Abstract
Pm21, originating from wheat wild relative Dasypyrum villosum, confers immunity to all known races of Blumeria graminis f. sp. tritici (Bgt) and has been widely utilized in wheat breeding. However, little is known on the genetic basis of the Pm21 locus. In the present study, four seedling-susceptible D. villosum lines (DvSus-1 ∼ DvSus-4) were identified from different natural populations. Based on the collinearity among genomes of Brachypodium distachyon, Oryza, and Triticeae, a set of 25 gene-derived markers were developed declaring the polymorphisms between DvRes-1 carrying Pm21 and DvSus-1. Fine genetic mapping of Pm21 was conducted by using an extremely large F2 segregation population derived from the cross DvSus-1/DvRes-1. Then Pm21 was narrowed to a 0.01-cM genetic interval defined by the markers 6VS-08.4b and 6VS-10b. Three DNA markers, including a resistance gene analog marker, were confirmed to co-segregate with Pm21. Moreover, based on the susceptible deletion line Y18-S6 induced by ethyl methanesulfonate treatment conducted on Yangmai 18, Pm21 was physically mapped into a similar interval. Comparative analysis revealed that the orthologous regions of the interval carrying Pm21 were narrowed to a 112.5 kb genomic region harboring 18 genes in Brachypodium, and a 23.2 kb region harboring two genes in rice, respectively. This study provides a high-density integrated map of the Pm21 locus, which will contribute to map-based cloning of Pm21.
Keywords: Dasypyrum villosum, Pm21, powdery mildew resistance, genetic mapping, physical mapping, comparative mapping
Introduction
Common wheat (Triticum aestivum L.) is the most widely grown cereal crop occupying ∼17% of all cultivated land of the world and provides ∼20% of the calories consumed by humankind (Fu et al., 2009). However, wheat production is seriously threatened by various diseases, such as head scab, rusts, and powdery mildew. Wheat powdery mildew caused by the obligate biotrophic fungal pathogen Blumeria graminis f. sp. tritici (Bgt) is one of the most important factors leading to yield losses. Development of resistant varieties using powdery mildew resistance (Pm) genes is an effective, economical and environmental-friendly way to reduce yield losses caused by Bgt. Up to now, 58 resistant genes have been formally designated (Pm1 ∼Pm58). Among them, some Pm genes were identified from the species in the tertiary gene pool, including Pm7, Pm8, Pm17, and Pm20 from Secale cereal, Pm40 and Pm43 from Thinopyrum intermedium, and Pm21 and Pm55 from Dasypyrum villosum (McIntosh et al., 2013; Wiersma et al., 2017).
Dasypyrum villosum Candargy (2n = 14, VV), a diploid wild relative of common wheat, has been an important resource for wheat improvement which provides resistance to multiple wheat diseases, such as powdery mildew, rusts, and eyespot (Chen et al., 2013). The powdery mildew resistance gene Pm21, located on the short arm of chromosome 6V (6VS) of D. villosum, confers immunity to all known Bgt races (Chen et al., 1995; Cao et al., 2011). After emergence of highly virulent Bgt isolates, such as Bgt YZ01 (He et al., 2016), wheat varieties carrying Pm2a or Pm4a broadly planted are gradually losing their resistance in the middle and lower reaches of the Yangtze River Valley, the most rampant area of powdery mildew in China. As an important replacement, varieties carrying the translocated chromosome T6AL.6VS are being planted more widely in this region (Bie et al., 2015). In the other major wheat regions in China, the number of the varieties integrating Pm21 is also growing fast. To better utilize Pm21, clarification of the genetic basis and functional mechanism of Pm21 becomes very important and urgent.
Previously, Pm21 has been physically mapped to the bin 6VS FL0.45–0.58 by using the resistant deletion line del.6VS-1 (FL0.58) and the susceptible deletion line del.6VS-2 (FL0.45) (Cao et al., 2011). Recently, several candidate genes located in the bin FL0.45–0.58 were found and verified to be required by Pm21-mediated resistance to powdery mildew (Cao et al., 2011; He et al., 2016). However, the exact genetic relationships of these genes and Pm21 remain unclear. To date, there are still two major obstacles for genetic mapping of Pm21. First, D. villosum chromosome 6VS cannot recombine with homoeologous wheat chromosomes. Second, there is neither natural nor artificial induced susceptible mutant of D. villosum that could be used for genetic mapping.
To fulfill genetic mapping of Pm21 at the diploid level, an attempt was made to obtain susceptible D. villosum line(s). Fortunately, by screening more than 100 D. villosum accessions collected from different germplasm resources organizations, we found four D. villosum accessions susceptible to wheat powdery mildew. Using an F2 population derived from the cross between a resistant D. villosum line and a susceptible one, fine genetic mapping of Pm21 was initiated. In combination with physical and comparative mapping, a high-density integrated map of Pm21 was successfully constructed. This study will contribute to map-based cloning of Pm21 and understanding of its functional mechanism of broad-spectrum resistance.
Materials and Methods
Plant Materials
A collection of 110 accessions of D. villosum were kindly provided by Germplasm Resources Information Network (GRIN) (51), GRIN Czech (16), Genebank Information System of the IPK Gatersleben (GBIS-IPK) (35), Nordic Genetic Resource Center (NordGen) (7), and the Cytogenetics Institute, Nanjing Agricultural University (CI-NAU) (1). The susceptible D. villosum line DvSus-1 was crossed with the resistant line DvRes-1 carrying Pm21 (Table 1), and the generated F2 population containing 10,536 individuals was used for genetic analysis. The powdery mildew resistant wheat variety Yangmai 18, carrying a pair of translocated chromosomes T6AL.6VS, and the susceptible variety Yangmai 9 were both developed in Yangzhou Academy of Agricultural Sciences (YAAS). About 2,000 seeds of Yangmai 18 were treated with 0.8% ethyl methanesulfonate (EMS), and 1,216 M2 families were generated to use for screening of mutants susceptible to powdery mildew.
Table 1.
Origin of Dasypyrum villosum lines used in this study.
| Line | Powdery mildew response | Origin | Original accession | Provider |
|---|---|---|---|---|
| DvRes-1 | Resistant | United Kingdom | Unknown | CI-NAU |
| DvSus-1 | Susceptible | Greece | GRA 2738 | GBIS-IPK |
| DvSus-2 | Susceptible | Unknown | GRA 962 | GBIS-IPK |
| DvSus-3 | Susceptible | Italy | GRA 1105 | GBIS-IPK |
| DvSus-4 | Susceptible | Former Soviet Union | PI 598390 | GRIN |
CI-NAU, the Cytogenetics Institute, Nanjing Agricultural University; GBIS-IPK, Genebank Information System of the IPK Gatersleben; GRIN, Germplasm Resources Information Network.
Evaluation of Powdery Mildew Resistance
The D. villosum accessions, F1 and F2 individuals of the cross DvSus-1/DvRes-1, and M2 individuals of EMS-induced Yangmai 18 at one-leaf stage were inoculated with Bgt isolate YZ01, a predominant race collected from Yangzhou (He et al., 2016), by dusting from sporulating susceptible variety Yangmai 9 and powdery mildew responses were assessed at 7 days post-inoculation in greenhouse. Due to that Pm21 confers immunity to Bgt isolate YZ01, powdery mildew responses can be simply divided into two types, resistant and susceptible.
Development of DNA Markers
Polymorphic DNA markers between the resistant line DvRes-1 and the susceptible line DvSus-1 were developed using the CISP-IS (conserved intron scanning primer combined with intron sequencing) strategy based on the collinearity relationship between Brachypodium, rice and Triticeae species (He et al., 2013). The common genes between Brachypodium and rice were used to search their homologous full-length cDNAs or ESTs of wheat, barley, Aegilops or other Triticeae crops deposited in GenBank database. After alignment of these sequences by the Clustalw tool, primers were designed according to the conserved extron sequences and used to amplify the corresponding intronic regions of the parents DvRes-1 and DvSus-1. After T/A cloning and sequencing of the target fragments, the flanking conserved sequences of the variant introns were further used to design primers and screen polymorphic markers. DNA markers used for physical mapping were also developed using the same strategy. The details of polymorphic markers used in this study were listed in Supplementary Tables S1, S2.
Genomic DNA Isolation and Marker Analysis
Genomic DNA with high-quality was isolated from fresh leaves of the seedlings by the CTAB method (Murray and Thompson, 1980). For high-throughput marker analysis, a simple and rapid boiling method for crude DNA extraction was newly developed in this study. A leaf segment (about 1 cm long) was grinded by glass rod in a 1.5-ml tube. Then, added 150 μl of DNA protection solution (50 mM Tris–HCl, 25 mM EDTA, pH 8.0) into the tube and boiled at 90°C for 10 min. After centrifugation at 10,000 rpm for 1 min, the supernatant was directly used as DNA template for PCR amplification. PCR amplification was performed in a Peltier thermal cycler (Bio-Rad, United States) in 25 μl volume containing 1 × PCR buffer (Mg2+ free), 2.7 mM of MgCl2, 0.2 mM of each dNTP, 2 μM of each primer, 1 unit of Taq DNA polymerase, and 1 μl of DNA template. PCR was carried out with an initial denaturation at 94°C for 3 min, 35 cycles of 20 s at 94°C, 30 s at 60°C, 1 min at 72°C, and a final extension for 5 min at 72°C. PCR products were separated in 6% ∼ 12% non-denaturing polyacrylamide gels, silver stained, and photographed.
Genetic and Deletion Mapping of Pm21
Genetic analysis of Pm21 was performed on an F2 population derived from the cross between the resistant line DvRes-1 carrying Pm21 and the seedling-susceptible line DvSus-1 of D. villosum. Chi-squared (χ2) test was used to determine the goodness-of-fit of the observed segregation ratio to theoretical Mendelian ratio. Chromosomal deletion analysis was carried out using the susceptible mutant line Y18-S6 obtained from an EMS-induced Yangmai 18 population.
Comparative Genomics Analysis
The genome sequences of Brachypodium, rice and wheat were obtained from the Brachypodium distachyon genome assemblies v2.01, the rice genome pseudomolecule release 72, and the IWGSC Sequence Repository3, respectively. Genes were predicted using the FGENESH tool4 and re-annotated using the BLAST program5 and the SMART program6.
Results
Reactions of D. villosum Accessions to Bgt Isolate YZ01
A total of 110 D. villosum accessions were collected and inoculated with Bgt YZ01 at one-leaf stage. Fortunately, susceptible individuals were identified from four different accessions, and all the other accessions were immune. The susceptible lines obtained were designated as DvSus-1, DvSus-2, DvSus-3, and DvSus-4, respectively (Table 1). At different growth stages, powdery mildew responses of the four susceptible D. villosum lines were further investigated. All individuals were susceptible at one-leaf stage, but interestingly, an unknown complex resistance gradually increased at two-leaf stage, and fully expressed from three-leaf stage to adult stage (Figures 1A–C).
FIGURE 1.
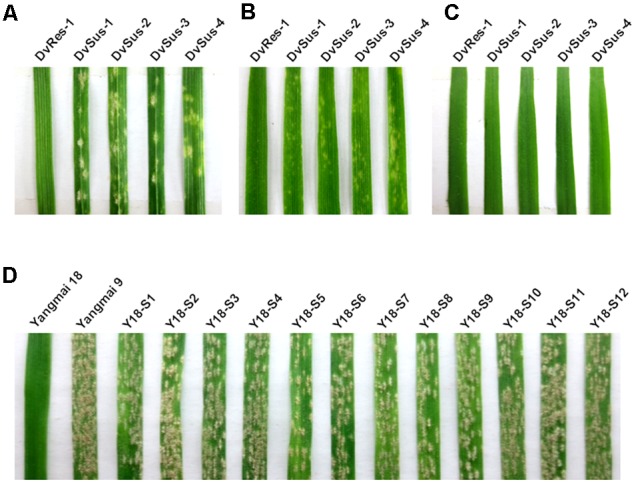
Powdery mildew responses of different Dasypyrum villosum lines and Yangmai 18 mutants. (A–C) Responses of the resistant D. villosum line DvRes-1 and four susceptible lines (DvSus-1 ∼ DvSus-4) to Bgt isolate YZ01 at one-leaf stage (A), two-leaf stage (B) and three-leaf stage (C), respectively. (D) Responses of 12 independent Yangmai 18 mutants (Y18-S1 ∼ Y18-S12) to Bgt isolate YZ01 at two-leaf stage, using Yangmai 18 and Yangmai 9 as the resistant and susceptible controls, respectively.
Development of Polymorphic Markers at Different Ploidy Levels
To develop polymorphic markers between the resistant line DvRes-1 and the susceptible line DvSus-1, a total of 54 6VS-specific markers reported previously (Qi et al., 2010; Chen et al., 2013; He et al., 2016) were tested. Among them, however, only four markers, Xcfe164, 6VS-11, 6VS-25, and 6VS-30, showed polymorphisms between DvRes-1 and DvSus-1. To get more markers for mapping Pm21, the CISP-IS strategy based on comparative genomics were applied and 21 polymorphic markers were further developed (Supplementary Table S1), among which, six were single nucleotide polymorphism (SNP) markers. Taken together, a total of 25 markers were recruited for genetic mapping of Pm21 in diploid D. villosum. Using the CISP-IS strategy, another seven 6VS-specific markers were newly developed here and used for physical mapping of Pm21 in hexaploid wheat (Supplementary Table S2).
Fine Genetic Mapping of Pm21
For genetic mapping of Pm21 gene, a total of 10,536 F2 individuals derived from the cross between the resistant DvRes-1 and the susceptible DvSus-1 were evaluated for their resistance to powdery mildew at one-leaf stage. The result showed that 8,147 were resistant while 2,389 were susceptible, resulting in a resistance-to-susceptible ratio as 3.41:1 that is significantly higher than 3:1, the theoretical Mendelian segregation ratio (χ2 = 30.500, P < 0.01). Genotyping of F2 individuals using the co-segregated marker 6VS-09.4b indicated that the ratio of 2,492 resistant homozygotes, 5,655 heterozygotes, and 2,389 susceptible homozygotes is 1.04:2.37:1, not in accordance with a 1:2:1 ratio (χ2 = 59.066, P < 0.01).
To accelerate screening for recombinants, a double-PCR system was conducted using two markers 6VS-00.1 and Xcfe164, respectively, located on the distal and paracentric regions of 6VS (Figure 2). Sixty-four recombinants were identified from the F2 population, indicating that the total genetic distance between the two markers was approximately 0.30 cM. Subsequently, a set of 25 markers were used to genotyping these recombinants. In combination with powdery mildew resistance evaluation, Pm21 was finely mapped into a 0.01-cM interval defined by the markers 6VS-08.4b and 6VS-10b (Figure 3A). Furthermore, three genic markers (6VS-08.8b, 6VS-09b and 6VS-09.4b) were found to co-segregate with Pm21 (Figure 4).
FIGURE 2.
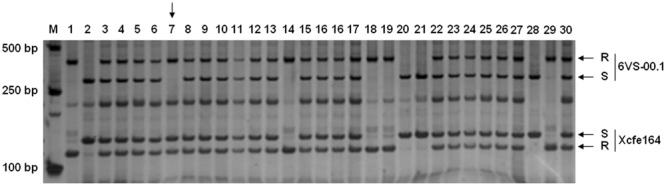
PCR amplification patterns in the F2 population using a double-PCR system with the markers 6VS-00.1 and Xcfe164. M: DNA marker DL2000; 1: the resistant parent DvRes-1; 2: the susceptible parent DvSus-1; 3–30: different F2 plants. R and S represent the DNA bands that were same to the resistant and susceptible parents, respectively. The vertical arrow indicates the recombinant individual.
FIGURE 3.
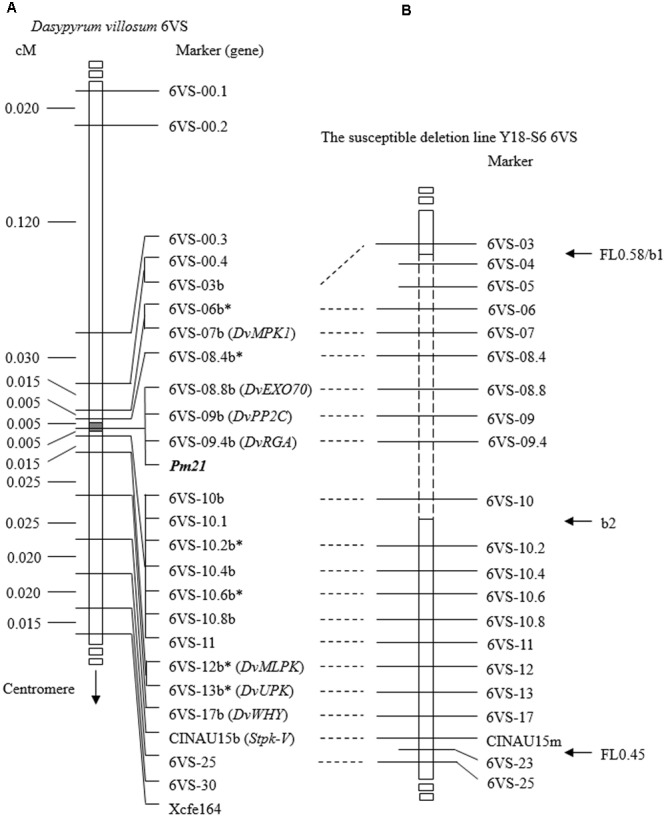
Genetic and physical mapping of Pm21. (A) Genetic map of Pm21 and 6VS markers obtained by using the F2 population derived from the cross between the resistant D. villosum DvRes-1 and the susceptible DvSus-1. The gray block on the chromosome 6VS denotes the genetic interval carrying Pm21. All markers used here were derived from genes based on comparative genomics among wheat crops and Brachypodium and designated according to the gene order in Brachypodium. The DNA markers marked by b and the corresponding ones were developed from the same gene. The asterisks indicate single nucleotide polymorphism (SNP) markers. The genes in brackets were reported previously (Cao et al., 2011; He et al., 2016) or first named in this study. (B) Physical map of Pm21 obtained by using the susceptible deletion line Y18-S6. CINAU15m was a co-dominant marker newly developed from Stpk-V gene in this study. The 6VS chromosome breakpoints b1 and b2 in Y18-S6 as well as the bin FL0.45–0.58 are pointed by horizontal arrows. The dashed box represents the chromosomal deletion region in Y18-S6. The vertical arrow shows the direction of the centromere of 6V.
FIGURE 4.
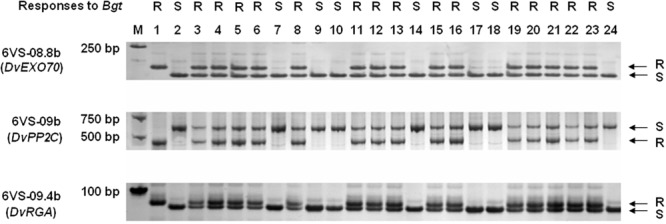
PCR amplification patterns of the co-segregated markers 6VS-08.8b, 6VS-09b, and 6VS-09.4b, corresponding to the genes DvEXO70, DvPP2C, and DvRGA, respectively. 1: the resistant parent DvRes-1; 2: the susceptible parent DvSus-1; 3–24: different recombinants. Powdery mildew responses were showed above the corresponding parents and recombinants. The arrows indicate the DNA bands that were same to the resistant (R) or susceptible (S) parents.
Physical Mapping of Pm21
From 1,216 EMS-induced M2 families of Yangmai 18 carrying Pm21, a total of 12 independent mutant lines highly susceptible to powdery mildew were screened out (Y18-S1 ∼ Y18-S12) (Figure 1D). Among them, the susceptible mutant line Y18-S6 was characterized as a chromosomal deletion involving the Pm21 locus. Molecular analysis revealed the chromosomal breakpoints b1 and b2 in Y18-S6 were closely flanked by the markers 6VS-03 and 6VS-04, 6VS-10 and 6VS-10.2, respectively (Figures 3B, 5). Interestingly, the breakpoint b1 in Y18-S6 was close to that in the deletion line del.6VS-1 (FL0.58) reported previously (He et al., 2016). Physical mapping also proved that the deleted chromosomal segment in Y18-S6 spanned the genetic interval carrying Pm21.
FIGURE 5.
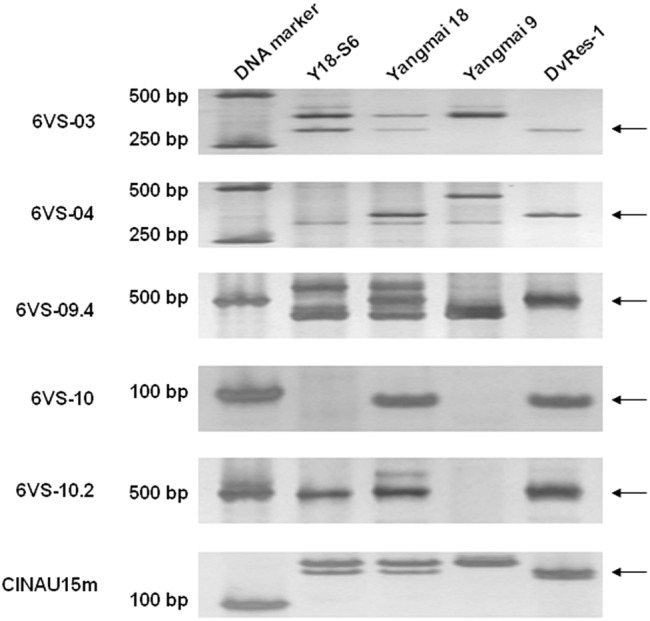
PCR amplification patterns of six representative markers (6VS-03, 6VS-04, 6VS-09.4, 6VS-10, 6VS-10.2, and CINAU15m) used for physical mapping of Pm21. The DNA templates used here are successively the susceptible deletion line Y18-S6, Yangmai 18, Yangmai 9, and the resistant D. villosum line DvRes-1. The 6VS-specific DNA bands are pointed by arrows.
Comparative Mapping of Pm21
The orthologs of the corresponding genes of the flanking markers 6VS-08.4b and 6VS-10b were Bradi3g03840 (2574344–2579921) and Bradi3g03970 (2692381–2693197) in Brachypodium, LOC_Os02g05610 (2728848–2734454) and LOC_Os02g05640 (2757693–2758714) in rice, respectively. Hence, the orthologous regions of the Pm21 locus were narrowed to a 112.5 kb genomic region harboring 18 predicted genes in Brachypodium, and a 23.2 kb region harboring two predicted genes in rice. In the orthologous regions, two genes were shared by Brachypodium and rice (Figure 6 and Table 2) and used to develop the DNA markers 6VS-08.8b and 6VS-09b. Both of the markers were tested to co-segregate with Pm21 in the F2 population.
FIGURE 6.
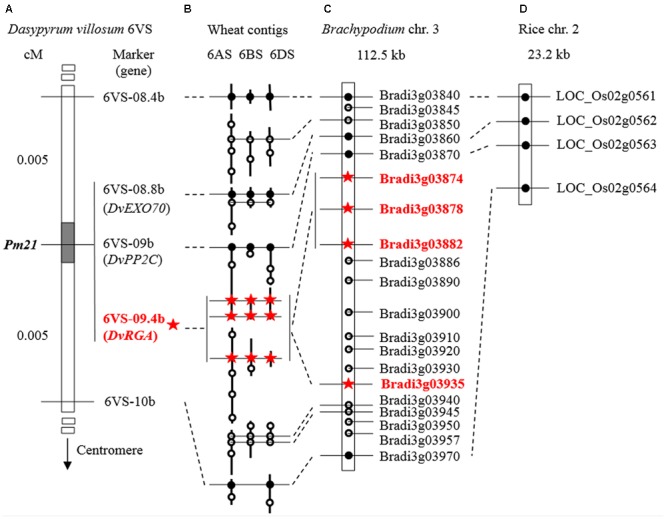
Comparative mapping of Pm21. (A) Genetic map (partial) using the F2 population derived from the cross between the resistant D. villosum line DvRes-1 and the susceptible line DvSus-1. The dark region on the chromosome 6VS demonstrates the genetic interval carrying Pm21. The vertical arrow shows the direction of the centromere of 6V. The complete genetic map can be found in Figure 3A. (B–D) Comparative maps of the Pm21 locus among D. villosum 6VS, wheat 6AS, 6BS, and 6DS (B), the short arms of Brachypodium chromosome 3 (C) and rice chromosome 2 (D). The homologous wheat contigs of genes of interest were obtained and annotated by using the BLAST, FGENESH, and SMART programs (Table 2). All genes from Brachypodium and rice are adopted according to the annotations of the corresponding genomes except the RGA Bradi3g03935 in Brachypodium that was re-annotated in this study. The solid circles and hollow circles indicate conserved genes and unconserved genes among different genomes, respectively. The orthologous RGAs among D. villosum, wheat and Brachypodium are marked by red stars.
Table 2.
Gene annotation in Brachypodium, rice and wheat orthologous regions of the Pm21 locus.
| Brachypodium | Rice | Wheat | Gene annotation |
|---|---|---|---|
| Bradi3g03845 | Retrotransposable element protein | ||
| Bradi3g03850 | 6AS_contigs_4399884 | Eukaryotic translation | |
| 6BS_contigs_2953789 | initiation factor | ||
| 6DS_contigs_2117578 | |||
| Bradi3g03860 | LOC_Os02g05620 | 6AS_contigs_4431592 | Exocyst complex subunit |
| 6BS_contigs_2953283 | EXO70-like protein | ||
| 6DS_contigs_2092656 | |||
| Bradi3g03870 | LOC_Os02g05630 | 6AS_contigs_4363243 | Serine/threonine protein |
| 6BS_contigs_2962596 | phosphatase 2C | ||
| 6DS_contigs_2093935 | |||
| Bradi3g03874a | Six contigsa | Disease resistance protein | |
| Bradi3g03878a | Six contigsa | Disease resistance protein | |
| Bradi3g03882a | Six contigsa | Disease resistance protein | |
| Bradi3g03886 | Unknown protein | ||
| Bradi3g03890 | Unknown protein | ||
| Bradi3g03900 | Unknown protein | ||
| Bradi3g03910 | Cytochrome P450 | ||
| Bradi3g03920 | Unknown protein | ||
| Bradi3g03930 | Poly(A) polymerase | ||
| Bradi3g03935a | Six contigsa | Disease resistance protein | |
| Bradi3g03940 | 6AS_contigs_4428294 | Photosystem II protein J | |
| 6BS_contigs_2926507 | |||
| 6DS_contigs_2114667 | |||
| Bradi3g03945 | 6AS_contigs_4428294 | Photosystem II | |
| 6BS_contigs_2926507 | cytochrome b559 alpha | ||
| 6DS_contigs_2114667 | subunit | ||
| Bradi3g03950 | Unknown protein | ||
| Bradi3g03957 | 6AS_contigs_4431958 | Polyubiquitin | |
| 6BS_contigs_2953283 | |||
| 6DS_contigs_2081863 |
aFour RGAs (Bradi3g03874, Bradi3g03878, Bradi3g03882, and Bradi3g03935) in Brachypodium are highly homologous with six contigs (6AS_contigs_4363243, 6AS_contigs_4428238, 6BS_contigs_3017519, 6BS_contigs_2958212, 6DS_contigs_2091261, and 6DS_contigs_1402211) on the short arms of wheat homologous group 6.
Comparative analysis also revealed a conserved resistance gene analog (RGA) locus between Brachypodium and wheat orthologous regions (Figure 6). In Brachypodium, four highly homologous RGAs, Bradi3g03874, Bradi3g03878, Bradi3g03882, and Bradi3g03935, were identified. Among them, Bradi3g03874, Bradi3g03878, and Bradi3g03882 were tandemly aligned as a cluster, and Bradi3g03935 were segregated by other six genes. In wheat, the orthologous RGA loci were also located in the contigs on 6AS, 6BS, and 6DS, and each locus harbored three RGAs at least. According to the conserved locus, the RGA markers 6VS-09.4 and 6VS-09.4b was developed. Physical mapping showed that 6VS-09.4 was lost in the susceptible deletion line Y18-S6, whereas genetic analysis demonstrated that 6VS-09.4b co-segregated with Pm21 in the F2 population.
Discussion
The powdery mildew resistance gene Pm21 was previously mapped to the physical bin FL0.45–0.58 of the chromosome 6VS (Cao et al., 2011; Chen et al., 2013). In our previous study, the orthologous regions of the bin FL0.45–0.58 in Brachypodium and rice were comparatively mapped (He et al., 2016). In the present study, we carried out physical mapping of Pm21 by using the susceptible deletion line Y18-S6 obtained from the Yangmai 18 population induced by EMS. We also successfully performed genetic mapping of Pm21 in diploid D. villosum, which was mainly attributed to the finding of D. villosum lines seedling-susceptible to wheat powdery mildew. Qi et al. (1998) observed that wheat-D. villosum disomic addition line DA6V#1 (Sears, 1953) was susceptible to powdery mildew. However, due to the utilization of colchicine in the incorporation of alien genome by Sears, it is difficult to explain whether the variation came from colchicine treatment or natural variation of the alien parent D. villosum. Nevertheless, it remains the possibility to mine susceptible D. villosum from natural populations. Thereafter, Qi et al. (1998) screened 46 D. villosum accessions but none was susceptible.
We also did not find any D. villosum accession susceptible to powdery mildew through the whole growth period. In this study, all the four susceptible D. villosum lines displayed complex resistance to wheat powdery mildew, which fully expressed since three-leaf stage. It’s now known to us that powdery mildew resistance of D. villosum comes from multi-gene effects, in which, the gene on 6VS carry out an immunologic mechanism different from the others, such as Pm55, an adult plant resistance (APR) gene newly found (Zhang et al., 2016). Therefore, to eliminate the background noise, we have to evaluate the Pm21-mediated resistance reaction at a very early stage (one-leaf stage). Furthermore, because Pm21 is immune to all Bgt races (Chen et al., 1995), problems probably resulted from race-specific resistance conferred by other genes in background should be avoided in resistance evaluation. Hence, the isolate Bgt YZ01 with high virulence was used, to which, only wheat varieties containing Pm21 or PmV (Li et al., 2005; Zeng et al., 2005) were immune. Additionally, D. villosum is a cross-pollinated species, which is adverse to forming homozygote of the recessive mutant gene (Hartfield and Glémin, 2014). So, the susceptible homozygotes account for a very small proportion naturally (2 ∼ 5%) as our observation. As a result, susceptible D. villosum is difficult to obtain. But fortunately, in this study, we successfully found susceptible individuals from 4 of 110 D. villosum accessions.
Based on the discovery of susceptible D. villosum resources, genetic mapping of Pm21 was fulfilled. By double-PCR program, 64 recombinants were identified from 10,536 F2 plants, indicating that the total genetic distance of the interval flanked by the markers 6VS-00.1 and Xcfe164 was only about 0.30 cM. It suggested that there exists recombination suppression between two 6VS chromosomes with different origins. The observation given by Qi et al. (1998) also showed that, in wheat background, the two alien 6V chromosomes could not pair normally in pollen mother cells of hybrids between the resistant DA6V#2 (Hyde, 1953) and the susceptible DA6V#1 (Sears, 1953). It is not clear whether recombination suppression observed here is related to low pairing of two different 6V chromosomes in D. villosum. In addition, the ratio of resistant and susceptible homozygotes (1.04:1) in the F2 population was fit for the ratio 1:1 (χ2 = 2.174, P > 0.01), suggesting that the male and female gametes have no obvious difference in viability and transmission. However, unexpectedly, the ratio of heterozygotes (53.6%) was higher than the theoretic ratio (50%), indicating that heterozygotes could survive more easily, which might be due to the heterosis in cross-pollinated D. villosum.
In the previous studies, eight genes, including Stpk-V, DvMPK1, DvMLPK, DvUPK, DvPSYR1, DvPP2C, DvGATA, and DvWHY, have been mapped to the bin FL0.45–0.58 carrying Pm21 and confirmed to be required by the Pm21 resistance. However, silencing of these genes mediated by barley stripe mosaic virus (BSMV) could not lead to macroscopic symptom of powdery mildew (Cao et al., 2011; He et al., 2016). It is suggested that these genes play roles in complex Pm21 resistance pathways but none of them is the Pm21 gene itself. Interestingly, overexpression of Stpk-V can confer high resistance to powdery mildew in transgenic wheat (Cao et al., 2011). Hence, these genes might have potential to be used to transgenic breeding for control of wheat powdery mildew. Furthermore, revealing their functions and mechanisms could contribute to understanding disease resistance pathways mediated by Pm21. Among the above genes, only DvPP2C (6VS-09b), encoding a serine/threonine protein phosphatase 2C, was confirmed to be located in the genetic interval carrying Pm21 by physical, genetic and comparative analyses in this study. In addition, another gene DvEXO70 (the corresponding markers 6VS-08.8 and 6VS-08.8b), encoding an exocyst complex subunit EXO70-like protein, appears in the Pm21 involved interval; however, whether it plays a role in the Pm21 resistance remains unclear.
Although disease resistance genes evolve rapidly and have limited collinearity in cereals (Keller et al., 2005), a conserved RGA locus was observed in Brachypodium and wheat orthologous regions of the genetic interval of Pm21. The members in this RGA locus encode typical coiled-coil, nucleotide-binding site, leucine-rich repeat (CC-NBS-LRR) proteins that are the major class of plant disease resistance proteins (Gururani et al., 2012). Both physical mapping and genetic mapping demonstrated that the corresponding RGA markers 6VS-09.4 and 6VS-09.4b were located in the Pm21 locus. It suggested that this co-segregated RGA locus could be considered as an important candidate of the Pm21 locus. However, due to large genome size, it would be still a great challenge to directly perform map-based cloning of Pm21 in D. villosum. Recently, a fast cloning method using mutagenesis and next-generation sequencing (NGS) has been successfully utilized to identify the stem rust resistance genes Sr22 and Sr45 (Steuernagel et al., 2016) and the powdery mildew resistance gene Pm2 (Sánchez-Martín et al., 2016) in wheat. In this study, 11 independent susceptible wheat mutants, not involving in chromosomal deletion, could be used to find the exact sequence of Pm21, and further researches are in progress.
Author Contributions
HH and TB conceived and designed the experiments. HH screened the resource of D. villosum. YJ and SZ developed DNA markers. YJ, BL, RZ, and ZJ performed genetic mapping. HH and TB performed physical mapping. HH and SZ analyzed the data and wrote the paper. HH and TB revised the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Germplasm Resources Information Network (GRIN), GRIN Czech, Genebank Information System of the IPK Gatersleben (GBIS-IPK), Nordic Genetic Resource Center (NordGen), and the Cytogenetics Institute, Nanjing Agricultural University (CI-NAU) for providing Dasypyrum villosum accessions.
Funding. This research was supported by Grants from the National Natural Science Foundation of China (31471497), the Natural Science Foundation of Jiangsu Province (BK20130503, BK20151319), the Innovation Foundation of Jiangsu Academy of Agricultural Sciences [ZX(17)2011] and the Foundation of Jiangsu University (13JDG103).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01914/full#supplementary-material
References
- Bie T., Zhao R., Jiang Z., Gao D., Zhang B., He H. (2015). Efficient marker-assisted screening of structural changes involving Haynaldia villosa chromosome 6V using a double-distal-marker strategy. Mol. Breed. 35 34 10.1007/s11032-015-0211-y [DOI] [Google Scholar]
- Cao A., Xing L., Wang X., Yang X., Wang W., Sun Y., et al. (2011). Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc. Natl. Acad. Sci. U.S.A. 108 7727–7732. 10.1073/pnas.1016981108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., You C., Hu Y., Chen S., Zhou B., Cao A., et al. (2013). Radiation-induced translocations with reduced Haynaldia villosa chromatin at the Pm21 locus for powdery mildew resistance in wheat. Mol. Breed. 31 477–484. 10.1007/s11032-012-9804-x [DOI] [Google Scholar]
- Chen P. D., Qi L. L., Zhou B., Zhang S. Z., Liu D. J. (1995). Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor. Appl. Genet. 91 1125–1128. 10.1007/BF00223930 [DOI] [PubMed] [Google Scholar]
- Fu D., Uauy C., Distelfeld A., Blechl A., Epstein L., Chen X., et al. (2009). A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323 1357–1360. 10.1126/science.1166289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururani M. A., Venkatesh J., Upadhyaya C. P., Nookaraju A., Pandey S. K., Park S. W. (2012). Plant disease resistance genes: current status and future directions. Physiol. Mol. Plant Pathol. 78 51–65. 10.1016/j.pmpp.2012.01.002 [DOI] [Google Scholar]
- Hartfield M., Glémin S. (2014). Hitchhiking of deleterious alleles and the cost of adaptation in partially selfing species. Genetics 196 281–293. 10.1534/genetics.113.158196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Zhu S., Jiang Z., Ji Y., Wang F., Zhao R., et al. (2016). Comparative mapping of powdery mildew resistance gene Pm21 and functional characterization of resistance-related genes in wheat. Theor. Appl. Genet. 129 819–829. 10.1007/s00122-016-2668-4 [DOI] [PubMed] [Google Scholar]
- He H., Zhu S., Sun W., Gao D., Bie T. (2013). Efficient development of Haynaldia villosa chromosome 6VS-specific DNA markers using a CISP-IS strategy. Plant Breed. 132 290–294. 10.1111/pbr.12035 [DOI] [Google Scholar]
- Hyde B. B. (1953). Addition of individual Haynaldia villosa chromosomes to hexaploid wheat. Am. J. Bot. 40 174–182. 10.2307/2438775 [DOI] [Google Scholar]
- Keller B., Feuillet C., Yahiaoui N. (2005). Map-based isolation of disease resistance genes from bread wheat: cloning in a supersize genome. Genet. Res. 85 93–100. 10.1017/S0016672305007391 [DOI] [PubMed] [Google Scholar]
- Li H., Chen X., Xin Z., Ma Y., Xu H., Chen X., et al. (2005). Development and identification of wheat–Haynaldia villosa T6DL.6VS chromosome translocation lines conferring resistance to powdery mildew. Plant Breed. 124 203–205. 10.1111/j.1439-0523.2004.01062.x [DOI] [Google Scholar]
- McIntosh R. A., Dubcovsky J., Rogers J., Morris C., Appels R., Xia X. C. (2013). Catalogue of Gene Symbols for Wheat: Komugi Integrated Wheat Science Database. Available at: http://www.shigen.nig.ac.jp/wheat/komugi/genes/download.jsp [Google Scholar]
- Murray M. G., Thompson Y. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8 4321–4325. 10.1093/nar/8.19.4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L. L., Wang S. L., Chen P. D., Liu D. J., Gill B. S. (1998). Identification and physical mapping of three Haynaldia villosa chromosome-6V deletion lines. Theor. Appl. Genet. 97 1042–1046. 10.1007/s001220050989 [DOI] [Google Scholar]
- Qi X. L., Cui F., Yu L., Ding A. M., Li J., Chen G. L., et al. (2010). Molecular tagging wheat powdery mildew resistance gene Pm21 by EST-SSR and STS markers. Mol. Plant Breed. 1 22–26. 10.5376/mpb.2010.01.0004 [DOI] [Google Scholar]
- Sánchez-Martín J., Steuernagel B., Ghosh S., Herren G., Hurni S., Adamski N., et al. (2016). Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 17 221. 10.1186/s13059-016-1082-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears E. R. (1953). Addition of the genome of Haynaldia villosa to Triticum aestivum. Am. J. Bot. 40 168–174. 10.2307/2438774 [DOI] [Google Scholar]
- Steuernagel B., Periyannan S. K., Hernández-Pinzón I., Witek K., Rouse M. N., Yu G., et al. (2016). Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 34 652–655. 10.1038/nbt.3543 [DOI] [PubMed] [Google Scholar]
- Wiersma A. T., Pulman J. A., Brown L. K., Cowger C., Olson E. L. (2017). Identification of Pm58 from Aegilops tauschii. Theor. Appl. Genet. 130 1123–1133. 10.1007/s00122-017-2874-8 [DOI] [PubMed] [Google Scholar]
- Zeng X. Y., Zhang Z. Y., Du L. P., Xin Z. Y., Chen X. (2005). Development of wheat germplasms with multi-resistance to powdery mildew, stripe rust and yellow dwarf virus by molecular marker-assisted selection (in Chinese). Sci. Agric. Sin. 38 2380–2386. [Google Scholar]
- Zhang R., Sun B., Chen J., Cao A., Xing L., Feng Y., et al. (2016). Pm55, a developmental-stage and tissue-specific powdery mildew resistance gene introgressed from Dasypyrum villosum into common wheat. Theor. Appl. Genet. 129 1975–1984. 10.1007/s00122-016-2753-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


