Abstract
We examine a gas-phase approach for converting a sub-set of amino acid residues in polypeptide cations to dehydroalanine (Dha). Subsequent activation of the modified polypeptide ions gives rise to specific cleavage N-terminal to the Dha residue. This process allows for the incorporation of selective cleavages in the structural characterization of polypeptide ions. An ion/ion reaction within the mass spectrometer between a multiply-protonated polypeptide and the sulfate radical anion introduces a radical site into the multiply-protonated polypeptide reactant. Subsequent collisional activation of the polypeptide radical cation gives rise to radical side-chain loss from one of several particular amino acid side-chains (e.g., leucine, asparagine, lysine, glutamine, and glutamic acid) to yield a Dha residue. The Dha residues facilitate preferential backbone cleavages to produce signature c- and z-ions, demonstrated with cations derived from melittin, mechano growth factor (MGF), and ubiquitin. The efficiencies for radical side-chain loss and for subsequent generation of specific c- and z-ions have been examined as functions of precursor ion charge state and activation conditions using cations of ubiquitin as a model for a small protein. It is noted that these efficiencies are not strongly dependent on ion trap collisional activation conditions but are sensitive to precursor ion charge state. Moderate to low charge states show the greatest overall yields for the specific Dha cleavages whereas small molecule losses (e.g., water/ammonia) dominate at the lowest charge states and proton catalyzed amide bond cleavages that five rise to b- and y-ions tend to dominate at high charge states.
Keywords: ion/ion reactions, sulfate radical anion, protein radical cation, dehydroalanine
INTRODUCTION
Tandem mass spectrometry is widely used to identify proteins based on the primary sequence information obtained by MS/MS experiments applied to intact protein ions or to digestion-derived peptide ions using various forms of activation and precursor ion types.1,2 The most common activation method, collision induced dissociation (CID), tends to lead to cleavages at amide linkages to produce b- and y-ions from protonated peptide or protein ions.3 Depending on the sequence and charge state of the precursor, some favorable backbone dissociation channels are observed, such as cleavages at aspartic acid4,5,6,7 and proline8,9,10 residues. The aspartic acid side chain can cyclize to cleave the peptide bond under a proton deficient environment, leading to b- and y-ions C-terminal to the aspartic acid.3–7 Conversely with the presence of a mobile proton, the higher basicity of the amide nitrogen of proline facilitates the association with a mobile proton to enhance the formation of b- and y-ions N-terminal to proline resiudes.3, 8–9 Each of these cleavages have been found to dominate spectra under favorable conditions.
Recently, ornithine, an amino acid produced in nature via deguanidination of arginine, was found to introduce a facile backbone cleavage channel C-terminal to ornithine via the side chain initiated formation of a stable six-membered lactam ring.11 This ornithine effect is found to be competitive with the aspartic acid effect and the proline effect. Dehydroalanine (Dha), another non-proteinogenic but naturally occurring amino acid with an unsaturated side chain, is found to promote N-Cα bond cleavage of Dha to produce specific c- and z-ions.12 Selective cleavages can provide useful information because they introduce a degree of specificity that is analogous to the use of selective enzymes in solution. For example, a novel top-down protein identification scoring scheme weighted on the frequency of preferential fragmentation occurring at proline and aspartic acid residues was described13 that improves the discriminatory utility of the scoring algorithm.
In this work, the Dha effect on protein ion fragmentation is further explored. Dha can form naturally14 or through solution phase derivatization from serine or cysteine residues.15,16 Multiple gas-phase processes have been shown to result in conversion of amino acids residues to Dha residues in peptide ions within the mass spectrometer,12 including loss of an alkyl sulfenic acid via activation of oxidized S-alkyl cysteine residues17,18,19 or oxidized methionine residues,20 and asymmetric cleavage of disulfide bonds upon activation of protein ions with limited proton mobility.21,22 A more general and efficient way to convert some amino acid residues to Dha is through a radical process that follows the ion/ion reaction of multiply protonated peptide ions with sulfate radical anions.12 The sulfate radical anion can extract a hydrogen atom from the protonated peptide within the electrostatic complex formed via ion/ion interaction, producing a hydrogen-deficient peptide radical cation and the loss of a neutral sulfuric acid molecule.12 There are multiple approaches to generate hydrogen-deficient peptide radical cations,23 sych as CID of a transition metal-ligand associated peptide ion,24,25 hemolytic dissociation of fragile bonds in peroxy-, azo-, tempo-based reagents upon CID,27,26,28 ultraviolet photodissociation (UVPD) of photolabile groups like carbon-iodine bond.29,30,31,32 However, these methods suffers from the requirement of a priori chemical derivatization, low efficiency, use of special instrument setup, etc. The ion/ion reaction with sulfate radical ion, on the other hand, is fast, low-cost and efficient in generating hydrogen-deficient peptide radical cations. Subsequent radical rearrangement leads to the loss of a neutral radical side chain, resulting in an even-electron Dha residue.23,33 It was demonstrated that arginine, methionine, and leucine residues can be converted to Dha on model peptides and melittin using these methods.12 In principle, all amino acids except for proline, glycine, isoleucine, and alanine can undergo this radical mechanism. However, only seven have been shown to form Dha in practice.33 The gas-phase approach described herein, therefore, emphasizes the intentional generation of Dha residues in polypeptide ions involving the amino acids observed to provide the highest conversion yields.
EXPERIMENTAL
Materials
Methanol and glacial acetic acid were purchased from Mallinckrodt (Phillipsburg, NJ). Sodium persulfate, melittin from honey bee venom, and bovine ubiquitin were obtained from Sigma-Aldrich (St. Louis, MO). PEGylated Mechano Growth Factor (PEG MGF) was purchased from Peptide Sciences. All materials were used without further purification. All peptide and protein solutions for positive nanoelectrospray (nESI) were prepared in 49.5/49.5/1 (v/v/v) methanol/water/acetic acid (~10 μM). Non-PEGylated MGF cations can be observed under this condition. The sodium persulfate solution was prepared in water at a concentration of ~0.1 mg/mL. Sulfate radical anions were observed via negative nESI of the sodium persulfate solution.
Mass Spectrometry
All experiments were performed using a TripleTOF (QTOF 5600, Sciex, Concord, ON, Canada), previously modified for ion/ion reactions34 with an alternately pulsed nESI source.35 Multiply protonated peptide/protein cations and reagent radical anions were sequentially isolated in the Q1-mass filter and injected into the q2 reaction cell that has been modified for mutual storage of ions of opposite polarity. After a defined mutual storage reaction time of 20–50 milliseconds, the product ions were then transferred back to Q1 with high energy to activate the complex, resulting in the sequential loss of neutral sulfuric acid and radical side chain. The resulting Dha-containing species were then isolated in Q1 prior to their second injection into q2 where they were activated by a low amplitude ion-trap CID for 10–1000 ms to promote fragmentation and enable sequence analysis. The product ions were then mass analyzed by TOF.
Data analysis
The product ion partitioning in the CID spectra was determined by using a program written in MATLAB to sum fragment peak areas from different dissociation channels. These summations were used to calculate the percentage of each channel.
RESULTS AND DISCUSSION
Generation of Dha via ion/ion radical chemistry
Several methods for generating radical peptide cations via ion/ion reactions have been described,12,32 the most efficient of which is ion/ion reactions with sulfate radial anion (SO4−•),36,12 A proposed mechanism for the generation of a peptide radical cation, subsequent generation of Dha, and the specific fragmentation to yield c-, z-ions is outlined in Scheme 1.
An electrostatic complex can be formed via ion/ion reaction between multiply protonated peptide ions and sulfate radical anions (m/z 95.95) derived from negative nESI of a sodium persulfate solution. The sulfate radical anion can abstract a hydrogen atom and a proton from the peptide, thus yielding a peptide radical cation and a neutral sulfuric acid molecule. The fragmentation pathways of peptide radical cations differ significantly from those of their even-electron counterparts, with amino acid side chain losses being more predominant in the former. There are two common side chain loss pathways from peptide radical cations.33,38 The first pathway is shown in Scheme 1 where the α-H atom is abstracted, resulting in the formation of a Dha residue and a loss of part of the side chain as a radical. The second pathway is the abstraction of the side chain γ-H that leads to the loss of the whole even-electron side chain, leaving the radical at the α-carbon. Table 1 lists the radical side chain losses of a few amino acid residues that give rise to Dha via the first pathway and their corresponding losses via the second pathway. With the exception of proline, glycine, isoleucine, and alanine, all other amino acid residues are theoretically capable of forming Dha via the first radical pathway because the mechanism in Scheme 1 requires a secondary γ-C (or two γ-H) in the amino acid residue. Experimentally, however, the losses from valine (15 Da), phenylalanine (77 Da), tyrosine (93 Da), histidine (67 Da), threonine (15 Da), serine (17 Da), and tryptophan (116 Da), for example, are never or rarely observed.33 The propensity for observing each side chain loss is related to the Bond Dissociation Energies (BDE) for abstracting the α-H of the amino acid residue.39 Glutamic acid, leucine, and methionine show the highest radical side chain loss while arginine and aspartic acid are among the lowest.33 Besides the BDE of the αC-H bond, other factors can also play roles in determining the likelihood for Dha formation. For example, the tyrosine hydroxyl side chain H can be abstracted and the loss of p-quinomethide (106 Da) is a more competitive side chain loss pathway. In the case of phenylalanine, side chain loss by the mechanism in Scheme 1 involves aromatic carbon radicals, rendering Dha formation unlikely.33 There are a very limited number of methionine residues in the sample ions in this study (i.e., ions derived from melittin, MGF, and ubiquitin). Therefore, the Dha effect is focused here on leucine, asparagine, lysine, glutamic acid, and glutamine residues.
Table 1.
| Amino acid residue | Radical loss to produce Dha | MW | Even-electron loss | MW |
|---|---|---|---|---|
| Leucine | C3H7• | 43.05 | C4H8 | 56.06 |
| Asparagine | CONH2• | 44.01 | CH2CONH2 | 58.03 |
| Lysine | C3H8N• | 58.07 | C4H9N | 71.07 |
| Glutamine | NH2COCH2• | 58.03 | NH2COC2H4 | 71.04 |
| Glutamic acid | CH3COO• | 59.01 | C2H4COO | 72.02 |
| Methionine | CH3S• | 61.01 | C3H6S | 74.02 |
| Arginine | C3H8N3• | 86.07 | C4H9N3 | 99.09 |
The [M+4H]4+ and [M+3H]3+ ions of melittin, a 26 amino acid polypeptide that is the active component of bee venom, are used to illustrate the Dha effect in Figure 1. Figure 1(a) shows the post-ion/ion reaction spectrum in positive mode that resulted from the reaction of melittin [M+4H]4+ ions with persulfate anions. The major peaks in the spectrum arise from residual cationic reactant ions and adduct ions from the attachment of one and two persulfate anions, respectively. The relative abundances of these products are sensitive to the number of anions and mutual storage time. Though it has been shown that multiple anion attachment can introduce more than one radical site on the peptide,36 only single radical species were investigated in this study. Therefore, ion/ion reaction conditions to maximize the single adduction complex were used here. Upon ion trap collisional activation of the isolated complex with one attached persulfate, the charge-reduced peptide radical cation, [M+2H]3+•, is produced as the nearly exclusive ionic product with the loss of a neutral H2SO4 molecule (Figure S1). Further activation of the radical cation species produces proton-directed backbone cleavage (b-, y-ions), radical side chain losses, and small neutral losses (H2O, NH3, CO2). In Figure 1(b), beam-type CID is used to generate the radical cation and the sequential side chain losses from the electrostatic complex in one step. Several highly abundant radical side chain losses are observed: •C2H5 (29.04 Da) from isoleucine to produce a dehydrobutyrine (Dhb) residue, as well as •C3H7 (43.05 Da) from leucine, •C3H8N (58.07 Da) from lysine or NH2COCH2• (58.03 Da) from glutamine to produce Dha residues. A small •C3H8N3 (86.07 Da) loss from arginine is also observed. Figure 1(c) shows the result of the MS3 experiment involving the ion trap CID of the isolated ion generated by 43 Da loss from the radical cation, (i.e., the [M+2H-43]3+ ion). The base peak in the product ion spectrum, z11-43 (z11▲2+), and its complementary c15 ion correspond to cleavage N-terminal to the leucine-16 residue. Peaks corresponding to c- and z-43 ions N-terminal to other leucine residues are also observed (shaded red). Non-specific b- and y-43 ions that originated from the proton-directed pathways are also in high abundance. Note that the isotopic peaks of the 43 Da loss are mixed with the 44 Da CO2 loss from the C-terminus. Those isomers with a CO2 loss contribute to the b- and y-43 ions as well. No c- or z-ions are observed in the product ion spectrum of [M+2H-29]3+, indicating the Dhb does not undergo the dissociation pathway shown in Scheme 1. The same MS3 process was repeated with a lower charge state of melittin precursor, [M+3H]3+. The ion trap CID spectrum of [M+H-43]2+ is shown in Figure 1(d). In this case, the c- and z-43 ions from Dha effect are dominant and only low abundances of b- and y-ions are observed. With one less proton in the system, the Dha pathway is favored over the mobile proton pathways, and is also more competitive than the proline effect, which gives rise to b13 and y13 ions.
Figure 1.
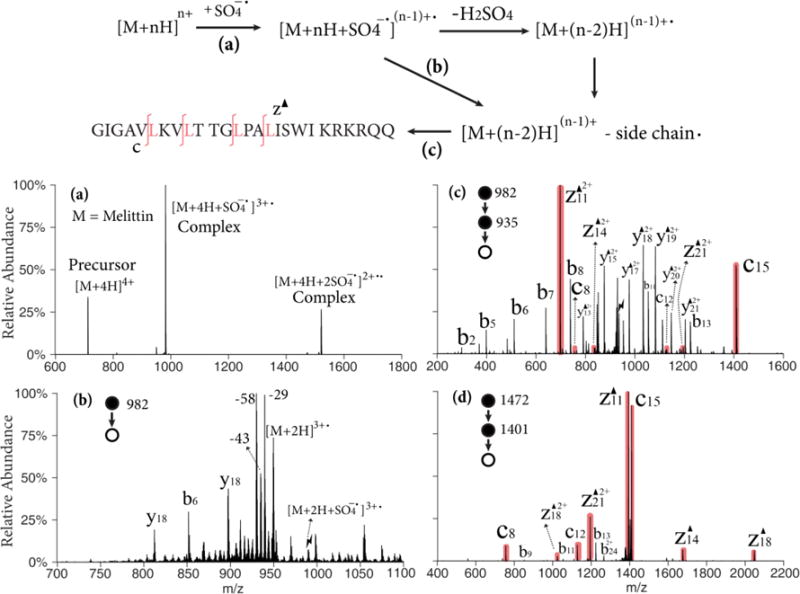
Upper: Workflow and sequence of melittin (M); Lower: (a) Ion/ion reaction between sulfate radical anion and [M+4H]4+; (b) Beam-type CID of singly adducted species; (c) CID of the 43 Da loss from the triply charged melittin radical cation, [M+2H-43]3+; (d) CID of the 43 Da loss from the doubly charged melittin radical cation, [M+H-43]2+ derived from ion/ion reaction with [M+3H]3+. Black triangles indicate fragment ions containing a Dha residue. Red shaded peaks indicate fragments resulted from Dha effect. Lightning bolts indicate species subjected to activation.
Scheme 1.
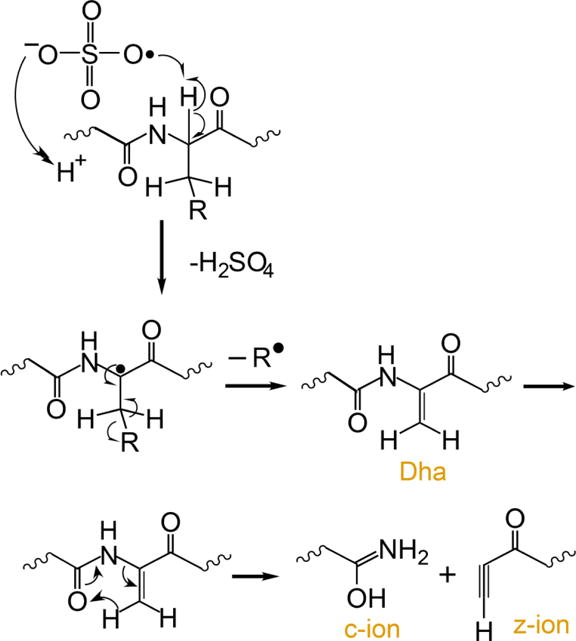
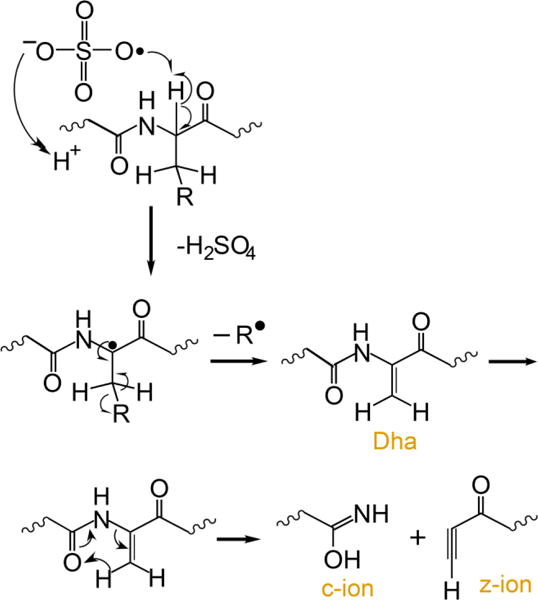
Proposed mechanism for the generation of dehydroalanine via ion/ion reaction between sulfate radical anion and multiply protonated protein ions and sequential generation of c- and z-ions N-terminal to dehydroalanine residues upon CID.37,12
Mechano growth factor (MGF), a 24-amino acid peptide involved in muscle tissue repair was examined to illustrate the process on a leucine-deficient peptide. The radical cation of MGF is obtained via attachment of sulfate radical anion and detachment of neutral sulfuric acid in the gas-phase. The 58/59 Da loss from the radical cation can be the radical side chain loss of lysine (C3H8N•, 58 Da), glutamine (NH2COCH2•, 58 Da) or glutamic acid (CH3COO•, 59 Da). As the isotopic envelopes of the 58 Da loss and the 59 Da loss overlap, it is difficult to avoid activating both populations. Figure 2 shows the MS3 product spectra of different charge states associated with the 58/59 Da losses from the radical cations generated from MGF, (i.e., [MGF+2H-58/59]3+ in Figure 2(a) and [MGF+H-58/59]2+ in Figure 2(b)). From [MGF+2H-58/59]3+, dominant c- and z-58/59 ions can be observed N-terminal to nearly every lysine, glutamine, and glutamic acid residue (shaded peaks). Triply protonated MGF, an even-electron species, was also subjected to CID as a comparison (Figure S2), in which backbone cleavages are observed at a very limited number of sites. At a lower charge state (Figure 2(b)), all the expected c- and z-ions are observed but the water loss channel becomes predominant thereby reducing the relative contributions of all other dissociation channels. In both Figure 2(a) and (b), the Dha effect on glutamine and glutamic acid residues are more favored that that on lysine residues though there are more lysine residues on MGF. This phenomenon cannot be generalized to other peptides or proteins because the reactivity of different residues are affected by the particular sequence of the peptide and gas-phase ion conformation.
Figure 2.
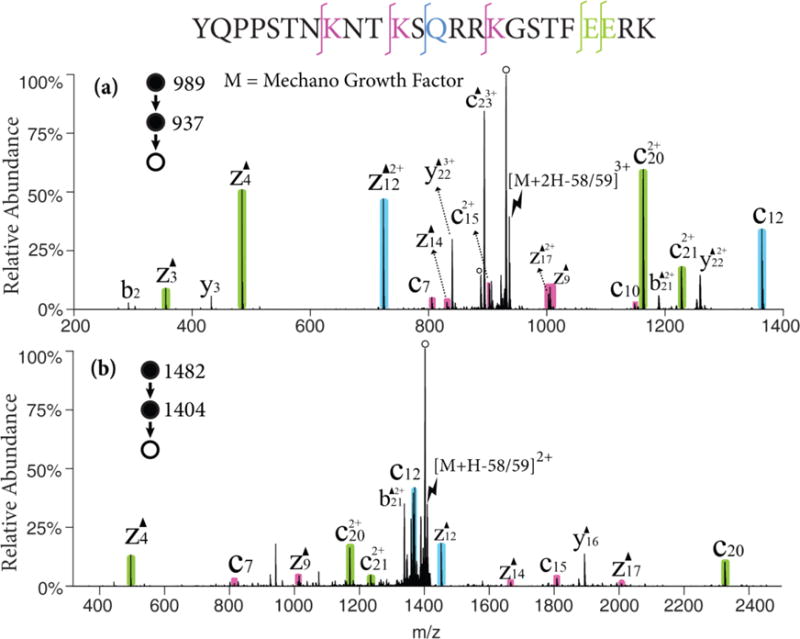
CID of the 58/59 Da loss from (a) the triply charged MGF radical cation, [MGF+2H]3+• and (b) the doubly charged MGF radical cation, [MGF+H]2+•. Black triangles indicate fragment ions containing a Dha residue and degree signs indicate water losses. Violet, green and blue shaded peaks indicate c- and z- fragments N-terminal to lysine, glutamic acid, and glutamine, respectively. Lightning bolts indicate species subjected to activation.
The loss of 44 Da from the MGF radical cations was also a significant channel fragmentation channel. This mass loss could arise from CO2 loss from the C-terminus, aspartic acid, or glutamic acid residues, as well as CONH2• loss from asparagine side chains. The ion trap CID spectrum of the [MGF+2H-44]2+ ion, for example, showed prominent c6/z18-44 and c8/z16-44 complementary pairs associated with the asparagine residues at positions 7 and 9, respectively (See Figure S3). This strong evidence for the Dha effect suggests that a significant fraction of the 44 Da loss was due to loss of CONH2•. The only clear evidence for contributions from CO2 loss arises from a prominent b232+ ion, which suggests a loss of CO2 from the C-terminus.
The generation of Dha in ubiquitin
Ubiquitin, a 76 residue polypeptide chain is used as a model system for a small protein, given that it contains nearly every residue able to lose the radical partial side chain and produce Dha. It also allows for a more comprehensive study of the role of charge state on the Dha effect relative to melittin and MGF. The ion/ion reaction of the [ubiquitin+6H]6+ ion was carried out with the sulfate radical anion to generate an electrostatic complex and subsequent CID generated the ubiquitin radical cation [ubiquitin+4H]5+•. As shown in Figure 3(a), side chain losses from the ubiquitin radical cations [ubiquitin +4H]5+• are predominant with only minimal abundances associated with NH3/H2O loss or b/y-ions from backbone cleavages. There are three main envelopes of side chain losses: (1) the 43/44 Da losses from leucine or asparagine, (2) the 58/59 Da losses from lysine, glutamine, or glutamic acid, and (3) the 86 Da loss from arginine. The ions generated from the 43/44 Da and the 58/59 Da losses were isolated and subjected to further activation with the results shown in Figures 3(b) and (c), respectively. Though abundant loss of neutral NH3 or H2O is observed, extensive c- and z-ions N-terminal to asparagine, leucine, lysine, glutamine, and glutamic acid residues are observed, as illustrated by the color-coded peaks. The b32, y24 and y18 ions C-terminal to aspartic acid residues are observed in lower abundances than the fragments generated from the Dha effect. These five reactive residues can be identified and localized in this method and more sequence information can be obtained than with traditional CID of the protonated species, [ubiquitin+5H]5+, which is dominated by fragments from the proline effect and aspartic acid effects.32
Figure 3.
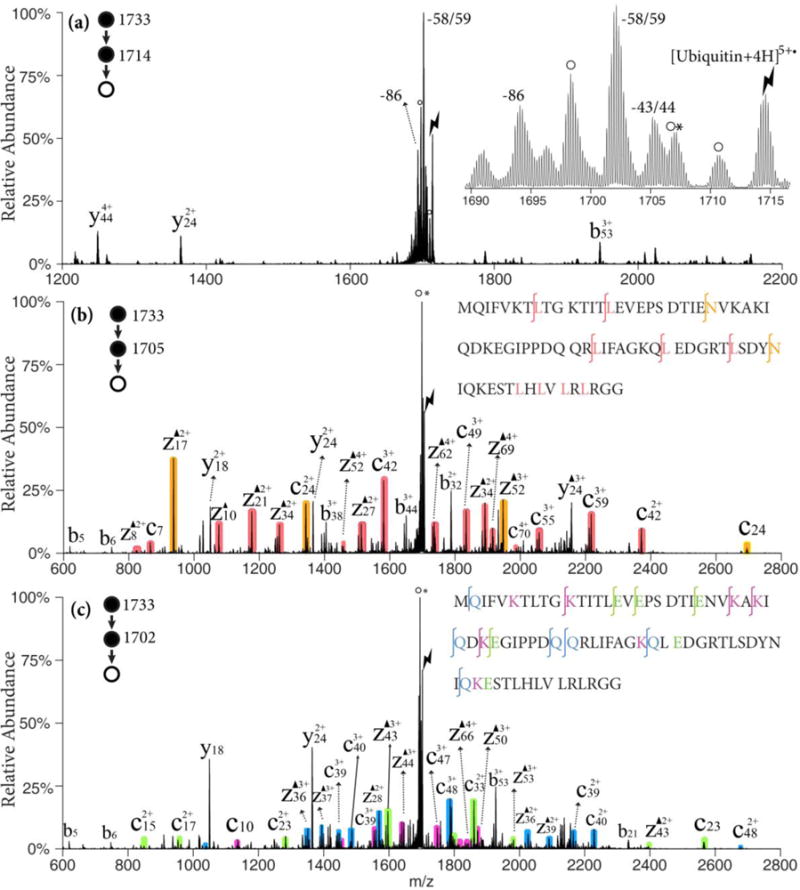
(a) CID of [ubiquitin +4H]5+• ion derived from ion/ion reaction between [ubiquitin +6H]6+ and SO4−•; Insert: zoom-in of the radical side chain loss region. (b) CID of the 43/44 Da loss from [ubiquitin +4H]5+•; (c) CID of the 58/59 Da loss from [ubiquitin +4H]5+•; Black triangles indicate fragment ions containing a Dha residue. Degree signs indicate water losses whereas asterisks indicate ammonia losses. Red, orange shaded peaks indicate c- and z-fragments N-terminal to leucine and asparagine. Violet, green and blue shaded peaks indicate c- and z-fragments N-terminal to lysine, glutamine and glutamic acid residues, respectively. Lightning bolts indicate species subjected to activation.
Optimization of Dha formation in ubiquitin
To maximize the overall yield of the Dha effect starting from the protein precursor, the efficiencies of (1) the loss of partial radical side chain from peptide radical cations, and (2) the formation of c- and z-ions N-terminal to Dha were investigated as functions of precursor ion charge state and activation conditions.
The yield of Dha formation from ubiquitin radical cations by side chain losses is first described. Upon CID ubiquitin radical cations can undergo a variety of fragmentation pathways, including radical-directed side chain losses, radical directed backbone cleavages, proton-directed backbone cleavages, charge-remote backbone cleavages and loss of small neutrals (NH3/H2O). Each of the pathways can dominate the CID spectrum under favorable conditions. Figures 4(a) and (b) show the ion-trap CID of two ubiquitin radical cations, [ubiquitin+8H]9+• and [ubiquitin+3H]4+•, respectively, under similar activation conditions. It was reported that extensive nonspecific fragmentation of the protonated ubiquitin was observed upon ion-trap CID at moderate charge states (+7 to +9), whereas at lower charge states, abundant loss of NH3 or H2O was observed.40 A similar trend is also observed here. Abundant b- and y-ions are present in the product spectrum of [ubiquitin+8H]9+•, while the H2O/NH3 loss peak is dominant from the [ubiquitin+3H]4+• ion. Some z-ions from the radical-directed backbone cleavages are also present in Figure 4(a). In both cases, the radical side chain losses, 43/44 Da and 58/59 Da are observed. To reveal the charge state where these side chain losses have the highest yield, the partitioning of the peak abundances between the backbone fragments, radical side chain losses, and H2O/NH3 losses are plotted versus the charge states of the precursor radical cations in Figure 4(c). As the charge state decreases, the relative intensity of the H2O/NH3 losses gradually increase and the backbone fragments decrease, with the exception of [ubiquitin+5H]6+• where abundant y18/b58 ions from the aspartic acid effect generate a high ratio of the backbone fragments. Nevertheless, the yield of radical side chain losses reaches its highest value of ~60% of the total fragments at the charge state of 5+.
Figure 4.
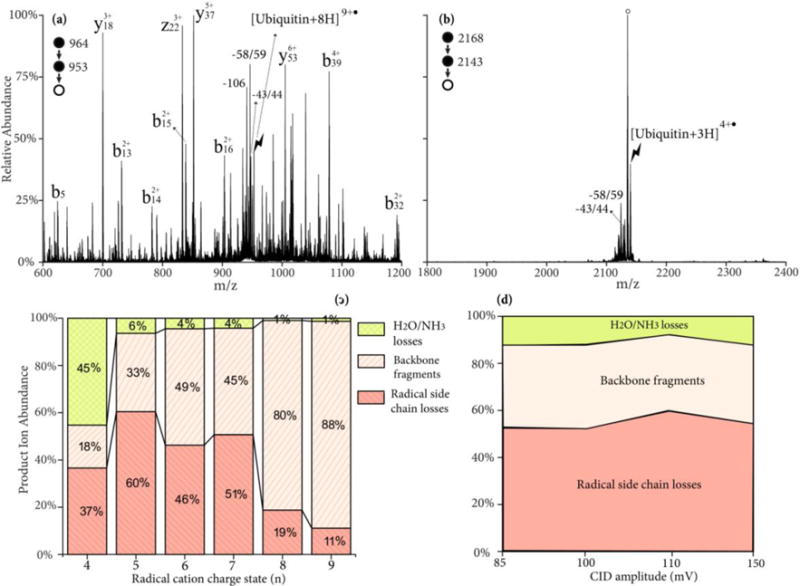
CID spectrum of ubiquitin radical cation (a) [ubiquitin+8H]9+• and (b) [ubiquitin+3H]4+• at 110mV for 100 ms; (c) Product ion partitioning at different charge states of radical cations; (d) Product ion partitioning of [ubiquitin+4H]5+• at different CID energies. Degree signs indicate water losses whereas lightning bolts indicate species subjected to activation.
The role of activation conditions on the relative contributions of the various types of fragmentation channels was studied with the [ubiquitin+4H]5+• ion. The ion-trap CID amplitude was varied from 85 mV to 150 mV and the activation time was varied (10–1000 ms) to ensure a >85% depletion of the precursor ions at each activation amplitude. The relative contributions of H2O/NH3 loss, backbone cleavages, and radical side chain losses are plotted in Figure 4(d), where a nearly constant ratio is observed across different energies. The similar yield of side chain losses (~50%) is also observed upon high-energy beam-type CID of the complex [ubiquitin+6H+SO4−•]5+• to generate the sequential H2SO4 and side chain losses.
Optimizing the relative contributions of Dha specific cleavages of ubiquitin
The roles of precursor charge state and activation conditions on fragment ion partitioning were also investigated for the Dha-containing species, [ubiquitin+(n-1)H-43/44]n+. Upon activation, there are three main dissociation channels: (1) fragmentation via Dha, giving rise to c- and z-ions N-terminal to leucine or asparagine residues, (2) charge-directed or charge remote backbone fragmentation, producing b- and y-ions,3 (3) and neutral H2O/NH3 losses via a variety of mechanisms.3 The trend from abundant nonspecific backbone cleavages to yield b/y-ions at high charge states to dominant small molecule neutral loss at low charge states, as noted for the radical cation of Figure 4, is also observed for the side-chain radical loss precursor ions (see Figures 5(a) and (b)). Interestingly, the tendency for the site of radical side chain loss to yield a Dha residue also appears to be sensitive to charge state. The Dha effect is almost exclusively observed at asparagine rather than leucine residues in the CID spectrum of [ubiquitin+7H-43/44]8+ depicted in Figure 5(a), while more Dha-specific cleavages N-terminal to leucine residues are observed from lower charge states, such as [ubiquitin+4H-43/44]5+ ion, as depicted in Figure 5(b). The partitioning of the product ions between the three classes of dissociative channels (viz., b- and y-ions from non-specific backbone cleavages, c- and z-ions from cleavages N-terminal to leucine and asparagine residues, and H2O/NH3 losses) are plotted as a function of the precursor charge state in Figure 5(c). Again, the yield of the Dha fragmentation channels reaches is highest (~ 30%) at the 5+ charge state. The energy dependence of Dha specific cleavages is shown in Figure 5(d). The activation time is varied (10–1000 ms) at each CID amplitude to allow >85% depletion of the precursor ions. The low energy H2O/NH3 loss channels (rearrangement) are preferred at lower CID amplitude, while the b- and y-ions channels are most competitive at higher CID amplitude. The specific c- and z-ions reach their maximum at ~110 mV.
Figure 5.
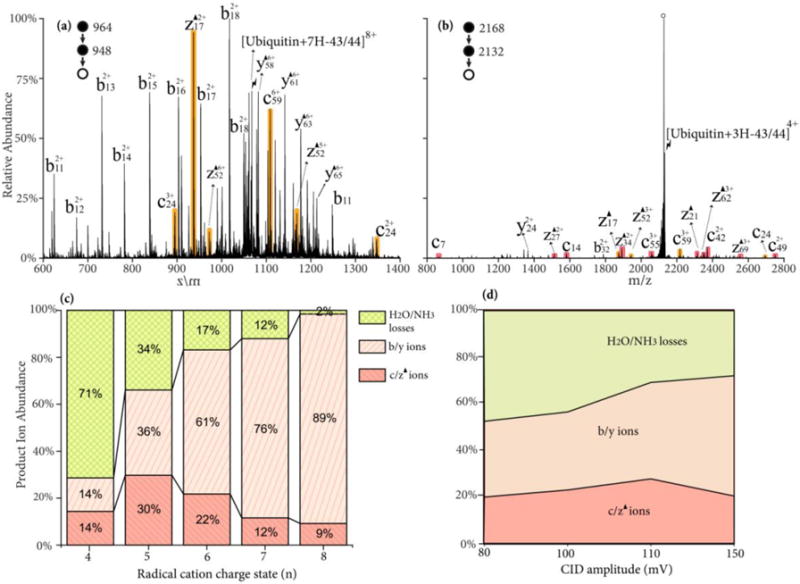
CID spectrum of radical side chain loss from ubiquitin radical cation (a) [ubiquitin+7H-43/44]8+; (b) [ubiquitin+3H-43/44]4+ at 110 mV for 100 ms; (c) Product ion partitioning at different charge states of radical cations; (d) Product ion partitioning of [Ubiquitin+4H-43/44]5+ at different CID energies. Red, orange shaded peaks indicate c- and z-fragments N-terminal to leucine and asparagine. Black triangles indicate fragment ions containing a Dha residue and degree signs indicate water losses. Lightning bolts indicate species subjected to activation.
The charge state and activation condition experiments for the ubiquitin ions suggest that the Dha effect is greatest at intermediate-to-low charge states and is most competitive at intermediate activation amplitudes, although the latter effect is not dramatic. The losses of small neutral molecules, which are generally low frequency/low critical energy processes, are known to be favored at low activation energies/long activation times and at the lowest charge states. Charge catalyzed processes, such as those favored by mobilized protons, are less favorable at low charge states due both to extensive charge solvation and localization at highly basic sites (e.g., arginine side chains). (At very high charge states, which were not examined here, proton mobility tends to be limited due to Coulombic repulsion, which limits the number of contributing channels.32) Over the charge state range examined here, diverse non-specific backbone cleavages become increasingly competitive as the charge state increases, as expected based on previous studies in which the Dha residues are absent.32 The mechanism indicated in Scheme 1 for the Dha effect that leads to N-Cα bond cleavage does not require a mobile proton, thus this pathway is not favored at higher charge states (+6 to +8) of ubiquitin compared to the charge-directed pathways to produce b- and y-ions. On the other hand, there is a relatively high tendency for the loss of small molecules at low charge states (+4) likely due to the compact gas-phase ion conformation at low charge states.41,42,43,44,45 The Dha effect can therefore be expected to be most competitive at moderate to low charge states.
CONCLUSIONS
Ion/ion reactions with sulfate radical anions to generate radical cations from multiply-protonated proteins followed by CID of the radical cation can be used to convert leucine, asparagine, lysine, glutamine, and glutamic acid residues to Dha residues in multiply protonated protein ions via the loss of radical side chains. As leucine, asparagine, lysine, glutamine, and glutamic acid are all frequently occurring amino acids, consisting of more than 28% of all amino acids in vertebrates,46,47 this radical cation-mediated approach to generating Dha residues is expected to have a high degree of generality. Furthermore, a degree of selectivity in choosing the residues in the modified protein ion that are converted to Dha in MS3 experiments is afforded via the characteristic masses of the radical side chain losses. The ‘Dha effect’ upon dissociation leads to specific c-/z-ions N-terminal the Dha residue. This readily recognizable process can give rise to significant contributions to the overall dissociation of the protein ions under favorable conditions. The favorable conditions for these specific cleavages were investigated here using ubiquitin ions as models. While the non-specific backbone cleavages and the small H2O/NH3 losses dominate at higher or lower charge states, respectively, the Dha effect is most prominent at an intermediate charge state of ubiquitin 5+. This gas-phase approach to convert selected residues in protein ions to Dha provides a novel means for introducing selective cleavages in the CID of gaseous protein ions. While beyond the scope of the present work, a database search approach that incorporates the replacement of specific residues with Dha, in conjunction with CID of gas-phase modified protein ions, could provide novel capabilities in top-down and middle-down protein characterization.
Supplementary Material
Figure S1. Ion-trap CID spectrum of electrostatic complex [melittin+4H+SO4−]3+ derived from ion/ion reaction between sulfate radical anion and [melittin +4H]4+. Lightning bolts indicate species subjected to activation.
Figure S2. Ion-trap CID spectrum of [MGF+3H]3+. Lightning bolts indicate species subjected to activation and degree signs indicate water losses.
Figure S3. Ion-trap CID spectrum of [MGF+H−44]2+. Lightning bolts indicate species subjected to activation. Black triangles indicate fragment ions containing a Dha residue. Degree signs indicate water losses. Orange shaded peaks indicate c− and z− fragments N-terminal to asparagine.
Acknowledgments
This work was supported by the National Institutes of Health under Grant GM R37-45372.
References
- 1.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 2.McLuckey SA, Mentinova M. Ion/neutral, ion/electron, ion/photon, and ion/ion interactions in tandem mass spectrometry: do we need them all? Are they enough? J Am Soc Mass Spectrom. 2011;22:3–12. doi: 10.1007/s13361-010-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paizs B, Suhai S. Fragmentation pathways of protonated peptides. Mass Spectrom Rev. 2005;24:508–548. doi: 10.1002/mas.20024. [DOI] [PubMed] [Google Scholar]
- 4.Yu W, Vath JE, Huberty MC, Martin SA. Identification of the facile gas-phase cleavage of the Asp-Pro and Asp-Xxx peptide bonds in matrix-assisted laser desorption time-of-flight mass spectrometry. Anal Chem. 1995;65:3015–3023. doi: 10.1021/ac00069a014. [DOI] [PubMed] [Google Scholar]
- 5.Tsaprailis G, Somogyi Á, Nikolaev EN, Wysocki VH. Refining the model for selective cleavage at acidic residues in arginine-containing protonated peptides. Int J Mass Spectrom. 2000;195/196:467–479. [Google Scholar]
- 6.Sullivan AG, Brancia FL, Tyldesley R, Bateman R, Sidhu K, Hubbard SJ, Oliver SG, Gaskell SJ. The exploitation of selective cleavage of singly protonated peptide ions adjacent to aspartic acid residues using a quadrupole orthogonal time-of-flight mass spectrometer equipped with a matrix-assisted laser desorption/ionization source. Int J Mass Spectrom. 2001;210/211:665–676. [Google Scholar]
- 7.Huang Y, Wysocki VH, Tabb DL, Yates JR., III The influence of histidine on cleavage C-terminal to acidic residues in doubly protonated tryptic peptides. Int J Mass Spectrom. 2002;219:233–244. [Google Scholar]
- 8.Schwartz BL, Bursey MM. Some proline substituent effects in the tandem mass spectrum of protonated pentaalanine. Biol Mass Spectrom. 1992;21:92–96. doi: 10.1002/bms.1200210206. [DOI] [PubMed] [Google Scholar]
- 9.Vaisar T, Urban J. Probing the proline effect in CID of protonated peptides. J Mass Spectrom. 1996;31:1185–1187. doi: 10.1002/(SICI)1096-9888(199610)31:10<1185::AID-JMS396>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 10.Bleiholder C, Suhai S, Harrison AG, Paizs B. Towards understanding the tandem mass spectra of protonated oligopeptides. 2: The proline effect in collision-induced dissociation of protonated Ala-Ala-Xxx-Pro-Ala (Xxx = Ala, Ser, Leu, Val, Phe, and Trp) J Am Soc Mass Spectrom. 2011;22:1032–1039. doi: 10.1007/s13361-011-0092-1. [DOI] [PubMed] [Google Scholar]
- 11.McGee WM, McLuckey SA. The ornithine effect in peptide cation dissociation. J Mass Spectrom. 2013;48:856–861. doi: 10.1002/jms.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pilo AL, Peng Z, McLuckey SA. The dehydroalanine effect in the fragmentation of ions derived from polypeptides. J Mass Spectrom. 2016;51:857–866. doi: 10.1002/jms.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reid GE, Shang H, Hogan JM, Lee GU, McLuckey SA. Gas-phase concentration, purification, and identification of whole proteins from complex mixtures. J Am Chem Soc. 2002;124:7353–7362. doi: 10.1021/ja025966k. [DOI] [PubMed] [Google Scholar]
- 14.Chatterjee A, Abeydeere ND, Bale S, Pai PJ, Dorrestein PC, Russell DH, Ealick SE, Begley TP. Saccharomyces cerevisiae THI4p is a suicide thiamine thiazole synthase. Nature. 2002;478:542–U146. doi: 10.1038/nature10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rich DH, Tam J, Mathiaparanam P, Grant JA, Mabuni C. General synthesis of didehydroalanine acids and peptides. J Chem Soc Chem Commun. 1974;21:879–898. [Google Scholar]
- 16.Ranganathan D, Shah K, Vaish N. An exceptionally mild and efficient route to dehydroalanine peptides. J Chem Soc Chem Commun. 1992;16:1145–1147. [Google Scholar]
- 17.Steen H, Mann M. Similarity between condensed phase and gas phase chemistry: fragmentation of peptides containing oxidized cysteine residues and its implications for proteomics. J Am Soc Mass Spectrom. 2001;12:228–232. doi: 10.1016/S1044-0305(00)00219-1. [DOI] [PubMed] [Google Scholar]
- 18.Froelich JM, Reid GE. Mechanisms for the proton mobility dependent gas-phase fragmentation reactions of S-alkyl cysteine sulfoxide-containing peptide ions. J Am Soc Mass Spectrom. 2007;18:1690–1705. doi: 10.1016/j.jasms.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 19.Pilo AL, Zhao FF, McLuckey SA. Selective gas-phase oxidation and localization of alkylated cysteine residues in polypeptide ions via ion/ion chemistry. J Proteome Res. 2016;15:3139–3146. doi: 10.1021/acs.jproteome.6b00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pilo AL, McLuckey SA. Oxidation of methionine residues in polypeptide ions via gas-phase ion/ion chemistry. J Am Soc Mass Spectrom. 2014;25:1049–1057. doi: 10.1007/s13361-014-0861-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wells JM, Stephenson JL, McLuckey SA. Charge dependence of protonated insulin decompositions. Int J Mass Spectrom. 2000;203:A1–A9. [Google Scholar]
- 22.Bilusich D, Bowie JH. Fragmentations of (M-H)− anions of underivatised peptides. Part 2: characteristic cleavages of Ser and Cys and of disulfides and other post-translational modifications, together with some unusual internal processes. Mass Spectrom Rev. 2009;28:20–34. doi: 10.1002/mas.20206. [DOI] [PubMed] [Google Scholar]
- 23.Tureček F, Julian RR. Peptide radicals and cation radicals in the gas phase. Chem Rev. 2013;113:6691–6733. doi: 10.1021/cr400043s. [DOI] [PubMed] [Google Scholar]
- 24.Chu IK, Rodriquez CF, Lau TC, Hopkinson AC, Siu KWM. Molecular radical cations of oligopeptides. J Phys Chem B. 2000;104:3393–3397. [Google Scholar]
- 25.Chu IK, Laskin J. Review formation of peptide radical ions through dissociative electron transfer in ternary metal-ligand-peptide complexes. Eur J Mass Spectrom. 2011;17:543–556. doi: 10.1255/ejms.1156. [DOI] [PubMed] [Google Scholar]
- 26.Wee S, Mortimer A, Moran D, Wright A, Barlow CK, O’Hair RAJ, Radom L, Easton CJ. Gas-phase regiocontrolled generation of charged amino acid and peptide radicals. Chem Commun. 2006;40:4233–4235. doi: 10.1039/b608724h. [DOI] [PubMed] [Google Scholar]
- 27.Laskin J, Yang Z, Ng CMD, Chu IK. Fragmentation of alpha-radical cations of arginine-containing peptides. J Am Soc Mass Spectrom. 2010;21:511–521. doi: 10.1016/j.jasms.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 28.Osburn S, O’Hair RAJ, Ryzhov V. Gas-phase reactivity of sulfur-based radical ions of cysteine derivatives and small peptides. Int J Mass Spectrom. 2012;316:133–139. [Google Scholar]
- 29.Cheng PY, Zhong D, Zewail AH. Kinetic-energy, femtosecond resolved reaction dynamics – modes of dissociation (in iodobenzene) from time-velocity correlations. Chem Phys Lett. 1995;237:399–405. [Google Scholar]
- 30.Ly T, Julian RR. Residue-specific radical-directed dissociation of whole proteins in the gas phase. J Am Chem Soc. 2008;130:351–358. doi: 10.1021/ja076535a. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Julian RR. Photoinitiated intramolecular diradical cross-linking of polyproline peptides in the gas-phase. Phys Chem Chem Phys. 2012;14:16243–16249. doi: 10.1039/c2cp42242e. [DOI] [PubMed] [Google Scholar]
- 32.Gilbert JD, Fisher CM, Prentice BM, Redwine JG, McLuckey SA. Strategies for generatin peptide radical cations via ion/ion reactions. J Mass Spectrum. 2015;50:418–428. doi: 10.1002/jms.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun Q, Nelsea H, Ly T, Stoltz BM, Julian RR. Side chain chemistry mediated backbone fragmentation in hydrogen deficient peptide radicals. J Proteome Res. 2009;8:958–966. doi: 10.1021/pr800592t. [DOI] [PubMed] [Google Scholar]
- 34.Xia Y, Wu J, Londry FA, Hager JW, McLuckey SA. Mutual storage mode ion/ion reactions in hybrid linear ion trap. J Am Soc Mass Spectrom. 2005;16:71–81. doi: 10.1016/j.jasms.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 35.Liang XR, Xia Y, McLuckey SA. Alternatively pulsed nanoelectrosptray ionization/atmospheric pressure chemical ionization for ion/ion reactions in an electrodynamic ion trap. Anal Chem. 2006;78:3208–3212. doi: 10.1021/ac052288m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pilo AL, Bu J, McLuckey SA. Transformation of [M+ 2H]2+ peptide cations to [M-H]+, [M+H+O]+, and M+• cations via ion/ion reactions: reagent anions derived from persulfate. J Am Soc Mass Spectrom. 2015;26:1103–1114. doi: 10.1007/s13361-015-1125-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chrisman PA, McLuckey SA. Dissociations of disulfide-linked gaseous polypeptide/protein anions: ion chemistry with implications for protein identification and characterization. J Proteome Res. 2002;1:549–557. doi: 10.1021/pr025561z. [DOI] [PubMed] [Google Scholar]
- 38.Barlow CK, McFadyen WD, O’Hair RAJ. Formation of cationic peptide radicals by gas-phase redox reactions with trivalent chromium, manganese, iron, and cobalt complexes. J Am Chem Soc. 2005;127:6109–6115. doi: 10.1021/ja043088f. [DOI] [PubMed] [Google Scholar]
- 39.Rauk A, Yu D, Armstrong DA. Toward site specificity of oxidative damage in proteins: C−H and C−C bond dissociation energies and reduction potentials of the radicals of alanine, serine, and threonine residues – An ab Initio study. J Am Chem Soc. 1997;119:208–217. [Google Scholar]
- 40.Reid GE, Wu J, Chrisman PA, Wells JM, McLuckey SA. Charge-state-dependent sequence analysis of protonated ubiquitin ions via ion trap tandem mass spectrometry. Anal Chem. 2001;73:3274–3281. doi: 10.1021/ac0101095. [DOI] [PubMed] [Google Scholar]
- 41.Valentine SJ, Counterman AE, Clemmer DE. Conformer-dependent proton-transfer reactions of ubiquitin ions. J Am Soc Mass Spectrom. 1997;8:954–961. [Google Scholar]
- 42.Li J, Taraszka JA, Counterman AE, Clemmer DE. Influence of solvent composition and capillary temperature on the conformations of electrosprayed ions: unfolding of compact ubiquitin conformers from pseudonative and denatured solutions Int. J Mass Spectrom. 1999;185:37–47. [Google Scholar]
- 43.Purves RW, Barnett DA, Guevremont R. Separation of protein conformers using electrospray-high field asymmetric waveform ion mobility spectrometry-mass spectrometry. Int J Mass Spectrom. 2000;197:163–177. [Google Scholar]
- 44.Purves RW, Barnett DA, Ellis B, Guevremont R. Investigation of bovine ubiquitin conformers separated by high-field asymmetric waveform ion mobility spectrometry: Cross section measurements using energy-loss experiments with a triple quadrupole mass spectrometer. J Am Soc Mass Spectrom. 2000;11:738–745. doi: 10.1016/S1044-0305(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 45.Freitas MA, Hendrickson CL, Emmett MR, Marshall AG. Gas-phase bovine ubiquitin cation conformations resolved by gas-phase hydrogen/deuterium exchange rate and extent. Int J Mass Spectrom. 1999;185:565–575. [Google Scholar]
- 46.King JL, Jukes TH. Non-Darwinian evolution. Science. 1969;164:788–798. doi: 10.1126/science.164.3881.788. [DOI] [PubMed] [Google Scholar]
- 47.Dyer KF. The quiet revolution: A new synthesis of biological knowledge. J Bio Edu. 1971;5:15–24. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Ion-trap CID spectrum of electrostatic complex [melittin+4H+SO4−]3+ derived from ion/ion reaction between sulfate radical anion and [melittin +4H]4+. Lightning bolts indicate species subjected to activation.
Figure S2. Ion-trap CID spectrum of [MGF+3H]3+. Lightning bolts indicate species subjected to activation and degree signs indicate water losses.
Figure S3. Ion-trap CID spectrum of [MGF+H−44]2+. Lightning bolts indicate species subjected to activation. Black triangles indicate fragment ions containing a Dha residue. Degree signs indicate water losses. Orange shaded peaks indicate c− and z− fragments N-terminal to asparagine.


