Abstract
Sequencing in all areas of the tree of life has produced more than 300,000 cytochrome P450 (CYP) sequences that have been mined and collected. Nomenclature has been assigned to more than 41,000 CYP sequences and the majority of the remainder has been sorted by BLAST searches into clans, families and subfamilies in preparation for naming. The P450 sequence space is being systematically explored and filled in. Well-studied groups like vertebrates are covered in greater depth while new insights are being added into uncharted territories like horseshoe crab (Limulus polyphemus), tardigrades (Hypsibius dujardini), velvet worm (Euperipatoides_rowelli), and basal land plants like hornworts, liverworts and mosses. CYPs from the fungi, one of the most diverse groups, are being explored and organized as nearly 800 fungal species are now sequenced. The CYP clan structure in fungi is emerging with 805 CYP families sorting into 32 CYP clans. More than 3000 bacterial sequences are named, mostly from terrestrial or freshwater sources. Of 18,379 bacterial sequences downloaded from the CYPED database, all are greater than 43% identical to named CYPs. Therefore, they fit in the 602 named P450 prokaryotic families. Diversity in this group is becoming saturated, however 25% of 3,305 seawater bacterial P450s did not match known P450 families, indicating marine bacterial CYPs are not as well sampled as land/freshwater based bacterial CYPs. Future sequencing plans of the Genome 10K project, i5k and GIGA (Global Invertebrate Genomics Alliance) are expected to produce more than one million cytochrome P450 sequences by 2020.
Keywords: Cytochrome P450, biodiversity, evolution, birds, insects, Limulus polyphemus, tardigrades, velvet worm, plants, fungi, bacteria
1. Introduction
The first full-length cytochrome P450 sequences rat CYP2B1 and CYP2B2 were published in 1982 by Fujii-Kuriyama, et al. [1] and also by Waxman and Walsh [2]. Earlier partial sequences were determined by protein chemical methods (Botelho, et al. 1979 [3] and Ozols, et al. 1981 [4]. Thirty-five years later we have more than 300,000 P450 sequences and this number is expected to reach one million in four years. Such riches were not anticipated in the 1980s. Early genome sequencing focused on model organisms. The first microorganisms were somewhat disappointing since E. coli had no P450s and baker’s yeast had only three (CYP51, CYP56 and CYP61). This rarefied status did not last long. Today we have >62,000 bacterial CYPs and >85,000 fungal CYPs identified in public and confidential databases. Currently, ~11,000 of these are named. Animals have more than 13,000 named P450s and plants have over 16,000. As the practicing member of the Standardized Cytochrome P450 Nomenclature Committee, it has been my task and privelege to assign names to this family of genes. Much of the resulting information is posted to the Cytochrome P450 Homepage [5], but a large part is treated as confidential and is not posted until published.
The breadth and depth of sequencing projects determines the coverage of P450 sequence space. A major contribution has come from the 1KP one-thousand plant transcriptome project [6–9]. More than half of the known CYP sequences (171,000) were produced from this project that has sequenced ~1200 plant transcriptomes from algae to angiosperms. Another “thousand” project is the One Thousand Fungal Genomes Project from JGI [10]. The project had 759 fungal genomes in the Mycocosm database on Feb. 21, 2017 [11]. Searching for the P450 Interpro code IPR001128 as a keyword for any genome in the set will find all the P450 sequences. Based on current findings ~100,000 P450s are expected in these fungal genomes once the 1000 genomes are complete (estimated by the end of 2017). Three additional sequencing projects are the i5k (5,000 arthropod genomes project), the Genome 10K (10,000 vertebrate genomes project) and GIGA (Global Invertebrate Genomics Alliance for 7,000 non-arthropod invertebrate genomes). The value of genome sequencing as a unifying biological principle is discussed by Richards [12].
For gene family studies these projects provide a window on the evolution of the family. In the case of P450s the evidence suggests one or two present-day P450s already existed in the earliest Eukaryote, probably a CYP51 and soon after a CYP61/CYP710. Deep branching eukaryotes like Trypanosoma, Leishmania and Euglena have from 2–4 CYPs, which include CYP51 and CYP710. These P450s are part of the sterol biosynthesis pathway that defines eukaryotic membranes. CYP61/CYP710 a sterol C22-destaurase is found later in the same pathway and has been lost in animals. From these primordial sequences and some that are not known today, all other eukaryotic P450s evolved (allowing for some lateral transfers like CYP55 from a bacterial CYP105 family member). Our task is to take all the available sequence data and reconstruct as much as feasible the evolutionary history of the family. There are approximately 60 eukaryotic lineages of which plants, animals and fungi are the best known (Margulis and Schwartz 1998 [13]). Each lineage may begin with the primordial P450(s) and run the tape anew along each line. So we may have 60 different billion-year experiments to examine. This paper presents new progress in a few limited areas of plants, animals, fungi and bacteria to see how the quest is coming along.
2. Materials and methods
2.1 Bird sequences
Sequences for five bird genomes (chicken, zebrafinch, turkey, flycatcher and duck) P450s were obtained from Ensembl using Biomart searching for the genes matching PFAM PF00067. Kiwi, chimney swift and medium ground finch P450s were obtained from GenBank by searching the protein section for “species[orgn] AND P450 NOT reductase”. Additional searches were done by BLAST searching for specific sequences not found by the more general method. Analysis of human P450s have identified a set of 66 loci where P450 genes and/or pseudogenes are found. These loci are highly conserved among birds. Each of these loci was checked on the UCSC genome browser for P450 content by obtaining the DNA sequence covering that region and blastx searching it against a large set of named vertebrate P450 protein sequences (currently 4,222 sequences). In this manner all P450 sequences including pseudogenes could be found for each species.
2.2 Naming of bird sequences
Once sequences were identified they were batch BLAST searched against the named P450s from vertebrates to identify the family and subfamily each sequence belonged to. The alignments were examined to make corrections to the sequences if they were missing exons or had insertions like retained introns. This was especially needed for the pseudogenes to account for frame shifts and stop codons in the sequences. The names were based on sequence relatedness to named vertebrate P450s.
2.3 Panarthropod sequences
I maintain a large collection of insect and other invertebrate P450s as part of my curatorial duties for naming P450s from all species. Recent progress has permitted the sequencing of deep evolutionary branches on the panarthropod tree including horseshoe crab (Limulus polyphemus), velvet worm (Euperipatoides rowelli) and a tardigrade (Hypsibius dujardini). Methods similar to those described for birds were used to obtain the P450s from Limulus. GenBank was searched for Limulus[orgn] AND P450 NOT reductase. Sequences recovered were batch BLAST searched against named insect sequences (currently 9,125 sequences) or invertebrate sequences (currently 2,206 sequences). Sequences were assigned to known families if they were >40% identical over full length or near full length. Other sequences were assigned to new families based on their clustering on trees and their percent identity to each other. Sequences alignments were examined to look for regions that were missing or were not matching well. These regions were improved by BLASTX searching the DNA against similar sequences. The list of P450s was compared against known lists from other arthropods. Any sequences that were missing but expected such as the Halloween genes and highly conserved genes like CYP4Gs and CYP15, CYP18, CYP20 etc. were searched for specifically by BLAST searching.
The tardigrade (Hypsibius dujardini) has been sequenced [14] (ensembl.tardigrades.org) and it has a searchable web site at http://badger.bio.ed.ac.uk/H_dujardini/search/all_searched#. This site was searched for P450 returning 720 hits (both nucleotide and protein) 358 protein sequences were recovered. These 358 were in duplicate giving 179 unique protein sequences. Each sequence was present in two entries such as snap_masked-nHd.2.3.scaf02184-processed-gene-0.7-mRNA-1 and nHd.2.3.1.t17128-RA presumably representing the gene and the transcribed version of the gene. These 179 were batch BLAST searched against named P450s. Bacterial contamination (two sequences) and fungal contamination (one sequence) was removed. One bacterial sequence was 43% identical to Ardenticatena strain Cfx-K (Bacteria; Chloroflexi), but only 27% identical to the best animal P450 match. The second bacterial sequence was 47% identical to CYP1214A8 Acidobacterium capsulatum and only 30% to an animal P450. A duplicated copy of NADPH cytochrome P450 reductase was removed and a methyltransferase-like protein was removed. Fused P450 sequences were split. Multiple sequences belonging to the same gene were joined leaving 124 sequences, 54 are less than 450 amino acids long. Sequences were revised based on BLAST alignments.
Velvet worm (Euperipatoides_rowelli) has a genome project but it is not very contiguous yet so P450s were analyzed using transcriptome sequences supplied by Lars Hering and Georg Mayer. The process for these sequences was different, The DNA sequences were formatted for BLAST searching on NCBI stand-alone BLAST software. A P450 search set was prepared that has good representation of P450 families in insects and other arthropods. This search set was used to batch BLAST search the transcriptome sequences for hits. Once hits were found these DNA sequences were translated and BLAST searched against the named set of P450s from insects and other invertebrates to identify family membership. Novel sequences <40% identical to named P450s were assigned to new families.
2.4 Spider sequences
Two spider genomes were analyzed for P450s: the wolf spider (Pardoas pseudoannulata, confidential, not included here) and the common house spider (a part of the i5k project). P450s available in the protein section of GenBank were obtained by the search Parasteatoda tepidariorum[orgn] AND P450 NOT reductase (334 sequences). Only four sequences seemed to be false positives. There were sequences from two Bioprojects, a transcriptome project PRJDB4182 and a genomic project PRJNA316108. Because of this there are some duplicate sequences of 97–100% identity between the two projects. There are ~137 unique P450s. Sequences were compared to the wolf spider sequences and named based on sequence similarity.
2.5 Alignments and trees
Protein sequences not including pseudogenes or short sequences were aligned using CLUSTAL Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). The NJ phylogenetic tree produced at the same site was used. The tree was midpoint rooted, drawn in Figtree v1.3.1 and labeled in Adobe Illustrator CC. After making hundreds of trees of cytochrome P450s for many years, the trees produced in this manner are very consistent. The same CYP clans are always found and sequences sort into CYP families that are reproducible when previously named sequences from other species are included as markers.
2.6 Plant sequences from the 1KP project
P450s were recovered from the 1KP data in two rounds. The first round used the 1KP Orthogroups Extractor, A web api available at http://iptol-api.iplantcollaborative.org/onekp/v1. MCL clustering had been performed previously on 22 annotated plant genomes to cluster the sequences into 53,136 orthogroups and 6,054 super orthogroups [15]. The 1KP transcriptome data were mapped onto these orthogroups as described [16]. The orthogroup extractor allowed queries of these orthogroups with seed sequences used in their construction. By querying the data with the seed sequence gene IDs orthogroups could be retrieved. Approximately 100 tomato P450 Soly (Solanum lycopersicum) IDs from all CYP plant families were used as seed sequences to recover P450s from 86 orthogroups. These 86 orthogroups contained 153,374 P450 sequences. The second round of P450 recovery used a different method. The MCL clustering data had been created by setting seven levels of stringency in the clustering algorithm[6]. The end result at the lowest stringency was super orthogroups. By searching the super orthogroup locations of the first 86 orthogroups two super orthogroups appeared to contain all the P450 sequences. The majority of P450s were in super orthogroup 9 and CYP74s that are highly divergent sequences were partly in super orthogroup 9 and also in super orthogroup 122. When these were downloaded a total of 490 orthogroups were obtained that had P450 sequences. There were an additional 36,061 sequences in this new data set. After combining the two sets and filtering to remove duplicates in more than one orthogroup and screening to detect and remove fungal, insect and other contamination 171,940 plant P450s remained. These sequences were sorted by their phylogeny into nine taxa for further analysis. The nine taxa are algae, liverworts, hornworts, mosses, lycophytes, ferns, gymnosperms, non-eudicot angiosperms and eudicots.
Analysis of 175 short eudicot P450s that were less than 40% identical to named P450s was performed. By BLAST searching the 1KP set, best matches were found to full length sequences that had already been assigned to a family. In some cases there was almost 100% identity. The majority of sequences could be assigned to families this way. The remaining sequences were mostly mid-region pieces. These were BLAST searched against the named P450 set and if all 40 top hits were to the same family it was assumed that the fragment belonged to that family. Three sequences were found to be contamination and they were removed.
To complete CYP family discovery and naming in this gymnosperms, 15,594 gymnosperm sequences were batch BLAST searched against the named plant P450s now including the families added from liverworts and hornworts. Only 714 were <40% identical to named P450s. The blastclust algorithm (part of the NCBI stand alone blast software) was used to cluster these sequence into seven groups. The largest group was assigned to CYP947. One hundred sequences from this family were named and added to the searchable plant BLAST file. The set was re-searched against the modified set to find any other members of CYP947. The procedure was repeated for each of the seven groups. Any singletons were searched at the end after adding all other named gymnosperm sequences to the searchable file.
1KP data is BLAST searchable at https://db.cngb.org/blast4onekp/blast with a login required.
2.7 Bacterial sequences
58,854 Bacterial RefSeq sequences were downloaded and batch BLAST searched against the 2,979 named bacterial and 64 archaeal sequences, using NCBI’s stand alone BLAST software.
3. Results and discussion
3.1 Birds
The complete sequencing of 48 bird genomes from 35/40 orders was accomplished in 2015 [17]. This was followed in the same year by deep sequencing of selected genomic regions in 198 birds covering all 40 bird orders [18]. Now, there is a bird 10K project B10K to sequence all 10,500 bird species [19]. Eight bird genomes from five orders have had their P450s analyzed. The deepest branching birds are the Palaeognathae, (includes kiwi, ostriches and relatives). The brown kiwi Apteryx australis has had its P450s named. The next branch on bird phylogeny is the Galloanserae or landfowl (Galliformes) and waterfowl (Anseriformes). Two Galliformes species chicken (Gallus gallus) and turkey (Meleagris gallopavo) as well as one Anseriformes species the mallard duck (Anas platyrhynchos) have been included. Most modern birds (33 orders) are in a large clade called Neoaves. Chaetura pellagica (chimney swift) is the only Apodiformes (swifts, hummingbirds, nightjars and relatives) species included here. Three Passeriformes or perching bird species: collared flycatcher (Fidcedula albicollis), zebrafinch (Taeniopygia guttata) and one of Darwin’s finches the medium ground finch (Geospiza fortis) have had their P450s named. Bird phylogeny is quite complex and the reader is referred to [18] for more details and a beautiful phylogenetic tree.
The birds are remarkably similar as seen in Suppl. Table S1 (Bird P450s compared). There is a core set of 46 CYPs shared by nearly all of the birds. This is a smaller CYPome than seen in mammals and there are almost no gene blooms. The one exception is in chicken that has five CYP2Js and one pseudogene. Turkey has three CYP2Js and nine pseudogenes. All other birds have CYP2J19. The Passeriformes birds the finches and the flycatcher also have a CYP2J40 not seen in other bird orders. The CYP2J19 gene was identified as the gene responsible for red colored plumage in red factor canaries and red beaks and legs in zebrafinches via production of ketocarotenoids [20, 21. The ortholog was also identified in turtles that produce red retinal oil droplets to aid in color vision [22].
An unexpected finding is the absence of three CYPs found in most other vertebrates. CYP8A, CYP11B and CYP27B are missing in 7/8 bird genomes except CYP27B is found in the chimney swift. CYP1C1 is also missing in Neoaves after Anseriformes, but only four species have been examined so far. Kiwi has pseudogenes of CYP8A and CYP27B and a functional CYP11B gene. These absences point to gene losses after other birds diverged from Palaeognathae. There might be concern that these genes are missing by chance. The contig N50 for each genome is published at NCBI for each genome project. Bird contig N50s range from 17,252 for kiwi to 410,964 for collared flycatcher. The fact that pseudogenes or full length sequences of these three genes were found in the kiwi with the lowest contig N50 reduces the likelihood of absence by chance. The mouse lemur Microcebus marinus assembly 1 had a contig N50 of 3,511. Assembly of P450 genes was fragmentary but 1:1 orthologs of P450s could be found even if they were not complete. These included CYP8A1, CYP11B1, CYP11B2 and CYP27B1. CYP8A1 is prostacyclin synthase also called PTGIS. Prostacyclin regulates blood hemostasis and acts in opposition to CYP5A1-derived thromboxane A2. It was noted in the 1980s that prostacyclin was not found in chicken aorta, a tissue normally a rich source of this molecule[23]. It has been noted that PGE1 and PGE2 have the ability to function like prostacyclin in birds[24]. Notably also in the glucocorticoid synthesis pathways, the CYP11B gene is found only in kiwi, but not in other birds. CYP11B1 is key in the biosynthesis of the glucocorticoids cortisol and corticosterone, while CYP11B2 is aldosterone synthase. Birds make glucocorticoids, so the absence of the CYP11B genes is puzzling. There must be an alternative enzyme to replace CYP11B [25]. CYP27B is a more complicated case since that gene is seen in chimney swift and Pseudopodoces humilis (Tibetan ground tit). CYP27B1 or 25-hydroxy vitamin D3 1-alpha hydroxylase, is missing from most of the birds. The 1, 25 dihydroxy vitamin D3 isoform is important for calcium homeostasis. The absences of these genes suggest bird physiology has some significant differences from mammalian physiology.
3.2 Panarthropods
Much progress had been made in insect P450s since the completion of the Drosophila genome in 2004 [26]. Insects are part of the arthropods (insects, crustaceans, myriapods (centipedes and millipedes) and chelicerates (spiders, ticks, mites and the horseshoe crab)). The phylogeny extends farther as panarthropods by adding in the sister group of velvet worms and finally tardigrades or waterbears (Figure 1). Eighty seven panarthropod genomes have had their P450s named (Table 1). At the base of this group the velvet worms and tardigrades are considered to be ‘living fossils’ [27, 28]. Comparison of these deepest branching members to each other and insects can give a window into what the node organism for this clade was like. One emerging feature is the loss of P450 clans along the lineage leading to insects. Note that tardigrades and velvet worms have seven P450 clans, including CYP51, a CYP20 and a CYP26 clan member. CYP51 is lost on the branch leading to the arthropods. CYP20 and CYP26 vanish between myriapods and crustaceans. Though the substrate for CYP20 is not known, CYP26 acts to inactivate retinoids by hydroxylation. Insects seem to have altered their use of retinoids by loss the CYP26 clan.
Figure 1.
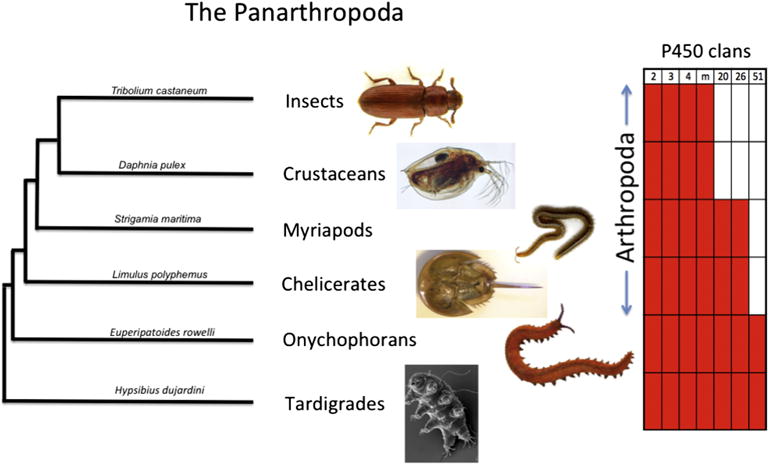
A schematic tree showing the relationships of the major taxa in the panarthropoda. The chart on the right shows the presence of clans in these groups.
Table 1.
Panarthropod genomes sequenced with CYPomes analyzed.
| Insects | CYPomes complete | Sequences | |
|---|---|---|---|
| Lepidoptera (moths, butterflies) | 20 | 1692 | |
| Hemiptera (bugs) | 15 | 1231 | |
| Coleoptera (beetles) | 14 | 1278 | |
| Diptera (flies, mosquitos) | 11 | 1390 | |
| Hymenoptera (bees, wasps, ants) | 8 | 678 | |
| Phthiraptera (body louse) | 1 | 36 | |
| total | 69 | 6305 | |
| Crustaceans | (copepods 5, amphipods 1, Daphnia 1) | 7 | 397 |
| Myriapods | (centipedes 1, millipedes 1) | 2 | 114 |
| Chelicerates | (spiders 2, mites 4, ticks 1, horseshoe crab 1) | 8 | 711 |
| Onychophorans | (velvet worm) | 1 | 94 |
| Tardigrades | (water bear) | 1 | 124 |
| total | 87 | 7745 |
To look in a little more detail Figure 2 is a tree that includes the CYPs from horseshoe crab Limulus polyphemus and a tardigrade Hypsibius dujardini. The tardigrade has only one member each in the CYP51 clan, the CYP26 clan, the CYP20 clan and the CYP4 clan. It has two sequences in the mito clan and one of these is CYP315, the Halloween gene shadow that is the final step on the path to ecdysone. CYP314, the shade gene that produces 20-hydroxyecdysone, was not found. Yet the tardigrade molts. No CYP family in Limulus has more than seven sequences. Though it is possible that Limulus experienced gene losses to achieve a low CYP count, I favor the opposite view that Limulus is closer to the node organism in having a lower number of P450 genes.
Figure 2.
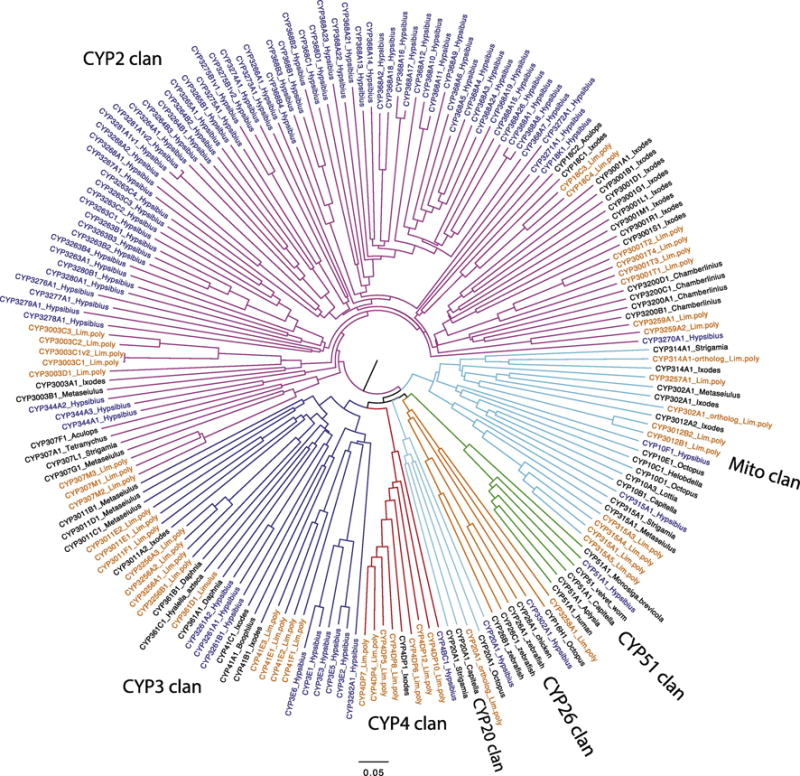
CYPs from the horseshoe crab Limulus polyphemus and a tardigrade Hypsibius dujardini. The spokes are colored to indicate CYP clan and the tip labels are colored blue for tardigrade and orange for the horseshoe crab Limulus. A few other sequences are included to clarify the branching pattern. This is an NJ tree made using CLUSTAL Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). The tree was drawn in Figtree v1.3.1 and labeled in Adobe Illustrator CC. Tardigrade gene model IDs are given in Suppl. Table S7. Sequences are included in Suppl. Sequence file 1.
Most of the CYP2 clan P450s in the tardigrade fall on two branches with 15 and 48 sequences, indicating large gene blooms. Only three other tardigrade branches are found in the CYP2 clan with only one, two or three sequences each. This may mean that the node organism had only a few CYP2 clan P450s that underwent dramatic expansion. The tardigrade CYP3 clan has nine sequences on this tree in three CYP families. The data supports a view that the node panarthropod genome had seven P450s clans and possibly only 1–4 P450s per clan. This organism may have had only a dozen P450s. Limulus on the other hand is more typical. It has the Halloween genes CYP302, CYP307, CYP314, CYP315 and CYP18 for 20-hydroxyecdysone catabolism.
The velvet worm P450s have been assembled from transcriptome data provided by Lars Hering and Georg Mayer [29]. A tree including Limulus, tardigrade and velvet worm sequences along with some additional sequences to define the families is included as Suppl. figure S1. In both figure 2 and Suppl. Figure S1 the CYP2 clan makes up more than half the sequences. Velvet worm has a 41 sequence gene bloom in the CYP2 clan and three smaller expansions in the CYP2, CYP3 and CYP4 clans with 9–11 sequences. Limulus does not have gene blooms.
The common house spider Parasteatoda tepidariorum was mined for P450s as part of the i5k project. These sequences are shown in Figure 3. No CYP20 or CYP26 sequences were found even though these are present in Limulus, a more primitive Chelicerate. The sequence diversity in this spider is somewhat limited since there are some large CYP families. The CYP2 clan has three of these families CYP3001 (29 CYPs), CYP3304 (40 CYPs) and CYP3306 (21 CYPs) in Figure 3. The CYP41 family in the CYP3 clan has 40 sequences in the tree. The CYP4 clan is made up entirely of subfamilies of the very divergent CYP4 family. Among the conserved CYP sequences house spider has CYP18, CYP302, CYP314 and CYP315. It may be significant that no CYP4G sequences were found. CYP4G codes for the genes used by insects to make cuticular hydrocarbons. It is thought that water proofing was an essential factor of emerging onto the land. Spiders are proposed to have made the water to land transition independently of insects and so the waterproofing biochemistry is probably different.
Figure 3.
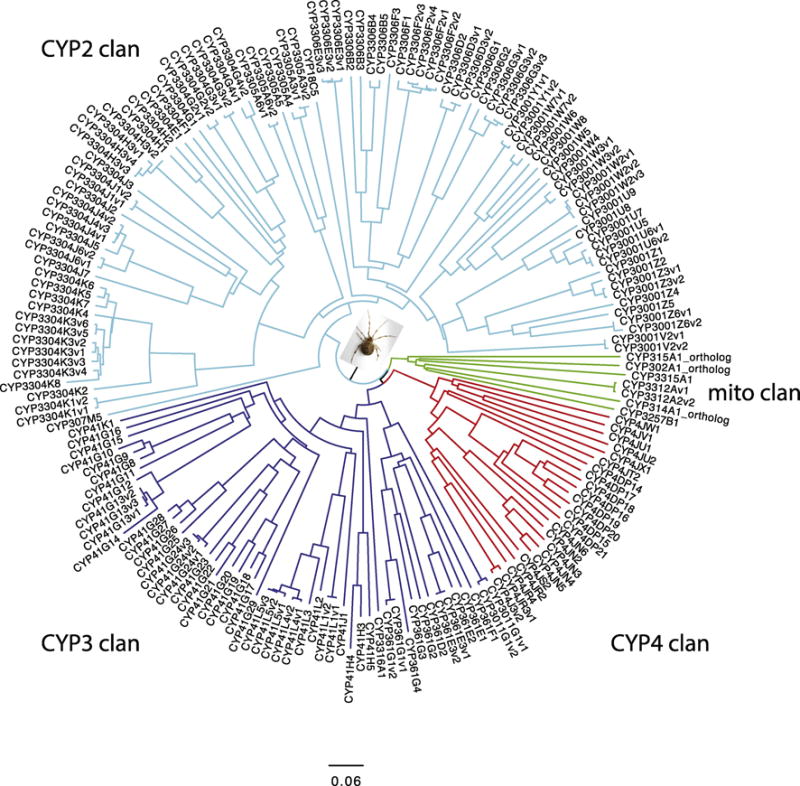
House spider tree. The spokes are colored to indicate the four CYP clans. This tree was made as described in Material and Methods. It is a midpoint rooted NJ tree. Sequences used are in Suppl. Sequence File 4. Accession numbers are in Suppl. Table S8.
Figure 4 shows the number of panarthropod CYP sequences per genome sorted by phylogeny. There is a saw tooth pattern. Each taxon has a range of P450 genes. The low end is generally 50 or less. Diptera exceed this and they also have the highest number of P450s observed in an animal (Stomoxys calcitrans the stable fly has 215 CYPs). The Ixodes scapularis tick is also an outlier with 206 CYPs. The body louse Pediculus humanus currently has the fewest at 36, but lower numbers are anticipated from unpublished genomes. The larger number of CYPs in some species is due to gene blooms but it is not clear what triggers a bloom. The Ixodes tick only eats blood meals, so the diet cannot be responsible for the high number of CYPs. This tick may be exposed to chemicals in its environment as it crawls through leaf litter and up the stalks and leaves of plants to find a host.
Figure 4.
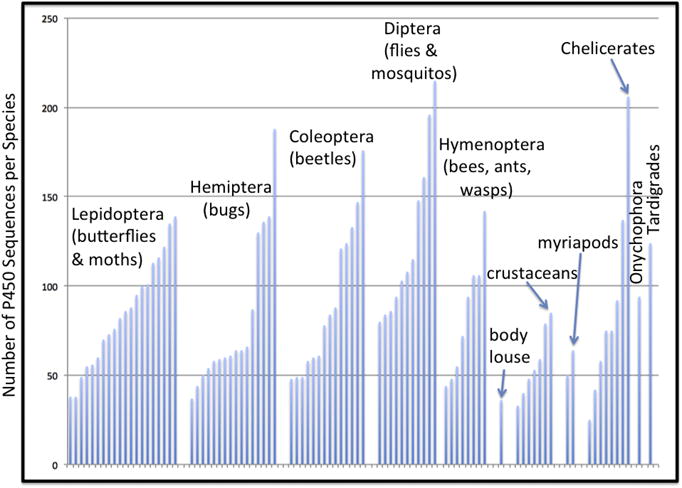
Distribution of numbers of P450s in 87 panarthropods by phylogeny
3.3 Plants: Liverworts, hornworts and gymnosperms
The 1KP thousand plant transcriptome project is headed by Gane Ka-Shu Wong in Alberta, Canada with sequencing done by BGI in Shenzhen, Guangdong, China. The sequence data included 1171 unique species in 916 genera and I added 19 additional species already annotated for P450s to the data (Suppl. Table S2). After following the protocols outlined in Materials and Methods more than 171,000 sequences were obtained (Table 2). Sequence sets from each taxon were batch BLAST searched against all named plant P450s (~7,500 sequences at the time of searching). The data was imported to a spreadsheet and sorted by percent identity (largest to smallest) to find those sequences <40% identical to named P450s. These would be the candidates for new P450 families or contaminants.
Table 2.
1KP P450 sequences in nine different plant taxa.
| Taxa | P450 sequences after removal of contamination and duplicates* | Fungal contaminant sequences | Insect contaminant sequences | |
|---|---|---|---|---|
| 1 | Algae | 4,837 | 17 (0.3%) | 0 |
| 2 | Liverworts | 2,422 | 540 (22%) | 12 (0.5%) |
| 3 | Mosses | 3,963 | 118 (3.0%) | 26 (0.7%) |
| 4 | Hornworts | 185 | 2 (1%) | 0 |
| 5 | Lycophytes | 2,561 | 49 (1.9%) | 4 (0.2%) |
| 6 | Ferns | 8,977 | 21 (0.2%) | 26 (0.3%) |
| 7 | Gymnosperms | 15,954 | 65 (0.4%) | 8 (<0.1%) |
| 8 | non-Eudicot Angiosperms | 21,023 | 20 (0.1%) | 56 (0.3%) |
| 9 | Eudicots | 112,940 | 170 (0.2%) | 222 (0.2%) |
| Total | 171,940 | 1002 (0.6%) | 354 (0.2%) |
duplicates means sequences with the same name found in more than one orthogroup.
The ultimate goal for categorizing all these sequences would be to name them all. That task is quite large. A simpler option would be to find all the novel CYP families in each taxon without necessarily naming all the sequences. The ranking by best hit percent identity as described above provides the starting point for this effort. Those sequences above 40% identity to a named sequence should belong to that CYP family. There are some caveats regarding shorter sequences that require some scrutiny when short sequences are between 40–55%. However, it is possible to take all sequences below 55% identity to a named P450 and Blast search them against each other to find family clusters of >40% sequence identity. This is essentially what the Blastclust algorithm does but the results are not so clean that total reliance on this algorithm will give satisfactory results. Some manual checking is needed.
To illustrate the procedure the eudicot group of 112,018 sequences was examined. Since most named P450s are from the eudicots only 175 sequences in this group were below 40% identity. Nearly all were short pieces from the N-terminal or middle region before the I-helix, a poorly conserved region. These sequences were analyzed as described in Materials and methods. The result was impressive in that there were no new families found in more than 112,000 new eudicot plant sequences. This same procedure could be applied to the eight remaining taxa, thus defining all the plant P450 families.
Liverworts and hornworts were the two smallest taxa, so they were chosen first to find all the new CYP families and then to name all the sequences. After binning the most diverse liverwort sequences by sorting them into family related groups 2,422 liverwort sequences from 22 different species were named. Some sequences had more than one transcript with different IDs. After subtracting the duplicate names 1,827 unique liverwort sequences remain. Figure 5 shows all eleven P450 clans and 39/69 CYP families found in liverworts. This tree features Marchantia polymorpha and Marchantia paleacea. 48 liverwort CYP families were not seen in other taxa at the time liverworts were named. Some of these families are expected to be found in other groups like moss, hornworts or lycophytes, since liverworts are the first of the lower plant taxa to be named. Liverworts are one of the earliest land plants so the distribution of P450s among the clans is of interest. Figure 6 shows a comparison between liverwort and named angiosperm CYPs by clan. They are highly similar except angiosperms have lost the CYP746 clan. The transition from water to land set the P450 composition and it has changed relatively little since.
Figure 5.
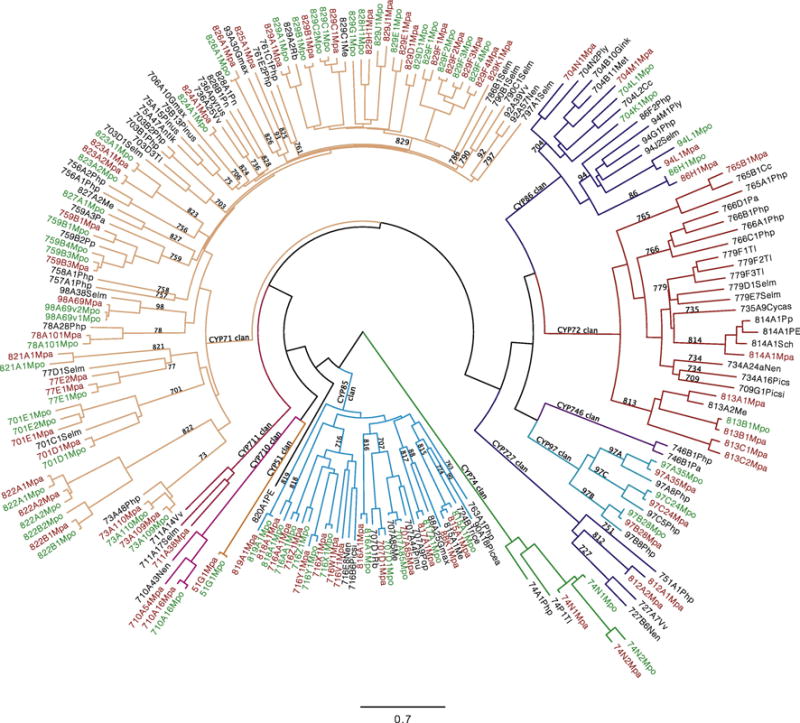
Liverwort tree showing representative sequences and featuring Marchantia polymorpha (green labels) and Marchantia paleacea (red labels). The spokes are colored to show CYP clans. Liverworts = Mpa Marchantia paleacea; Mpo Marchantia polymorpha; Me Marchantia emarginata; Pn Porella navicularis; Pp Ptilidium pulcherrimum; Pa Plagiochila asplenoides; Rb Riccia berychiana; Cc Conochephalus conicum; Ply Pallavicinia lyellii; Tl Treubia lacunose; Sch Schistochila sp.; PE Pellia cf. Epiphylla; Other species = Php Physcomitrella patens (moss); Selm Selaginella moellendorffii (lycophyte); Nen Nelumbo nucifera (sacred lotus); Cycas Cycas rumphii (cycad); Met Medicago trucatula; Pinus or Pinu Pinus taeda; Picea Picea glauca; Picsi or Pics Picea sitchensis; Gmax Glycine max (soybeans); rice Oryza sativa; Pop Populus trichocarpa; Antik Antirrhinum kelloggii; Vv Vitis vinifera; Pyrus Pyrus communis (Pear); Gink Ginkgo biloba. Sequences used are in Suppl. Sequence File 5.
Figure 6.
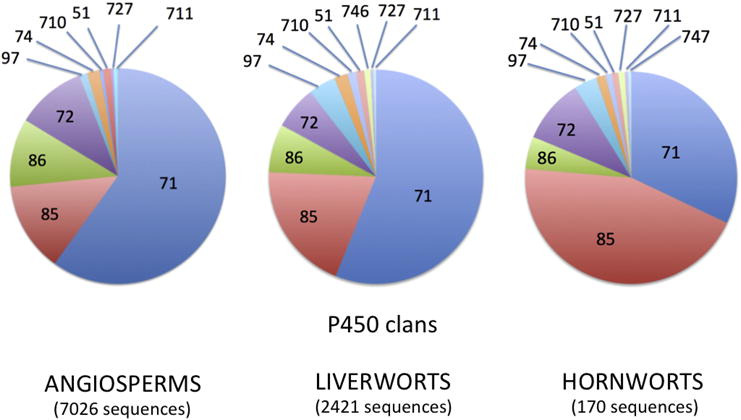
Pie chart showing the size of the P450 clans in Liverworts compared to Angiosperms and Hornworts
Hornworts initially had only 185 sequences, but the 1KP project added six more hornwort species later in the project. An exhaustive BLAST approach was used to find all the P450s from these eight species of hornworts. This yielded 2,165 P450 sequences. An approach like the one used for liverworts was able to bin these into families and subfamilies, though not all of them have been named. Thirty eight new CYP families were found in hornworts, demonstrating that it is significantly different from other plant CYPomes. The CYP85 clan is expanded compared to liverworts and the CYP71 clan is reduced (Figure 6). Hornworts have retained the CYP747 family seen in green algae.
The methods used in liverworts, hornworts and eudicots can be applied to each plant taxa in turn to define all the CYP families in plants. Algae will be particularly challenging due to the high biodiversity. Land plants have mosses, lycophytes, ferns, gymnosperms and non-eudicot angiosperms remaining. Considerable progress has already been made with gymnosperms from sequencing pine and spruce genomes. The details of the approach used for gymnosperms is described in Materials and Methods. Since two large conifer genomes had been named already, only 13 new CYP families were found. To be confident that no gymnosperm families were missed a total of 1,030 lowest percent identity sequences were named.
3.4 Fungi
Cytochrome P450s in fungi are extremely diverse, possibly more so than in any other phylum. Even though Saccharomyces cerevisiae has only three P450s and four Schizosaccharomyces species have only two, other fungi have >1% of their genomes as P450s. The saw tooth pattern observed before in Panarthropoda (Figure 4) is also characteristic of fungi (Figure 7). Many species have over 200 CYPs and at least one Sphaerolobus stellata the cannonball fungus [30] has 601 cytochrome P450 genes, with 564 found in Dendrothele bispora (both in Agaricomycotina). The number of CYP families is also quite large (currently 805), more than seen in bacteria. The saw tooth pattern conveys two points about P450s in fungal genomes. These genes are luxury genes with the exception of CYP51 and CYP61 in sterol biosynthesis. Even Agaricomycotina has some members with very few P450s (as low as 4–5 CYPs). Yet some Agaricomycotina species explode their P450 content. In contrast, Saccharomycotina is definitely frugal with their P450s, usually less than 10 but never more than 26. There appears to be a brake on gene expansion in Saccharomycotina. This has been noted by others [31].
Figure 7.
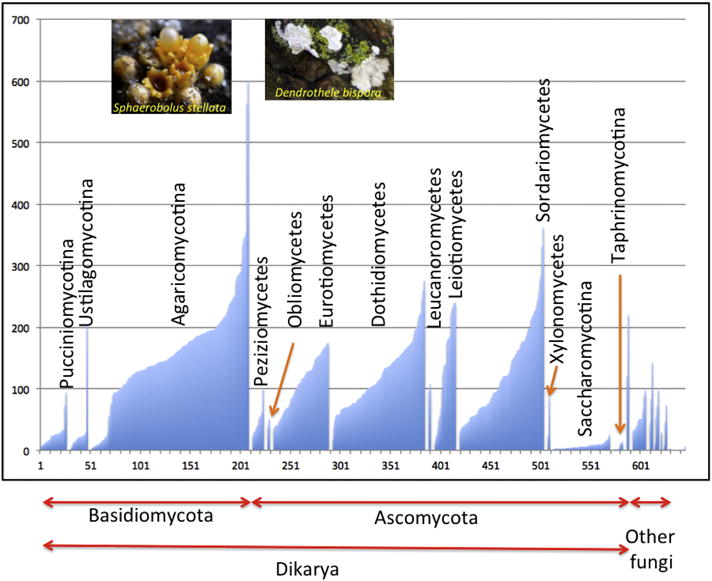
Distribution of the numbers of P450s in 570 fungi by phylogeny
Sphaerolobus stellatus seems so unusual that the sequences were checked for fragmented or incomplete genes. Figure 8 shows the 601 sequence lengths in a histogram. Nine are fusion proteins that belong to the known fused P450 family CYP505 (P450 on the N-terminal) and four sequences belong to the P450 fusion family CYP6005 (P450 on the C-terminal). These are legitimate longer P450s. One long sequence is actually two P450s fused together. About 300 are complete or nearly complete sequences. The graph has a long tail tapering to about 50 amino acids. Examination of the 216 sequences shorter than 250 amino acids shows 78 are N-terminal up to about the C-helix, 63 are mid region up to about the PKG motif and 89 are C-terminal from about PKG to the end. None were false positives. The shorter sequences may be pseudogenes or some of them might be partial genes that could be combined with others to make complete sequences. There may be about 400 full-length sequences after such combination. The S. stellatus genome is not highly fragmented. The contig N50 is 19,443, long enough to cover fungal P450 genes (1,500–3,000bp). so the high gene count is not an artifact.
Figure 8.
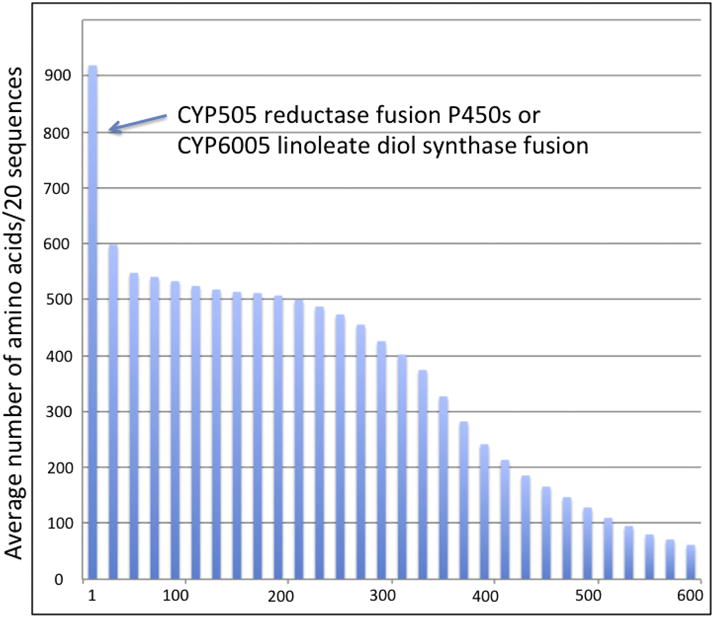
Sphaerobolus stellatus P450 length of sequences in decreasing order. Sequence lengths were averaged in groups of 20.
Diversity in fungal genomes is being explored by JGI in the 1KFG the One Thousand Fungal Genomes Project [10,11]. This project is now more than three quarters complete with 771 genomes sequenced. Seventy five of these genomes and an additional 28 fungal genomes have had their P450s named (103 species). P450s from 796 genomes have been downloaded (Suppl. Table S3) giving a total of 85,103 fungal P450 sequences, of which 7,925 are named. BLAST searching of 52,582 sequences from 571 genomes has been performed against ~7,900 named fungal P450s. The BLAST output from these first 571 genomes showed an unexpected result. Only 14% of these sequences (7,513) were below 40% identical to named P450s. This is still a large number but 86% belonged to existing CYP families. Only 0.5% were false positive hits that were not P450 sequences. To define the total number of P450 families in fungi these 7,513 sequences will have to be analyzed as described earlier for plant taxa. Then the newer genome sequences will also have to be examined. It is probable that the number of fungal P450 families will exceed 1000.
P450 clans are deep branching clades on phylogenetic trees that form natural divisions among the P450 sequences from one kingdom. Members of a clan always cluster together on trees even though the relative position of branching of the clans can vary. Animals have only 11 clans and multicellular land plants also have 11. A few more plant clans (CYP745, CYP747) will be added as green algae are included. Plant and animal clans only overlap in CYP51 and CYP74 clans and the CYP74 overlap is probably due to a lateral transfer, not to a common ancestral sequence in the node organism. In fact all Eukaryotes are expected to share the CYP51 clan, allowing for some losses in a few groups like arthropods, nematodes, tunicates, etc. Fungi make up the third large eukaryotic phylum to be well sampled by genome sequencing. The fungal CYP clan structure is more complex and these branches have not all been defined yet. An early attempt to define fungal clades was conducted by Deng, et al., 2007 [32]. They used 376 P450s from diverse fungal species and made a solid attempt to define families. Their families differ from the P450 nomenclature families, but there is some strong congruence. However, with tens of thousands of sequences another approach needs to be made besides making a single master tree. A more recent attempt was produced by Moktali et al., 2012 [33]. These authors made use of a fungal P450 database and algorithms to define clans. There is significant overlap in their clans and my own set but they are not exactly the same.
Figure 9 shows a base set of 184 P450s to begin to define the fungal P450 clans. This tree has 17 clans and one lone sequence CYP5346A1, normally in the CYP54 clan. These 17 clans can be considered as a base to which new clans can be added. Sequences that are <40% identical can be added to the tree in batches of about 100 sequences at a time to see where they sort. Sequences that fall in new deep branches between existing clans are candidates for new clans. Clans cannot be defined by percent identity cutoff as has been done for families and subfamilies. The percent identity begins to lose defining power as it dips below 30%. For example CYP4B1 from mammals can be 28–30% identical to some plant sequences and some fungal sequences, though these sequences are not in the same clans. How can one define a clan? The clans are a reproducible outcome of clustering algorithms that use scoring matrices to align sequences and then produce a hierarchical tree based on a difference matrix representing the similarities between all of the sequences. The fact that the same clusters are seen in hundreds of trees indicates that the clustering algorithms are extracting a pattern from the sequence data that is reproducible. What is not reproducible is the final connectivity of the deepest branches. This is variable from tree to tree. Where the reproducible clusters end and the variablity begins in the deepest branches is the de facto definition of a clan.
Figure 9.
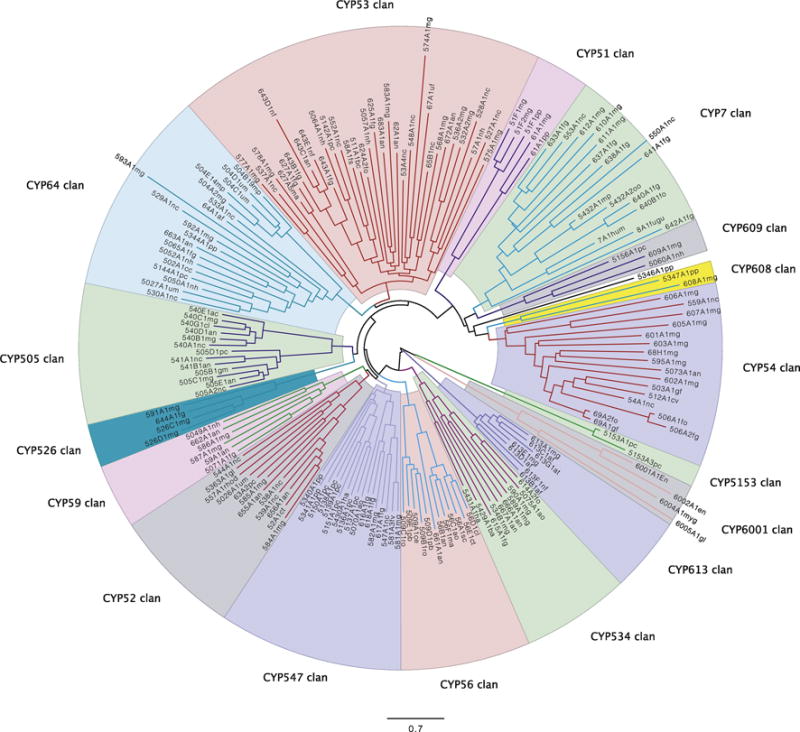
Fungal clan tree. The sequences used in this tree are provided in Suppl. Sequence File 2.
Looking at the Figure 9 tree in more detail, note in clan 7 that there are two animal CYP7 sequences from human and fugu. These sequences fall inside this clan and give it their name. Clans are named for the lowest numbered family member in this case CYP7.
Opisthokonta is made from the sister groups animals and fungi. Therefore, it might be possible that the Opisthokonta common ancestor had a CYP7-like sequence that gave rise to both animal and fungal CYP7 clans. Alternatively, there could have been a lateral transfer to move the clan between species. Lateral transfers are known to happen in fungal P450s since the CYP55 fungal sequences are derived from bacterial CYP105 sequences [34]. The CYP55 clan is not included in this tree but it is on a distinct branch of its own.
A 2013 compilation made by adding sequences to the base tree in Figure 9 included 611 families in 26 fungal clans (Suppl. Table S4). As further documentation, A 296 sequence tree with 20 clans including the CYP5108, CYP5224, CYP5225 and CYP5226 clans is provided as Suppl. Figure S2. Note in this tree the CYP51 and CYP61 clans are split though they were joined in Figure 9. Five clans of the original 26 are not shown in either of these two trees. These are CYP55 (1 family), CYP645 (2 families), CYP5189 (2 families), CYP5241 (1 family) and CYP6003 (1 family). In March 2017, 177 new fungal families were added to the tree in Suppl. Figure S4 in two batches to assign clans. A total of 168 new fungal families were assigned to clans (shown as red font in Suppl. Table S4). Currently, 779/805 fungal CYP families are assigned clan membership. 26 families were represented by pseudogenes or short sequences only and these could not be assigned. Thirty two CYP clans were defined, including six new clans but three of these are singletons and do not have strong evidence to support them (Suppl. Table S4). As the number of fungal CYP families begins to plateau as the sequence space is more fully sampled the absolute number of fungal CYP clans will become known.
3.5 Bacteria
Bacterial P450s were among the first to be sequenced and the first to have crystal structures determined. Some bacteria have no cytochrome P450s and some have 50 or more (Frankia sp. EAN1pec 50 CYPs, Mycobacterium vanbaalenii PYR-1 51 CYPs, Streptomyces RK95-74 52 CYPs), Streptomyces clavuligerus ATCC 27064 58 CYPs and Streptomyces rapamycinicus NRRL_5491 61 CYPs). For comparison humans have only 57 CYPs. The diversity among bacteria is legendary so the diversity of P450s was expected to be extremely high, even higher than that of fungi. A total of 2,979 bacterial and 64 archaeal CYP sequences are named in 602 prokaryotic CYP families (Suppl. Table S5) but sequencing is much faster with bacteria so many more sequences are in the databases. The CYPED (Cytochrome P450 Engineering Database) has 18,379 bacterial CYPs. Some effort has gone into naming of metagenome samples to cover the biodiversity of bacterial CYPs. Because of that all of the CYPED sequences were found to fit in existing P450 families, defying expectations of high diversity. The majority of these sequences were from land or freshwater samples, so to test the diversity of marine bacteria, 3,305 P450 sequences from the Sorcerer II Global Ocean Sampling Expedition were compared to the named set. The result was 25% were <40% identical, indicating that the diversity in the marine bacteria is much higher or less well sampled than the CYPED sequences. Even so, there appears to be a trend toward saturation as most new bacterial sequences seem to fit in existing families.
A search of GenBank’s protein section with the query “bacteria[orgn] AND P450 NOT reductase” returned 223,600 hits. Experience with this database suggests that both RefSeq and original entries are included so that would reduce this number by half. Still that would be over 100,000 sequences. Even the RefSeq number given in the Source Databases link on the left of the page shows 58,854 hits. Of these 14,454 are from Mycobacterium and 13,530 are from Streptomyces and these are already well sampled in the named P450 set. A total of 2,181 of the RefSeq sequences were 100% identical to named P450s and 5,094 sequences were >99% identical to named P450s. Less than 1% or 437 sequences had no BLAST hits and were considered false positives. After removal of those and some additional false positives 58,336 sequences remained. The sequences and their best BLAST hit are presented in Suppl. Table S6 Only six % (2,268 sequences) were below 40% identical to named P450 sequences. In this group 209 are also <100 amino acids long and may be false positives. Ninety four % of the sequences are >40% identical to named P450s. This supports the earlier conclusion drawn from the CYPED set of sequences that the diversity of bacterial P450s has been well sampled excepting the marine bacteria.
3.6 P450 statistics
Table 3 shows the number of cytochrome P450 sequences currently named and the number in my collection but not yet named. The number of CYP families in each taxon is also given. Due to some families being present in more than one taxon, the total number of 2,252 CYP families is less than the sum of the nine taxons. Over 41,000 sequences are named. A total of nearly 350,000 sequences have been mined from various databases described earlier. The Genome 10K project claims that it will sequence 10,000 vertebrates by 2020 [35]. Assuming ~75 P450s per vertebrate genome will add 750,000 more sequences, easily exceeding one million.
Table 3.
Cytochrome P450 statistics including CYP family counts.
| Cytochrome P450 Statistics | |||||
|---|---|---|---|---|---|
| Taxon | |||||
| named P450s | Unnamed but in my possession | total | CYP families | ||
| Animals | |||||
| Insects | 7,426 | 105 | 7,531 | 208 | |
| Non-insect Invertebrates | 1,925 | 0 | 1,925 | 311 | |
| Mammals | 2,419 | 1,334 | 3,753 | 18 | |
| Other Vertebrates | 1,461
|
83 | 1,544 | 19 | |
| Total | 13,231 | ||||
| Plants | 16,219 | 168,303 | 184,522 | 277 | |
| Fungi | 7,925 | 77,178 | 85,103 | 805 | |
| Protozoa | 602 | 0 | 602 | 63 | |
| Bacteria | 2,979 | 59,620 | 62,606 | 591 | |
| Archaea | 64 | 84 | 148 | 14 | |
| Viruses | 28 | 0 | 28 | 6 | |
| Total | 41,048 | 306,707 | 347,762 | 2252* | |
60 Families present in more than one taxon are included only once
4.0 Conclusions
The annotation of cytochrome P450 sequences is a project I have undertaken for 30 years [36]. At first, the task was open-ended and of unknown scale. Today the framework for a comprehensive nomenclature is in place. Clans define the deepest branching clades of P450s followed by CYP families. Clan nomenclature was introduced in 1998 [37] and 1999 [38] to “indicate relationships that lie outside the family designations.” In practical terms this means that some CYP families shared a common ancestor recently enough that they still cluster with each other on phylogenetic trees. These relationships are exclusive in that members of one clan do not cluster with members of a different clan when both are present. The clan concept works well within kingdoms. Ruggiero et al., 2015 recognize 34 animal phyla. Animal CYP clans cross these boundaries [39]. The clans from different kingdoms when mixed still sort into consistent distinct clusters on trees. Mixing 314 sequences from 9 plant clans and 10 animal clans produced 18 distinct clans in a single tree (data not shown). CYP51 from plants merged with CYP51 from animals to make one clan. None of the other clans merged. Animals and fungi (termed Opisthokonta) are closer relatives than plants but they do not appear to share clans either, with exception of CYP51 and a possible lateral transfer of CYP7. The lack of shared clans between animals and fungi, even though they are sister groups speaks to the timing of clan formation. Clans presumably formed after these kingdoms separated. Berbee and Taylor, 2010 date the animal fungal divergence at about 1 billion years, so clans seem to be the product of gene divergence within the sub-billion year time scale. Any branch on the tree of life that is older than animal/fungi should have independently evolved clans of P450s derived from whatever P450s existed in the ancestor of each group.
The number of vertebrate and even animal clans is fixed at 11 and is unlikely to change. The last animal clan was CYP74 added in 2008 [40]. The number of vertebrate families is also stable at 19, with the last family detected CYP16 being added in 2010. The land plants appear to have 11 clans as well but more families. The last land plant clan identified was CYP746 found in moss in 2006 but first observed in Chlamydomonas [41]. The discovery of all these plant groups is moving forward thanks to the 1KP project. Soon all the clans and all the common families should be known in plants. There is always room for new genus specific families, but these should not change the overall landscape of the sequence space. Even fungi and bacteria are yielding to the sequence onslaught.
The dark matter of P450 is currently in the protozoa. Only 602 P450 sequences have been named in protozoa. Partly this is due to a lack of sequencing in these organisms and partly it is due to few P450s being found in some species. More than half of the known sequences (356) were analyzed from 13 oomyctes (Stramenopiles) [42]. Studies on the eukaryotic branching pattern at the deepest nodes are resolving the evolutionary history of eukaryotes [43–46]. Five Supergroups have been defined and refined: Opisthikonta, Amoebozoa, SAR (Stramenopiles, Alveolates, and Rhizaria), Archaeplastida and Excavata. Opisthokonta (animals and fungi) are united with Amoebozoa. SAR has now joined with Archaeplastida (green plants, red algae and gluacophytes). Together with Excavata and a set of lesser known enigmatic orphan taxa, these three megagroups are being examined further to define their ultimate branching order and the root to the tree of eukaryotes. Two groups claim to have a fully resolved tree and their root positions agree [47,48 ] Since these branches are ancient it is expected that each deep branch has evolved P450s independently from a core set of P450s in the earliest node organism. How many P450s were in these ancestors? To quote Feyereisen “The number and identity of common CYP ancestors is unknown, and in fact, unknowable, and only an estimate of the minimal number can be made…” [49]. Our ability to recognize ancient P450s over newer (post-kingdom) P450s depends on their conservation of sequence. CYP51 has preserved its function and sequence over very large time periods so that distant protist branches such as Euglenozoa have clear CYP51 members. Another candidate for this ancient status is CYP710/CYP61. This is the C22-desaturase enzyme of sterol biosynthesis whose function is experimentally known in fungi as CYP61 and in plants as CYP710. Some protists have CYP710 as well (Euglenozoa: Leishmania, Trypanosoma, Euglena; Amoebozoa: Acanthamoeba). Since this gene is downstream of CYP51 in sterol biosynthesis in can be argued that it evolved later and probably from a CYP51 duplication. The fungal clans CYP51 and CYP61 often group together. Barring multiple lateral transfers, these two genes were present in the last ancestors of plants, fungi and some protists. How many others could have been present? There may have been more than two but their sequences have not been preserved to the present day. Their lineages may have been lost or their sequences could have changed to the point that they can no longer be recognized across kingdoms.
In some cases evolution resulted in gene loss leading to no P450s in lines like Giardia intestinalis, Plasmodium falciparum (malaria) and Encephalitozoon cuniculi (all parasitic). In other cases expansions have occurred as in the ciliates Tetrahymena thermophila (48 CYPs) and Paramecium tetraurelia (22 CYPs). Since ciliate organisms do not use sterols in their membranes except when available from the diet they have lost CYP51 and make hopanes like tetrahymenol instead [50]. Their P450s are unique to their own line. This will probably be the case for most eukaryotic deep lineages. Of the original LECA (Last Eukaryotic Common Ancestor) CYPome, only CYP51 and CYP61/CYP710 are recognizable in modern times. Any other CYPs that may have been present in the LECA are now extinct or evolved into new families that are not traceable back to the Eukaryotic root.
A method has been described here to systematically find all the P450 families in large sets of P450 sequences from a single taxon like gymnosperms or liverworts. Application of this method to algae, moss, lycophytes, ferns and non-eudicot angiosperms in the near future should complete the discovery of all P450 families in plants. Another process for finding and defining new CYP clans in fungi with more than 85,000 sequences is being applied. Even this most diverse group of P450s is showing the trend toward saturation, with relatively few sequences belonging to the <40% sequence identical group. This number will fall toward zero as the data from the one thousand fungal genomes project is analyzed. Insects are moving forward with the i5k project and nearly 70 species have been analyzed for their P450s. With more than one million species the insects will be a rich vein to mine in the future. One can expect saturation of the sequence space as the insect biodiversity is sampled. Non-insect invertebrates are only beginning to be explored [51 Goldstone, this issue] and much work remains to be done in this area. The GIGA compilation of genomes should be quite helpful as raw material for future progress. Bacteria are surprisingly well sampled with 94% of GenBank RefSeq P450s falling in named P450 families. More work will need to be done on marine bacteria and the microbiomes from inside animal’s guts. Much diversity is still expected as the sources of bacteria broaden beyond soil and freshwater. If P450 diversity is like a jigsaw puzzle being assembled, the picture has become quite clear in some areas, while it is still terra incognita in a few less frequently visited regions. Of course, this refers to the sequences only. The function of P450s is more difficult to determine and much less is known about function. At least in 2017 we know the scope of the project and can perhaps see the border pieces being inserted for parts of the sequence diversity puzzle.
Supplementary Material
Figure 10.
Figure 11.
Highlights.
~350,000 cytochrome P450 sequences are collected from public and private sources.
P450s from panarthropods horseshoe crab, velvet worm and a tardigrade are anlayzed.
The P450s from a first spider genome are named.
Progress is presented on naming P450 clans in fungi.
Methods are described for finding all P450 families in any taxa (gymnosperms, etc).
Acknowledgments
Velvet worm transcriptome data were provided by Lars Hering lars.hering@uni-kassel.de, and Georg Mayer gmayer@onychophora.com. The 1000 Plants (1KP) initiative led by Gane Ka-Shu Wong was funded by the Alberta Ministry of Advanced Education, Alberta Innovates Technology Futures (AITF), Innovates Centres of Research Excellence (iCORE), Musea Ventures, and China National Genebank (CNGB). The BGI-Shenzhen group led by Yong Zhang was funded by the Shenzhen Supporting Projects Program under grant CXZZ20140421112021913.
Funding
Vertebrate studies were supported by NIHGR U41-HG003345
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The author declares that no conflict of interest exists.
References
- 1.Fujii-Kuriyama Y, Mizukami Y, Kawajiri K, Sogawa K, Muramatsu M. Primary structure of a cytochrome P-450: coding nucleotide sequence of phenobarbital-inducible cytochrome P-450 cDNA from rat liver. Proc Natl Acad Sci U S A. 1982;79(1982):2793–2797. doi: 10.1073/pnas.79.9.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waxman DJ, Walsh C. Phenobarbital-induced rat liver cytochrome P-450. Purification and characterization of two closely related isozymic forms. J Biol Chem. 1982;257:10446–10457. [PubMed] [Google Scholar]
- 3.Botelho LH, Ryan DE, Levin W. Amino acid compositions and partial amino acid sequences of three highly purified forms of liver microsomal cytochrome P-450 from rats treated with polychlorinated biphenyls, phenobarbital, or 3-methylcholanthrene. J Biol Chem. 1979;254:5635–5640. [PubMed] [Google Scholar]
- 4.Ozols J, Heinemann FS, Johnson EF. Amino acid sequence of an analogous peptide from two forms of cytochrome P-450. J Biol Chem. 1981;256:11405–11408. [PubMed] [Google Scholar]
- 5.Nelson DR. The Cytochrome P450 Homepage. Human Genomics. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matasci N, Hung LH, Yan Z, Carpenter EJ, Wickett NJ, Mirarab S, Nguyen N, Warnow T, Ayyampalayam S, Barker M, Burleigh JG, Gitzendanner MA, Wafula E, Der JP, dePamphilis CW, Roure B, Philippe H, Ruhfel BR, Miles NW, Graham SW, Mathews S, Surek B, Melkonian M, Soltis DE, Soltis PS, Rothfels C, Pokorny L, Shaw JA, DeGironimo L, Stevenson DW, Villarreal JC, Chen T, Kutchan TM, Rolf M, Baucom RS, Deyholos MK, Samudrala R, Tian Z, Wu X, Sun X, Zhang Y, Wang J, Leebens-Mack J, Wong GK. Data access for the 1,000 Plants (1KP) project. Gigascience. 2014 Oct 27;3:17. doi: 10.1186/2047-217X-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA, Ruhfel BR, Wafula E, Der JP, Graham SW, Mathews S, Melkonian M, Soltis DE, Soltis PS, Miles NW, Rothfels CJ, Pokorny L, Shaw AJ, DeGironimo L, Stevenson DW, Surek B, Villarreal JC, Roure B, Philippe H, dePamphilis CW, Chen T, Deyholos MK, Baucom RS, Kutchan TM, Augustin MM, Wang J, Zhang Y, Tian Z, Yan Z, Wu X, Sun X, Wong GK, Leebens-Mack J. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci U S A. 2014;111:E4859–4868. doi: 10.1073/pnas.1323926111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie Y, Wu GJ, Tang R, Luo J, Patterson S, Liu W, Huang G, He S, Gu S, Li S, Zhou X, Lam TW, Li Y Y, Xu X, Wong GK, Wang J. SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics. 2014;30:1660–1666. doi: 10.1093/bioinformatics/btu077. [DOI] [PubMed] [Google Scholar]
- 9.Johnson MT, Carpenter EJ, Tian Z, Bruskiewich R, Burris JN, Carrigan CT, Chase MW, Clarke ND, Covshoff S, dePamphilis CW, Wong GK. Evaluating methods for isolating total RNA and predicting the success of sequencing phylogenetically diverse plant transcriptomes. PLoS One. 2012 Nov;7:e50226. doi: 10.1371/journal.pone.0050226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, Smirnova T, Nordberg H, Dubchak I, Shabalov I. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42:D699–704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.http://genome.jgi.doe.gov/programs/fungi/index.jsf (accessed March 21, 2017)
- 12.Richards S. It’s More Than Stamp Collecting: How Genome Sequencing Can Unify Biological Research. Trends Genet. 2015;31:411–421. doi: 10.1016/j.tig.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Margulis L, Schwartz KV. An illustrated Guide to the Phyla of Life on Earth. third. W.H Freeman; New York: 1998. Five Kingdoms. [Google Scholar]
- 14.Koutsovoulos G, Kumar S, Laetsch DR, Stevens L, Daub J, Conlon C, Maroon H, Thomas F, Aboobaker AA, Blaxter M. No evidence for extensive horizontal gene transfer in the genome of the tardigrade Hypsibius dujardini. Proc Natl Acad Sci U S A. 2016;113:5053–5058. doi: 10.1073/pnas.1600338113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amborella Genome Project. The Amborella genome and the evolution of flowering plants. Science. 2013;342:1241089. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- 16.https://pods.iplantcollaborative.org/wiki/display/iptol/Access+to+OneKP+data+set
- 17.Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, Ho SY, Faircloth BC, Nabholz B, Howard JT, Suh A, Weber CC, da Fonseca RR, Li J, Zhang F, Li H, Zhou L, Narula N, Liu L, Ganapathy G, Boussau B, Bayzid MS, Zavidovych V, Subramanian S, Gabaldón T, Capella-Gutiérrez S, Huerta-Cepas J, Rekepalli B, Munch K, Schierup M, Lindow B, Warren WC, Ray D, Green RE, Bruford MW, Zhan X, Dixon A, Li S, Li N, Huang Y, Derryberry EP, Bertelsen MF, Sheldon FH, Brumfield RT, Mello CV, Lovell PV, Wirthlin M, Schneider MP, Prosdocimi F, Samaniego JA, Vargas Velazquez AM, Alfaro-Núñez A, Campos PF, Petersen B, Sicheritz-Ponten T, Pas A, Bailey T, Scofield P, Bunce M, Lambert DM, Zhou Q, Perelman P, Driskell AC, Shapiro B, Xiong Z, Zeng Y, Liu S, Li Z, Liu B, Wu K, Xiao J, Yinqi X, Zheng Q, Zhang Y, Yang H, Wang J, Smeds L, Rheindt FE, Braun M, Fjeldsa J, Orlando L, Barker FK, Jønsson KA, Johnson W, Koepfli KP, O’Brien S, Haussler D, Ryder OA, Rahbek C, Willerslev E, Graves GR, Glenn TC, McCormack J, Burt D, Ellegren H, Alström P, Edwards SV, Stamatakis A, Mindell DP, Cracraft J, Braun EL, Warnow T, Jun W, Gilbert MT, Zhang G. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014;346:1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prum RO, Berv JS, Dornburg A, Field DJ, Townsend JP, Lemmon EM, Lemmon AR. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature. 2015;526:569–573. doi: 10.1038/nature15697. [DOI] [PubMed] [Google Scholar]
- 19.http://www.sciencemag.org/news/2017/02/biologists-propose-sequence-dna-all-life-earth
- 20.Mundy NI, Stapley J, Bennison C, Tucker R, Twyman H, Kim KW, Burke T, Birkhead TR, Andersson S, Slate J. Red Carotenoid Coloration in the Zebra Finch Is Controlled by a Cytochrome P450 Gene Cluster. Curr Biol. 2016;26:1435–1440. doi: 10.1016/j.cub.2016.04.047. [DOI] [PubMed] [Google Scholar]
- 21.Lopes RJ, Johnson JD, Toomey MB, Ferreira MS, Araujo PM, Melo-Ferreira J, Andersson L, Hill GE, Corbo JC, Carneiro M. Genetic Basis for Red Coloration in Birds. Curr Biol. 2016;26:1427–1434. doi: 10.1016/j.cub.2016.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Twyman H, Valenzuela N, Literman R, Andersson S, Mundy NI. Seeing red to being red: conserved genetic mechanism for red cone oil droplets and co-option for red coloration in birds and turtles. Proc Biol Sci. 2016;283(1836):20161208. doi: 10.1098/rspb.2016.1208. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Claeys M, Wechsung E, Herman AG, Nugteren DH. Lack of prostacyclin biosynthesis by aortic tissue of the chicken. Prostaglandins 1981. 1981;21:739–749. doi: 10.1016/0090-6980(81)90231-8. [DOI] [PubMed] [Google Scholar]
- 24.Bult H, Wechsung E, Houvenaghel A, Herman AG. Prostanoids and hemostasis in chickens: anti-aggregating activity of prostaglandins E1 and E2, but not of prostacyclin and prostaglandin D2. Prostaglandins. 1981;21:1045–1058. doi: 10.1016/0090-6980(81)90172-6. [DOI] [PubMed] [Google Scholar]
- 25.de Matos R. Adrenal steroid metabolism in birds: anatomy, physiology, and clinical considerations, Vet. Clin North Am Exot Anim Pract. 2008;11:35–57. vi. doi: 10.1016/j.cvex.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 26.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 27.Fortey R. Horseshoe Crabs and Velvet Worms: The Story of the Animals and Plants That Time Has Left Behind, Alfred A Knopf. 2012 [Google Scholar]
- 28.Maas A, Mayer G, Kristensen RM, Waloszek D. A Cambrian micro-lobopodian and the evolution of arthropod locomotion and reproduction. Chinese Science Bulletin. 2007;52:3385–3392. [Google Scholar]
- 29.Hering L, Henze MJ, Kohler M, Kelber A, Bleidorn C, Leschke M, Nickel B, Meyer M, Kircher M, Sunnucks P, Mayer G. Opsins in onychophora (velvet worms) suggest a single origin and subsequent diversification of visual pigments in arthropods. Mol Biol Evol. 2012;29:3451–3458. doi: 10.1093/molbev/mss148. [DOI] [PubMed] [Google Scholar]
- 30.Kohler A, Kuo A, Nagy LG, Morin E, Barry KW, Buscot F, Canback B, Choi C, Cichocki N, Clum A, Colpaert J, Copeland A, Costa MD, Dore J, Floudas D, Gay G, Girlanda M, Henrissat B, Herrmann S, Hess J, Hogberg N, Johansson T, Khouja HR, LaButti K, Lahrmann U, Levasseur A, Lindquist EA, Lipzen A, Marmeisse R, Martino E, Murat C, Ngan CY, Nehls U, Plett JM, Pringle A, Ohm RA, Perotto S, Peter M, Riley R, Rineau F, Ruytinx J, Salamov A, Shah F, Sun H, Tarkka M, Tritt A, Veneault-Fourrey C, Zuccaro A, Tunlid A, Grigoriev IV, Hibbett DS, Martin F. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet. 2015;47:410–415. doi: 10.1038/ng.3223. Epub 2015 Feb 23. [DOI] [PubMed] [Google Scholar]
- 31.Arvas M, Kivioja T, Mitchell A, Saloheimo M, Ussery D, Penttila M, Oliver S. Comparison of protein coding gene contents of the fungal phyla Pezizomycotina and Saccharomycotina, ( BMC Genomics. 2007;20078:325. doi: 10.1186/1471-2164-8-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng J, Carbone I, Dean RA. The evolutionary history of cytochrome P450 genes in four filamentous Ascomycetes. BMC Evol Biol. 2007 Feb 26;7:30. doi: 10.1186/1471-2148-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moktali V, Park J, Fedorova-Abrams ND, Park B, Choi J, Lee YH, Kang S. Systematic and searchable classification of cytochrome P450 proteins encoded by fungal and oomycete genomes. BMC Genomics. 2012 Oct 4;13:525. doi: 10.1186/1471-2164-13-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson D, Werck-Reichhart D. A P450-centric view of plant evolution. Plant J. 2011;66:194–211. doi: 10.1111/j.1365-313X.2011.04529.x. [DOI] [PubMed] [Google Scholar]
- 35.https://genome10k.soe.ucsc.edu/news/article/27 (Accessed March 21, 2017)
- 36.Nelson DR, Strobel HW. Evolution of cytochrome P-450 proteins. Mol Biol Evol. 1987;4:572–593. doi: 10.1093/oxfordjournals.molbev.a040471. [DOI] [PubMed] [Google Scholar]
- 37.Nelson DR. Metazoan Cytochrome P450 Evolution. Comparative Biochemistry and Physiology Part C. 1998;121:15–22. doi: 10.1016/s0742-8413(98)10027-0. [DOI] [PubMed] [Google Scholar]
- 38.Nelson DR. Cytochrome P450 and the individuality of species, Arch. Biochem Biophys. 1999;369:1–10. doi: 10.1006/abbi.1999.1352. [DOI] [PubMed] [Google Scholar]
- 39.Nelson DR, Goldstone JV, Stegeman JJ. The cytochrome P450 genesis locus and the origin of animal cytochrome P450s. Philosophical Transactions of the Royal Society Part B Biological Sciences. 2012;368:20120474. doi: 10.1098/rstb.2012.0474. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee DS, Nioche P, Hamberg M, Raman CS. Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature. 2008;455:363–368. doi: 10.1038/nature07307. [DOI] [PubMed] [Google Scholar]
- 41.Nelson DR. Plant Cytochrome P450s from Moss to Poplar. Phytochemistry Reviews. 2006;5:193–204. doi: 10.1007/s11101-006-9015-3. [DOI] [Google Scholar]
- 42.Sello MM, Jafta N, Nelson DR, Chen W, Yu JH, Parvez M, Kgosiemang IK, Monyaki R, Raselemane SC, Qhanya LB, Mthakathi NT, Sitheni Mashele S, Syed K. Diversity and evolution of cytochrome P450 monooxygenases in Oomycetes. Sci Rep. 2015 Jul 1;5:11572. doi: 10.1038/srep11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren R, Sun Y, Zhao Y, Geiser D, Ma H, Zhou X. Phylogenetic Resolution of Deep Eukaryotic and Fungal Relationships Using Highly Conserved Low-Copy Nuclear Genes. Genome Biol Evol. 2016;8:2683–2701. doi: 10.1093/gbe/evw196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burki F, Kaplan M, Tikhonenkov DV, Zlatogursky V, Minh BQ, Radaykina LV, Smirnov A, Mylnikov AP, Keeling PJ. Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista. Proc Biol, Sci. 2016 Jan 27;283(1823):20152802. doi: 10.1098/rspb.2015.2802. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katz LA, Grant JR. Taxon-rich phylogenomic analyses resolve the eukaryotic tree of life and reveal the power of subsampling by sites. Syst Biol. 2015;64:406–415. doi: 10.1093/sysbio/syu126. [DOI] [PubMed] [Google Scholar]
- 46.Adl SM, Simpson AG, Lane CE, Lukeš J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, Heiss A, Hoppenrath M, Lara E, Le Gall L, Lynn DH, McManus H, Mitchell EA, Mozley-Stanridge SE, Parfrey LW, Pawlowski J, Rueckert S, Shadwick L, Schoch CL, Smirnov A, Spiegel FW. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Derelle R, Torruella G, Klimeš V, Brinkmann H, Kim E, Vlček Č, Lang BF, Eliáš M. Bacterial proteins pinpoint a single eukaryotic root. Proc Natl Acad Sci U S A. 2015;112:E693–699. doi: 10.1073/pnas.1420657112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burki F, Kaplan M, Tikhonenkov DV, Zlatogursky V, Minh BQ, Radaykina LV, Smirnov A, Mylnikov AP, Keeling PJ. Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista. Proc Biol Sci. 2016;283:20152802. doi: 10.1098/rspb.2015.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feyereisen R. Arthropod CYPomes illustrate the tempo and mode in P450 evolution. Biochim Biophys Acta. 2011;1814:19–28. doi: 10.1016/j.bbapap.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 50.Tomazic ML, Poklepovich TJ, Nudel CB, Nusblat AD. Incomplete sterols and hopanoids pathways in ciliates: gene loss and acquisition during evolution as a source of biosynthetic genes. Mol Phylogenet Evol. 2014;74:122–134. doi: 10.1016/j.ympev.2014.01.026. [DOI] [PubMed] [Google Scholar]
- 51.Goldstone (mollusks) this issue
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


