Abstract
Epigenetic signaling pathways are implicated in tumorigenesis and therefore histone deacetylases (HDACs) represent novel therapeutic targets for cancers including multiple myeloma (MM). Although non-selective HDAC inhibitors show anti-MM activities, unfavorable side effects limit their clinical efficacy. Isoform- and/or class-selective HDAC inhibition offers the possibility to maintain clinical activity while avoiding adverse events attendant to broad non-selective HDAC inhibition. We have previously reported that HDAC3 inhibition, either by genetic knockdown or selective inhibitor BG45, abrogates MM cell proliferation. Here we show that knockdown of HDAC3, but not HDAC1 or HDAC2, as well as BG45, downregulate expression of DNA methyltranseferase 1 (DNMT1) mediating MM cell proliferation. DNMT1 expression was also regulated by c-Myc, and HDAC3 inhibition triggers degradation of c-Myc protein. Moreover, HDAC3 inhibition results in hyperacetylation of DNMT1, thereby reducing the stability of DNMT1 protein. Combined inhibition of HDAC3 and DNMT1 with BG45 and DNMT1 inhibitor 5-azacytidine (AZA), respectively, triggers synergistic downregulation of DNMT1, growth inhibition, and apoptosis in both MM cell lines and patient MM cells. Efficacy of this combination treatment was confirmed in a murine xenograft MM model. Our results therefore provide the rationale for combination treatment using HDAC3 inhibitor with DNMT1 inhibitor to improve patient outcome in MM.
Introduction
Histone deacetylases (HDACs)-mediated posttranslational modifications represent novel therapeutic targets in various types of cancers, including multiple myeloma (MM).1–4 Recently, a non-selective HDAC inhibitor panobinostat was approved by the US Food and Drug Administration (FDA) for the treatment of patients with MM. However, adverse side effects attendant to broad non-selective HDAC inhibitors, such as thrombocytopenia, fatigue and diarrhea, limit their clinical application.5, 6 To increase tolerability and exploit anti-cancer activity of HDAC inhibitors, isoform- or class-selective HDAC inhibitors are under development. Specifically, we have previously reported that selective genetic or pharmacologic HDAC3 inhibition shows remarkable anti-MM activities in vitro and in vivo in a xenograft mouse model of human MM.7 However, the molecular mechanisms of action have not yet been delineated.
c-Myc regulates a large number of genes related to cell proliferation and differentiation and is a potent oncogene.8, 9 MM can develop from a premalignant stage of monoclonal gammopathy of undetermined significance (MGUS),10–12 and during the progression from MGUS to MM, MYC activation plays a crucial role.13–15 Indeed MM cells have been reported to be addicted to c-Myc, which therefore represents a promising therapeutic target in MM.16
Recent studies have shown that HDACs regulate deacetylation not only of histones, but also of non-histone proteins such as p53 and signal transducer and activator of transcription (STAT3).17, 18 In terms of posttransrational modification of c-Myc, ubiquitination leads to degradation of the protein.19 In addition, acetylation of c-Myc by a histone acetyltranferase p300 also triggers proteasomal degradation of c-Myc protein through its ubiquitination.20 Moreover, HDAC3 interacts with p300 in neuron cells,21 and HDAC inhibitors, especially class I HDAC inhibitors, can target c-Myc in MM cells.22, 23 DNA methyltransferase 1 (DNMT1) maintains DNA methylation and is implicated in tumorigenesis.1, 24, 25 Interestingly, previous studies show that HDAC1 forms a complex with DNMT1,26 and that the protein stability of DNMT1 is regulated by posttranslational modifications of acetylation and ubiquitination.27, 28 Importantly, the binding of DNMT1 with ubiquitin specific peptidase 7 (USP7, also known as HAUSP) is regulated by the acetylation of DNMT1.28 However, it is not yet known which HDAC isoform specificity mediates regulation of c-Myc deacetylation or regulates interaction between DNMT1 and USP7.
In the present study, we first demonstrate that HDAC3 inhibition leads to downregulation of c-Myc, in turn resulting in downregulation of DNMT1 mRNA expression. Furthermore, we demonstrate that HDAC3 forms a complex with DNMT1, and that HDAC3 inhibition results in an increased acetylation of DNMT1 and leads to degradation of DNMT1. Finally, combination inhibition of DNMT1 and HDAC3 triggers synergistic MM growth inhibition in vitro and in in vivo in a murine xenograft model of human MM, providing the framework for clinical evaluation of this combination therapy.
Materials and Methods
No statistical analysis was used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment. For a more detailed description of the methods used, see supplemental Materials and Methods.
Results
HDAC3 regulates c-Myc
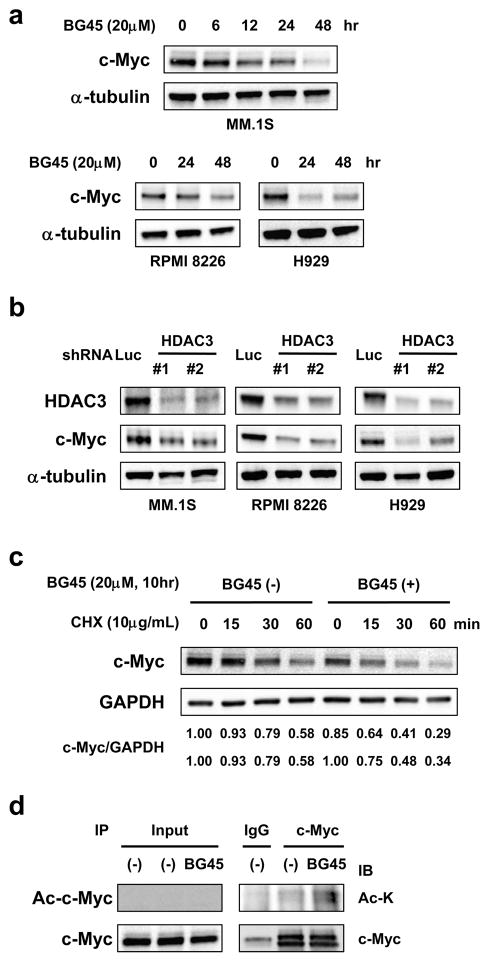
Since prior studies show that HDAC inhibitors, especially class I HDAC inhibitors, downregulate c-Myc,22, 23 we first examined whether HDAC3-selective inhibition triggers downregulation of c-Myc expression using an HDAC3-selective small molecule inhibitor BG45. As expected, BG45 treatment is associated with decreased c-Myc in MM.1S, RPMI 8226, and NCI-H929 (H929) cells in a time-dependent manner (Figure 1a). c-Myc expression was downregulated by BG45 in a dose dependent fashion; however, mRNA levels were not altered by the treatment (Figure S1a). To confirm whether c-Myc downregulation was solely due to HDAC3 inhibition, we carried out HDAC3 knockdown in MM.1S, RPMI 8226, and H929 cells using lentiviral short hairpin RNAs (shRNAs) targeting HDAC3 (shHDAC3 #1 and #2) or control shRNA targeting luciferase (shLuc). Consistent with BG45 treatment, c-Myc was markedly downregulated in HDAC3-knockdown MM cells (Figure 1b), without significant alteration of MYC mRNA (Figure S1b). Moreover, c-Myc was significantly decreased in BG45-treated MM.1S cells in the presence of cycloheximide (CHX), further suggesting that downregulation of c-Myc by HDAC3 inhibition is not due to inhibition of transcription (Figure 1c). Importantly, we observed that hyperacetylated c-Myc was detected in MM.1S cells treated with BG45 (Figure 1d). A previous study shows that treatment with non-selective HDAC inhibitors lead to acetylation of c-Myc due to inhibition of deacetylases, resulting in degradation of c-Myc by proteasome.20 Our results therefore suggest HDAC3 inhibition plays an important role in hyperacetylation of c-Myc, followed by its degradation. Taken together, these results suggest that HDAC3 regulates c-Myc protein levels, rather than modulating transcription.
Figure 1. HDAC3 inhibition by gene silencing or by the HDAC3 selective inhibitor BG45 downregulates c-Myc.
(a) MM.1S, RPMI 8226, and NCI-H929 (H929) cells were treated with or without the HDAC3 selective inhibitor BG45 (20 μM) for the indicated time periods. (b) MM.1S, RPMI 8226, and H929 cells were transduced with either HDAC3-specific shRNA (shHDAC3 #1 and #2) or luciferase (shLuc) by lentivirus. (c) MM.1S cells were treated with or without BG45 (20 μM) for 10 hours, further cultured with cycloheximide (CHX) (10 μg/mL), and then harvested at the indicated time courses. Relative expression levels of c-Myc (compared to GAPDH expression) are shown by densitometric analysis using the ImageJ software. The upper baseline ratio defined the condition of CHX 0 min without BG45 treatment as 1, and the lower baseline ratio defined each condition of CHX 0 min as 1. (d) MM.1S cells were treated with or without BG45 (20 μM) for 10 hours. Whole cell lysates were extracted from the treated cells, followed by immunoprecipitation of c-Myc or IgG as a negative control. Acetylated c-Myc (Ac-c-Myc) was detected by immunoblotting with Ac-lysine (K). As input, whole cell lysates were analyzed by immunoblotting using the indicated antibodies. (a – c) Whole cell lysates were extracted from the treated or transduced cells and were subjected to immunoblot analysis. α-tubulin (a, b), GAPDH (c), or c-Myc (d) served as a loading control. Data are representative of at least two independent experiments.
HDAC3 regulates DNMT1
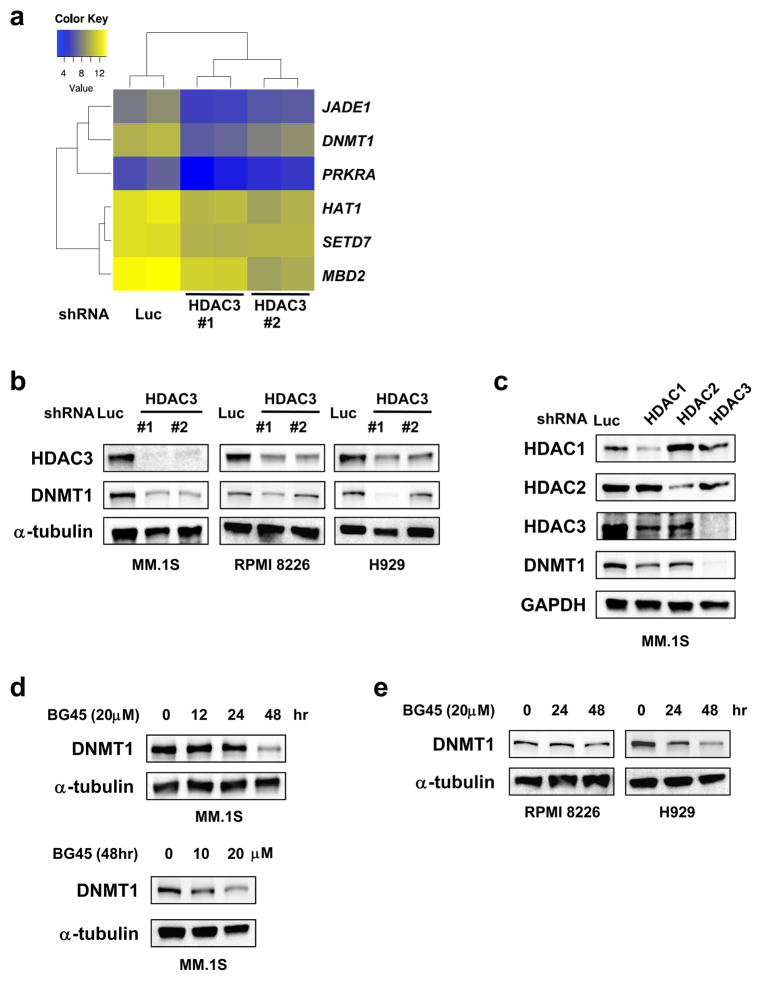
To identify the target molecules of HDAC3, we next carried out gene expression profiling using MM.1S cells transduced with shHDAC3 (#1 and #2) or shLuc. A total of 397 genes were down or up-regulated (adjusted P-value < 0.001, log fold change > 0.5) in HDAC3-knockdown cells compared to control cells (Table S5). We previously reported that HDAC3 knockdown induces apoptosis in MM cells.7 Consistent with this observation, knockdown of HDAC3 markedly decreased expression of XIAP and Bcl-2 members of the inhibitor of apoptosis family of proteins (Table S5 and Figure S2). Since HDAC plays a crucial role in epigenetic modulation, we here focused on epigenetic regulators implicated in tumor pathogenesis.1, 2 Of the 423 epigenetic regulators defined by Gene Ontology, DNMT1 was significantly downregulated in HDAC3-knockdown cells (Figure 2a).
Figure 2. HDAC3 inhibition by gene silencing or by BG45 downregulates DNMT1, which is a critical regulator in MM cells.
(a – c) MM.1S, RPMI 8226, and H929 cells were transduced with HDAC1-specific shRNA, HDAC2-specific shRNA, shHDAC3 (#1, #2), or shLuc. (a) Heatmap of the expression of genes participating in DNA methylation, histone acetylation, and histone methylation in MM.1S cells transduced with either shHDAC3 (#1, #2) or shLuc (control). Genes were differentially expressed relative to control (adjusted P-value <0.001). (d, e) MM.1S, RPMI 8226, and H929 cells were treated with or without BG45 for the indicated time periods and concentrations. (f) MM.1S cells were treated with Merck60, MS-275, or LBH-589 for the indicated time periods and concentrations. (b – f) Whole cell lysates extracted from the transduced or treated cells were subjected to immunoblot analysis. α-tubulin (b, d, e), GAPDH (c, f) served as a loading control. Data are representative of at least two independent experiments. NS, not significant, *P < 0.05, ***P < 0.001 compared with control; Student’s t-test.
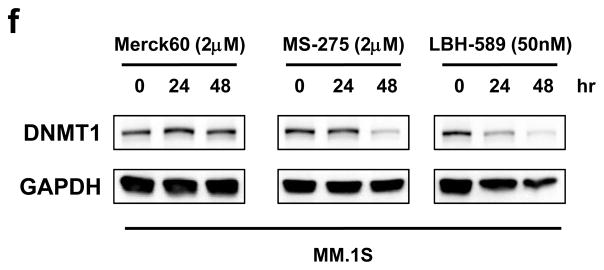
Downregulation of DNMT1 by HDAC3 knockdown was confirmed by quantitative real-time PCR (Q-PCR) (Figure S3a). We further confirmed downregulation of DNMT1 protein expression in HDAC3-knockdown MM.1S, RPMI 8226, and H929 cells with immunoblotting (Figure 2b). Importantly, HDAC3 knockdown, but not HDAC1 or HDAC2 knockdown, significantly downregulated DNMT1 expression (Figure 2c). Consistent with the results of HDAC3-knockdown, DNMT1 was downregulated after BG45 treatment in MM.1S, RPMI 8226, and H929 cells (Figure 2d, 2e, and S3b). DNMT1 expression was also reduced after treatment with non-selective HDAC inhibitor LBH-589, or with HDAC1 and 3 selective inhibitor MS-275, but was not affected by treatment with HDAC1 and 2 selective inhibitor Merck60 (Figure 2f), further confirming that both genetic and pharmacological inhibition of HDAC3 trigger downregulation of DNMT1 expression in MM cells.
We next examined the effect of DNMT1 downregulation on MM cell growth. Growth of MM.1S, RPMI 8226, and H929 MM cell lines transduced with shRNAs targeting DNMT1 (shDNMT1 #1 and #2), but not shLuc, was significantly inhibited, associated with downregulation of DNMT1 expression (Figure S4a). We observed cleavage of caspase-3 and poly (ADP-ribose) polymerase (PARP), suggesting that DNMT1 knockdown induces apoptotic cell death (Figure S4b). Taken together, these results suggest that DNMT1 downregulation mediates, at least in part, HDAC3 inhibition-induced MM cell growth inhibition.
c-Myc regulates DNMT1
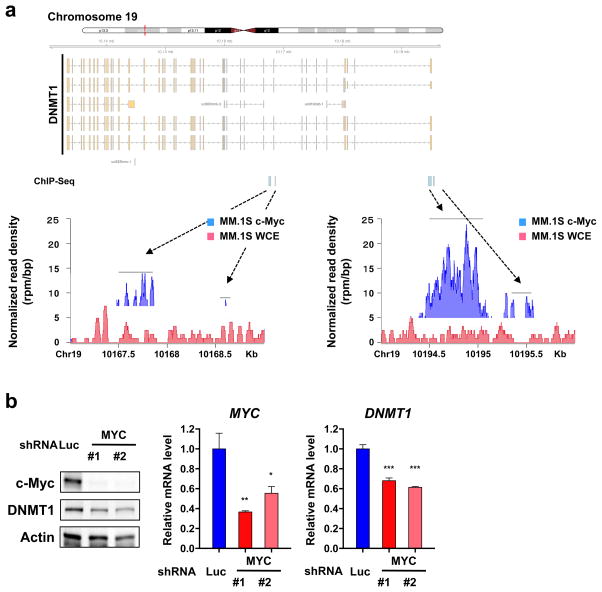
Since HDAC3 inhibition downregulates both c-Myc and DNMT1, we hypothesized that c-Myc may regulate DNMT1 expression. We therefore next analyzed the occupancy of c-Myc at DNMT1 locus using publicly available ChIP-Seq data (GSE36354) (Figure 3a).29 As expected, there was a significant occupancy of c-Myc at DNMT1 locus, and the majority of c-Myc binding sites were localized around intron 1 of DNMT1 (Figure 3a). We next validated ChIP-seq results by ChIP-Q-PCR. Among the four enriched regions at the DNMT1 locus by ChIP-seq, sequences of regions at introns 1 and 9 were recovered at a high level in the c-Myc antibody immunoprecipitate compared to that of control IgG (Figure S5a). Furthermore, these ChIP signals were reduced in MYC-knockdown MM.1S cells (Figure S5a), confirming c-Myc binding at these regions. We further confirmed downregulation of DNMT1 mRNA level and protein expression in MM.1S and RPMI 8226 cells transduced with shRNAs targeting MYC (Figure 3b and S5b). Consistent with these results, there is a positive correlation between expression of MYC and DNMT1 in primary myeloma samples in publicly available data (GSE6477 and GSE19784) (Figure S5c). Taken together, these results indicate that c-Myc transcriptionally regulates DNMT1 in MM cells.
Figure 3. c-Myc regulates DNMT1 in MM cells.
(a) Publicly available ChIP-Seq data (GSE36354) were analyzed for the distribution of c-Myc binding at the DNMT1 locus in MM.1S cells. The x axis shows genomic position. The y axis shows signal strength of c-Myc binding (rpm/bp). WCE; whole cell extract (b) MM.1S cells were transduced with either MYC-specific shRNA (shMYC #1 and #2) or shLuc by lentivirus. Whole cell lysates and total RNAs were extracted from the transduced cells. The lysates were subjected to immunoblot analysis. A served as a loading control. Q-PCR of MYC and DNMT1 was performed. Values represent the amount of mRNA relative to shLuc, defined as 1. Error bars represent s.d. of triplicate measurements. Data are representative of at least two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 compared with control; Student’s t-test.
HDAC3 regulates DNMT1 protein stability through its acetylation
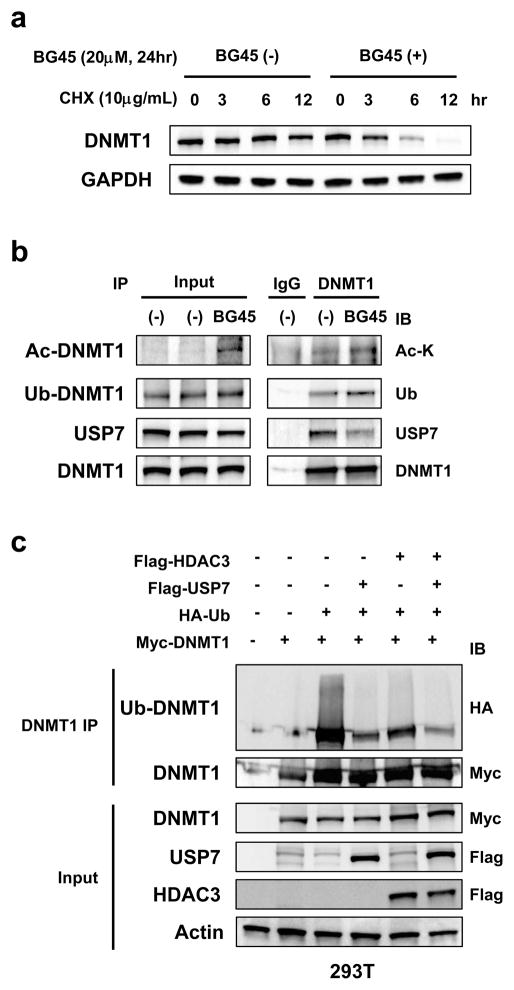
Since previous studies show that protein stability of DNMT1 is regulated through its acetylation and subsequent ubiquitination, 27, 28 we next examined whether HDAC3 inhibition triggers acetylation and regulates protein stability of DNMT1. We first showed that protein stability of DNMT1 in MM.1S cells was significantly decreased after treatment with BG45 in the presence of cycloheximide (CHX) (Figure 4a). Since previous studies have shown that DNMT1 forms a complex with HDAC1 or USP7,26–28 we next examined whether HDAC3 physically interacts with DNMT1. Importantly, we observed that DNMT1 co-immunoprecipitated not only with USP7, but also with HDAC3 (Figure S6), suggesting that HDAC3, DNMT1, and USP7 form a complex. We also assessed whether HDAC3 inhibition modulated this complex formation: acetylation and ubiquitination of DNMT1 was increased in BG45-treated MM.1S cells, associated with decreased binding between DNMT1 and USP7 (Figure 4b).
Figure 4. HDAC3 inhibition reduces the protein stability of DNMT1.
(a) MM.1S cells were treated with or without BG45 (20 μM) for 24 hours, further cultured with CHX (10 μg/mL), and then harvested at the indicated time intervals. The cell lysates were subjected to immunoblot analysis. (b) MM.1S cells were treated with or without BG45 (20 μM) for 24 hours; whole cell lysates were then extracted, followed by immunoprecipitation of DNMT1 or IgG as a negative control. Acetylated DNMT1 (Ac-DNMT1), ubiquitinated DNMT1 (Ub-DNMT1), and USP7 were detected by immunoblotting with Ac-K, ubiquitin, and USP7 antibody, respectively. DNMT1 served as a loading control. (c, d) 293T cells were transfected with indicated Myc-DNMT1 (WT), acetylation-dead Myc-DNMT1 (4KQ), HA-ubiquitin (Ub), Flag-USP7, and/or Flag-HDAC3 for 48 hours. In vivo ubiquitination of DNMT1 proteins was assessed by immunoprecipitation. Polyubiquitination was detected by immunoblotting with HA antibody. (b – d) As input, the whole cell lysates were analyzed by immunoblotting using the indicated antibodies. GAPDH (a), DNMT1 (b), or Actin (c, d) served as a loading control. Data are representative of at least two independent experiments. (e) Schema of regulation of DNMT1 expression by HDAC3 in MM cells. HDAC3 deacetylates c-Myc, and maintains the protein stability of c-Myc. c-Myc regulates DNMT1 mRNA expression. HDAC3 deacetylates DNMT1, and maintains the protein stability of DNMT1.
We further examined the effect of HDAC3 on ubiquitination of DNMT1 using a deubiquitination assay in 293T cells. Importantly, both HDAC3 and USP7 decreased DNMT1 ubiquitination (Figure 4c, DNMT1 IP). In contrast, DNMT1 protein level was increased in the presence of HDAC3 and USP7 (Figure 4c Input). A previous report showed that acetylation of Lysine (K1111/K1113/K1115/K1117) is required for ubiquitination of DNMT1.28 Consistent with this report, mutations of these acetylation sites abrogated ubiquitination of DNMT1 (Figure 4d). These results suggest that HDAC3 promotes stability of DNMT1, whereas HDAC3 inhibition induces DNMT1 ubiquitination, followed by its degradation by proteasomes.
Taken together, HDAC3 modulates DNMT1 mRNA expression through the regulation of c-Myc protein stability. Furthermore, HDAC3 also deacetylates and stabilizes DNMT1 protein. Our results indicate that HDAC3 regulates DNMT1 expression by modulating transcription and protein stability (Figure 4e).
Pharmaceutical inhibition of HDAC3 and DNMT1 synergistically inhibits MM cell growth in vitro as well as in vivo
5-Azacytidine (AZA) is used as a DNMT1 inhibitor in the treatment of acute myeloid leukemia and myelodysplastic syndrome.38 Since previous reports have also shown anti-MM activity of AZA,39, 40 we examined whether HDAC3 inhibitor with AZA induces synergistic DNMT1 inhibition and MM cell growth inhibition. We first evaluated the effect of AZA on DNMT1 protein expression in MM cells. Consistent with the previous reports, AZA downregulated DNMT1 expression in dose- and time-dependent manners (Figure S7a). Importantly, BG45 with AZA markedly reduced DNMT1 in MM.1S and RPMI8226 cells (Figure S7b), associated with synergistic cell growth inhibition associated with increased cleavage of caspase-3, −8, −9 and PARP (Figure S7c and S7d). Similar results were observed in primary tumor cells from MM patients (Figure S7e and S7f). Taken together, these results suggest that cell growth inhibition induced by combination treatment is due to apoptotic cell death.
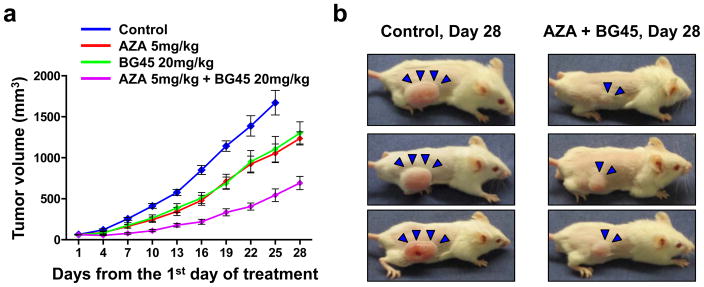
We next evaluated this combination treatment in vivo using a murine xenograft model of human MM. At day 25 of treatment, significant tumor growth inhibition was observed in mice treated with BG45 or AZA alone compared to the vehicle control group (P = 0.021 and P = 0.005, respectively) (Figure 5a). Importantly, BG45 with AZA enhanced tumor growth inhibition compared to monotherapy cohorts (P = 0.001 in AZA versus the combination, and P = 0.003 in BG45 versus the combination), without significant body weight loss (Figure S8a). Representative images of tumor growth inhibition by the combination treatment are shown in Figure 5b. The median OS of the combination treated cohort (45 days) was significantly prolonged compared to vehicle or monotherapy treated cohorts (P = 0.0002 in vehicle versus the combination, P = 0.0006 in AZA versus the combination, and P = 0.0002 in BG45 versus the combination) (Figure S8b). Consistent with our in vitro experiments, c-Myc and DNMT1 was decreased in tumor harvested from the BG45-treated or combination treated cohort (Figure S8c and S8d). These results indicate that AZA and BG45 in combination enhanced in vivo cell growth inhibition through targeting DNMT1.
Figure 5. AZA and BG45 in combination inhibit human MM cell growth in vivo through the downregulation of DNMT1.
(a, b) After development of measurable tumors (> 50 mm3), cohorts were treated for 3 weeks with vehicle control (n= 9; blue line), 5mg/kg AZA once a week (n= 9; red line), 20 mg/kg BG45 5 days a week (n= 9; green line), or 20 mg/kg BG45 5 days a week with 5 mg/kg AZA once a week (n= 9; purple line). Data represent mean ± s.e.m. (b) Images show representative whole-body images from vehicle control (left panel) and AZA and BG45 combination group (right panel) after 28 days treatment. Arrows indicate tumors.
Discussion
HDACs have numerous target molecules, which are not only histones but also non-histone proteins including transcription factors.17, 18 We have previously reported that HDAC3 targets STAT3, and that inhibition of HDAC3 dephospholylates STAT3, thereby reducing MM cell growth.7 In this study, we further examined the mechanisms whereby HDAC3 inhibition abrogates MM cell growth, and show that HDAC3 targets c-Myc and DNMT1. c-Myc is a potent oncoprotein which has a critical role in progression of MGUS to MM.13–15 Previous reports show that HDAC inhibitors downregulate c-Myc expression22, 23; however, the effect of HDAC isoform-selective inhibition on c-Myc modulation has not yet been elucidated. Here we show that HDAC3 inhibition triggers downregulation of c-Myc protein level, without alteration of mRNA level.
Previous reports have shown that HDAC inhibitors target DNMT1 in various cancer cell types27, 28, 30; however, which HDAC isoform (s) regulate DNMT1 expression has not been defined. In addition, prior studies suggest that HDACs regulate protein stability, but not transcription, of DNMT1.27, 28, 30 In this study, our microarray analysis and Q-PCR showed that HDAC3 regulates DNMT1 mRNA expression in MM cells. Importantly, we demonstrate that c-Myc regulates DNMT1 expression, confirming that HDAC3 regulates DNMT1 mRNA expression via its effects on protein stability of c-Myc. Of note, DNMT1 expression is not solely mediated by c-Myc and/or HDAC3. Indeed, DNMT1 expression can be regulated by other signaling pathways. For example, it is also modulated via Rb/E2F or p53/SP1 cascade at the transcriptional level.31, 32 We therefore carried out knockdown of E2F1 and SP1, but noted no significant change in DNMT1 protein expression in MM.1S cells (Figure S9). These results suggest that c-Myc is a key factor maintaining DNMT1 expression in MM cells.
DNMT1 protein expression is regulated by its protein stability via its acetylation and ubiquitination status.27, 28, 33 For example, USP7 has been shown to deubiquitinate DNMT1,27, 28 and a recent study by Cheng and colleagues demonstrates that protein stability of DNMT1 is mediated by binding of USP7, a physical interaction regulated by DNMT1 acetylation.28 However, this study did not identify which HDAC isoform impacts DNMT1 deacetylation. In our study, we demonstrate that HDAC3 is associated with deacetylation of DNMT1, followed by decreased ubiquitination and degradation of DNMT1, indicating that HDAC3 regulates DNMT1 protein stability via its deacetylation.
A recent study has shown that DNMT1 is an essential factor not only for mammary, but also cancer stem cell maintenance and tumorigenesis.34 Our current study demonstrated that DNMT1 is also essential for MM cell survival. To reveal the downstream targets of DNMT1 in MM cells, we performed gene expression profiling after knockdown of DNMT1 in MM.1S cells (Table S7). Gene set enrichment analysis revealed that genes encoding regulators of cell cycle were significantly reduced after knockdown of DNMT1 (Figure S10a). We then confirmed that knockdown of DNMT1 leads to S-phase cell cycle arrest in MM.1S cells (Figure S10b). These data suggest that DNMT1 is an important cell cycle regulator in MM cells.
Class I or II HDACs are attractive therapeutic targets in MM. In particular, a large number of HDAC inhibitors target mainly class I HDACs (especially HDAC1, 2, 3) and IIb HDACs (HDAC6).35 Our previous report validates genetic knockdown or pharmacologic inhibition of HDAC3 as a novel potential therapeutic strategy in MM.7 Although non-selective HDAC inhibitors induce anti-MM activity, they also have attendant unfavorable side effects.5, 6 Moreover, HDAC inhibitor monotherapy induces only modest anti-MM clinical activity.36, 37 To reduce unfavorable toxicities and augment the anti-tumor effect of HDAC inhibitors, we therefore need to develop more tolerable HDAC inhibitors that can be used in combination. Our previous study demonstrated that the combination of BG45 and a proteasome inhibitor bortezomib induces synergistic cytotoxicity against MM cells.7 Here we show that knockdown of HDAC3, as well as HDAC3 inhibition by BG45, markedly decreased the expression of members of the inhibitor of apoptosis family of proteins XIAP and Bcl-2 (Figure S2). Thus, the combination of BG45 with bortezomib may synergistically augment apoptotic signaling pathways. Regarding DNMT1, previous studies show that the combination of non-selective HDAC inhibitors with DNMT inhibitors are effective treatment of cancers.38, 39 Importantly, our study shows that BG45 in combination with AZA markedly downregulates DNMT1, triggers synergistic anti-MM activity in vitro, as well as significantly inhibits in vivo tumor cell growth in a murine xenograft mouse model of human MM.
Taken together, we demonstrate that HDAC3 inhibition reduces DNMT1 expression via regulation of mRNA and protein levels; and importantly, that HDAC3 inhibitor and DNMT1 inhibitor induces synergistic anti-MM activity. Our results therefore provide the rationale for combination treatment with HDAC3 inhibitor and DNMT1 inhibitor to improve patient outcome in MM.
Supplementary Material
Acknowledgments
This study was supported by the National Institute of Health Grants SPORE-P50100707 (KCA), P01-CA078378 (KCA), R01-CA050947 (K.C.A.), and R01-CA178264 (TH and KCA). K.C.A. is an American Cancer Society Clinical Research Professor.
Footnotes
Conflict of interest
All of the authors have no potential conflicts of interest.
Authorship contributions
T.H., H.O., and T.H. designed the research. T.H. performed experiments. Y.G performed the bioinformatics analysis. R.M. synthesized BG45. T.H., H.O., S.K, M.S., T.H, and K.C.A analyzed the data. Y.T and K.C.A. provided MM patient samples. H.O., T.H., and K.C.A. supervised the study. T.H., H.O., Y.G., T.H. and K.C.A. wrote the manuscript.
Supplementary information accompanies at this paper on the Leukemia website (http://nature.com/leu)
References
- 1.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012 Jul 6;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nature reviews Drug discovery. 2012 May;11(5):384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 3.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nature reviews Drug discovery. 2014 Sep;13(9):673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 4.Harada T, Hideshima T, Anderson KC. Histone deacetylase inhibitors in multiple myeloma: from bench to bedside. International journal of hematology. 2016 Sep;104(3):300–309. doi: 10.1007/s12185-016-2008-0. [DOI] [PubMed] [Google Scholar]
- 5.Dimopoulos M, Siegel DS, Lonial S, Qi J, Hajek R, Facon T, et al. Vorinostat or placebo in combination with bortezomib in patients with multiple myeloma (VANTAGE 088): a multicentre, randomised, double-blind study. The lancet oncology. 2013 Oct;14(11):1129–1140. doi: 10.1016/S1470-2045(13)70398-X. [DOI] [PubMed] [Google Scholar]
- 6.San-Miguel JF, Hungria VT, Yoon SS, Beksac M, Dimopoulos MA, Elghandour A, et al. Panobinostat plus bortezomib and dexamethasone versus placebo plus bortezomib and dexamethasone in patients with relapsed or relapsed and refractory multiple myeloma: a multicentre, randomised, double-blind phase 3 trial. The lancet oncology. 2014 Oct;15(11):1195–1206. doi: 10.1016/S1470-2045(14)70440-1. [DOI] [PubMed] [Google Scholar]
- 7.Minami J, Suzuki R, Mazitschek R, Gorgun G, Ghosh B, Cirstea D, et al. Histone deacetylase 3 as a novel therapeutic target in multiple myeloma. Leukemia. 2014 Mar;28(3):680–689. doi: 10.1038/leu.2013.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dang CV. MYC on the path to cancer. Cell. 2012 Mar 30;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, Metabolism, and Cancer. Cancer discovery. 2015 Oct;5(10):1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palumbo A, Anderson K. Multiple myeloma. The New England journal of medicine. 2011 Mar 17;364(11):1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 11.Landgren O, Kyle RA, Pfeiffer RM, Katzmann JA, Caporaso NE, Hayes RB, et al. Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study. Blood. 2009 May 28;113(22):5412–5417. doi: 10.1182/blood-2008-12-194241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiss BM, Abadie J, Verma P, Howard RS, Kuehl WM. A monoclonal gammopathy precedes multiple myeloma in most patients. Blood. 2009 May 28;113(22):5418–5422. doi: 10.1182/blood-2008-12-195008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chesi M, Robbiani DF, Sebag M, Chng WJ, Affer M, Tiedemann R, et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer cell. 2008 Feb;13(2):167–180. doi: 10.1016/j.ccr.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anguiano A, Tuchman SA, Acharya C, Salter K, Gasparetto C, Zhan F, et al. Gene expression profiles of tumor biology provide a novel approach to prognosis and may guide the selection of therapeutic targets in multiple myeloma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009 Sep 1;27(25):4197–4203. doi: 10.1200/JCO.2008.19.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chng WJ, Huang GF, Chung TH, Ng SB, Gonzalez-Paz N, Troska-Price T, et al. Clinical and biological implications of MYC activation: a common difference between MGUS and newly diagnosed multiple myeloma. Leukemia. 2011 Jun;25(6):1026–1035. doi: 10.1038/leu.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holien T, Vatsveen TK, Hella H, Waage A, Sundan A. Addiction to c-MYC in multiple myeloma. Blood. 2012 Sep 20;120(12):2450–2453. doi: 10.1182/blood-2011-08-371567. [DOI] [PubMed] [Google Scholar]
- 17.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science (New York, NY) 2009 Aug 14;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 18.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. The Journal of clinical investigation. 2014 Jan 2;124(1):30–39. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gregory MA, Hann SR. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt’s lymphoma cells. Molecular and cellular biology. 2000 Apr;20(7):2423–2435. doi: 10.1128/mcb.20.7.2423-2435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faiola F, Liu X, Lo S, Pan S, Zhang K, Lymar E, et al. Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Molecular and cellular biology. 2005 Dec;25(23):10220–10234. doi: 10.1128/MCB.25.23.10220-10234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, He X, Liu L, Jiang M, Zhao C, Wang H, et al. Hdac3 Interaction with p300 Histone Acetyltransferase Regulates the Oligodendrocyte and Astrocyte Lineage Fate Switch. Developmental cell. 2016 Feb 8;36(3):316–330. doi: 10.1016/j.devcel.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hideshima T, Cottini F, Ohguchi H, Jakubikova J, Gorgun G, Mimura N, et al. Rational combination treatment with histone deacetylase inhibitors and immunomodulatory drugs in multiple myeloma. Blood cancer journal. 2015;5:e312. doi: 10.1038/bcj.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angela N, Carafa V, Conte M, Tambaro FP, Abbondanza C, Martens JH, et al. c-Myc modulation & acetylation is a key HDAC inhibitor target in cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016 Jun 29; doi: 10.1158/1078-0432.CCR-15-2388. [DOI] [PubMed] [Google Scholar]
- 24.Yang L, Rau R, Goodell MA. DNMT3A in haematological malignancies. Nature reviews Cancer. 2015 Mar;15(3):152–165. doi: 10.1038/nrc3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, et al. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nature genetics. 2007 Mar;39(3):391–396. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- 26.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nature genetics. 2000 Jul;25(3):338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 27.Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Science signaling. 2010;3(146):ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng J, Yang H, Fang J, Ma L, Gong R, Wang P, et al. Molecular mechanism for USP7-mediated DNMT1 stabilization by acetylation. Nature communications. 2015;6:7023. doi: 10.1038/ncomms8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012 Sep 28;151(1):56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Q, Agoston AT, Atadja P, Nelson WG, Davidson NE. Inhibition of histone deacetylases promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells. Molecular cancer research : MCR. 2008 May;6(5):873–883. doi: 10.1158/1541-7786.MCR-07-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura H, Nakamura T, Ogawa T, Tanaka S, Shiota K. Transcription of mouse DNA methyltransferase 1 (Dnmt1) is regulated by both E2F-Rb-HDAC-dependent and -independent pathways. Nucleic acids research. 2003 Jun 15;31(12):3101–3113. doi: 10.1093/nar/gkg406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin RK, Wu CY, Chang JW, Juan LJ, Hsu HS, Chen CY, et al. Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer research. 2010 Jul 15;70(14):5807–5817. doi: 10.1158/0008-5472.CAN-09-4161. [DOI] [PubMed] [Google Scholar]
- 33.Denis H, Ndlovu MN, Fuks F. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO reports. 2011 Jul;12(7):647–656. doi: 10.1038/embor.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pathania R, Ramachandran S, Elangovan S, Padia R, Yang P, Cinghu S, et al. DNMT1 is essential for mammary and cancer stem cell maintenance and tumorigenesis. Nature communications. 2015;6:6910. doi: 10.1038/ncomms7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, et al. Chemical phylogenetics of histone deacetylases. Nature chemical biology. 2010 Mar;6(3):238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richardson P, Mitsiades C, Colson K, Reilly E, McBride L, Chiao J, et al. Phase I trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) in patients with advanced multiple myeloma. Leukemia & lymphoma. 2008 Mar;49(3):502–507. doi: 10.1080/10428190701817258. [DOI] [PubMed] [Google Scholar]
- 37.Wolf JL, Siegel D, Goldschmidt H, Hazell K, Bourquelot PM, Bengoudifa BR, et al. Phase II trial of the pan-deacetylase inhibitor panobinostat as a single agent in advanced relapsed/refractory multiple myeloma. Leukemia & lymphoma. 2012 Sep;53(9):1820–1823. doi: 10.3109/10428194.2012.661175. [DOI] [PubMed] [Google Scholar]
- 38.Kalac M, Scotto L, Marchi E, Amengual J, Seshan VE, Bhagat G, et al. HDAC inhibitors and decitabine are highly synergistic and associated with unique gene-expression and epigenetic profiles in models of DLBCL. Blood. 2011 Nov 17;118(20):5506–5516. doi: 10.1182/blood-2011-02-336891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pathania R, Ramachandran S, Mariappan G, Thakur P, Shi H, Choi JH, et al. Combined Inhibition of DNMT and HDAC Blocks the Tumorigenicity of Cancer Stem-like Cells and Attenuates Mammary Tumor Growth. Cancer research. 2016 Jun 1;76(11):3224–3235. doi: 10.1158/0008-5472.CAN-15-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silverman LR, McKenzie DR, Peterson BL, Holland JF, Backstrom JT, Beach CL, et al. Further analysis of trials with azacitidine in patients with myelodysplastic syndrome: studies 8421, 8921, and 9221 by the Cancer and Leukemia Group B. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006 Aug 20;24(24):3895–3903. doi: 10.1200/JCO.2005.05.4346. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.