggest that Lnc-IL7R might contribute to regulating SASP factor production [87].
Lethe
The pseudogene lncRNA Lethe was shown to be downregulated in aging tissues. It was also reported that aging tissues have highly active NF-κB which could induce several pro-inflammatory genes [88]. Lethe interacts with the NF-κB subunit RelA to inhibit the DNA-binding activity of NF-κB, leading to less production of pro-inflammatory cytokines [89]. The age-associated reduction in Lethe may be one of the reasons for the increased NF-κB activity in older individuals.
THRIL
In THP1 macrophages, the lncRNA THRIL (TNF- and hnRNPL-related immunoregulatory lincRNA) interacts with the RBP hnRNP L to form a ribonucleoprotein complex that promotes TNF transcription through binding to the TNF gene promoter [90].
4. CircRNAs
Circular RNAs (circRNAs) have attracted much interest in recent years. Although circRNAs were first identified decades earlier, extensive investigation was not performed due to poor knowledge of their function and rudimentary detection techniques [91–93]. Recent work has uncovered that thousands of circRNAs are expressed in mammalian cells [94, 95], generally arising from the joining of 5′ and 3′ ends of exonic or intronic sequences during splicing, a process known as ‘backsplicing’. Due to the lack of free ends, circRNAs are resistant to cellular exonucleases. Recent studies have described various functions of circRNA in cells including sponging of microRNAs and RBPs, as reviewed recently [96]. Although knowledge of circRNAs in SASP is still at early stages, we anticipate that their contribution to SASP will become more apparent as our understanding of circRNAs progresses.
Circ-Foxo3
Highly expressed in old organs (heart, intestines, lung, and skin) relative to young organs, Circ-Foxo3 is predominantly localized in the cytoplasm and interacts with senescence-associated proteins FAK, HIF1A, ID1, and E2F1, helping to elicit senescence [97]. In turn, Circ-Foxo3 increases the production of SASP factors, likely through indirect mechanisms.
CircPVT1
Hundreds of senescence-associated circRNAs (SAC-RNAs) differentially expressed in senescent WI-38 human diploid fibroblasts were recently reported [98]. CircPVT1, a SAC-RNA generated from the lncRNA PVT1, was identified as being markedly reduced in senescent fibroblasts. The high levels of CircPVT1 in dividing cells resulted in the sequestration of let-7 and enabling of a proliferative phenotype [98], suppressing senescence and SASP.
5. Concluding remarks and perspectives
We have discussed the current knowledge and possible roles of ncRNAs (microRNAs, lncRNAs, circRNAs) in one of the major facets of senescence, SASP. As in other areas of senescence-associated gene regulation [6, 7], it is interesting that the various levels of control of SASP protein production rely on the actions of ncRNAs of all types. We propose that this extensive network of regulatory mechanisms underscores the critical role of SASP in senescence, and highlight the joint roles of RNA regulators in conjunction with protein and DNA to elicit a highly precise control of the SASP program. The reliance on regulatory ncRNA for the control of cellular processes may be particularly important in processes such as senescence and SASP in which cells experience cumulative damage to protein and DNA and thus transcriptional regulation alone may not control gene expression patterns with sufficient accuracy. Alternatively, ncRNAs may provide the ‘checks and balances’ that characterize gene regulation driving critical cellular processes. One additional possibility is that SASP-regulatory ncRNAs might be shared between cells via extracellular vesicles originating from neighboring cells or from distant tissues [99, 100] to ensure that this important phenotype is maintained even if cells have impaired transcriptional programs.
As discussed, mounting evidence supports functions for ncRNAs as regulators of the production and secretion of SASP factors by modulating the transcription of SASP genes, the stability of SASP mRNAs, and the translation and/or secretion of SASP proteins. microRNAs target large fractions of mRNAs, and each microRNA can influence hundreds of target mRNAs [101]. Given their pleiotropic actions, inhibition of one microRNA using a stable antisense RNA molecule (an ‘antagomiR’) that binds to the mature microRNA of interest and inhibits its function [102] could have a broad clinical impact. Such approaches have been successfully used in cancer therapy [103–105]. In a similar way, microRNA-based therapeutics can be used to inhibit SASP-associated microRNAs in senescent cells. LncRNAs can interact with DNA, RNA, and protein to alter the expression of specific set of genes implicated in various diseases [106, 107]. Accordingly, lncRNAs are also emerging as therapeutic targets; an interfering lncRNA was recently shown to suppress carcinogenesis by blocking multiple oncogenic microRNAs [108] and might similarly be envisioned to affect SASP. Nonetheless, our knowledge of the roles of lncRNAs in cellular senescence is still very limited, and this knowledge is even more limited for circRNAs. As our understanding of ncRNAs regulating SASP expands and deepens, their potential therapeutic value in age-associated diseases will come into view.
Acknowledgments
This research was supported in full by the National Institute on Aging Intramural Research Program, NIH.
Abbreviations
- TNF
Tumor necrosis factor alpha
- IL
Interleukin
- LPS
lipopolysaccharide
- SASP
Senescence-associated secretory phenotype
- TLR
Toll-like receptors
- NF-κB
nuclear factor-κB
- lncRNA
long noncoding RNA
- miRNA
microRNA
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hayflick L. The Limited in Vitro Lifetime of Human Diploid Cell Strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 2.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24:2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldstein S. Replicative senescence: the human fibroblast comes of age. Science. 1990;249:1129–1133. doi: 10.1126/science.2204114. [DOI] [PubMed] [Google Scholar]
- 4.Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc. 2009;4:1798–806. doi: 10.1038/nprot.2009.191. [DOI] [PubMed] [Google Scholar]
- 5.Dimri GP1, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, Peacocke M, Campisi J. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cristofalo VJ, Volker C, Francis MK, Tresini M. Age-dependent modifications of gene expression in human fibroblasts. Crit Rev Eukaryot Gene Expr. 1998;8:43–80. doi: 10.1615/critreveukargeneexpr.v8.i1.30. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, Pan KH, Cohen SN. Senescence-specific gene expression fingerprints reveal cell-type-dependent physical clustering of up-regulated chromosomal loci. Proc Natl Acad Sci USA. 2003;100:3251–3256. doi: 10.1073/pnas.2627983100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coppé JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev Cancer. 2009;9:81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- 10.Acosta JC, Banito A, Wuestefeld T, Georgilis A, Janich P, Morton JP, Athineos D, Kang TW, Lasitschka F, Andrulis M, Pascual G, Morris KJ, Khan S, Jin H, Dharmalingam G, Snijders AP, Carroll T, Capper D, Pritchard C, Inman GJ, Longerich T, Sansom OJ, Benitah SA, Zender L, Gil J. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat Cell Biol. 2013;15:978–990. doi: 10.1038/ncb2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Acosta JC, O’Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, Takatsu Y, Melamed J, d’Adda di Fagagna F, Bernard D, Hernando E, Gil J. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 12.Storer M, Mas A, Robert-Moreno A, Pecoraro M, Ortells MC, Di Giacomo V, Yosef R, Pilpel N, Krizhanovsky V, Sharpe J, Keyes WM. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell. 2013;155:1119–1130. doi: 10.1016/j.cell.2013.10.041. [DOI] [PubMed] [Google Scholar]
- 13.Muñoz-Espín D, Cañamero M, Maraver A, Gómez-López G, Contreras J, Murillo-Cuesta S, Rodríguez-Baeza A, Varela-Nieto I, Ruberte J, Collado M, Serrano M. Programmed cell senescence during mammalian embryonic development. Cell. 2013;155:1104–1118. doi: 10.1016/j.cell.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 14.Demaria M, Ohtani N, Youssef SA, Rodier F, Toussaint W, Mitchell JR, Laberge RM, Vijg J, Van Steeg H, Dollé ME, Hoeijmakers JH, de Bruin A, Hara E, Campisi J. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev Cell. 2014;31:722–733. doi: 10.1016/j.devcel.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munoz-Espin D, Serrano M. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol. 2014;15:482–496. doi: 10.1038/nrm3823. [DOI] [PubMed] [Google Scholar]
- 16.Lujambio A, Akkari L, Simon J, Grace D, Tschaharganeh DF, Bolden JE, Zhao Z, Thapar V, Joyce JA, Krizhanovsky V, Lowe SW. Non-cell-autonomous tumor suppression by p53. Cell. 2013;153:449–460. doi: 10.1016/j.cell.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang TW, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D, Hohmeyer A, Gereke M, Rudalska R, Potapova A, Iken M, Vucur M, Weiss S, Heikenwalder M, Khan S, Gil J, Bruder D, Manns M, Schirmacher P, Tacke F, Ott M, Luedde T, Longerich T, Kubicka S, Zender L. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479:547–551. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- 18.Krtolica A, Campisi J. Cancer and aging: a model for the cancer promoting effects of the aging stroma. Int J Biochem Cell Biol. 2002;34:1401–1414. doi: 10.1016/s1357-2725(02)00053-5. [DOI] [PubMed] [Google Scholar]
- 19.Prieur A, Peeper DS. Cellular senescence in vivo: a barrier to tumorigenesis. Curr Opin Cell Biol. 2008;20:150–155. doi: 10.1016/j.ceb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Ovadya Y, Krizhanovsky V. Senescent cells: SASPected drivers of age-related pathologies. Biogerontology. 2014;15:627–642. doi: 10.1007/s10522-014-9529-9. [DOI] [PubMed] [Google Scholar]
- 21.Sikora E, Bielak-Zmijewska A, Mosieniak G. Cellular senescence in ageing, age-related disease and longevity. Curr Vasc Pharmacol. 2014;12:698–706. doi: 10.2174/1570161111666131219094045. [DOI] [PubMed] [Google Scholar]
- 22.Baker DJ1, Perez-Terzic C, Jin F, Pitel KS, Niederländer NJ, Jeganathan K, Yamada S, Reyes S, Rowe L, Hiddinga HJ, Eberhardt NL, Terzic A, van Deursen JM. Opposing roles for p16Ink4a and p19Arf in senescence and ageing caused by BubR1 insufficiency. Nat Cell Biol. 2008;10:825–36. doi: 10.1038/ncb1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, van Deursen JM. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coppé JP, Patil CK, Rodier F, Sun Y, Muñoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci USA. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu D, Hornsby PJ. Senescent human fibroblasts increase the early growth of xenograft tumors via matrix metalloproteinase secretion. Cancer Res. 2007;67:3117–3126. doi: 10.1158/0008-5472.CAN-06-3452. [DOI] [PubMed] [Google Scholar]
- 27.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 29.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 30.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yi R1, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3106. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 34.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 35.Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- 36.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 37.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 38.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 39.Panda AC, Sahu I, Kulkarni SD, Martindale JL, Abdelmohsen K, Vindu A, Joseph J, Gorospe M, Seshadri V. miR-196b-mediated translation regulation of mouse insulin2 via the 5′UTR. PLoS ONE. 2014;9:e101084. doi: 10.1371/journal.pone.0101084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 41.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–6. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 42.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 43.Chang S, Johnston RJ, Jr, Frøkjaer-Jensen C, Lockery S, Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430:785–789. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- 44.Plasterk RH. Micro RNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 45.Lee KP, Shin YJ, Panda AC, Abdelmohsen K, Kim JY, Lee SM, Bahn YJ, Choi JY, Kwon ES, Baek SJ, Kim SY, Gorospe M, Kwon KS. miR-431 promotes differentiation and regeneration of old skeletal muscle by targeting Smad4. Genes Dev. 2015;29:1605–1617. doi: 10.1101/gad.263574.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He X, He L, Hannon GJ. The guardian’s little helper: microRNAs in the p53 tumor suppressor network. Cancer Res. 2007;67:11099–11101. doi: 10.1158/0008-5472.CAN-07-2672. [DOI] [PubMed] [Google Scholar]
- 47.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226–30. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, Kowdley KV. MicroRNAs in common human diseases. Genomics Proteomics Bioinformatics. 2012;10:246–253. doi: 10.1016/j.gpb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boehm M, Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310:1954–1957. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- 50.Gorospe M, Abdelmohsen K. MicroRegulators come of age in senescence. Trends Genet. 2011;27:233–241. doi: 10.1016/j.tig.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harries LW. MicroRNAs as Mediators of the Ageing Process. Genes (Basel) 2014;5:656–670. doi: 10.3390/genes5030656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Orjalo AV, Rodier F, Lithgow GJ, Campisi J. MicroRNAs miR-146a/b negatively modulate the senescence-associated inflammatory mediators IL-6 and IL-8. Aging (Albany NY) 2009;1:402–411. doi: 10.18632/aging.100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bonifacio LN, Jarstfer MB. MiRNA profile associated with replicative senescence, extended cell culture, and ectopic telomerase expression in human foreskin fibroblasts. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0012519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gysler SM, Mulla MJ, Guerra M, Brosens JJ, Salmon JE, Chamley LW, Abrahams VM. Antiphospholipid antibody-induced miR-146a3p drives trophoblast interleukin-8 secretion through activation of Toll-like receptor 8. Mol Hum Reprod. 2016;22:465–474. doi: 10.1093/molehr/gaw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kabir TD, Leigh RJ, Tasena H, Mellone M, Coletta RD, Parkinson EK, Prime SS, Thomas GJ, Paterson IC, Zhou D, McCall J, Speight PM, Lambert DW. A miR-335/COX-2/PTEN axis regulates the secretory phenotype of senescent cancer-associated fibroblasts. Aging (Albany NY) 2016 doi: 10.18632/aging.100987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lang A, Grether-Beck S, Singh M, Kuck F, Jakob S, Kefalas A, Altinoluk-Hambüchen S, Graffmann N, Schneider M, Lindecke A, Brenden H, Felsner I, Ezzahoini H, Marini A, Weinhold S, Vierkötter A, Tigges J, Schmidt S, Stühler K, Köhrer K, Uhrberg M, Haendeler J, Krutmann J, Piekorz RP, et al. MicroRNA-15b regulates mitochondrial ROS production and the senescence-associated secretory phenotype through sirtuin 4/SIRT4. Aging. 2016;8:484–505. doi: 10.18632/aging.100905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rossato M, Curtale G, Tamassia N, Castellucci M, Mori L, Gasperini S, Mariotti B, De Luca M, Mirolo M, Cassatella MA, Locati M, Bazzoni F. IL-10-induced microRNA-187 negatively regulates TNF-alpha, IL-6, and IL-12p40 production in TLR4-stimulated monocytes. Proc Natl Acad Sci U S A. 2012;109:E3101–3110. doi: 10.1073/pnas.1209100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J1,2, Jia J1, Zhao L3, Li X1, Xie Q4, Chen X1, Wang J3, Lu F1. Down-regulation of microRNA-9 leads to activation of IL-6/Jak/STAT3 pathway through directly targeting IL-6 in HeLa cell. Mol Carcinog. 2016;55:732–742. doi: 10.1002/mc.22317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O’Leary JJ, Ruan Q, Johnson DS, Chen Y, O’Neill LA. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–7. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 61.Liu X, Yu J, Jiang L, Wang A, Shi F, Ye H, Zhou X. MicroRNA-222 regulates cell invasion by targeting matrix metalloproteinase 1 (MMP1) and manganese superoxide dismutase 2 (SOD2) in tongue squamous cell carcinoma cell lines. Cancer Genomics Proteomics. 2009;6:131–139. [PMC free article] [PubMed] [Google Scholar]
- 62.Badi I, Burba I, Ruggeri C, Zeni F, Bertolotti M, Scopece A, Pompilio G, Raucci A. MicroRNA-34a Induces Vascular Smooth Muscle Cells Senescence by SIRT1 Downregulation and Promotes the Expression of Age-Associated Pro-inflammatory Secretory Factors. J Gerontol A Biol Sci Med Sci. 2015;70:1304–1311. doi: 10.1093/gerona/glu180. [DOI] [PubMed] [Google Scholar]
- 63.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 64.Xu N, Zhang L, Meisgen F, Harada M, Heilborn J, Homey B, Grandér D, Ståhle M, Sonkoly E, Pivarcsi A. MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion. J Biol Chem. 2012;287:29899–29908. doi: 10.1074/jbc.M112.391243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng X, Chopp M, Lu Y, Buller B, Jiang F. MiR-15b and miR-152 reduce glioma cell invasion and angiogenesis via NRP-2 and MMP-3. Cancer Lett. 2013;329:146–154. doi: 10.1016/j.canlet.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu G, Friggeri A, Yang Y, Park YJ, Tsuruta Y, Abraham E. miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc Natl Acad Sci U S A. 2009;106:15819–24. doi: 10.1073/pnas.0901216106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen R, Alvero AB, Silasi DA, Kelly MG, Fest S, Visintin I, Leiser A, Schwartz PE, Rutherford T, Mor G. Regulation of IKKbeta by miR-199a affects NF-kappaB activity in ovarian cancer cells. Oncogene. 2008;27:4712–23. doi: 10.1038/onc.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Röder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigó R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 71.Abdelmohsen K, Panda AC, Kang MJ, Guo R, Kim J, Grammatikakis I, Yoon JH, Dudekula DB, Noh JH, Yang X, Martindale JL, Gorospe M. 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Res. 2014;42:10099–10111. doi: 10.1093/nar/gku686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bergmann JH, Spector DL. Long non-coding RNAs: modulators of nuclear structure and function. Curr Opin Cell Biol. 2014;26:10–18. doi: 10.1016/j.ceb.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–8. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, White EJ, Orjalo AV, Rinn JL, Kreft SG, Wilson GM, Gorospe M. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Luo Q, Chen Y. Long noncoding RNAs and Alzheimer’s disease. Clin Interv Aging. 2016;11:867–872. doi: 10.2147/CIA.S107037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grammatikakis I, Panda AC, Abdelmohsen K, Gorospe M. Long noncoding RNAs(lncRNAs) and the molecular hallmarks of aging. Aging. 2014;6:992–1009. doi: 10.18632/aging.100710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abdelmohsen K, Panda A, Kang MJ, Xu J, Selimyan R, Yoon JH, Martindale JL, De S, Wood WH, 3rd, Becker KG, Gorospe M. Senescence-associated lncRNAs: senescence-associated long noncoding RNAs. Aging Cell. 2013;12:890–900. doi: 10.1111/acel.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 82.Tominaga-Yamanaka K, Abdelmohsen K, Martindale JL, Yang X, Taub DD, Gorospe M. NF90 coordinately represses the senescence-associated secretory phenotype. Aging. 2012;4:695–708. doi: 10.18632/aging.100497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, O’Neill LA, Moore MJ, Caffrey DR, Fitzgerald KA. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Naganuma T, Hirose T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013;10:456–461. doi: 10.4161/rna.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, Kai C, Yada T, Suzuki Y, Yamada T, Ozawa T, Kaneki K, Inoue T, Kobayashi M, Kodama T, Wada Y, Sekimizu K, Akimitsu N. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 86.Cui H, Xie N, Tan Z, Banerjee S, Thannickal VJ, Abraham E, Liu G. The human long noncoding RNA lnc-IL7R regulates the inflammatory response. Eur J Immunol. 2014;44:2085–2095. doi: 10.1002/eji.201344126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Freund A, Orjalo AV, Desprez PY, Campisi J. Inflammatory networks during cellular senescence: causes and consequences. Trends Mol Med. 2010;16:238–246. doi: 10.1016/j.molmed.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Adler AS, Sinha S, Kawahara TL, Zhang JY, Segal E, Chang HY. Motif module map reveals enforcement of aging by continual NF-kappaB activity. Genes Dev. 2007;21:3244–3257. doi: 10.1101/gad.1588507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, Head SR, Burns JC, Rana TM. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A. 2014;111:1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cocquerelle C, Daubersies P, Majérus MA, Kerckaert JP, Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992;11:1095–1098. doi: 10.1002/j.1460-2075.1992.tb05148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 93.Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 94.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–57. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 96.Panda AC, Grammatikakis I, Munk R, Gorospe M, Abdelmohsen K. Emerging roles and context of circular RNAs. Wiley Interdiscip Rev RNA. 2016 doi: 10.1002/wrna.1386. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, Li X, Yang BB. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2016 doi: 10.1093/eurheartj/ehw001. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 98.Panda AC, Grammatikakis I, Kim KM, De S, Martindale JL, Munk R, Yang X, Abdelmohsen K, Gorospe M. Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic Acids Res. doi: 10.1093/nar/gkw1201. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ramachandran S1, Palanisamy V. Horizontal transfer of RNAs: exosomes as mediators of intercellular communication. Wiley Interdiscip Rev RNA. 2012;3:286–293. doi: 10.1002/wrna.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.John B, Sander C, Marks DS. Prediction of human microRNA targets. Methods Mol Biol. 2006;342:101–13. doi: 10.1385/1-59745-123-1:101. [DOI] [PubMed] [Google Scholar]
- 102.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13:622–38. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 103.Taylor MA, Schiemann WP. Therapeutic Opportunities for Targeting microRNAs in Cancer. Mol Cell Ther. 2014;2:1–13. doi: 10.1186/2052-8426-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Karagonlar ZF, Korhan P, Atabey N. Targeting c-Met in Cancer by MicroRNAs: Potential Therapeutic Applications in Hepatocellular Carcinoma. Drug Dev Res. 2015;76:357–367. doi: 10.1002/ddr.21274. [DOI] [PubMed] [Google Scholar]
- 105.Young DD, Connelly CM, Grohmann C, Deiters A. Small molecule modifiers of microRNA miR-122 function for the treatment of hepatitis C virus infection and hepatocellular carcinoma. J Am Chem Soc. 2010;132:7976–7981. doi: 10.1021/ja910275u. [DOI] [PubMed] [Google Scholar]
- 106.Chen X, Yan CC, Zhang X, You ZH. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinform. 2016 doi: 10.1093/bib/bbw060. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fatima R, Akhade VS, Pal D, Rao SM. Long noncoding RNAs in development and cancer: potential biomarkers and therapeutic targets. Mol Cell Ther. 2015;3:5. doi: 10.1186/s40591-015-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Su Y, Sun B, Lin X, Zhao X, Ji W, He M, Qian H, Song X, Yang J, Wang J, Chen J. Therapeutic strategy with artificially-designed i-lncRNA targeting multiple oncogenic microRNAs exhibits effective antitumor activity in diffuse large B-cell lymphoma. Oncotarget. 2016 doi: 10.18632/oncotarget.9237. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]


