Summary
Microsatellite expansion disorders are pathologically characterized by RNA foci formation and repeat-associated non-AUG (RAN) translation. However, their underlying pathomechanisms and regulation of RAN translation remain unknown. We report that expression of expanded UGGAA (UGGAAexp) repeats, responsible for spinocerebellar ataxia type 31 (SCA31) in Drosophila, causes neurodegeneration accompanied by accumulation of UGGAAexp RNA foci and translation of repeat-associated pentapeptide repeat (PPR) proteins, consistent with observations in SCA31 patient brains. We revealed that motor-neuron disease (MND)-linked RNA-binding proteins (RBPs), TDP-43, FUS and hnRNPA2B1, bind to and induce structural alteration of UGGAAexp. These RBPs suppress UGGAAexp-mediated toxicity in Drosophila by functioning as RNA chaperones for proper UGGAAexp folding and regulation of PPR translation. Furthermore, nontoxic short UGGAA repeat RNA suppressed mutated RBP aggregation and toxicity in MND Drosophila models. Thus, functional crosstalk of the RNA/RBP network regulates their own quality and balance, suggesting convergence of pathomechanisms in microsatellite expansion disorders and RBP proteinopathies.
Keywords: Microsatellite repeat expansion diseases, SCA31, ALS, RNA foci, RAN translation, TDP-43, ribonucleoprotein, RNA chaperone, Drosophila melanogaster
Introduction
Microsatellite repeat expansion disorders, such as myotonic dystrophy (DM), fragile X-associated tremor/ataxia syndrome (FXTAS), C9orf72-linked amyotrophic lateral sclerosis (ALS)/frontotemporal dementia (FTD), spinocerebellar ataxia type 31 (SCA31) and other several types of SCA, are a group of neurological and neuromuscular diseases caused by the expansion of microsatellite repeat sequences within the human genome (DeJesus-Hernandez et al., 2011; La Spada and Taylor, 2010; Renton et al., 2011). A pathological cellular hallmark of these diseases is the aggregation of expanded repeat RNA, termed RNA foci, which disrupt the function of specific RNA-binding proteins in trans (Cooper et al., 2009; Todd and Paulson, 2010). In addition, aberrant proteins generated by repeat-associated, non-AUG (RAN) translation accumulate in patient brains, potentially promoting neurodegeneration (Cleary and Ranum, 2014, Zu et al., 2011). Despite these advances in our understanding, it still remains a challenge to disentangle their individual contributions and to regulate their pathomechanisms of toxicity.
In SCA31, disease-causing microsatellites consist of pentanucleotide repeat complexes, including (TGGAA)n, (TAGAA)n, (TAAAATAGAA)n and (TAAAA)n, located in an intron shared by two different genes: brain expressed associated with NEDD4-1 (BEAN1) and thymidine kinase 2 (TK2) (Sato et al., 2009). SCA31 is one of the most common types of autosomal-dominant cerebellar ataxia in Japan. Among the repeat sequences associated with this condition, (TGGAA)n in the BEAN1 strand and (TTCCA)n in the TK2 strand are disease-specific repeats found exclusively in affected individuals. In addition, RNA foci containing UGGAA repeats were observed in Purkinje cell nuclei of SCA31 patients, but not in control individuals (Niimi et al., 2013). Therefore, (TGGAA)n is considered the critical motif of SCA31 pathogenesis, supporting a gain-of-toxic-function hypothesis of pathogenesis.
To gain insights into the mechanisms by which UGGAAexp causes neurodegeneration, we generated Drosophila models expressing UGGAAexp. First, we obtained evidence that the expression of UGGAAexp induced toxicity in vivo in a manner dependent on repeat length and gene dosage, accompanied by the accumulation of RNA foci. Second, we performed a screen for potential UGGAAexp-binding proteins, and identified the motor neuron disease (MND)-linked RBPs, TAR DNA-binding protein 43 kDa (TDP-43), Fused in Sarcoma (FUS) and heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNPA2B1), which exhibit RNA chaperone activity by inhibiting UGGAAexp RNA misfolding. Third, we observed an accumulation of UGGAAexp-encoded pentapeptide (Trp-Asn-Gly-Met-Glu) repeat (PPR) proteins in Drosophila and SCA31 human brains. Interestingly, upregulation of RBPs suppressed the translation of PPR proteins in vivo, indicating that RBPs are key molecules for both aggregate-prone repeat RNA quality control and regulation of repeat-associated translation. Finally, we demonstrated that the expression of nontoxic short UGGAA repeats mitigates MND-linked RBP (TDP-43, FUS and hnRNPA2B1) toxicities, suggesting that RNA species also contribute to RBP quality control. This study highlights the mechanistic link between RNA and RBP quality control in ribonucleoprotein (RNP) complex homeostasis and suggests potential therapeutic strategies for microsatellite expansion disorders and RBP proteinopathies.
Results
Expression of UGGAAexp in Drosophila exhibits RNA foci and results in toxicity
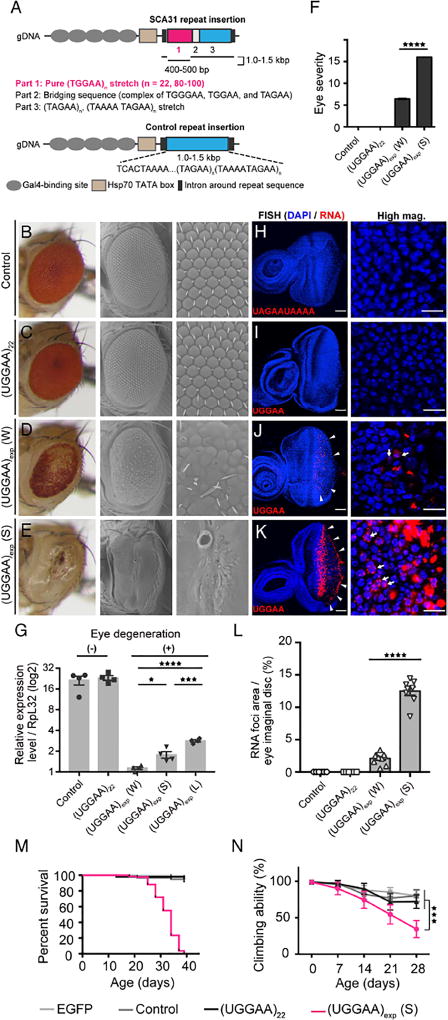
To define the toxic features of UGGAAexp that might be responsible for neurodegeneration in SCA31 in vivo, we expressed a human SCA31 repeat tract that consists of four repeat components, (TGGAA)n, (TAGAA)n, (TAAAA)n and (TAAAATAGAA)n, or a control repeat tract observed in the normal population lacking (TGGAA)n, in Drosophila using the UAS/GAL4 system (Figure 1A). Note that only (TGGAA)n is specific to affected SCA31 patients, whereas the other three repeat components are also observed in the unaffected Japanese population. We successfully established five Drosophila lines (lines #1, #2, #4, #5 and #6) carrying approximately 80–100 TGGAA repeats (0.4–0.5 kb), and one line (line #3) in which spontaneous contraction to 22 TGGAA repeats had occurred (UGGAA22 line), confirmed by long PCR and TGGAA-specific PCR for transgenes in the fly genome (Figures S1A–C). The repeat lengths of control lines were also evaluated by long PCR, and control line #1 was found to carry a longer repeat tract of 1.0–1.5 kb than the SCA31 lines (Figure S1D). To evaluate the effect of UGGAAexp expression, we expressed SCA31 or control repeat in the compound eyes of flies using the GMR-GAL4 driver. Although expression of the control repeat (all nine control lines) and short UGGAA22 had no significant effect on eye morphology (Figures 1B, 1C, 1F, S1E and S1F), strong expression of UGGAAexp (S line, line #1) caused severe disruption to eye morphology with reduced eye size and ommatidial abnormalities, whereas weak expression of UGGAAexp (W line, line #2) led to a milder rough-eye phenotype (Figures 1D–1F and S1E). A pupal lethal line (L line, line #4) had the highest level of UGGAAexp expression among the SCA31 fly lines, revealed by quantitative reverse-transcription PCR (RT-PCR) (Figure 1G). Given that the GMR-GAL4 driver is reported to be expressed in additional tissues including larval wing imaginal disc and neuronal cells in larval ventral and cerebral ganglia, besides eye imaginal disc, the leaky expression of UGGAAexp RNA in these tissues may be a cause of the pupal lethality (Ray and Lakhotia, 2015).
Figure 1. Expression of UGGAAexp in Drosophila Results in RNA Foci Formation and Toxicity.
(A) Schematic of constructs used to express UGGAAexp and control repeat.
(B–E) Light microscopy (LM; left) and scanning electron microscopy (SEM; middle and right) images of compound eyes from Drosophila females expressing control repeats (B), UGGAA22 (C), UGGAAexp (W) (D) or UGGAAexp (S) (E) using GMR-Gal4 (See also Figure S1 and Table S1).
(F) Eye degeneration scores from the flies of the indicated genotypes.
(G) Quantitative real-time PCR analysis of transgene expression in the indicated Drosophila lines (four independent experiments, n = 80 flies per genotype).
(H–K) RNA FISH analyses in eye imaginal discs of L3 expressing the control repeat (H), UGGAA22 (I), UGGAAexp (W) (J) or UGGAAexp (S) (K) under GMR-GAL4 using LNA probes (red) and DAPI (blue). Scale bars = 50 or 10 µm (high mag.). RNA foci (arrowheads) are observed in (J) and (K). RNA foci are present in nuclei (arrows) and cytoplasm.
(L) Quantification of area of RNA foci per larva. n = 8 (Control), 8(UGGAA22), 10 (W), 8 (S) respectively.
(M) Lifespan analysis using ELAV-GeneSwitch. UGGAAexp (S) flies had a shorter lifespan than flies expressing EGFP, control repeats or UGGAA22 (p < 0.0001, log-rank test; n = 100–120 flies per group).
(N) Changes in motor performance with age. UGGAAexp (S) flies showed more severe locomotor defects compared with flies expressing EGFP, control repeats or UGGAA22 (mean ± SD, n = 100–120; ***p < 0.001, two-way ANOVA). Data are presented as mean ± SEM. In (F), (G) and (L), p < 0.0001, one-way ANOVA; *p = 0.0186, ***p = 0.0008 (G), ****p < 0.0001, as assessed by Tukey’s post hoc analysis.
Notably, homozygous transgenic UGGAAexp flies had more severe degenerative eye morphologies than heterozygous flies, indicating that the effects of UGGAAexp are dosage-dependent (Figure S1H). RT-PCR using primers spanning the whole repeat configuration (“long PCR” in Figure S1A) showed that the entire transgene was transcribed in all lines (Figure S1G). To determine the localization of UGGAAexp transcripts in eye imaginal discs of these flies, we performed RNA fluorescence in situ hybridization (FISH) on UGGAAexp, UGGAA22 and a control repeat line using digoxigenin (DIG)-conjugated locked nucleic acid (LNA) probes (Figures 1H–1K, S1I and S1J). Extensive accumulation of UGGAAexp as RNA foci in eye imaginal discs of the UGGAAexp (S) line and, to a lesser extent, the (W) line (Figures 1J and 1K, arrowheads) was observed, reminiscent of the pathology of human SCA31 (Niimi et al., 2013), whereas RNA foci were not detected in lines expressing control repeat or UGGAA22 (Figures 1H and 1I). RNA foci appeared to be present in the nucleus (arrows) and cytoplasm; however, their precise localization was not clear because of severe degeneration. Importantly, the levels of RNA foci correlated with UGGAAexp expression levels and the severity of eye degeneration (Figure 1L). Foci were abolished when FISH was performed after pretreatment with RNase (Figure S1J). Thus, UGGAAexp-induced toxicity accompanied by the formation of abnormally aggregated structures is dependent on RNA dosage and length.
Next, we expressed UGGAAexp in the Drosophila nervous system under the control of ELAV-GAL4. Its continuous expression throughout development caused early pupal lethality in the UGGAAexp (S) line (Table S1). To determine whether UGGAAexp-mediated neurodegeneration progresses with age in Drosophila, we expressed UGGAAexp after adult eclosion using the inducible ELAV-GeneSwitch expression system. UGGAAexp (S) flies exhibited a shorter lifespan (median: 34 days; p < 0.001, log-rank test) (Figure 1M) and progressive locomotor defects with age compared with flies expressing enhanced green fluorescent protein (EGFP), control repeats and UGGAA22, as revealed by climbing assay (at 28 days, UGGAAexp (S): 34.4 ± 11.7%, EGFP: 80.2 ± 6.9%, control repeat: 79.1 ± 8.1% and UGGAA22: 72.1 ± 9.2%; ***p < 0.001, two-way ANOVA) (Figure 1N). Overall, UGGAAexp-expressing flies successfully reproduced several key aspects of human SCA31.
RNA-binding protein TDP-43 interacts with UGGAA RNA in vitro and in vivo
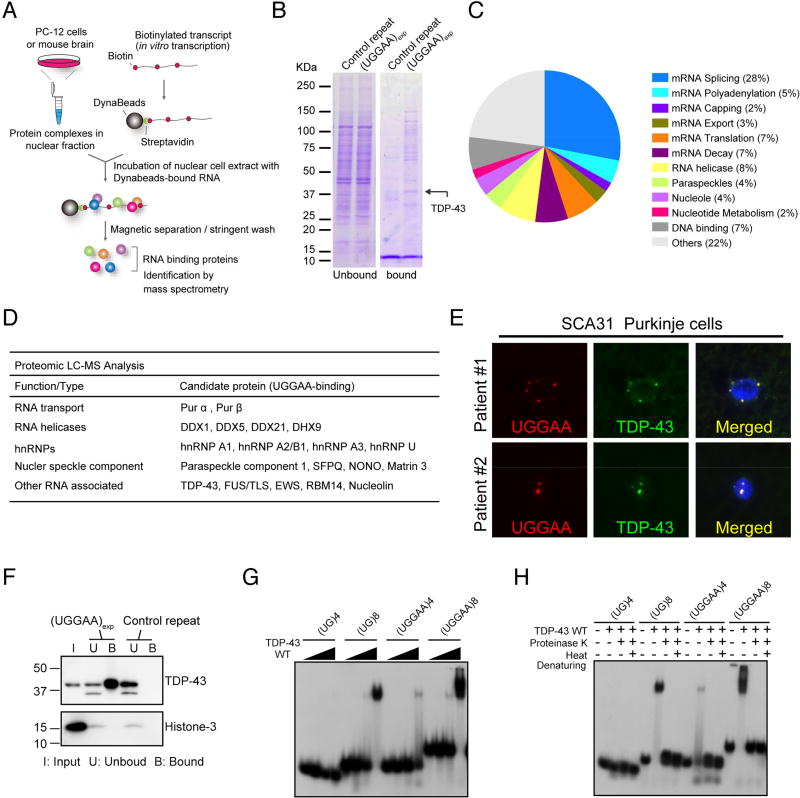
Because proteins sequestered in RNA foci are important in the pathogenesis of noncoding microsatellite expansion disorders (Echeverria and Cooper, 2012), we performed two independent screens for potential UGGAAexp-binding partners by in vitro RNA pull-down assay followed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and liquid chromatography–mass spectrometry in two different laboratories (Figure 2A). In the first screen, six UGGAA-binding proteins were found to be abundant on Coomassie-stained gels, and they were identified as TDP-43, NONO, SFPQ, DHX9, DDX5 and DDX17 (Figure 2B). These six proteins were also included among the 88 candidate proteins identified by the second mass spectrometry screen using mouse brain lysates. Eighty-eight proteins that potentially bind to (UGGAA)n were classified by their functions into 12 categories (Figure 2C). Among them, we noted several proteins involved in RNA transport, splicing and editing, which are components of nuclear speckles (Figure 2D and Table S2). Many ALS-linked RBPs, such as TDP-43, FUS, hnRNPs and matrin-3 (Johnson et al., 2014; Kim et al., 2013), were also found. Note that TDP-43 was one of the most abundant proteins identified in our screen (Figure 2B, arrow). We next found that TDP-43 binds to in vitro transcribed (UGGAA)n sequence (Figure 2E), consistent with previous studies indicating that TDP-43 has high binding affinity to TG- and UG-rich sequence clusters (Buratti et al., 2001; Tollervey et al., 2011). Furthermore, TDP-43 clearly colocalized with RNA foci in human SCA31 Purkinje cells (Figure 2F). Therefore, we focused on TDP-43 to analyze its role in SCA31 pathology. We assessed whether the length of UGGAA affects TDP-43-binding affinity. Band-shift analysis using radiolabeled oligonucleotide RNAs revealed length-dependent affinity of recombinant TDP-43 for (UGGAA)n, with increasing repeat size being associated with stronger affinity for TDP-43, as evidenced by the stronger binding of TDP-43 to (UGGAA)8 than to (UGGAA)4 (Figure 2G). This same length dependence was confirmed in the positive control oligonucleotide sequences, (UG)8 and (UG)4. Binding to (UGGAA)8 was similar to that to (UG)8 (Figure 2G). Binding was also shown to be TDP-43-dependent as heat denaturation and/or proteinase K abolished the shifted complex and the released RNA comigrated with unbound RNAs (Figure 2H). This suggests that the alterations, at least for these short oligos, were not permanent. Similar binding was observed for the full length and ΔC forms of recombinant TDP-43 (Figures S2A–C).
Figure 2. RNA-Binding Protein TDP-43 Interacts with UGGAA RNA In Vitro and In Vivo.
(A) Schematic of method to identify RBPs in PC-12 cells and mouse brain.
(B) Representative Coomassie-stained gels showing proteins pulled down by UGGAAexp repeat and control repeat. Arrow indicates TDP-43.
(C) A pie-chart showing all 88 hits categorized by GO terms.
(D) Protein candidates identified by binding to UGGAAexp RNA (See also Table S2).
(E) Immunoblots confirming specific binding of TDP-43 to UGGAAexp RNA.
(F) Combined RNA FISH and immunostaining in human SCA31 brains.
(G) Binding of RNA oligonucleotides to increasing amounts of TDP-43 WT (See also Figure S2).
(H) Heat denaturation and/or proteinase K treatment abolishes TDP-43 WT binding to RNA oligos, which then comigrate with unbound RNA.
TDP-43 suppresses UGGAAexp-mediated toxicity, and RNA recognition motifs in TDP-43 are required for its rescuing effects
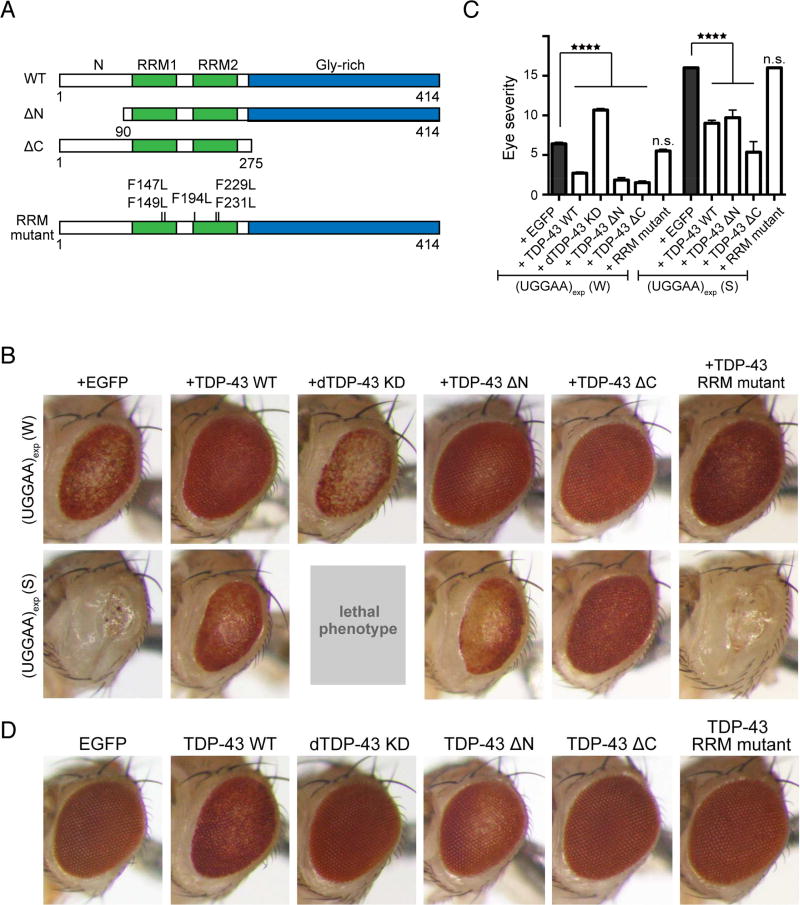
To investigate the potential roles of TDP-43 in UGGAAexp-mediated toxicity, we crossed flies expressing UGGAAexp and human wild-type TDP-43 (TDP-43 WT) (Figures 3A and 3D). Surprisingly, upon concomitant expression of TDP-43 WT, eyes of UGGAAexp (S) flies showed dramatic restoration of pigmentation and ommatidial formation, and pigmentation in UGGAAexp (W) eyes also significantly improved (Figures 3B and 3C). Thus, the coexpression of TDP-43 WT suppressed UGGAAexp-mediated toxicity in flies, even though upregulation of TDP-43 WT alone induced eye degeneration and a slight rough-eye phenotype (Figure 3D). In contrast, RNA interference (RNAi)-mediated knockdown of endogenous Drosophila TDP-43 (dTDP-43) (Lin et al., 2011) strikingly enhanced eye degeneration in the UGGAAexp (W) line and induced a lethal phenotype in the UGGAAexp (S) line (Figures 3B and 3C). Thus, TDP-43 might play a crucial role in mediating UGGAAexp toxicity in vivo. To determine which functions of TDP-43 are required to suppress UGGAAexp toxicity, we generated TDP-43 mutant transgenic flies lacking the N-terminal (ΔN) or C-terminal (ΔC) region. Previous reports suggested that the TDP-43 RNA recognition motif (RRM) contains at least twelve or nine residues that interact with nucleic acids (Buratti and Baralle, 2001; Kuo et al., 2014). We generated fly lines carrying five amino acid substitutions in RRM (RRM mutant), resulting in reduced binding to its RNA targets (Figure 3A). The expression of TDP-43 mutant proteins in flies was confirmed by immunoblotting with anti-TDP-43 antibody (Figure S2D). Consistent with previous findings (Zhang et al., 2013), we also found that TDP-43 WT and ΔC mutant, but not ΔN, in fly tissues treated with disuccinimidyl glutarate (DSG), a homobifunctional crosslinker based on the amine-reactive N-hydroxysuccinimide ester group widely used to study native protein structures, preserved the formation of homodimers (Figure S2E). Note that RRM mutations in TDP-43 led to a reduction of the binding to UGGAAexp RNA (Figure S2F). Coexpression of either TDP-43 deletion mutant markedly mitigated the phenotypes of UGGAAexp-expressing flies (Figures 3B and 3C), suggesting that dimerization of TDP-43 via its N-terminus (Zhang et al., 2013) and interaction of TDP-43 with other proteins via its C-terminus required for splicing regulation and microRNA (miRNA) biogenesis/silencing (Buratti et al., 2005; Kawahara and Mieda-Sato, 2012) may not be involved in suppressing UGGAAexp toxicity. Interestingly, coexpression of the TDP-43 RRM mutant failed to rescue UGGAAexp-mediated degeneration (Figures 3B and 3C), indicating that the RNA-binding ability of TDP-43 is essential for its suppression of UGGAAexp-mediated toxicity. Expression of the TDP-43 RRM mutant did not cause eye degeneration in Drosophila, consistent with previous reports (Ihara et al., 2013; Voigt et al., 2010), whereas flies expressing TDP-43 WT or deletion mutant alone exhibited slight eye degeneration (Figure 3D). Therefore, the direct interaction of TDP-43 and UGGAAexp might be essential for alleviating UGGAAexp-mediated toxicity.
Figure 3. TDP-43 Suppresses UGGAAexp-Mediated Toxicity, and RNA Recognition Motifs in TDP-43 Are Required for its Rescuing Effects.
(A) Schematic diagram of wild-type (WT) TDP-43, N- or C-terminus-deleted TDP43 (ΔN or ΔC, respectively), and RRM mutants used.
(B) LM images of compound eyes from Drosophila females co-expressing TDP-43 variants or endogenous dTDP43 RNAi with UGGAAexp (W) and (S) using GMR-Gal4 (See also Figure S2).
(C) Eye degeneration scores from the flies of the indicated genotypes (n = 10 per genotype). Data are presented as mean ± SEM, p < 0.0001, one-way ANOVA, ****p < 0.0001, p = 0.2353 (W line with EGFP versus W line with TDP-43 RRM mutant), p = 0.9999 (S line with EGFP versus S line with TDP-43 RRM mutant), Dunnett’s post hoc analysis; n.s. indicates no significant difference.
(D) LM images of compound eyes from Drosophila females expressing TDP-43 variants using GMR-GAL4. TDP-43 WT and ΔN exhibited a slight rough-eye phenotype in Drosophila, whereas EGFP-expressing flies had a normal eye phenotype.
TDP-43 functions as an RNA chaperone for UGGAAexp RNA misfolding
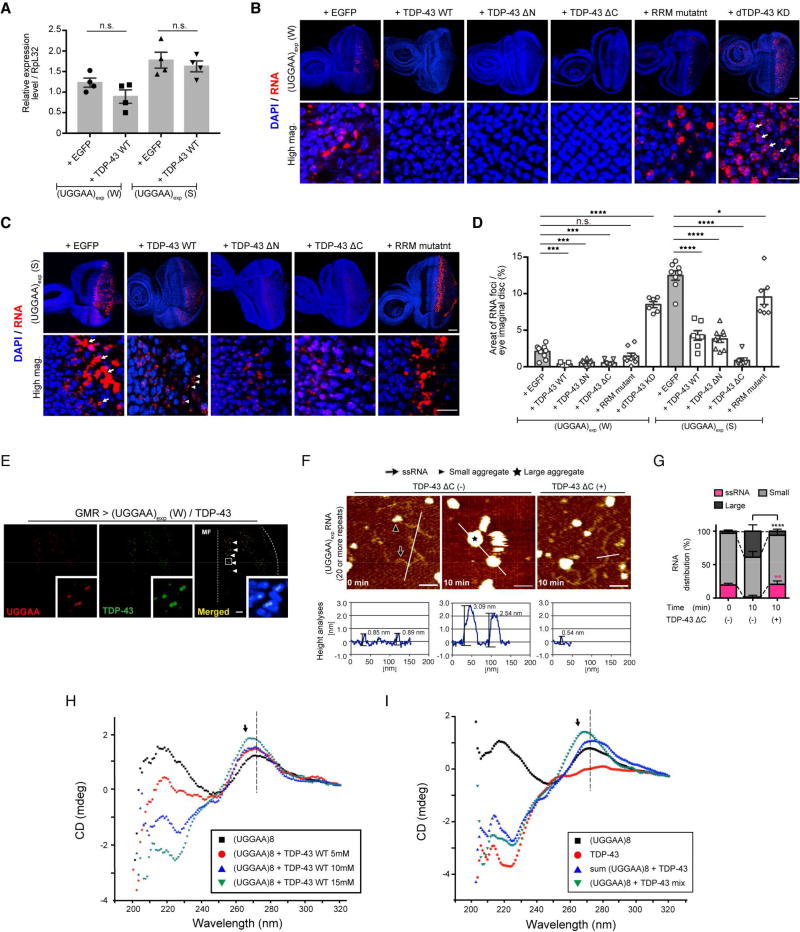
To explore the mechanisms by which TDP-43 suppresses UGGAAexp-mediated toxicity through its RNA-binding ability, we measured UGGAAexp transcript expression levels and the accumulation of RNA foci in flies coexpressing UGGAAexp and TDP-43. Coexpression of TDP-43 WT had no significant effect on UGGAAexp transcript expression levels (Figure 4A). However, coexpression of TDP-43 WT in UGGAAexp (W) flies resulted in a statistically significant 81.95% reduction in the area of RNA foci formation compared with that in UGGAAexp (W) flies expressing EGFP (Figure 4B). This suppression was even more dramatic in UGGAAexp (S) flies, where large RNA foci (Figure 4C, arrows) disappeared and were replaced by small granular RNA foci (Figure 4C, arrowheads). Similar results were observed when either TDP-43 deletion mutant (ΔN or ΔC) was coexpressed. Upon quantification, the total area of RNA foci was greatly suppressed by UGGAAexp–TDP-43 interactions (Figure 4D), whereas coexpression with the TDP-43 RRM mutant failed to suppress the accumulation of RNA foci in UGGAAexp-expressing flies (Figures 4B–4D). Consistent with the deterioration of eye morphology, dTDP-43 silencing dramatically enhanced the accumulation of RNA foci in UGGAAexp (W) flies (Figures 4B and 4D). Moreover, simultaneous RNA FISH and immunofluorescence analyses in eye imaginal discs of the UGGAAexp (W) line revealed that TDP-43 associated with UGGAAexp RNA in the nucleus (Figure 4E). Thus, TDP-43 does not promote the degradation of UGGAAexp transcripts, but disperses RNA foci, resulting in the neutralization of toxic RNAs through interactions of RNA and TDP-43 in vivo.
Figure 4. TDP-43 Functions as an RNA Chaperone for UGGAAexp RNA Misfolding.
(A) Quantitative real-time PCR analysis of transgene expression in the indicated Drosophila lines (four independent experiments, n = 80 flies per genotype). Data are presented as mean ± SEM, p = 0.139 (W line with EGFP versus W line with TDP-43 WT), p = 0.5514 (S line with EGFP versus S line with TDP-43 WT), two-tailed unpaired Student’s t-test; n.s.: not significant.
(B) RNA FISH analyses of eye imaginal discs of UGGAAexp (W) L3 coexpressing TDP-43, RRM mutant or dTDP-43 RNAi. Arrows indicate increased numbers of nuclear RNA foci. Scale bars = 50 or 10 µm (high mag.).
(C) RNA FISH analyses of eye imaginal discs of L3 of the UGGAAexp (S) coexpressing TDP-43 or RRM mutant. Arrows indicate abnormally large RNA foci, and arrowheads indicate granular RNA foci. Scale bars = 50 or 10 µm (high mag.).
(D) Quantification of the area of RNA foci in flies of each genotype (n = 6–10). Data are presented as mean ± SEM, p < 0.0001 (W line group), p < 0.0001 (S line group), one-way ANOVA; ***p < 0.001, ****p < 0.0001, p = 0.2577 (W line with EGFP versus W line with TDP-43 RRM), *p = 0.0125 (S line with EGFP versus S line with TDP-43 RRM), Dunnett’s post hoc analysis; n.s.: not significant.
(E) Immunostaining of TDP-43 in eye imaginal disc of L3 expressing UGGAAexp (W) and TDP-43. TDP-43 colocalized with UGGAAexp RNA transcripts in cell nuclei (arrowheads). Dotted line represents morphogenetic furrow (MF).
(F and G) AFM images (F) and distribution column (G) of RNAs UGGAAexp having approximately 20 repeat units or more, the RNA solution showing decreased UGGAAexp aggregation by TDP-43 ΔC. In (F), scale bars = 50 nm. (G) Relative amount of RNA species between TDP43 ΔC (−) and (+) at 10 min; n > 900 molecules counted. Data are presented as mean ± SD, p < 0.0001(ssRNA), p = 0.0001 (small aggregates), p < 0.0001 (large aggregates), one –way ANOVA. **p = 0.0016, ****p < 0.0001, Tukey’s post hoc analysis (See also Figure S3). (H) Titration experiments [0 mM (black), 5 mM (red), 10 mM (blue) and 15 mM (green)] of GST–TDP-43 WT were used with a fixed concentration of (UGGAA)8 RNAs (5 µM) and assessed by CD spectroscopy. (I) CD spectra of (UGGAA)8 alone (black), TDP-43 alone (red), the sum of spectrum obtained from TDP-43 and (UGGAA)8 separately (blue) and the spectra obtained from incubating them together, corresponding to (UGGAA)8 + TDP-43 binding (green).
These findings suggest that TDP-43 may directly interact with UGGAAexp to prevent aberrant RNAs from being trapped in RNA foci in vivo. Therefore, we examined whether TDP-43 directly suppressed UGGAAexp aggregation in vitro using atomic force microscopy (AFM). In vitro-transcribed UGGAAexp RNAs having approximately 20 repeat units or more were mostly present as single-stranded RNAs (ssRNAs, arrow; mean ± S.D.: 19.5% ± 2.6%) or small aggregates (arrowhead; 77.3% ± 3.4%) immediately after denaturation in solution (Figures 4F, 4G and S3A). After 10-min incubation at room temperature, the proportion of ssRNAs was markedly reduced (1.5% ± 0.9%, ****p < 0.0001) and many large aggregates appeared (Figure S3B, Figures 4F and 4G, star, 38.9% ± 9.5%, ****p < 0.0001; Figure S3B). However, coincubation with recombinant TDP-43 ΔC, which had lost its aggregation ability (Wang et al., 2012), resulted in significant prevention of the formation of large RNA aggregates (6.1% ± 3.1%, ****p < 0.0001), thus maintaining UGGAAexp RNA as single strands (20.6% ± 5.0%, **p = 0.0016) compared with that in RNA-only conditions (Figures 4F, 4G and S3C). We used TDP-43 ΔC instead of TDP-43 WT for the high aggregation ability of full-length TDP-43 (Wang et al., 2012). These results show that RBP TDP-43 prevented the aggregation of a target UGGAAexp RNA by directly binding to it, consequently abrogating UGGAAexp toxicity.
The above AFM results suggest that TDP-43 has RNA chaperone activity, structurally altering RNAs. We thus addressed whether TDP-43 could change the conformation of (UGGAA)n in a length-dependent manner. In vitro TDP-43 binding experiments (band-shifts) with the (UG)8 and (UGGAA)8 oligos revealed specific binding (Figures S2A and S2B). To further assess structural changes in the RNA and TDP-43 protein upon binding, we used circular dichroism (CD) spectroscopy to monitor such RNA changes with increasing TDP-43 WT protein. The CD spectrum of (UGGAA)8 RNA only has positive peaks at 270 and 220 nm, along with a negative peak at 250 nm (Figure 4H), which are characteristic of RNA molecules adopting the A-form structure (Vorlickova et al., 2012). As the TDP-43 concentration increased, the intensity of the 220-nm band decreased, becoming a negative peak, and the 270-nm peak shifted slightly left to a lower wavelength (Figure 4H, see arrow). The effect of TDP-43 on the CD spectrum of (UGGAA)8 is shown more clearly in Figure 4I, where the spectrum of the (UGGAA)8 + TDP-43 mixture does not overlap with the sum of the spectra for each alone (compare green and blue spectra, respectively). Notably, the spectrum of the mixture showed a less broad and shifted peak centered around 270 nm, indicating of that TDP-43 binding changed the (UGGAA)8 RNA structure. However, it seems that the structure of the protein does not change markedly once bound to (UGGAA)8, as seen from the near-superimposed profiles of the 220-nm peaks that can be attributed to the α-helix structures in the protein, for the (UGGAA)8 + TDP-43 mixture spectrum and the sum of the spectra for each alone (compare green and blue spectra, respectively) (Figure 4I). Interestingly, the structural changes in (UGGAA)4 are much less pronounced than for the (UGGAA)8 (Figures S3F and 4I; compare 270-nm peaks in green spectra between panels F and I), suggesting that TDP-43 induced changes in the overall folding pattern of (UGGAA)8, but only affected the base stacking interactions in (UGGAA)4. The shapes of the CD spectra of (UG)4 and (UG)8 remained the same upon TDP-43 titration, but they exhibited changes in intensity most likely due to an increased protein concentration (Figures S3G and S3H). While their CD profiles predominantly overlapped, there were slight increases in the breadth of the 280-nm peaks, suggesting subtle structural changes in the (UG)n RNA conformation (Figures S3I and S3J). Therefore, we can conclude that TDP43 binding induced structural alterations in both the (UGGAA)n and the (UG)n RNAs, where the latter were subtler than the former (Figures S3G–J). This structural alteration of RNAs by TDP-43 did not require adenosine triphosphate (ATP) or other proteins, thus conforming to the definition of an RNA chaperone (Rajkowitsch et al., 2007) and indicating a novel function of TDP-43 for the maintenance of RNA quality control.
The UGGAA repeat is translated into PPR proteins in SCA31 patient brains and in a Drosophila model of SCA31
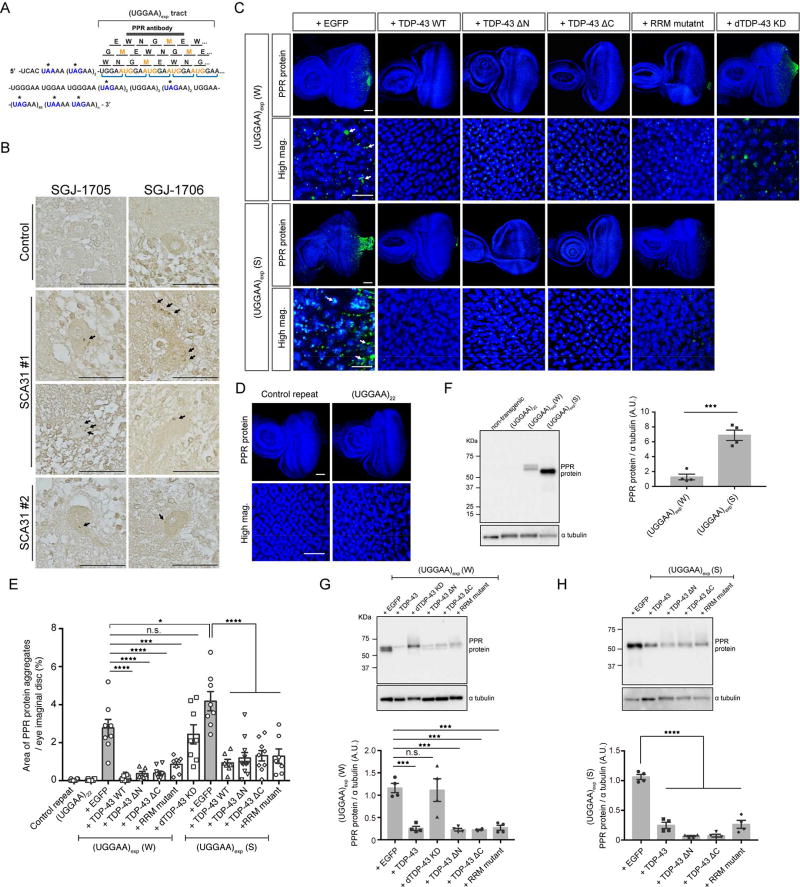
Several lines of evidence indicate that RAN translated proteins are involved in a growing number of diseases, including Huntington’s disease (HD), SCA8, DM1, FXTAS and C9ORF72-linked ALS/FTD (Banez-Coronel et al., 2015; Cleary and Ranum, 2014). Unlike other microsatellite expansion disorders, it is important to note that, in SCA31, the TGGAA repetitive sequence itself includes an AUG translation-initiation codon and both the TAAAA and the TAGAA sequences flanking the TGGAA repetitive sequence should include UAA and UAG translation stop codons (Figure 5A). Translation of the UGGAA repeat in all frames would thus result in the production of an identical PPR protein, poly-WNGME (Figure 5A). To examine whether intronic UGGAA repeats were aberrantly translated into PPR proteins, by either RAN translation mechanisms or AUG-initiated translation, we developed rabbit polyclonal antibodies (anti-PPR antibodies SGJ-1705 and SGJ-1706) against MEWNGMEWNGMEWNG peptides. Immunoblotting confirmed that affinity-purified PPR antibodies successfully detected recombinant poly-WNGME peptides generated in bacteria as Glutathione S-transferase (GST) fusions (Figure S4A).
Figure 5. TDP-43 Negatively Regulates the Synthesis of PPR Proteins.
(A) Schematic of PPR proteins. Amino acids are shown as single letters. Asterisk (*) indicates a stop codon in the sequence surrounding the UGGAA repeat.
(B) Immunostaining for PPR proteins with SGJ-1705 (top) and SGJ-176 (bottom) antibodies in human brain tissues. PPR proteins are detected in cell bodies and dendrites of SCA31 Purkinje cells as dot-like granules (arrows), but not in controls. Scale bars = 50 µm (See also Figure S4).
(C) Immunofluorescence of PPR proteins in eye imaginal discs of the UGGAAexp (W) or (S) line coexpressing TDP-43 WT, RRM mutant or dTDP-43 knockdown (RNAi). Scale bars = 50 or 10 µm (high mag.).
(D) PPR proteins are not observed in eye imaginal discs of flies expressing control repeat or UGGAA22 using GMR-GAL4 by immunostaining with an anti-PPR antibody. Scale bars = 50 or 10 µm (high mag.).
(E) Quantification of PPR protein area in flies of the indicated genotypes (n = 6–10).
(F) Immunoblotting and quantification of PPR proteins in lysates of eye imaginal discs (four independent experiments, n = 40 flies per each genotype). Data are presented as mean ± SEM, p = 0.0004, two-tailed unpaired Student’s t-test. Note that the molecular weights of PPR proteins are slightly different between the W and S lines or between experiments because of their difference in the sizes of UGGAA repeats in our SCA31 flies due to their instability.
(G–H) Immunoblotting and quantification of PPR proteins in lysates of eye imaginal discs (four independent experiments, n = 40 flies per each genotype) (See also Figure S5). In (E), data are presented as mean ± SEM, p < 0.0001 (W line group), p < 0.0001 (S line group), one-way ANOVA; *p = 0.0313, ***p = 0.0001, ****p < 0.0001, p = 0.8703 (W line with EGFP versus W line with dTBPH-KD), Dunnett’s post hoc analysis, n.s.: not significant. In (G–H), p < 0.0001, one-way ANOVA, ***p <0.001, ****p < 0.0001, p = 0.9984 (W line with EGFP versus W line with dTDP-43 KD), Dunnett’s post hoc analysis, n.s.: not significant.
To assess whether PPR proteins were present in SCA31 patient brains, we performed immunohistochemical analyses on human cerebellar tissue sections. In SCA31 patient tissue sections, anti-PPR antibodies detected granular structures (Figure 5B, filled arrows) in cell bodies and dendrites of Purkinje cells, but not in controls, suggesting that PPR proteins are expressed in SCA31 brains (Figure 5B). There were no striking differences in the staining properties of the two anti-PPR antibodies for PPR proteins in SCA31 brain tissue. Whether the PPR protein was generated through RAN translation or conventional AUG translation is currently unclear.
Next, we determined whether PPR proteins are also expressed in flies expressing UGGAAexp. Immunostaining and immunoblotting with anti-PPR antibodies revealed that PPR proteins are expressed in eye imaginal discs of both UGGAAexp (W) and (S) lines, whereas no PPR production was observed in nontransgenic and UGGAA22 fly lines (Figures 5C and 5F). Granular PPR proteins were detected mainly in the cytoplasm of cells in eye imaginal discs of both UGGAAexp (W) and (S) lines (Figure 5C, arrows). Furthermore, PPR protein levels correlated with the severity of eye degeneration (Figure 5E). Indeed, PPR proteins were also detected by anti-PPR antibodies in lysates from UGGAAexp fly lines by immunoblotting and PPR proteins were expressed at a higher level in the UGGAAexp (S) line than in the UGGAAexp (W) line (Figure 5F). The specificity of the affinity-purified PPR antibodies was confirmed by the treatment of cerebellar tissue sections and fly tissues with synthesized poly-WNGME peptides, resulting in no detection of PPR proteins (Figures S4B and S4C). There were also no differences in the staining properties of the two anti-PPR antibodies for PPR proteins in Drosophila tissue (Figure S4D).
TDP-43 negatively regulates PPR protein synthesis
Because TDP-43 acts as an RNA chaperone and affects UGGAAexp-mediated toxicity and RNA foci formation, we evaluated whether TDP-43 affected the production of PPR proteins in flies. Coexpression of TDP-43 significantly reduced the production of PPR proteins in UGGAAexp (W) (2.9% ± 0.53% to 0.1% ± 0.02%, **p < 0.01) and (S) lines (4.2% ± 0.51% to 0.9% ± 0.23%, **p < 0.01) (Figures 5C and 5E), as for RNA foci formation. Immunoblotting of UGGAAexp-expressing fly lysates revealed WNGME-immunoreactive bands and confirmed that TDP-43 markedly inhibited PPR production in vivo (Figures 5G and 5H). In contrast, dTDP-43 silencing, which caused a significant increase in RNA aggregation in the nucleus, did not significantly alter PPR protein levels compared with that in the UGGAAexp (W) flies expressing EGFP (2.9% ± 0.53% to 2.9% ± 0.73%) (Figures 5C, 5E and 5G). Furthermore, coexpression of the TDP-43 RRM mutant, which failed to suppress the accumulation of RNA foci and UGGAAexp-mediated toxicity, still suppressed PPR protein synthesis in UGGAAexp-expressing flies, as revealed by immunohistochemistry [UGGAAexp (W): 2.9% ± 0.53% to 0.9% ± 0.52%; UGGAAexp (S): 4.2% ± 0.51% to 1.0% ± 0.57%, **p < 0.01] (Figures 5C and 5E) or immunoblotting (Figures 5G and 5H). These data suggest that loss of RNA-binding ability of TDP-43 may have negative impact on suppression of RNA foci formation than that of PPR synthesis (Figure 4D and 5E). Taking these findings together, TDP-43 negatively regulates PPR protein levels in vivo; however, other factors such as unknown molecules that interact with TDP-43 or RNA folding states such as RNA foci and hairpin formation may also affect PPR protein levels.
FUS and hnRNPA2B1 modify UGGAAexp-mediated toxicity by suppressing RNA foci formation and PPR protein synthesis
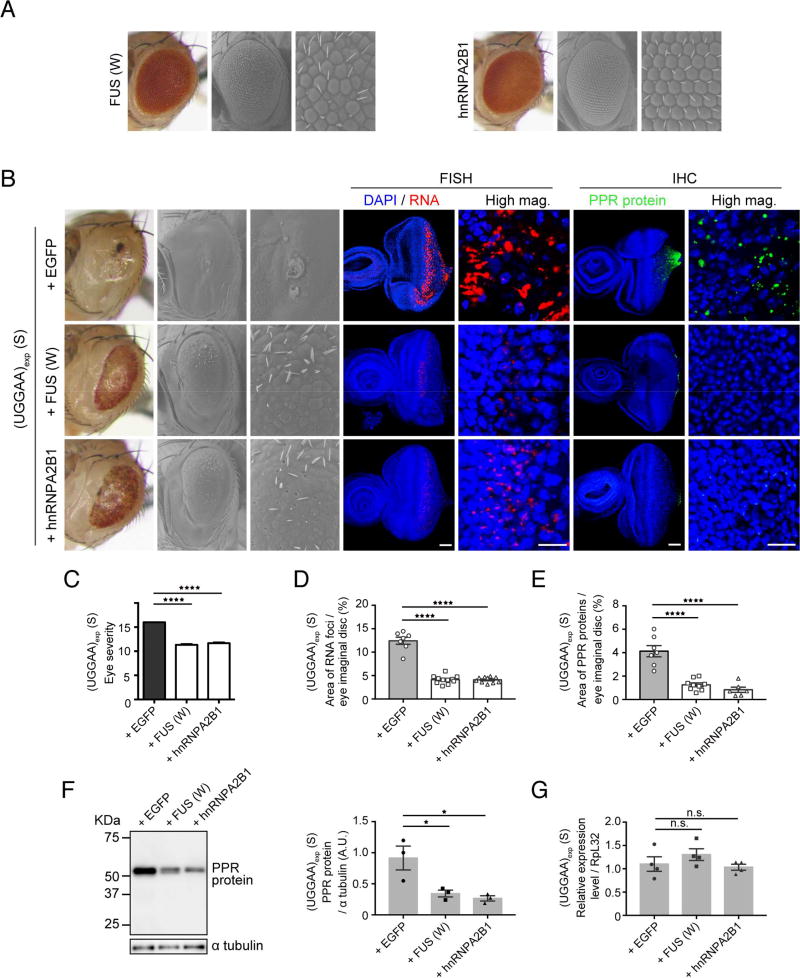
Our screen for potential UGGAAexp-binding proteins using in vitro RNA pull-down assays identified several RNA-associated proteins besides TDP-43, including FUS and hnRNPA2B1 (Figure 2). FUS and hnRNPA2B1 are disease-linked RBPs that share functional and structural similarities, harboring a canonical RRM and a putative prion domain that functions in transcriptional regulation and RNA processing in neurons. Therefore, we evaluated the effects of FUS and hnRNPA2B1 on UGGAAexp-mediated toxicity, using fly lines expressing FUS or hnRNPA2B1 (Figure 6A). While human FUS and hnRNPA2B1 flies displayed a slight rough-eye phenotype (Figure 6A), upregulation of FUS or hnRNPA2B1 dramatically mitigated compound eye degeneration in UGGAAexp (S) flies, as human TDP-43 WT did (Figures 6B and 6C). Furthermore, both FUS and hnRNPA2B1 suppressed the accumulation of RNA foci (4.1% ± 0.26% and 4.1% ± 0.20%, respectively, vs. 12.4% ± 0.76%; both ****p < 0.0001) and the production of PPR proteins (1.3% ± 0.16% and 0.8% ± 0.22%, respectively, vs. 4.1% ± 0.48%; ****p < 0.0001) in flies expressing UGGAAexp (Figures 6B–E). Immunoblot analyses confirmed that FUS and hnRNPA2B1 significantly reduced the amount of PPR proteins (Figure 6F). Coexpression of FUS or hnRNPA2B1 with UGGAAexp did not affect the expression levels of UGGAAexp transcript (Figure 6G). Notably, the effects of TDP-43, FUS and hnRNPA2B1 against RNA aggregation were contrary to those of other RBPs, such as RNA-specific adenosine deaminase B2 in C9orf72-linked ALS/FTD and MBNL1 in DM1/DM2, which were reported to promote RNA foci formation (Donnelly et al., 2013; Querido et al., 2011). Instead, the protective roles of TDP-43, FUS and hnRNPA2B1 to prevent aggregation of their target RNA are reminiscent of the roles of molecular chaperones that prevent the aggregation of mutant proteins, leading to the alleviation of neurodegenerative disease phenotypes. Furthermore, molecular chaperones, such as HSP70 or HSP40, did not mitigate the degenerative phenotypes of flies expressing UGGAAexp (Figure S5A), but did so in flies expressing CGG repeats linked to FXTAS (Jin et al., 2003).
Figure 6. FUS and hnRNPA2B1 Suppress UGGAAexp-Mediated Toxicity, RNA Foci Formation and PPR Protein Synthesis.
(A) LM (left) and SEM (middle and right) images of the compound eyes of Drosophila females expressing FUS (upper) or hnRNPA2B1 (lower). The expression of FUS or hnRNPA2B1 alone caused slight compound eye degeneration in Drosophila.
(B) Upregulation of FUS and hnRNPA2B1 mitigates compound eye degeneration of Drosophila expressing UGGAAexp (S) (See also Figure S6). FUS and hnRNPA2B1 reduce RNA foci and PPR protein accumulation in eye imaginal discs of L3. FUS (W) has weak FUS expression. Scale bars = 50 or 10 µm (high mag.).
(C) Eye degeneration scores from the flies of the indicated genotypes (n = 10).
(D) Quantification of the area of RNA foci in each Drosophila line (n = 7–11).
(E) Quantification of PPR proteins in each Drosophila line (n = 6–10).
(F) Immunoblotting and quantification of PPR protein in lysates of eye imaginal discs (three independent experiments, n =30 flies per each genotype).
(G) Quantitative real-time PCR analysis of transgene expression in each indicated Drosophila line (four independent experiments, n = 80 flies per each genotype). In (C–E), data are presented as mean ± SEM, p < 0.0001, one-way ANOVA; ****p =0.0001, as assessed by Dunnett’s post hoc analysis. In (F), p = 0.0155 (F), one-way ANOVA, *p = 0.0249 (S line with EGFP versus S line with FUS W), *p = 0.0143 (S line with EGFP versus S line with hnRNPA2B1), Dunnett’s post hoc analysis. In (G), p = 0.3073, one-way ANOVA, n.s.: not significant.
Numerous RBPs are implicated in a wide range of cellular functions, including transcription, alternative splicing, mRNA stability and RNA–protein complex formation (D’Ambrogio et al., 2009; Ling et al., 2013). Previous reports suggested that TDP-43 suppression of CGG repeat toxicity is dependent on hnRNPA2B1 orthologues in an FXTAS fly model (He et al., 2014). On the other hand, hnRNPA2B1 suppression of CGG repeat toxicity was reported to be independent of a TDP-43 orthologue (He et al., 2014). To determine whether these RBPs cooperate to achieve the optimal RNP homeostasis or play redundant roles, we knocked down Drosophila FUS (Caz) and Drosophila hnRNPA2B1 (Hrb87F or Hrb98DE) by utilizing fly lines with RNAi targeting these RBPs (Figure S6C). We also tested whether the Drosophila TDP-43 orthologue TBPH is important for FUS- or hnRNPA2B1-mediated suppression of UGGAAexp-induced toxicity using fly lines with RNAi targeting TBPH. Unlike the silencing effects of Hrb87F or Hrb98DE on TDP-43-mediated rescue in the FXTAS fly model, knockdown of Caz, Hrb87F or Hrb98DE did not prevent the TDP-43-mediated suppression of UGGAAexp-induced toxicity (Figures S6A, S6D and S6E). Likewise, TBPH silencing did not affect the FUS- or hnRNPA2B1-mediated suppression of UGGAAexp-induced toxicity in Drosophila (Figures S6B, S6F and S6G). These results suggest that TDP-43, FUS and hnRNPA2B1 could act independently to suppress UGGAAexp-induced toxicity. The effects of the complete absence of TBPH, FUS or hnRNPA2B1 were not investigated because the simultaneous loss of these RBPs has a lethal effect.
Nontoxic short UGGAA repeat RNAs mitigate ALS-linked TDP-43 toxicity
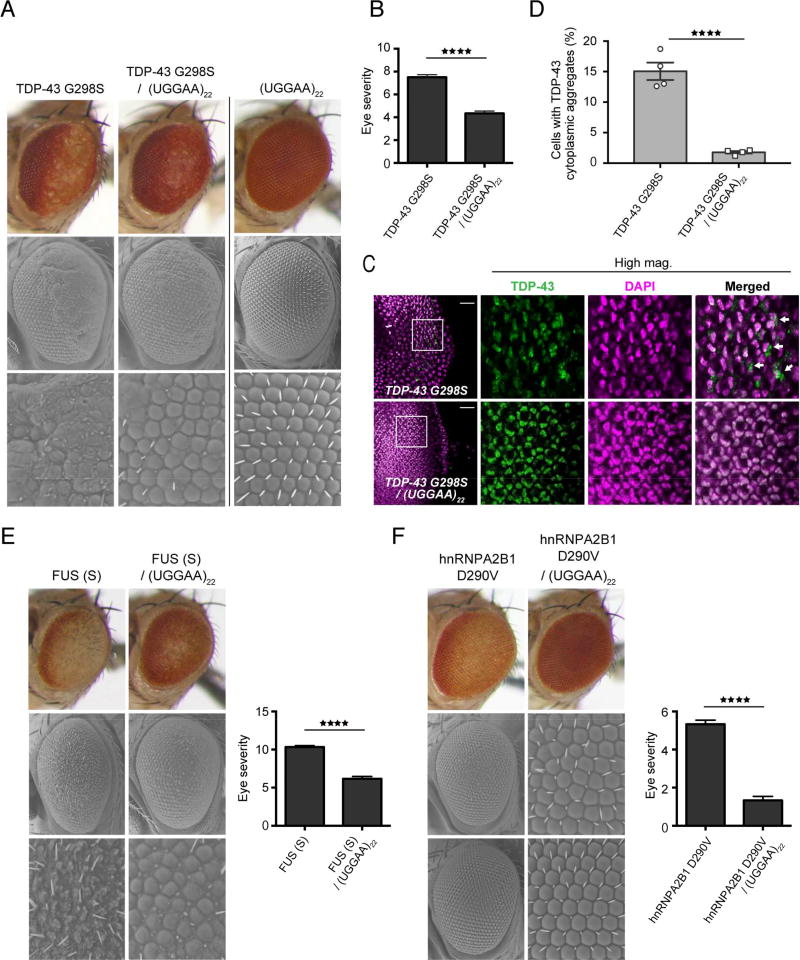
The aggregation of TDP-43 is commonly associated with neurodegenerative conditions, including sporadic and familial forms of ALS and FTD (Ling et al., 2013). Overexpression of TDP-43 in experimental animals recapitulated some of the pathological features of human ALS and FTD. Consistent with previous reports, we confirmed that our fly lines expressing human TDP-43, which has a protective role against UGGAAexp-mediated toxicity (Figure 3B), exhibited degenerative phenotypes (Figure 3C). To our surprise, flies coexpressing toxic RNA UGGAAexp (W line) and toxic RBP TDP-43 exhibited a modest but definite improvement in eye morphology compared with flies expressing either UGGAAexp (W line) or TDP-43 alone (Figures 3B and S7). TDP-43 WT expression in Drosophila by the eye-specific GMR-GAL4 driver was found to cause thinning of the retina. However, coexpression of nontoxic short UGGAA, UGGAA22 or relatively low-level expression of long UGGAA with TDP-43 WT led to restoration of retinal thickness in compound eyes of TDP-43 WT flies (Figure S7). To clarify whether the RNA-TDP-43 interaction modified TDP-43 toxicity in vivo, we assessed the potential effect of a nontoxic short repeat RNA (UGGAA22) on mutant TDP-43 toxicity. ALS-linked mutant TDP-43 G298S-expressing flies exhibited retinal degeneration with loss of pigmentation and ommatidial disruption, whereas coexpression of UGGAA22 significantly mitigated this degeneration (Figures 7A and 7B). Intriguingly, the coexpression of UGGAA22 with TDP-43 remarkably reduced the cytoplasmic aggregates of mutant TDP-43 and led to their retention in the nucleus in eye imaginal discs of flies compared with that in flies expressing mutant TDP-43 alone (1.8% ± 0.20% vs. 15.1% ± 1.42%, ****p < 0.0001) (Figures 7C and 7D). Because TDP-43 participates in multiple pathways of RNA metabolism via its RNA-binding activity (Ling et al., 2013) and TDP-43 RRM mutant flies did not show compound eye degeneration (Figure 3D), RNA–TDP-43 interactions might be crucial for TDP-43 toxicity (Ihara et al., 2013; Voigt et al., 2010). This result is consistent with a previous study showing that ssRNA/DNA binding inhibited TDP-43 aggregation in vitro (Huang et al., 2013). We next examined the effect of UGGAA22 on the toxicities of FUS and hnRNPA2B1, using flies expressing FUS and the MND-linked mutant hnRNPA2B1 D290V. Interestingly, coexpression of UGGAA22 also suppressed compound eye degeneration of flies expressing either FUS or mutant hnRNPA2B1 D290V (Figures 7E and 7F). Thus, nontoxic UGGAA22 exerts protective effects against RBP toxicities by suppressing RBP aggregation and mislocalization. A previous study showed that intronic lariat RNAs inhibited TDP-43 toxicity in yeast, by acting as a decoy competing with endogenous essential RNAs for TDP-43 binding (Armakola et al., 2012). Thus, some RNA species may affect RBP quality control through RNA–RBP interactions in RNP homeostasis.
Figure 7. Nontoxic Short UGGAA Repeat RNAs Mitigate ALS-Linked TDP-43 Toxicity.
(A) LM (upper) and SEM (middle and lower) images of compound eyes from Drosophila females expressing TDP-43 G298S mutant only, UGGGAA22 only and TDP-43 G298S mutant with UGGAA22 (See also Figure S7).
(B) Eye degeneration scores from the flies of the indicated genotypes (n = 7).
(C) Immunohistochemistry of eye imaginal discs detected TDP-43 (green) and DAPI staining (magenta). Scale bars = 20 µm.
(D) Quantification of proportion of cells with TDP-43 cytoplasmic inclusions in TDP-43 G298S and TDP-43 G298S/UGGAA22 fly lines.
(E–F) LM and SEM images with eye degeneration scores of compound eyes from Drosophila females in each genotype (n = 7). In (B–F), data are presented as mean ± SEM, ****p < 0.0001, two-tailed unpaired Student’s t-test.
Discussion
In this study, we elucidated the novel role of RBPs in neurodegeneration, based on analyses of SCA31-associated UGGAAexp-mediated toxicity in vivo. We found that TDP-43 directly binds to UGGAAexp to suppress its neurotoxicity. We demonstrated that TDP-43, FUS and hnRNPA2B1 act as RNA chaperones, suppressing RNA foci formation and PPR translation. Therefore, the facilitation of proper RNA folding by RNA chaperones might play crucial roles in RNA quality control pathways and regulation of RAN translation in vivo.
RNA folding is an essential step required for correct RNA metabolism, such as the processing of tRNAs and mRNAs, the translation of mRNAs and epigenetic regulation by long noncoding RNAs (lncRNAs) (Mercer and Mattick, 2013; Rajkowitsch et al., 2007; Russell, 2008). Several RBPs, including hnRNPs, viral nucleocapsid proteins, ribosomal proteins, cold-shock proteins, histone-like proteins and Sm-like protein Hfq, demonstrated RNA chaperone activity, facilitating conformational rearrangements of RNA from misfolded to functional structures (Rajkowitsch et al., 2007; Russell, 2008). RBPs are thought to bind ssRNA and stabilize RNA folding structures to prevent the misfolding of RNAs. Thus, our data showing that RRM is an essential region in TDP-43 for the suppression of UGGAAexp-mediated toxicity and RNA aggregation (Figures 3 and 4F–4G) suggest that TDP-43–UGGAAexp interactions might prevent aberrant RNA–RNA interactions, resulting in the maintenance of proper RNA folding. Indeed, TDP-43 belongs to the hnRNP family of proteins that shares structural and functional similarities with RNA chaperones (Lagier-Tourenne et al., 2010). Our in vitro binding data demonstrate the specific repeat-length-sensitive binding of TDP-43 to SCA31 (UGGAA)n RNA, similar to that for the common TDP-43 target (UG)n (Figures 2G and 2H). Our analysis clearly shows that TDP-43 binding induced structural alteration of (UGGAA)n repeats and, more subtly, that of (UG)n repeats. Note that structural changes within (UG)6 have been reported (Bhardwaj et al., 2013). We observed only limited changes to the TDP-43 structure upon RNA binding. Notably, a previous CD study also reported an absence of TDP-43 structural change upon its binding to (UG)6 (Bhardwaj et al., 2013); however, structural changes upon RNA binding were observed by both isothermal calorimetry (ITC) and nuclear magnetic resonance (NMR) spectroscopy (Lukavsky et al., 2013). Our AFM data from analyses of longer (UGGAA)20 repeat tracts suggest that TDP-43 can prevent the aggregation of RNA complexes to ssRNAs. Collectively, our AFM data, CD data and in vivo data reveal that TDP-43 may contribute to RNA folding as an RNA chaperone. Similarly, the upregulation of FUS and hnRNPA2B1 also inhibited UGGAAexp-mediated toxicity and RNA foci formation. Interestingly, these proteins could bind to expanded GGGGCC repeats responsible for ALS/FTD (Mori et al., 2013), and in particular, the overexpression of hnRNPA2B1, a CGG-binding protein identified in postmortem brain tissues of FXTAS patients, suppressed expanded CGG-mediated toxicity in a Drosophila model of FXTAS (Sofola et al., 2007). Taking these findings together, FUS, hnRNPA2B1 and TDP-43 might have protective roles against repeat-mediated toxicity as RNA chaperones, which might generally contribute to RNA quality control pathways by suppressing RNA misfolding in neurodegeneration.
Although RNA chaperone functions are ATP-independent, we also found that ATP-dependent RNA helicases such as DDX5, DDX17 and DHX9 bind to UGGAAexp transcripts and, in particular, that knockdown of endogenous DHX9 enhanced UGGAAexp-mediated toxicity in Drosophila (Figure S5B). A recent study also reported that the DEAD-box RNA helicase DDX6 binds to expanded CUG repeats and unwinds CUG-repeat duplexes in vitro in an ATP-dependent manner (Pettersson et al., 2014). These results suggest that ATP-dependent RNA helicases regulate RNA folding, and that dysregulated RNA folding might contribute to the pathomechanisms of repeat RNA diseases.
The precise mechanisms by which RBPs affect the translation of repeat-encoded PPR proteins in the SCA31 Drosophila model are still unclear. However, depletion of TDP-43 did not alter PPR protein levels, but caused marked accumulation of RNA foci in nuclei, suggesting the existence of other factors including RNA folding state and unknown molecules that might affect PPR protein levels. We propose three potential models of regulatory mechanisms for repeat-associated translation by TDP-43: i) the direct binding of RBP complex to UGGAAexp impedes the initiation of translation by blocking ribosomal scanning; ii) RNA chaperone activity alters UGGAAexp RNA conformation essential for aberrant repeat-associated translation; and iii) binding of RBPs to UGGAAexp repeats in the nucleus interferes with cytosolic transport of UGGAAexp transcripts, resulting in reduced PPR translation. Indeed, a recent study suggested that MBNL1 repressed the aberrant translation of repeat RNAs by the nuclear retention of repetitive CUG/CAG RNA (Kino et al., 2014).
Although we demonstrated that TDP-43, FUS and hnRNPA2B1 are potent modifiers of UGGAAexp-mediated neurodegeneration, the mechanisms leading to such neurodegeneration remain obscure. Regarding an RNA gain-of-toxic-function mechanism, the consequence of UGGAAexp RNA toxicity may be the sequestration of RBPs, resulting in their functional insufficiency leading to other noncoding repeat RNA diseases. Indeed, RNA foci in human postmortem tissues were positively immunostained for TDP-43, further suggesting that RNA chaperones are involved in SCA31 pathogenesis. Notably, silencing of dTDP-43 alone induced only slight compound eye degeneration compared with silencing of dTDP-43 and UGGAAexp expression in Drosophila (Figures 3B and 3C). Because knockdown of potential UGGAAexp-binding proteins, such as Drosophila DHX9 (maleless) and Drosophila NONO/SFPQ (nonA), exacerbated degeneration in flies expressing UGGAAexp (Figure S5B), we speculate that UGGAAexp-mediated toxicity may involve the sequestration of TDP-43 and additional UGGAAexp-binding proteins into RNA foci. However, the downstream effects of this sequestration on TDP-43 and other RBP targets remain to be determined.
Previous studies reported that aberrant RNAs caused neurodegeneration through innate immune activation (Tarallo et al., 2012), oxidative stress-induced p53 activation (Hanada et al., 2013) and gene silencing mediated by direct binding between repeat RNAs and the genome (Colak et al., 2014), providing mechanistic explanations for the numerous gains of toxic function of aberrant RNAs themselves. It is important to understand the contributions of repeat RNA and PPR proteins to disease pathogenesis. Interestingly, the number of RNA foci clearly correlated with disease severity in the SCA31 Drosophila model (Figures 4B–D). These findings expand the growing list of microsatellite expansion disorders in which repeat-associated translation occurs, and support a dual-toxicity mechanism of repeat RNAs and PPR proteins in neurodegeneration.
Finally, we discovered that UGGAA22, a nontoxic short repetitive RNA itself, conversely has a protective role against TDP-43-, FUS- and hnRNPA2B1-induced toxicities (Figure 7). Because RNA-binding activity mediates neurotoxicity in transgenic Drosophila models of TDP-43 and FUS proteinopathies (Daigle et al., 2013; Ihara et al., 2013; Voigt et al., 2010), we speculate that nontoxic RNAs may contribute to the control of RBP proteins, suppressing their aggregation and mislocalization. Indeed, the UGUGU pentamers are highly enriched in introns, lncRNAs and intergenic RNAs, which contain many putative TDP-43-binding sequences (Polymenidou et al., 2011; Tollervey et al., 2011). Given that microsatellites within the promoters and other cis-regulatory regions can greatly increase the divergence of gene expression profiles across the great apes (Bilgin Sonay et al., 2015; Gymrek et al., 2016), microsatellites located in untranslated regions or introns may also have a physiological role for proteotoxicity as disease modifiers.
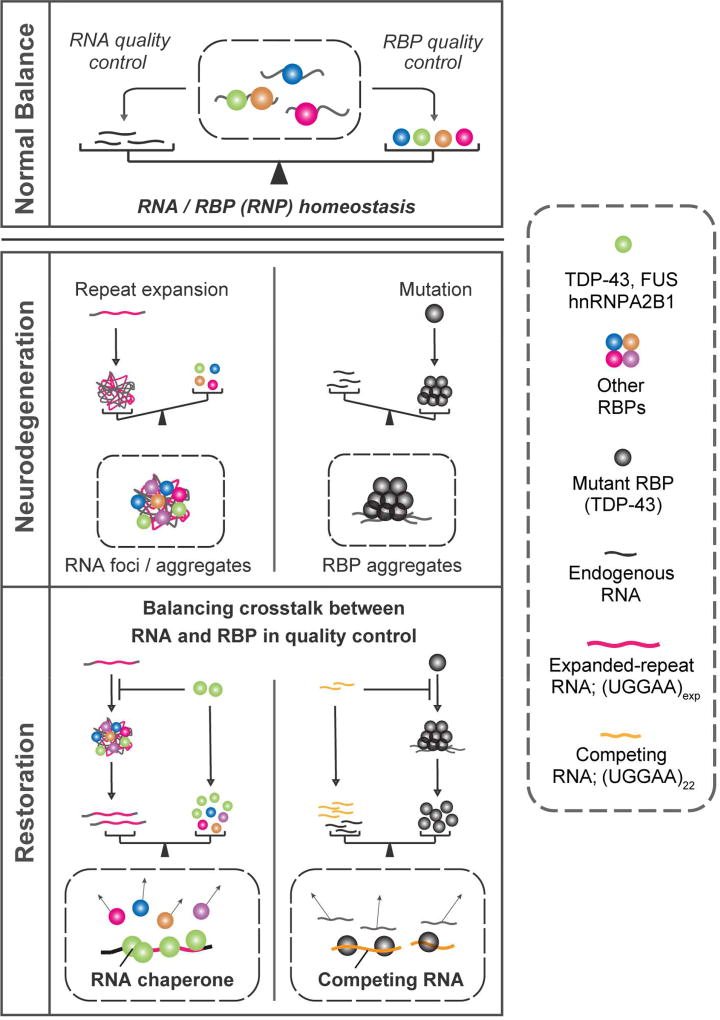
Thus, we hypothesize that both RNA and RBP quality control are inextricably linked and balanced RNA and RBP crosstalk contributes to RNP homeostasis (Figure 8). Therefore, mutations in either RNP component causing their aggregation initiate an imbalance in crosstalk between RNAs and RBPs, leading to neurodegeneration. Previous studies showed that TDP-43 binds to pre-mRNAs with long introns or to lncRNAs, helping to maintain transcript levels (Polymenidou et al., 2011; Tollervey et al., 2011), whereas lncRNAs function as a sponge or scaffold in a broad epigenetic regulatory network for RBPs including multiple protein complexes and miRNAs (Mercer and Mattick, 2013). Considering the widespread involvement of RNA–RBP interacting networks in neurons, our present findings may provide a novel concept towards understanding the mechanistic link between RNA and RBP metabolism in both physiological and pathological processes (Figure 8). Balancing RNA–RBP crosstalk is a potential therapeutic strategy for both noncoding microsatellite expansion disorders and RBP proteinopathies.
Figure 8. Model of Pathogenic Imbalance in RNP Homeostasis in Microsatellite Expansion Disorders or MND.
Our in vivo and in vitro data suggest that not only TDP-43 regulate RNA toxicity, but the opposite can also occur; namely, nontoxic short RNA can regulate TDP-43 toxicity, suggesting that balanced crosstalk of RNA and RBP contributes to maintaining RNP homeostasis.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yoshitaka Nagai (nagai@neurother.med.osaka-u.ac.jp).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly stocks
Transgenic RNAi fly lines bearing UAS-IR-TBPH #1 (#38379), UAS-IR-TBPH #2 (#38377), UAS-IR-Maleless (#19691), UAS-IR-nonA (#26442), UAS-IR-DmRH1 (#46933), UAS-IR-Caz (100291), UAS-IR-Hrb87F (#100732) and UAS-IR-Hrb98DE (#29523) were obtained from Vienna Drosophila RNAi Center (VDRC) and the Bloomington Drosophila Stock Center at Indiana University (BDSC). The level of knockdown of TBPH in the #38379 line was reported to be an approximately 23% reduction compared with that in wild-type flies (Lin et al., 2011). Transgenic fly lines bearing UAS-EGFP (#6658, #6874), elav-GAL4 (#458) and UAS-IR-GFP (#9330) were obtained from BDSC. The transgenic fly line bearing GMR-GAL4 has been described previously (Yamaguchi et al., 1999). The genotypes of the fly lines used are described in Supplemental Table 3. All experiments were performed at 25 °C unless otherwise stated. Eye phenotypes of 1–2-day-old flies were evaluated with the stereoscopic microscope model SZX10 (Olympus) and a scanning electron microscope (Miniscope TM1000; Hitachi High-Technologies Corporation.).
The fly lines were allocated to experimental groups based on genotype, equivalent age and gender. Where multiple groups of the same genotype were required, these were allocated randomly to the particular experimental conditions. Sample sizes were estimated using pilot experiments.
Human Tissues
All human tissues were collected at Tokyo Medical and Dental University. Brain autopsy was performed on each patient under the family’s written consent. The investigation was approved by the Institutional Ethics Committee of the Tokyo Medical and Dental University. The penta-nucleotide repeat insertion was tested by PCR in all SCA31 individuals, and controls. Two SCA31 individuals were both in a mild stage of the disease. Note that the lengths of the mutation are 2.81 kilo-base-pairs (kb) in Patient 1(age at death: 80 years/male) and 2.86 kb in Patient 2 (age at death: 74 years/male), roughly corresponding to 562 and 572 penta-nucleotide repeats respectively. We also studied four control brains which included two patients with sporadic ALS (age at death: 61 years/male and 73 years/male), cerebral infarction (age at death: 83 years/female), Parkinson’s disease (age at death: 88 years/male) and SCA6 (age at death: 75 years/female). The data represented in Figure 5 is one of each with cerebral infarction. The clinical summaries of SCA31 individuals are previously described (Niimi et al., 2013). Sample sizes were estimated using pilot experiments.
Cell Lines
PC12 cells (a pheochromocytoma of the rat adrenal medulla, ATCC® CRL-1721™) were cultured in DMEM + supplemented with 10% (v/v) HS, 5% (v/v) FBS and Penicillin/Streptomycin at 37C in a humidified atmosphere at 5% CO2 unless otherwise indicated in figure legends or method details.
METHOD DETAILS
Generation of fly lines
Transgenic flies expressing UGGAAexp, human wild-type TDP-43 and FUS were generated by standard techniques using a pUAST vector. Transformant lines were generated by standard procedures, using the y,w strain (BestGene Inc., Chino Hills, CA, USA). The transgenic fly line expressing human hnRNPA2B1 was previously described (Kim et al., 2013). SCA31 or control repeat insertion DNA was amplified by PCR from genomic DNA, as previously described (Sato et al., 2009). The PCR amplicons were digested with HaeIII, and purified and cloned into the pBluescript KS (+) vector (Strategene). Fly expression constructs were created by shuttling the DNA fragments from pBluescript KS (+) into the Drosophila transformation vector pUAST, between the NotI and XhoI cloning sites. These constructs include either a TAG or a TAA translation stop codon immediately upstream of the repeat sequences. The pUAST-TDP-43 ΔN and pUAST-TDP-43 ΔC constructs were generated by deleting the regions encoding amino acids 1–89 and amino acids 276–414, respectively. The TDP-43 RRM mutant construct was generated by mutating the phenylalanine residues 147, 149, 194, 229 and 231 to leucine, with the QuikChange Multi Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA), as previously described (Elden et al., 2010).
Repeat size estimation in fly strains
SCA31 and control repeats were amplified from fly genomic DNA by PCR. For amplification of the entire repeat, primers were designed to amplify the transcript region of the pUAST vectors. The forward primer was 5′-TCTACGGAGCGACAATTCAA-3′ and the reverse primer was 5′-TGCTCCCATTCATGAG-3′. The PCR protocol was as follows: denaturation (5 min at 94 °C) followed by 30 cycles of amplification (30 s at 94 °C and 6 min at 68 °C). For the PCR amplification of the UGGAAexp region (Part 1), the forward primer used was 5′-AGGGAATTGGGAATTCGTTAACA-3′ and the reverse primer was 5′-TCCATTCCATTCTATTCTATTCCCAT-3′. The PCR protocol (four-step PCR) was as follows: 5 min at 94 °C, 3 cycles (30 s at 94 °C, 1 min at 68 °C and 4 min at 72 °C), 3 cycles (30 s at 94 °C, 1 min at 66 °C and 4 min at 72 °C), 3 cycles (30 s at 94 °C, 1 min at 64 °C and 4 min at 72 °C), 3 cycles (30 s at 94 °C, 1 min at 62 °C and 4 min at 72 °C), 25 cycles (30 s at 94 °C, 1 min at 60 °C and 4 min at 72 °C) and 5 min at 72 °C. Subsequently, the approximate sizes of the repeats amplified by PCR were estimated by gel electrophoresis. To identify the exact repeat length of line 3, PCR products of line 3 were resolved on a 1% agarose gel by electrophoresis, purified and cloned into the PCRII TA vector (Invitrogen), and their repeat lengths were determined by DNA sequencing.
Fly experiment
Flies were crossed in the absence of RU486 (mifepristone) on standard fly food. One-day-old adult females were transferred and aged in food treated with either RU486 (100 µg) or ethanol for the periods indicated. Climbing and lifespan analyses were performed as described previously, with some modifications (Feany and Bender, 2000). For the climbing assay, 20 flies were placed in a plastic vial which was gently tapped to ensure their descent to the bottom of it. After 15 s of climbing, the number of flies at the top of the vial (a distance greater than 10 cm) was counted. Five trials were repeated for each time point. More than 100 flies were tested per genotype. Phenotype severity was quantified using a validated scale (Wen et al., 2014). Briefly, eye phenotypes were scored based on 4 criteria: reduction in size of the eye, loss of pigmentation in the eye, disruption of the ommatidial array; altered hair growth between the ommatidia. Each criterion was scored 1 to 4 depending on the severity.
RNA pull-down assay
The RNA pull-down assay was used to screen the UGGAAexp RNA-binding proteins (screen 1, performed in the laboratory of K.I.; screen 2, performed in the laboratory of N.C.B.). All manipulations were performed at 4 °C or on ice. The nuclear fraction of PC12 cells or mouse brain was analyzed in screen 1 or screen 2. PC12 cells were washed twice with ice-cold phosphate-buffered saline (PBS). For subcellular fractionation, cells were lysed in NE-PER extraction reagent (Pierce), in accordance with the manufacturer’s protocol. Biotinylated repeat RNA transcripts were synthesized in vitro with T7 RNA polymerase, in accordance with the manufacturer’s instructions (Roche). The genomic DNA of the patients or control individuals with normal repeats was amplified by PCR as previously described (Sato et al., 2009), and the purified PCR products were used as template DNA for in vitro transcription. For the binding reaction, biotinylated RNAs (3 µg) were captured with washed Dynabeads M-280 Streptavidin (Invitrogen), incubated with the nuclear fraction of PC12 cells in 1× binding buffer [10 mM HEPES (pH 7.6), 100 mM KCl, 5 mM MgCl2, 5 mM DTT and salmon sperm DNA) for 5 h and then washed twice with 1× binding buffer. For the competition experiments, nuclear extracts were incubated with a 10-fold excess of nonbiotinylated competitor RNA before addition of the biotinylated probe. The same amounts of biotin and lysates were used in the control binding reactions. The captured RNA–protein complexes were separated by SDS-PAGE and stained with Coomassie Blue. The lanes of interest were excised from the gel and subjected to LC-MS analysis. In screen 2, nuclear extract preparation and RNA pull-down assay were performed, as described previously (Sellier et.al. 2013). Briefly, a total of 300 mg of nuclear extract was passed over an in vitro-transcribed and -biotinylated RNA (Biotin 11 CTP; PerkinElmer) bound to streptavidin-coated magnetic beads (Dynabeads M-280 streptavidin; Thermo Fisher Scientific) in the presence of 20 mM HEPES, 300 mM NaCl, 8 mM MgCl2, 0.01% NP40, 1 mM DTT and protease inhibitor cocktail (PIC; Roche). The magnetic beads with immobilized RNA and proteins bound to it were washed and the bound proteins were eluted by boiling for 3 min in the sample buffer. The proteins were identified using a NanoESI_Ion Trap (LTQ XL; Thermo Fisher Scientific).
In situ hybridization in human samples
Preparation of human brain samples (patient 1: an 80-year-old man; patient 2, a 74-year-old man) and detection of the repeat transcripts in Purkinje cells by FISH were performed with digoxigenin (DIG)-conjugated locked nucleic acid (LNA) probes (5 nM; Exiqon) [(TTCCA)5 or (TTTTATTCTA)2.5], as described previously with some modifications (Sato et al.). Human brain sections were deparaffinized two times for 20 min in Histosol Plus (Shandon) and dehydrated as follows: twice in 100% ethanol (5 min), twice in 95% ethanol (5 min), once in 80% ethanol (5 min) and once in 70% ethanol (5 min), followed by rinsing in PBS before RNA FISH. Glass coverslips containing brain sections were fixed in 4% PFA for 20 min and washed three times with PBS. The slides were incubated for 10 min in PBS with 0.5% Triton X-100 and washed three times with PBS before prehybridization in 40% dimethyl sulfoxide (DMSO), 40% formamide, 10% bovine serum albumin (BSA) (10 mg/mL) and 2× saline-sodium citrate (SSC) buffer for 30 min. The slides were then hybridized for 2 h in 40% formamide, 10% DMSO, 2× SSC, 2 mM vanadyl ribonucleoside, 60 µg/mL tRNA and 30 µg/mL BSA with 0.75 µg of (TTCCA)×8-Cy3 or (TGGAA)×8-Cy3 DNA oligonucleotide probe (Sigma). Next, the slides were washed twice in 2× SSC/50% formamide and twice in 2× SSC. Following FISH, the slides were washed twice successively in 2× SSC/50% formamide, and then in 2× SSC and in PBS. The slides were subsequently incubated for 2 h with a primary antibody against TDP-43 (1/100 dilution; Abcam AB41972). The slides were then washed twice with PBS before incubation with a goat anti-rabbit secondary antibody conjugated with Alexa-Fluor 488 (1/500 dilution, Fisher) for 60 min, and incubated for 10 min in 2× SSC/DAPI (1/10,000 dilution) and rinsed twice in 2× SSC before mounting in Pro-Long media (Molecular Probes). The slides were finally examined using a fluorescence microscope (Leica). This experiment was repeated with two independent samples from patient by two independent laboratories (Ishikawa lab and Charlet-Berguerand lab).
In situ hybridization in fly samples
Eye imaginal discs from crawling third-instar larvae were fixed in 4% formaldehyde/PBS for 30 min at 4 °C and then incubated in methanol. Fixed samples were rehydrated by successive treatments with a graded methanol series (75%, 50% and 25% v/v in H2O), and then treated with HCl/H2O solution (0.2 N). Next, the samples were permeabilized in 0.2% Triton X-100/PBS at room temperature, fixed again in 4% formaldehyde/PBS for 5 min, washed with PBS and then incubated in glycine/PBS (2 mg/mL). After the acetylation steps, prehybridization was performed with hybridization buffer [40% formamide, 2× SSC, yeast tRNA (0.2 mg/mL), salmon sperm DNA (1 mg/mL), heparin (0.5 mg/mL)] for 30 min at 45 °C, and hybridized for 2 h (or overnight) at 45 °C with DIG-conjugated LNA probes (5 nM) in hybridization buffer. After washing, the samples were incubated in a solution containing the anti-DIG antibody Fab conjugated with alkaline phosphatase (Roche), and developed by HNPP/FAST TR (Roche). For nuclear counterstaining with DAPI, the samples were mounted with an aqueous mounting kit. For the RNase experiments, after fixation, the samples were incubated with 50 µg/mL RNase A for 30–60 min at 37 °C. The LNA-oligonucleotide probes (Exiqon) were (TTCCA)5 and (TTTTATTCTA)2.5. Images were taken using a confocal laser scanning microscope model LSM510 Meta (Carl Zeiss). For quantification assay of RNA foci area, data were collected from 6–10 independent fly samples in each genotype repeated with at least two independent experiments. Experiments were conducted in a blinded fashion.
Human tissue immunohistochemistry
The rabbit polyclonal antibodies SGJ-1705 and SGJ-1706 (anti-PPR) against the [H]CMEWNGMEWNGMEWNG[OH] peptide, predicted to be a UGGAA repeat-encoded pentapeptide expressed in SCA31 patients and Drosophila lines, were developed commercially (Sigma-Aldrich, Tokyo, Japan). For antigen retrieval, paraffin-embedded human sections were deparaffinized with xylene, washed with distilled water, boiled for 1 min three times in 10 mM sodium citrate (pH 7.4) and immersed in formic acid (99.5%) for 5 min. The sections were immunostained with an anti-PPR antibody at 1/500 dilution as the primary antibody. For western blotting analyses to confirm antibody specificity against PPR proteins, GST-fused PPR proteins were harvested by a modified version of a previously described method (Sato et al., 2009).
Immunohistochemical analysis in fly samples
For analysis of retinal tissues in one day-old female flies, fly heads were fixed in Carnoy’s fixative solution and embedded in paraffin. The serial 3 µm sections were stained with hematoxylin and eosin. For immunohistochemical analysis, the eye imaginal discs of third instar larvae were immunostained with primary antibodies including anti-PPR antibodies (SGJ-1705 and SGJ-1706) at 1/1,000 dilution or an anti-TDP-43 antibody (Proteintech) at 1/1,000 dilution and Alexa 488-conjugated anti-rabbit IgG antibody (Invitrogen) at 1/2,000 dilution as the secondary antibody. For quantification assay of PPR protein area, data were collected from 5–10 independent fly samples in each genotype repeated with at least two independent experiments. The numbers of cells with cytoplasmic TDP-43 were calculated using the National Institutes of Health Image J software and at least five discs were analyzed for each genotype. Primary antibodies are listed in Key Resource Table. Experiments were conducted in a blinded fashion.
Immunoblotting
Western blotting of fly tissue was performed following standard protocols. To assess the expression levels of the TDP-43 protein, five heads of flies expressing TDP-43 or TDP-43 deletion mutants were collected, frozen and lysed in 1× Laemmli sample buffer. To assess the expression levels of the PPR protein, 10 eye discs of third-instar larvae were collected, frozen and lysed in 1× Laemmli sample buffer. Lysates were incubated at 99 °C for 5 min and then centrifuged at 17,000 g for 10 min at 25 °C. The resultant supernatants were then subjected to SDS-PAGE (5%–20% gradient polyacrylamide gel; ATTO) and transferred onto polyvinylidene fluoride membranes (Thermo Fisher Scientific, Waltham, MA USA). These membranes were blocked for 1 h in 5% nonfat dry milk at room temperature (25 °C) and then incubated overnight (16–18 h) with the primary antibody at 4 °C. Membranes were washed four times in Tris-buffered saline (TBS), incubated in horseradish peroxidase (HRP)-conjugated secondary antibody (1/5,000 dilution) for 1 h and then washed four times in TBST (0.1% Tween20 in TBS). Proteins were visualized using ECL plus (GE Healthcare UK Ltd., Buckinghamshire, England). The primary antibodies used were as follows: anti-TDP-43 rabbit polyclonal (1/5,000 dilution; Proteintech), anti-tubulin (E7, 1/10,000 dilution; Developmental Studies Hybridoma Bank) and anti-PPR antibodies (SGJ-1705 and SGJ-1706; 1/5,000 dilution). Immunoblot data are representative of at least two independent experiments.
Quantitative real-time PCR
Total RNA from 20 male or female third-instar larvae was extracted using the RNeasy kit (Qiagen). All experiments were repeated four times using independent RNA preparations (total 80 fly larvae in each genotype). cDNAs were obtained using PrimeScript™ RT Master Mix (Takara Bio). Before PCR, cDNA was treated with DNase I (Invitrogen) to remove genomic DNA. Real-time PCR was performed in triplicate using a LightCycler 480 (Roche). Data were analyzed using either the ΔΔCt method or the standard curve method (for Rp49 levels). The primers used for SCA31 or the control insertion were as follows: forward primer 5′-AGGGAATTGGGAATTCGTTAACA-3′ and reverse primer 5′-CTGGAGTGCAGTGACGTGATCT-3′. The quantities measured by these analyses were adjusted by predesigned, gene-specific primers and probe sets of Rp49 named RpL32 in FlyBase (Applied Biosystems).
Electrophoretic Mobility Shift Assays
RNA oligonucleotides were purchased as purified from Sigma-Aldrich, Toronto, Canada, and end-labeled using [γ-32P]ATP. Reactions were carried out in a 10 µL volume containing ~2 pmol of RNA with purified TDP-43 WT or TDP-43 ΔC. Binding reactions contained a final buffer concentration of 10 mM Tris pH8, 10 mM NaCl, 2mM MgCl2, 5% glycerol, and 1mM DTT. RNA was incubated with TDP-43 and the buffer described above for 30 minutes at room temperature prior to loading on a 6% native polyacrylamide gel. The gel was then electrophoresed at 120 Volts for 1 hour and 45 minutes in 1X TBE, dried and then autoradiographed.
CD spectroscopy
CD experiments were performed at room temperature using an Aviv model 62 DS spectropolarimeter using indicated concentrations of RNA samples. An average of three CD spectra, over the wavelength range of 320–220 nm, was recorded in a 1-mm path length cuvette at a scan rate of 20 nm/min. For the titrations, aliquots of TDP-43 in the same buffer as the RNA were added to the sample and mixed before recording the spectrum.
RNA preparation and AFM imaging
RNA was prepared by in vitro transcription with T7 RNA polymerase, in accordance with a standard protocol (Roche). The transcripts were subjected to DNase I (Invitrogen) digestion for 30 min at 37 °C. Then, synthesized RNAs were purified with the RNeasy Mini Elute Cleanup kit (Qiagen) and diluted with sterile purified water to a concentration of 1 ng/µL. The RNA solution was then heat-denatured (90 °C) and incubated on ice. Next, it was incubated with or without recombinant TDP-43 ΔC (Abcam) for 10 min at room temperature, followed by the addition of MgCl2 (final concentrations: 0.7 ng/µL RNA, 9 ng/µL TDP-43 ΔC, 10 mM MgCl2). We observed the RNA solutions under in-liquid conditions using High-speed AFM (HS-AFM, NanoExplorer, Research Institute of Biomolecule Metrology) with BL-AC10DS probes (Olympus) on which sharp electron beam-deposited tips had been grown. The buffer composition and denaturation conditions for the reaction were optimized further for taking AFM images. AFM images were taken using a 2-µL drop of RNA solution or protein–RNA solution spotted onto freshly cleaved mica (1.5 nm in diameter) for 5 min, and rinsed with water. The scanning rate was 0.5 frames/s. Fields of a 500 × 500 nm square area were scanned (total area, 1,050,000 nm2, n > 900 molecules counted). Image processing and measurement of heights and widths were performed using Scanning Probe Image Processor (SPIP™, Image Metrology) and ImageJ. The amount of UGGAAexp RNAs was quantified with delineations for single-stranded RNAs (height < 0.9 nm), small aggregates (height > 1.5 nm; width > 15 nm and < 40 nm) or large aggregates (height > 1.5 nm, width > 40 nm) (Fig. 4g). Statistical analyses were performed using one-way ANOVA with Tukey’s post hoc analysis.
Filter binding assay
Preparation of recombinant TDP-43 proteins and binding analysis of TDP-43 with nucleotides were performed as described previously (Furukawa et al., 2011). Briefly, TDP-43 dissolved in a buffer containing 6 M guanidine hydrochloride was first refolded by 40-fold dilution with 100 mM Na-Pi, 100 mM NaCl, 5 mM EDTA, 5 mM DTT and 10% glycerol at pH 8.0, followed by centrifugation at 20,000 g for 30 min at 4 °C. Soluble and refolded TDP-43 in supernatant fraction was then incubated with nucleotides biotinylated at the 5f-end: 0.1 µM (TG)12 and (AC)12 single-stranded DNA, and 6 ng/µL in vitro-transcribed UGGAA repeat RNA. After 1 h at room temperature, the mixture was filtered through a nitrocellulose membrane (PROTRAN, 0.2 µm; Schleicher & Schuell) overlaid on a nylon membrane (Hybond-N+, 0.45 µm; Amersham Biosciences) in a 96-well dot-blot apparatus (ATTO). The bound DNA/RNA was crosslinked to the membranes by UV Stratalinker (Stratagene) at an energy level of 0.12J. The membranes were blocked with 3% bovine serum albumin in PBS with 0.1% Tween 20, and the biotin-labeled nucleotides were detected with streptavidin-HRP (1:10,000; Nakalai Tesque) and Pierce Western Blotting Substrate (Pierce).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical parameters including the definitions and exact values of n (e.g., number of experiments, number of flies, number of eye imaginal discs, etc.), distributions and deviations are reported in the Figures and corresponding Figure Legends. Data of eye degeneration score, quantitative real-time PCR analysis, quantification of RNA foci / PPR protein area of eye imaginal discs and quantification of band intensities in immunoblots were presented as mean ± SEM using one-way ANOVA with Tukey’s post hoc analysis or Dunnett’s post hoc analysis (Figure 1–6, S2D, S5 and S6). Lifespan analyses were performed with log-rank test (Figure 1M). Climbing analyses were presented as mean ± SD using two-way ANOVA (Figure 1N). Quantification of RNA species in AFM images were presented as mean ± SD using one-way ANOVA with Tukey’s post hoc analysis (Figure 4G). Data of eye degeneration score and quantification of TDP-43 aggregates were presented as mean ± SEM using two-tailed unpaired Student’s t-test (Figure 7). Differences in means were considered statistically significant at p < 0.05. Significance levels are: * p < 0.05; ** p < 0.01; *** p < 0.001; **** p <0.0001; n.s., non-significant. Statistical analysis was performed in GraphPad PRISM 7.0 software.
Supplementary Material
Highlights.
UGGAA repeats form RNA foci and elicit repeat-associated translation in SCA31.
TDP-43 binds UGGAA repeats and suppresses their toxicity.
TDP-43 regulates UGGAA RNA folding and PPR synthesis, acting as an RNA chaperone.
Exogenous RNA that binds to TDP-43 suppressed TDP-43 aggregation and toxicity.
Acknowledgments
We thank Hisae Kikuchi and Drs. Takeshi Kasama, Michi Okita, Helena Akiko Popiel for their advice and technical support. We thank Drs. Francisco E. Baralle and Emanuele Buratti for the kind gifts of the TDP-43 bacterial expression constructs and for the proteins. We also thank Dr. Leonard Petrucelli for critical reading of the manuscript and helpful discussions. This work was supported in part by a grant from Core Research for Evolutional Science and Technology (H.M.) from the Japan Science and Technology Agency; by a Grant-in-Aid for Scientific Research on Innovative Areas (Synapse and Neurocircuit Pathology) (Y.N.) and the Strategic Research Program for Brain Sciences (Integrated Research on Neuropsychiatric Disorders) (Y.N.) from the Ministry of Education, Culture, Sports, Science and Technology, Japan; by Grants-in-Aid for Scientific Research (A) (H.M. and K.I.) and for Scientific Research (C) (K.I.), and for Challenging Exploratory Research (Y.N. and T.I.) from the Japan Society for the Promotion of Science; by Grants-in-Aid as a Health and Labour Sciences Research Grant (Y.N.), and from the Research Committee for Ataxic Diseases (H.M. and Y.N.) from the Ministry of Health, Labour and Welfare; and finally by a grant for Practical Research Projects for Rare/Intractable Diseases (Y.N) from the Japan Agency for Medical Research and Development.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
Conceptualization, T.I., K.I., Y.N.; Methodology, T.I., N.F.; Investigation, T.I., N.S., M.U., N.F., C.S., A.K. T.G.D., M.M., B.Z., C.E.P., E.T. and Y.F, N.C.B.; Writing – Original Draft, T.I., K.I. and Y.N.; Writing – Review & Editing, T.I., C.E.P., K.I. and Y.N.; Resources, J.P.T.; Supervision, T.Y., K.W. and H.M.
References
- Armakola M, Higgins MJ, Figley MD, Barmada SJ, Scarborough EA, Diaz Z, Fang X, Shorter J, Krogan NJ, Finkbeiner S, et al. Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nat. Genet. 2012;44:1302–1309. doi: 10.1038/ng.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banez-Coronel M, Ayhan F, Tarabochia AD, Zu T, Perez BA, Tusi SK, Pletnikova O, Borchelt DR, Ross CA, Margolis RL, et al. RAN translation in Huntington disease. Neuron. 2015;88:667–677. doi: 10.1016/j.neuron.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj A, Myers MP, Buratti E, Baralle FE. Characterizing TDP-43 interaction with its RNA targets. Nucleic Acids Res. 2013;41:5062–5074. doi: 10.1093/nar/gkt189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgin Sonay T, Carvalho T, Robinson MD, Greminger MP, Krutzen M, Comas D, Highnam G, Mittelman D, Sharp A, Marques-Bonet T, Wagner A. Tandem repeat variation in human and great ape populations and its impact on gene expression divergence. Genome Res. 2015;25:1591–1599. doi: 10.1101/gr.190868.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary JD, Ranum LP. Repeat associated non-ATG (RAN) translation: new starts in microsatellite expansion disorders. Curr. Opin. Genet. Dev. 2014;26C:6–15. doi: 10.1016/j.gde.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colak D, Zaninovic N, Cohen MS, Rosenwaks Z, Yang WY, Gerhardt J, Disney MD, Jaffrey SR. Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science. 2014;343:1002–1005. doi: 10.1126/science.1245831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle JG, Lanson NA, Jr, Smith RB, Casci I, Maltare A, Monaghan J, Nichols CD, Kryndushkin D, Shewmaker F, Pandey UB. RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Hum. Mol. Genet. 2013;22:1193–1205. doi: 10.1093/hmg/dds526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrogio A, Buratti E, Stuani C, Guarnaccia C, Romano M, Ayala YM, Baralle FE. Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res. 2009;37:4116–4126. doi: 10.1093/nar/gkp342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Heusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverria GV, Cooper TA. RNA-binding proteins in microsatellite expansion disorders: mediators of RNA toxicity. Brain Res. 2012;1462:100–111. doi: 10.1016/j.brainres.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feany MB, Bender WW. A Drosophila model of Parkinson's disease. Nature. 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- Furukawa Y, Kaneko K, Watanabe S, Yamanaka K, Nukina N. A seeding reaction recapitulates intracellular formation of Sarkosyl-insoluble transactivation response element (TAR) DNA-binding protein-43 inclusions. J Biol Chem. 2011;286:18664–18672. doi: 10.1074/jbc.M111.231209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymrek M, Willems T, Guilmatre A, Zeng H, Markus B, Georgiev S, Daly MJ, Price AL, Pritchard JK, Sharp AJ, Erlich Y. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat. Genet. 2016;48:22–29. doi: 10.1038/ng.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J, Hanada R, Orthofer M, Cronin SJ, Komnenovic V, et al. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature. 2013;495:474–480. doi: 10.1038/nature11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Krans A, Freibaum BD, Taylor JP, Todd PK. TDP-43 suppresses CGG repeat-induced neurotoxicity through interactions with HnRNP A2/B1. Hum. Mol. Genet. 2014;23:5036–5051. doi: 10.1093/hmg/ddu216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YC, Lin KF, He RY, Tu PH, Koubek J, Hsu YC, Huang JJ. Inhibition of TDP-43 aggregation by nucleic acid binding. PLoS One. 2013;8:e64002. doi: 10.1371/journal.pone.0064002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara R, Matsukawa K, Nagata Y, Kunugi H, Tsuji S, Chihara T, Kuranaga E, Miura M, Wakabayashi T, Hashimoto T, et al. RNA binding mediates neurotoxicity in the transgenic Drosophila model of TDP-43 proteinopathy. Hum. Mol. Genet. 2013;22:4474–4484. doi: 10.1093/hmg/ddt296. [DOI] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Zhang F, Pearson CE, Lucchesi JC, Moses K, Warren ST. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron. 2003;39:739–747. doi: 10.1016/s0896-6273(03)00533-6. [DOI] [PubMed] [Google Scholar]
- Johnson JO, Pioro EP, Boehringer A, Chia R, Feit H, Renton AE, Pliner HA, Abramzon Y, Marangi G, Winborn BJ, et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat. Neurosci. 2014;17:664–666. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl. Acad. Sci. USA. 2012;109:3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino Y, Washizu C, Kurosawa M, Oma Y, Hattori N, Ishiura S, Nukina N. Nuclear localization of MBNL1: splicing-mediated autoregulation and repression of repeat-derived aberrant proteins. Hum. Mol. Genet. 2015;24:740–756. doi: 10.1093/hmg/ddu492. [DOI] [PubMed] [Google Scholar]
- Kuo PH, Chiang CH, Wang YT, Doudeva LG, Yuan HS. The crystal structure of TDP-43 RRM1-DNA complex reveals the specific recognition for UG- and TG-rich nucleic acids. Nucleic Acids Res. 2014;42:4712–4722. doi: 10.1093/nar/gkt1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat. Rev. Genet. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 2010;19:R46–64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MJ, Cheng CW, Shen CK. Neuronal function and dysfunction of Drosophila dTDP. PLoS One. 2011;6:e20371. doi: 10.1371/journal.pone.0020371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky PJ, Daujotyte D, Tollervey JR, Ule J, Stuani C, Buratti E, Baralle FE, Damberger FF, Allain FH. Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat. Struct. Mol. Biol. 2013;20:1443–1449. doi: 10.1038/nsmb.2698. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- Mori K, Lammich S, Mackenzie IR, Forne I, Zilow S, Kretzschmar H, Edbauer D, Janssens J, Kleinberger G, Cruts M, et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 2013;125:413–423. doi: 10.1007/s00401-013-1088-7. [DOI] [PubMed] [Google Scholar]
- Niimi Y, Takahashi M, Sugawara E, Umeda S, Obayashi M, Sato N, Ishiguro T, Higashi M, Eishi Y, Mizusawa H, et al. Abnormal RNA structures (RNA foci) containing a penta-nucleotide repeat (UGGAA) in the Purkinje cell nucleus is associated with spinocerebellar ataxia type 31 pathogenesis. Neuropathology. 2013;33:600–611. doi: 10.1111/neup.12032. [DOI] [PubMed] [Google Scholar]
- Pettersson OJ, Aagaard L, Andrejeva D, Thomsen R, Jensen TG, Damgaard CK. DDX6 regulates sequestered nuclear CUG-expanded DMPK-mRNA in dystrophia myotonica type 1. Nucleic Acids Res. 2014;42:7186–7200. doi: 10.1093/nar/gku352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querido E, Gallardo F, Beaudoin M, Menard C, Chartrand P. Stochastic and reversible aggregation of mRNA with expanded CUG-triplet repeats. J. Cell Sci. 2011;124:1703–1714. doi: 10.1242/jcs.073270. [DOI] [PubMed] [Google Scholar]
- Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, Schroeder R. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- Ray M, Lakhotia SC. The commonly used eye-specific sev-GAL4 and GMR-GAL4 drivers in Drosophila melanogaster are expressed in tissues other than eyes also. J. Genet. 2015;94:407–416. doi: 10.1007/s12041-015-0535-8. [DOI] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R. RNA misfolding and the action of chaperones. Front. Biosci. 2008;13:1–20. doi: 10.2741/2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Amino T, Kobayashi K, Asakawa S, Ishiguro T, Tsunemi T, Takahashi M, Matsuura T, Flanigan KM, Iwasaki S, et al. Spinocerebellar ataxia type 31 is associated with "inserted" penta-nucleotide repeats containing (TGGAA)n. Am. J. Hum. Genet. 2009;85:544–557. doi: 10.1016/j.ajhg.2009.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellier C, Freyermuth F, Tabet R, Tran T, He F, Ruffenach F, Alunni V, Moine H, Thibault C, Page A, et al. Sequestration of DROSHA and DGCR8 by expanded CGG RNA repeats alters microRNA processing in fragile X-associated tremor/ataxia syndrome. Cell Rep. 2013;3:869–880. doi: 10.1016/j.celrep.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofola OA, Jin P, Qin Y, Duan R, Liu H, de Haro M, Nelson DL, Botas J. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarallo V, Hirano Y, Gelfand BD, Dridi S, Kerur N, Kim Y, Cho WG, Kaneko H, Fowler BJ, Bogdanovich S, et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012;149:847–859. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Paulson HL. RNA-mediated neurodegeneration in repeat expansion disorders. Ann. Neurol. 2010;67:291–300. doi: 10.1002/ana.21948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt A, Herholz D, Fiesel FC, Kaur K, Muller D, Karsten P, Weber SS, Kahle PJ, Marquardt T, Schulz JB. TDP-43-mediated neuron loss in vivo requires RNA-binding activity. PloS One. 2010;5:e12247. doi: 10.1371/journal.pone.0012247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorlickova M, Kejnovska I, Bednarova K, Renciuk D, Kypr J. Circular dichroism spectroscopy of DNA: from duplexes to quadruplexes. Chirality. 2012;24:691–698. doi: 10.1002/chir.22064. [DOI] [PubMed] [Google Scholar]
- Wang IF, Chang HY, Hou SC, Liou GG, Way TD, James Shen CK. The self-interaction of native TDP-43 C terminus inhibits its degradation and contributes to early proteinopathies. Nat. Commun. 2012;3:766. doi: 10.1038/ncomms1766. [DOI] [PubMed] [Google Scholar]
- Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB, et al. Antisense Proline-Arginine RAN Dipeptides Linked to C9ORF72-ALS/FTD Form Toxic Nuclear Aggregates that Initiate In Vitro and In Vivo Neuronal Death. Neuron. 2014;84:1213–1225. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Hirose F, Inoue YH, Shiraki M, Hayashi Y, Nishi Y, Matsukage A. Ectopic expression of human p53 inhibits entry into S phase and induces apoptosis in the Drosophila eye imaginal disc. Oncogene. 1999;18:6767–6775. doi: 10.1038/sj.onc.1203113. [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Caulfield T, Xu YF, Gendron TF, Hubbard J, Stetler C, Sasaguri H, Whitelaw EC, Cai S, Lee WC, et al. The dual functions of the extreme N-terminus of TDP-43 in regulating its biological activity and inclusion formation. Hum. Mol. Genet. 2013;22:3112–3122. doi: 10.1093/hmg/ddt166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. USA. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.