Abstract
Background: Selective serotonin reuptake inhibitor (SSRI) antidepressant drugs are the first-line of treatment for major depressive disorder (MDD) but are effective in <70% of patients. Our earlier genome-wide studies indicated that two genes encoding for cell adhesion proteins, close homolog of L1 (CHL1) and integrin beta-3 (ITGB3), and microRNAs, miR-151a-3p and miR-221/222, are implicated in the variable sensitivity and response of human lymphoblastoid cell lines (LCL) from unrelated individuals to SSRI drugs.
Methods: The microRNAs miR-221, miR-222, and miR-151-a-3p, along with their target gene binding sites, were explored in silico using miRBase, TargetScan, microRNAviewer, and the UCSC Genome Browser. Luciferase reporter assays were conducted for demonstrating the direct functional regulation of ITGB3 and CHL1 expression by miR-221/222 and miR-151a-3p, respectively. A human LCL exhibiting low sensitivity to paroxetine was utilized for studying the phenotypic effect of CHL1 regulation by miR-151a-3p on SSRI response.
Results: By showing direct regulation of CHL1 and ITGB3 by miR-151a-3p and miR-221/222, respectively, we link these microRNAs and genes with cellular SSRI sensitivity phenotypes. We report that miR-151a-3p increases cell sensitivity to paroxetine via down-regulating CHL1 expression.
Conclusions: miR-151a-3p, miR-221/222 and their (here confirmed) respective target-genes, CHL1 and ITGB3, are implicated in SSRI responsiveness, and possibly in the clinical response to antidepressant drugs.
Keywords: ITGB3, CHL1, miR-221, miR-222, miR-151a-3p, selective serotonin reuptake inhibitors
Introduction
Major depressive disorder (MDD) is a complex, relatively common, costly, and recurrent mental disorder that, according to the World Health Organization, is the leading cause of disability worldwide and is among the leading causes of disease burden globally (Centers for Disease Control Prevention (CDC), 2010; Greenberg et al., 2015)1. One of the major risks of MDD is suicide, which has been reportedly linked to psychiatric disorders in general and to MDD specifically (Zweig and Hinrichsen, 1993; Lesage et al., 1994). Improving the treatment for MDD would have huge financial consequences and would greatly improve the quality of life for millions of patients globally. Selective serotonin reuptake inhibitor (SSRI) antidepressant drugs, which block serotonin uptake via binding directly to the serotonin transporter (SERT) (Sangkuhl et al., 2009), have remained for the past three decades as the first-line treatment for MDD (Thaler et al., 2012). However, about 30–40% of MDD patients fail to reach sufficient remission with SSRI treatment (Souery et al., 2007). In such cases, clinicians often increase the dosage of the same SSRI drug, switch to another antidepressant of the same or a different class [such as serotonin-norepinephrine reuptake inhibitors (Girardi et al., 2009), tricyclic antidepressants, or serotonin ligands (Ruhé et al., 2006)], or augment the antidepressant by adding the mood-stabilizing drug lithium (Price et al., 1990; Bauer et al., 2003).
Clinical guidelines recommend waiting at least 4–6 weeks before switching to an alternative drug (Kato and Serretti, 2010). Meanwhile, patients may experience long periods of depressive symptoms and an increased risk of suicide with no benefit from their first-line SSRI treatment. However, to date no diagnostic tools for predicting patient response to specific antidepressants are available (Dale et al., 2015).
Biomarkers for predicting antidepressant drug response, in particular for SSRIs as the first-line antidepressants, are therefore needed for aiding clinicians in drug and dosage choice in order to decrease the time from diagnosis to remission for the SSRI non-responder patient population. Additionally, reliable diagnostics are needed for early identification of treatment-resistant depression (TRD, patients who do not respond to any of the approved antidepressants). Several genome-wide association studies (GWAS) have searched for common single nucleotide polymorphisms (SNPs) associated with SSRI drug response (Ising et al., 2009; Garriock et al., 2010; Uher et al., 2010; Ji et al., 2013; Biernacka et al., 2015). However, none of these GWAS findings could be replicated, and several meta-analysis studies have been published (Tansey et al., 2012; GENDEP Investigators, MARS Investigators, STAR*D Investigators, 2013; Biernacka et al., 2015), which concluded that none of the identified SNPs had genome-wide significance.
A major landmark in mRNA regulation and protein expression levels came about with the discovery of microRNAs (miRNAs). miRNAs are short (22 nucleotides on average) endogenous non-coding RNAs that down-regulate gene expression at the post-transcriptional level (Bartel, 2009; Friedman et al., 2009; Shomron, 2009; Rukov and Shomron, 2011). Thousands of miRNAs are encoded within the human genome; they are prevalent in all cells, tissues (Liang et al., 2007) and body fluids (Weber et al., 2010; Gurwitz, 2015). At least half of all human gene transcripts are estimated to be targets of evolutionarily conserved miRNA regulation (Lewis et al., 2005; Friedman et al., 2009). Many miRNAs were implicated in various diseases including those involved in brain disorders (Mor et al., 2013; Serafini et al., 2014; Modai and Shomron, 2016).
Given that miRNA can potentially target dozens of genes, they have been recognized as master regulators of gene expression in multicellular organisms. Thus, changes in miRNA levels can affect and might even predict changes in global gene expression (Lim et al., 2005). Consequently, miRNAs are being studied as diagnostics, prognostics, therapeutics, or as pharmacogenomic biomarkers. Indeed, in recognition of their important role in health, disease, and drug response, a new trend in molecular medicine, termed “miRNA pharmacogenomics,” has emerged (Shomron, 2010; Rukov and Shomron, 2011; Rukov et al., 2011, 2014).
Genome-wide expression profiling for identifying SSRI response biomarkers
We recently utilized human lymphoblastoid cell lines (LCLs) from unrelated healthy individuals for conducting a genome-wide transcriptomic microarray-based search for SSRI sensitivity and response to chronic treatment biomarkers (Morag et al., 2011; Oved et al., 2012, 2013). After applying this genome-wide, hypothesis-free approach, we reported several genes and miRNAs as tentative SSRI response biomarkers potentially implicated in the mode of action of SSRI antidepressants (Morag et al., 2011; Oved et al., 2012, 2013). Among these, the expression levels of CHL1 (close homolog of L1) and miR-151a-3p, predicted by bioinformatics tools to target CHL1, were found to be associated with SSRI sensitivity (Morag et al., 2011; Oved et al., 2012). In a separate study, ITGB3 (coding for integrin beta-3, also known as platelet glycoprotein IIIa and CD61) as well as miR-221 and the closely related miR-222, both predicted by bioinformatics tools to target ITGB3, exhibited the most consistent expression level changes following chronic (21 days) paroxetine exposure of human LCLs (Oved et al., 2013). Additionally, ITGB3 and miR-221/miR-222 exhibited opposite expression level changes (Oved et al., 2013). Both CHL1 and ITGB3 code for cell adhesion proteins implicated in neurogenesis and synaptogenesis, and therefore seem to be promising SSRI response biomarkers. Notably, CHL1 knockout mice exhibit mood-related neurological deficits as well as a defective organization of the limbic serotonergic neurons, projecting from thalamic nuclei to the visual rather than prefrontal cortex (Buhusi et al., 2003; Montag-Sallaz et al., 2003; Demyanenko et al., 2004, 2010, 2011; Carneiro et al., 2008; Cingolani and Goda, 2008; Cingolani et al., 2008; Schlatter et al., 2008; Carter et al., 2011; Huang et al., 2011; Katic et al., 2014; Kleene et al., 2015; Mazalouskas et al., 2015). Indeed, cell adhesion proteins, including those coded by CHL1 and ITGB3, were shown to play key roles in neurogenesis and synaptogenesis, which in turn, are crucial for remission from depression (Thomas and Peterson, 2008; Hanson et al., 2011; Danzer, 2012; Eisch and Petrik, 2012; Eyre and Baune, 2012; Bambico and Belzung, 2013; Mateus-Pinheiro et al., 2013; Duman, 2014; Duman and Duman, 2015).
In order to link the genes and miRNAs (Morag et al., 2011; Oved et al., 2012, 2013) to a single set cohort of potential SSRI response biomarkers, we integrated our findings and identified five candidate miRNA-target gene pairs (see Methods). miR-221/222, miR-151a-3p, and their predicted target genes, ITGB3 and CHL1, respectively, were chosen for further studies, since they were implicated in our proposed model regarding the mode of action of SSRI drugs (Oved et al., 2013). This model depicts the cell membrane proteins encoded by CHL1 and SLC6A4 (coding for the serotonin transporter), competing on a limited cell membrane protein pool of integrin beta-3 (encoded by ITGB3) (Oved et al., 2013).
The aim of the current study was to show the direct regulation of CHL1 and ITGB3 by their proposed regulators, miR-151a-3p and miR-221/222, respectively. In addition, we examined the phenotypic effects of altering the expression of these genes on the in vitro SSRI sensitivity of cultured human cells. Furthermore, this study also explored the expression levels of candidate genes and miRNAs that we previously reported as associated with SSRI response and that are known to be implicated in cell adhesion, in LCLs cultured in serum-free (SF) compared with serum-supplemented media. The adhesion of cultured cells to the matrix is known to be enhanced in the absence of serum (Thirumala et al., 2007; Audiffred et al., 2010; Nakayama et al., 2014).
Our new findings link miR-151a-3p and miR-221/222 with SSRI sensitivity phenotypes in human cells via direct regulation of CHL1 and ITGB3. Importantly, we show that human miR-151a-3p and miR-221/222 and their respective target genes, CHL1 and ITGB3, may be implicated in the response of human LCLs to SSRI antidepressant drugs and may tentatively serve as novel MDD drug targets, following validation by additional studies using clinical blood samples and/or studies with brain tissues or blood samples from animal MDD models.
Methods
Data integration analysis for miRNAs and their target sites
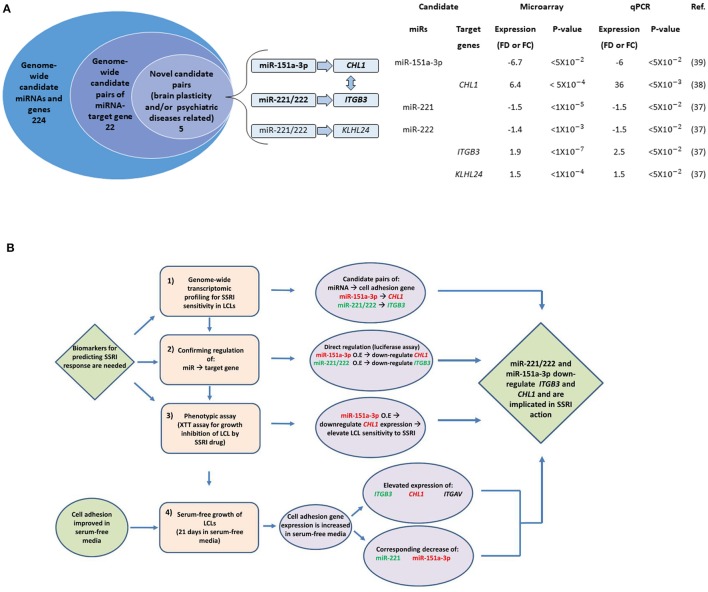
The microRNAs miR-221, miR-222, and miR-151-a-3p, along with their binding sites, were explored in silico using miRBase (Griffiths-Jones et al., 2008)3, TargetScan4, microRNAviewer (Kiezun et al., 2012)5, and the UCSC Genome Browser (Multiz Alignment of 100 Vertebrates)6 In compiling the data in Figure 1, we first compiled a list of 224 candidate biomarker SSRI response genes and miRNAs identified in our earlier studies using genome-wide searches (Morag et al., 2011; Oved et al., 2012, 2013). We then created a list of the top 22 predicted miRNA-target gene pairs using TargetScan and additional software tools, based on miRNA-binding site conservation and a high level of agreement between different software (Oved et al., 2012, 2013). Next, we screened for novel miRNA-target gene pairs for which, both miRNA and the target gene, were reportedly expressed in neuronal cells and were associated with brain plasticity (synaptogenesis/neurogenesis) or with psychiatric diseases; we identified five such pairs.
Figure 1.
The study design. (A) Data integration findings from our previously published genome-wide transcriptomic studies (Morag et al., 2011; Oved et al., 2012, 2013). On the left: A stacked Venn diagram. The outer circle represents the number of candidate miRNAs and genes showing a fold-difference>1.4 and a statistical significance of p < 0.05. The second inner circle represents the number of predicted miRNA-target gene pairs using TargetScan and additional software tools (see Oved et al., 2012, 2013). The third inner circle represents the number of miRNA-target gene pairs for which both miRNA and the target gene were reportedly expressed in neuronal cells and were associated with brain plasticity (synaptogenesis/neurogenesis) or with psychiatric diseases (note that the five pairs include both miR-221 and miR-222). The horizontal arrows denote the relationship between miRs and target genes. The vertical arrow denotes a mutual physical interaction between ITGB3 and CHL1 taking place in the cell membrane (Katic et al., 2014). On the right: Microarray and qPCR data for candidate miRNA-target gene pairs. Fold-difference represents basal expression levels in LCLs with low vs. high paroxetine sensitivity. Fold-change represents expression levels in LCLs following paroxetine exposure compared with controls grown and studied in parallel, as measured by microarray and qPCR experiments. (B) A flow chart presenting the study design. Phase 1 represents the results generated from previous studies described in Figure 1A. Abbreviation: FD, Fold difference. FC, Fold change. O.E., Over-Expression.
Cell culture
Human LCLs were obtained from the National Laboratory for the Genetics of Israeli Populations (NLGIP) at Tel-Aviv University as described in Morag et al. (2010, 2011); Oved et al. (2012, 2013)7. The cell lines were immortalized from the peripheral blood lymphocytes of consenting healthy adult donors. Cells were maintained in RPMI medium supplemented with 10% FBS and antibiotics (100 U ml−1 penicillin; 100 μg ml−1 streptomycin) and kept at a temperature of 37°C, with 6% CO2 and 100% humidity. The human cell lines MCF-7 (adenocarcinoma breast cell line), HEK-293T (transformed human embryonic kidney cells), and SH-SY5Y neuroblastoma cells were maintained in DMEM medium supplemented with 10% FBS and antibiotics (100 U ml−1 penicillin; 100 μg ml−1 streptomycin) under similar conditions.
Serum-free growth of LCLs
LCLs previously maintained in 10% FBS-containing medium were washed in PBS and then resuspended in serum-free RPMI medium containing 4% BIOGRO-2 (Biological Industries, Israel) commercial serum supplement. This BIOGRO-2 concentration was previously reported to be optimal for long-term serum-free growth of LCLs (Milanesi et al., 2015). Control cultures were grown in parallel with 10% FBS.
miRNA constructs
The miRNA expression vectors, miRVec-221 and miRVec-222, which contain the genomic regions of human pre-miR-221 and pre-miR-222, respectively, were provided by Prof. Reuven Agami (Voorhoeve et al., 2006). The genomic region of the human pre-miR-151a was cloned into the BamHI–EcoRI restriction sites of the miRNA expression vector miRVec. miRVec-151a-3p was prepared with the genomic loci ~70 bp upstream and downstream of the pre-miR by PCR-amplification from human genomic DNA (gDNA). BamHI–EcoRI restriction sites were added (indicated by uppercase letters) to the primers:
miRVec-151a-3p forward: gcGGATCCgctaaactaaccctcctgtcagccc
miRVec-151a-3p reverse: gccttGAATTCagtgcctgggtgactcttcctg
Dual luciferase reporter assays
Fragments of ~500 bp from CHL1 and ITGB3 3′UTR, spanning the miRNA-binding sites, were cloned into the XhoI–NotI restriction site downstream of the Renilla luciferase reporter of the psiCHECK-2 plasmid (Promega, USA) that contains a Firefly luciferase reporter (used as a control) under a different promoter. For this purpose, the 3′ UTR fragments were PCR amplified using Phusion High-Fidelity DNA Polymerase (Finnzymes) from gDNA of LCL, and XhoI–NotI restriction sites were added. The miRNA binding sites were mutated using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent, USA). For luciferase assays, HEK-293T, MCF-7, and SH-SY5Y cells were transfected using Lipofectamine 2000 transfection reagent. Next, the cells were transfected with 5 ng of psiCHECK-2 plasmid containing the desired 3′ UTR, with or without site-directed mutations, and 485 ng miRVec containing the desired pre-miRNA or an empty vector. At 24 and 48 h after transfection, firefly and Renilla luciferase activities were measured using the Dual Luciferase reporter assay system kit (Promega, USA) and the LUMIstar Omega Luminometer (BMG LabTech, Germany), according to the manufacturer's recommendations. Renilla luciferase results were normalized to the values of the firefly luciferase. Results represent 3–4 biological replicates. Transfection efficiencies were measured by Green Fluorescent Protein (GFP) fluorescence measurements in all cells, indicating a reproducible transfection efficiency of at least 20%.
miRNA transfections
For miRNA transfection experiments, LCL code #5000 was seeded in 12-well plates at a concentration of 9.6*105 cells/well and transfected with 2 μg of miRVec-151a-3p or an empty vector (for 24 h), or in 24-well plates at a concentration of 4.8*105 cells/well and transfected with 1 μg of the indicated vectors (for 6 and 12 h). Transfections were performed in triplicate using Lipofectamine 2000 transfection reagent (Invitrogen, USA) according to the manufacturer's instructions. Following 6, 12, or 24 h transfection, RNA was extracted. Results of miR-151a-3p over-expression and CHL1 expression down-regulation represent three technical replicates. Transfection efficiencies were measured by GFP fluorescence in all cells, indicating a transfection efficiency of at least 20%.
RNA extraction
Total RNA purification was achieved using phenol-chloroform extraction: cells were centrifuged and then lysed using TRIzol Reagent (Thermo Fisher Scientific, USA), followed by RNA separation using chloroform and isopropanol precipitation. The final RNA concentration and purity were measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Thermo Fisher Scientific, USA).
Cell proliferation assays
LCL code #5000 was seeded in 96-well plates at a concentration of 2*105 cells/well. Following 12 h transfection with 200 ng of miRvec-151a-3p or empty vector, growth inhibition of LCL was examined by exposure to 10 μM paroxetine for 24 h. Following 24 h, XTT cell proliferation assays (Biological Industries, Israel) were carried out as previously described (Morag et al., 2010, 2011). Results represent 4 biological replicates.
Real-time RT-PCR
Reverse transcription reactions for mRNA and for specific mature miRNAs were performed using the High-Capacity cDNA Reverse-Transcription Kit with random primers or TaqMan miRNA assays, respectively, according to the manufacturer's recommendations (Life Technologies, USA). The expression of single miRNA or mRNA was tested similarly using TaqMan Universal PCR Master Mix (Life Technologies, USA) or Solaris qPCR Gene Expression Master Mix (Thermo Scientific, USA), respectively as described (Oved et al., 2012, 2013), and the specific Solaris quantitative PCR gene-expression assay (Thermo Scientific, MA, USA) or the ABI TaqMan Assay probe (ABI, USA). PCR amplification and analysis were performed using the Step-One Detection System (ABI, USA). Comparative critical threshold (Ct) values, obtained by real-time PCR analysis, were used for relative quantification of gene or miRNA expression and determination of the fold-change of expression. Fold changes were obtained by using the formula: 2−ΔΔCt (Schmittgen and Livak, 2008). Individual forward and reverse primer sequences are detailed next:
| Gene | Forward primer | Reverse primer |
|---|---|---|
| GUSB (control) | CTGCTGGCTACTACTTGAAGATG | GAGTTGCTCACAAAGGTCAC |
| ITGB3 | ACCAGTAACCTGCGGATTG | CAGGTGGTCTTCATATCATAGC |
| CHL1 | GCACAGCCAGCAATTTCTTG | TCTTTGTCCAGCGAGGA |
| ITGAV | ABI assay ID - Hs00233808 | |
| SLC6A4 | ABI assay ID - Hs00984349 | |
| miRNA | Mature miRNA sequence | ABI assay ID |
| U6 snRNA (control) | GTGCTCGCTTCGGCAG CACATATACTAAAATTG GAACGATACAGAGAAG ATTAGCATGGCCCCTG CGCAAGGATGACACGC AAATTCGTGAAGCGTT CCATATTTT |
1973 |
| miR-221 (miR-221-3p) | AGCUACAUUGUCUGCUGGGUUUC | 524 |
| miR-222 (miR-222-3p) | AGCUACAUCUGGCUACUGGGU | 2276 |
| miR-151a-3p (miR-151-3p) | CUAGACUGAAGCUCCUUGAGG | 2254 |
Results
Regulation of ITGB3 and CHL1 transcription by miRNAs
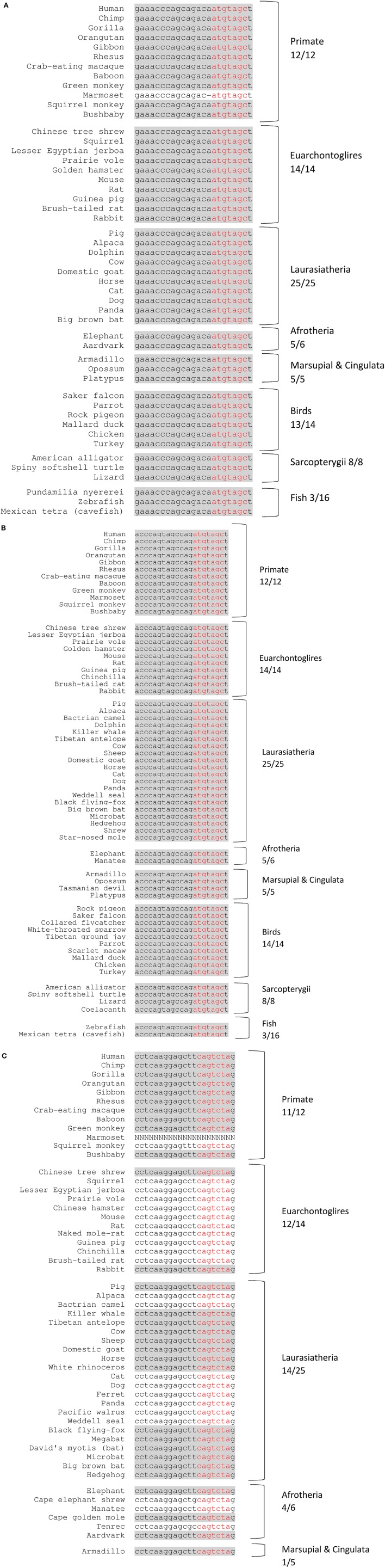
MicroRNAs, miR-221 and miR-222, which are clustered genes located in an intergenic region on the X chromosome in humans and have the same seed sequence, were found to be broadly conserved among vertebrates (89/100 with the conserved seed region), and highly conserved among primates (12/12 with the conserved mature miRNA region) (Figures 2A,B, and Supplementary Figures S1A,B). miR-151a-3p, which is located within intron-22 of the host gene PTK2 (also known as FAK) on chromosome 8 in humans, was found to be conserved only among mammals (52/62 with the conserved seed region), and especially among primates (11/12 with the conserved seed region) (Figure 2C and Supplementary Figure S1C).
Figure 2.
Conserved miRNAs across representative vertebrate species. Shading represents conserved fully mature miRNA. Red nucleotides represent the conserved seed region. x/y represents the number of species out of the total subset with a conserved seed region. (A) miR-221 (chr. X position 45746180–45746202 +strand. (B) miR-222 (chr. X position 45747036–45747056 +strand). (C) miR-151a-3p (chr. 8 position 140732587–140732607 +strand).
For both human miR-221/222 and miR-151a-3p, the binding sites are of the 8-mer type, defined as an exact match to positions 2–8 of the mature miRNA, the seed region + position 8 and are followed by an adenosine residue (Figures 3, 4A). Having a canonical 8-mer site was shown to be important for recognition and increased down-regulation of the targeted gene (Brennecke et al., 2005; Grimson et al., 2007). Furthermore, these miRNA binding sites are well conserved across mammals (33/62 mammals for the miR-221/222-ITGB3 binding site; 32/62 mammals for the miR-151a-3p-CHL1 binding site), particularly primates (10/12 primates for the miR-221/222-ITGB3 binding site and for the miR-151a-3p-CHL1 binding site) (Figure 3 and Supplementary Figure S2).
Figure 3.
Conserved miRNA binding sites across representative mammal species. Shading represents conserved nucleotides – note: for the primate species, the binding sites are of the 8-mer type (the seed region + position 8), and are followed by an adenosine residue. x/y represents the number of species out of the total subset with the conserved miR binding site (seed region). (A) miR-221/222 target site at ITGB3 3′UTR (chr. 17 position 45389536-45389550). (B) miR-151a-3p target site at CHL1 3′UTR (chr. 3 position 447894-447908).
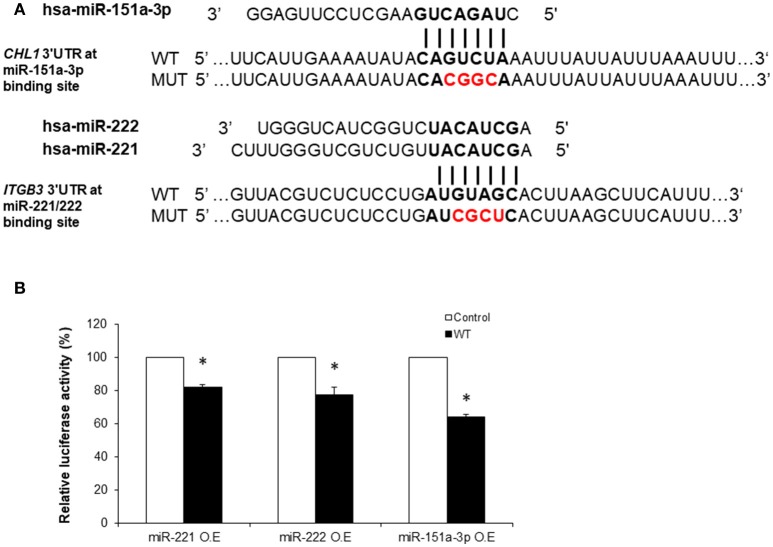
Figure 4.
Luciferase reporter assays demonstrating the in vitro regulation of ITGB3 by miR-221/222 and of CHL1 by miR-151a-3p. (A) Sequences of Renilla/Firefly Luciferase psiCHECK2 constructs under the regulation of ITGB3 and CHL1 3′UTRs that were used for transient reporter assay experiments. WT and Mutant alleles for miR-221/222 and miR-151a-3p binding sites are presented. (B) Luciferase activity 24 h following co-transfection with miR-221, miR-222 or miR-151a-3p combined with either of the ITGB3 or CHL1 3′UTR constructs (WT vs. mutant) using the HEK-293T cell line. Values are presented as the % mean ± SEM (n = 3–4; *p < 0.05). Bold nucleotides at the ITGB3 and CHL1 3′UTR constructs represent the miRNA binding sites. Red nucleotides represent the 4 mutated nucleotides at the seed miR-221/222 or miR-151a-3p binding sites. O.E., Over-Expression.
In order to demonstrate the direct functional regulation of ITGB3 and CHL1 expression by miR-221/222 and miR-151a-3p, respectively, luciferase reporter assays were conducted as follows: a region of ~500 bp from the 3′UTRs of human ITGB3 and CHL1 genes, containing a tentative miRNA binding site, was cloned into the Renilla/Firefly Luciferase psiCHECK2 construct (see Methods). Next, negative controls for the transfection assays were generated by performing site-directed mutagenesis reactions that resulted in changes of four nucleotides of the respective 3′UTR miRNA-binding sites of ITGB3 and CHL1 in the “seed” region (presented in Figure 4A).
ITGB3 was down-regulated by miR-221/222 in HEK-293T cells 24 h following co-transfection with either miR-221 or miR-222 in combination with the consensus ITGB3 3′UTR construct; this was compared to the mutant ITGB3 3′UTR construct co-transfection. The Renilla Luciferase-ITGB3 activity was reduced to 82 and 77%, respectively, of the negative control Renilla Luciferase activity (Figure 4B). Additional time points and human cell lines were tested (see Supplementary Figure S3).
CHL1 expression regulation by miR-151a-3p was similarly tested in HEK-293T human cell line. CHL1 expression was directly down-regulated by miR-151a-3p 24 h following co-transfection with miR-151a-3p in combination with the consensus CHL1 3′UTR construct and compared with the mutant CHL1 3′UTR construct co-transfection. The Renilla Luciferase-CHL1 activity was reduced to 64% of the negative control Renilla Luciferas activity (Figure 4B). Additional time points and human cell lines were tested (see Supplementary Figure S3).
miR-151a-3p increases cell sensitivity to paroxetine via down-regulating CHL1 expression
For determining how CHL1 regulation by miR-151a-3p affects the cellular response to SSRI drugs, a human LCL (healthy female donor, code #5000, see Methods) exhibiting low sensitivity to growth inhibition by paroxetine was analyzed (Morag et al., 2011; Oved et al., 2012). This particular LCL was chosen for transfection, since it was previously found to express relatively lower levels of miR-151a-3p (Oved et al., 2012), along with higher levels of CHL1 (Morag et al., 2011), compared with several other LCLs examined from healthy donors.
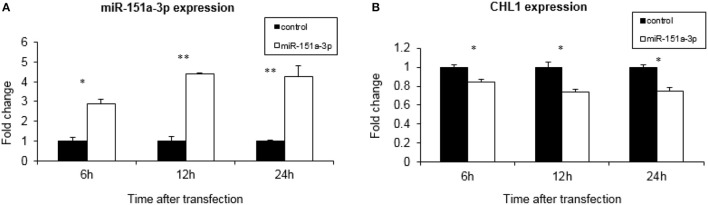
We over-expressed miR-151a-3p in this selected LCL and evaluated its expression at different time points following the transfection. miR-151a-3p was significantly elevated by 3–4.5-fold at 6, 12, and 24 h following miR-151a-3p transfection (Figure 5A), along with a corresponding reduction in CHL1 expression to 74–84% of control transfection values at 6, 12, and 24 h post-transfection (Figure 5B). Moreover, at 12 h following miR-151a-3p transfection, the cell sensitivity to growth inhibition by paroxetine (10 μM) was 24% higher than that of the control transfection (Figure 6).
Figure 5.
CHL1 expression affected by miR-151a-3p levels. Real-time PCR analysis of miRNA and gene expression following 6, 12, and 24 h of miR-151a-3p over-expression relative to control plasmid in LCL #5000. The data show the normalized fold-change of (A) miR-151a-3p and (B) CHL1. Values are presented as the mean ± SEM (n = 3; *p < 0.05 **p < 0.005).
Figure 6.
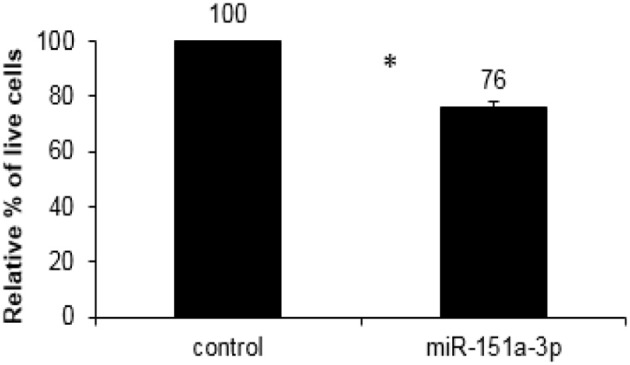
XTT cell viability assay for growth inhibition by paroxetine (10 μM; 24 h exposure) in LCL #5000 following 12 h of over-expression of miR-151a-3p. Values are presented as the mean ± SEM of the relative % of live miR-151a-3p transfected vs. control transfected cells (n = 4; *p < 0.05).
Cell adhesion gene expression is increased in serum-free media
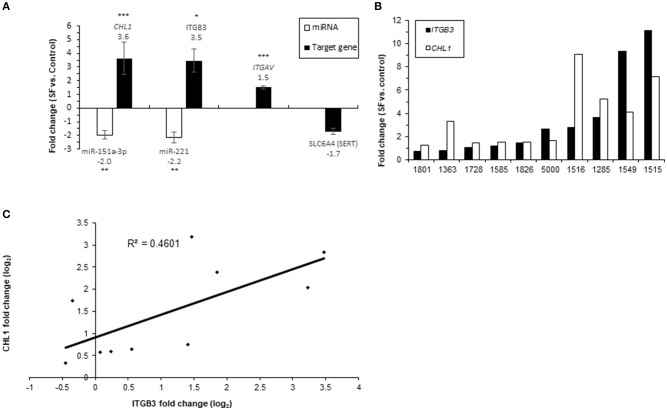
The expression levels of SERT (encoded by SLC6A4), ITGAV; ITGB3, CHL1, and their regulating miRNAs, miR-221 and miR-151a-3p, were compared following 21-day growth of human LCLs from 10 unrelated individuals in serum-free (SF) media supplemented with 4% Biogro-2 (see Methods). LCLs grown in SF media had growth rates and viabilities similar to control cultures grown in serum-supplemented (10% FBS) media, although they divided at a slightly lower rate (not shown). The expression levels of ITGAV, ITGB3, and CHL1 were elevated (average fold-change ± SD of 1.5 ± 0.38, 3.5 ± 2.74, and 3.6 ± 3.71, respectively) following 21-day growth in SF media (Figure 7A). No significant change was observed for SLC6A4 (SERT) expression levels. Notably, large inter-individual variations between human LCLs from unrelated individuals were observed regarding the extent of up-regulated expression of both ITGB3 and CHL1 following 21 days in SF vs. serum-supplemented media, ranging from relatively no change to 11-fold and 9-fold increased expression for ITGB3 and CHL1, respectively (Figure 7B). Moreover, a positive correlation (R = 0.68; P = 0.031) was observed between the elevated expression levels of these genes following 21 days in SF medium vs. serum-supplemented media (Figure 7C). The respective miRNAs, miR-221, which targets ITGB3, and miR-151a-3p, which targets CHL1, were down-regulated accordingly following 21 days in SF vs. serum-supplemented media (an average fold change ± SD of −2.2 ± 1.23-fold and −2 ± 1-fold for miR-221 and miR-151a-3p, respectively) (Figure 7A).
Figure 7.
Real-time PCR analysis of miRNAs and gene expression following 21-day growth in SF media vs. serum-supplemented media in 10 LCLs from individual donors. The data show the normalized fold-change of (A) miRNAs and their target genes; values are presented as the mean ± SEM (*p < 0.05, **p < 0.001, ***p < 0.005). (B) ITGB3 and CHL1 expression for each LCL. (C) Correlation plot for CHL1 fold change (log2) vs. ITGB3 fold change (log2). The numbers on the X axis are LCL donor codes.
Discussion
Accumulating evidence suggests that the expression of miRNAs is altered during chronic stress, and that dysregulation of miRNAs during such stress may contribute to the etiology and pathophysiology of MDD (Dwivedi, 2011; Mouillet-Richard et al., 2012; Issler et al., 2014; Lopez et al., 2014; Garbett et al., 2015). Moreover, miRNAs may be involved in antidepressant response and may serve as novel antidepressant targets, since different studies have demonstrated altered miRNA expression levels following antidepressant treatment (Bocchio-Chiavetto et al., 2013; O'Connor et al., 2013). Owing to the above considerations and because a single miRNA can potentially regulate scores of mRNAs, miRNAs have the potential to serve as pharmacogenomic biomarkers for SSRI response (Rukov and Shomron, 2011; Labermaier et al., 2013).
Affective disorders, including MDD, which seem to uniquely affect humans (or at least only primates) and are not easily modeled by rodent studies, are related to defects in higher human cognitive processes and to the complexity of the human brain compared with the rodent brain (Geschwind and Rakic, 2013; Czéh et al., 2016). MDD is a complex mood disorder with high comorbidity with anxiety disorders, characterized by psychiatric diagnoses of human behaviors such as sadness, guilt, and feeling of worthlessness, among others. Regulation of gene expression (e.g., by miRNAs) has been suggested to mediate stable adaptations in the brain (Tsankova et al., 2007; Labermaier et al., 2013). Thus, the high level of miRNA conservation among primates observed for our three candidate miRNAs and their respective target gene miRNA-binding sites is intriguing and suggests that an important (and thus evolutionary conserved) role exists for the regulation of ITGB3 and CHL1 by miR-221/222 and miR-151a-3p, respectively, in primates (especially in humans) vs. non-primate vertebrates. Moreover, miR-221/222 (to which we will refer together owing to their very close similarity) and miR-151a-3p have a canonical 8-mer site on the 3′UTR region of their targeted genes. Having an 8-mer site was shown to be important for recognition and down-regulation of the target gene (Brennecke et al., 2005; Grimson et al., 2007).
ITGB3, coding for the integrin beta-3 subunit, together with integrin alpha-v or with the integrin alpha-IIb, form the vitronectin receptor (αvβ3), and the fibrinogen receptor (αIIbβ3), respectively. Human ITGB3 has been shown to be regulated by different human miRNAs including miR-98 (Ni et al., 2015), miR-320a (Sun et al., 2015), let-7c (Zhao et al., 2014), and let-7a (Müller and Bosserhoff, 2008). However, in a previous study we did not observe changes in the expression levels of the latter miRNAs following chronic in vitro SSRI treatment of human cells (Oved et al., 2013), suggesting that miR-221/222 play a role in the mode of action of SSRI antidepressants.
Using luciferase assays, our present study confirms the direct regulation by miR-221/222 of their potential target-gene, ITGB3. In addition, the expression of ITGB3 was dramatically increased (3.5-fold) following the 21-day growth of human LCLs in SF media, whereas under these conditions, miR-221 was down-regulated in the same cells by 2.2-fold (Figure 7A).
miR-221/222 and their target-gene, ITGB3, are expressed in the human brain and are implicated in synaptogenesis/neurogenesis (Cingolani and Goda, 2008; Cingolani et al., 2008; Terasawa et al., 2009; Shao et al., 2010; Hamada et al., 2012; Pozo et al., 2012; Cheng et al., 2014). Using Ilumina sequencing, miR-221 and miR-222 were identified among the top 40 highly expressed miRNAs in post-mortem human brains (Shao et al., 2010); both were induced by nerve growth factor in rat pheochromocytoma PC12 cells (Terasawa et al., 2009; Hamada et al., 2012) and were shown to play a role in neurite guidance (Cheng et al., 2014). Moreover, miR-221 potentiated the formation of neurite networks in these cells (Hamada et al., 2012). ITGB3 is involved in synaptic plasticity in mouse neuronal hippocampal cultures (Cingolani and Goda, 2008) and regulates excitatory synaptic strength (Cingolani et al., 2008) and GluA2 AMPA receptor expression in mouse hippocampi (Pozo et al., 2012).
Importantly, ITGB3 is crucial for maintaining the activity of the serotonin transporter (SERT; encoded in humans by SLC6A4), the well-established drug target of SSRI drugs that block serotonin uptake via binding directly to SERT (Sangkuhl et al., 2009). This role of ITGB3 is evident from the diminished SERT activity in platelets (Carneiro et al., 2008), as well as in the midbrain (Whyte et al., 2014) and raphe nuclei synaptosomes (Mazalouskas et al., 2015) of Itgb3-deficient mice, either knockout (Itgb3−/−) (Carneiro et al., 2008) or heterozygous (Itgb3−/−) (Whyte et al., 2014; Mazalouskas et al., 2015). Itgb3-knockout mice exhibit altered social and repetitive behavior, e.g., behavior relevant for autism spectrum disorder (Carter et al., 2011). Neuroanatomical assessment of these mice indicated significantly different relative tissue volumes in several brain regions, among them reduced volume of the lateral wings of the dorsal raphe nuclei—a brain region important for the development of the CNS serotonergic system (Ellegood et al., 2012). Moreover, integrins, including ITGB3, interact with CHL1 in the plasma membrane and promote CHL1-induced neuronal migration and neurite outgrowth (Buhusi et al., 2003; Demyanenko et al., 2004; Schlatter et al., 2008; Katic et al., 2014). Considering the above, these findings suggest that ITGB3 plays an important role in correct neuroanatomical development of the CNS, a property it shares with CHL1.
Interestingly, in a pilot clinical study, miR-221 was found to be down-regulated in the plasma of MDD patients treated with the SSRI antidepressant escitalopram (Enatescu et al., 2016). In another study, miR-221 was found to be up-regulated in CSF and serum samples of MDD patients vs. control subjects (Wan et al., 2015). These observations correspond to the decreased expression of miR-221/222 we observed following chronic SSRI treatment in human LCLs (Oved et al., 2013); thus they demonstrate that human LCLs can serve as a legitimate research tool for searching biomarkers for SSRI response in neurons. Whereas in a study that examined blood mononuclear cell proteomes, several members of the integrin signaling pathway, including ITGB3, were found to be differentially expressed between responder and non-responder MDD patients (Martins-de-Souza et al., 2014). In the latter study, ITGB3 expression was found to be increased in responder vs. the non-responder MDD patients.
The identification of ITGB3 as a potential SSRI response biomarker was further supported by our recent study in which lower ITGB3 expression levels (~50% vs. healthy controls) were observed in peripheral blood mononuclear cells (PBMCs) obtained from MDD patients (Rzezniczek et al., 2016). The lower ITGB3 expression levels observed in the PBMCs obtained from the MDD patients is noteworthy and suggests that increased ITGB3 expression upon chronic SSRI treatment plays a role in the therapeutic action of these drugs in MDD.
Human CHL1 (close homolog of L1; also known as CALL or L1CAM2) has also been shown to be regulated by additional miRNAs such as miR-10a, miR-590, miR-182, and miR-21 (Long et al., 2012; Chu et al., 2014; Zhu et al., 2014; Li et al., 2016). However, in a previous study we did not observe changes in the expression levels of the latter miRNAs (Oved et al., 2012), suggesting that miR-151a-3p plays a role in the mode of action of SSRI antidepressants.
Using luciferase assays in cells transfected with miR-151a-3p, we confirmed the direct regulation of CHL1 transcription by this human miRNA, for which the expression levels were observed to be significantly correlated with the in vitro paroxetine sensitivity phenotype of LCLs from unrelated healthy individuals (Oved et al., 2012). In addition, the expression of CHL1 was dramatically increased (3.6-fold) following the 21-day growth of human LCLs in SF media, whereas under these conditions miR-151a-3p, which targets CHL1, was down-regulated in the same cells by ~2-fold (Figure 7A). Moreover, following over-expression of miR-151a-3p in human LCLs, we observed a strong reduction of CHL1 expression (Figure 5B), along with higher in vitro sensitivity to paroxetine (Figure 6), as expected from our previous genome-wide transcriptomic study reporting lower CHL1 expression in human LCLs exhibiting higher in vitro paroxetine sensitivity (Morag et al., 2011). Notably, plasma miR-151a-3p was down-regulated in escitalopram-medicated MDD patients (Enatescu et al., 2016).
CHL1 codes for a cell adhesion protein which plays central roles in neural cell proliferation, migration, differentiation, signal transduction and axon guidance (Maness and Schachner, 2007; Huang et al., 2011). It is implicated in correct brain circuitry (Montag-Sallaz et al., 2002, 2003) and in mental disorders (Frints et al., 2003; Chen et al., 2005). Moreover, CHL1 expression was downregulated in the hippocampus of mice exposed to early post-natal stress, a known aggravator of mood disorders (Desarnaud et al., 2008). Furthermore, CHL1/L1 double knockout (CHL1/L1−/−) mice have misguided neuronal circuitry from the limbic system to the cerebral cortex (Demyanenko et al., 2010, 2011). In addition, CHL1 was reported to interact with the serotonin 2c receptor and thereby act as a modulator of the serotonergic system (Kleene et al., 2015).
MDD and bipolar disorder are related affective disorders, with BD-II typified mostly by depressive episodes (Baldessarini et al., 2013; Carvalho et al., 2014). Lithium, which is used as a first-line treatment for bipolar disorder, is also employed for augmenting antidepressant therapy in treatment-resistant MDD (Price et al., 1990; Bauer et al., 2003). Notably, lithium was reported to down-regulate miR-221 in the hippocampi of chronically treated rats (Zhou et al., 2009). Moreover, miR-221/222 were down-regulated in the hippocampi of juvenile rats following the induction of lithium-pilocarpine status epilepticus with lithium chloride injections (Ashhab et al., 2013). Taken together, these findings suggest that lithium augmentation of SSRI efficacy in MDD (Price et al., 1990; Bauer et al., 2003) could be related to ITGB3 up-regulation secondary to miR-221/222 down-regulation.
A recent study by Milanesi et al. reported that the in vitro lithium sensitivity of LCLs derived from bipolar disorder patients is affected by IGF-1, an effect that could be observed only in the absence of serum (Milanesi et al., 2015). Notably, IGF-1 was also implicated in MDD (Kopczak et al., 2015). IGF-1 binds directly to integrin αvβ3, and this interaction was found to be essential for IGF-1 signaling through the IGF1R receptor (Saegusa et al., 2009). In addition, IGF-1 assembles the formation of a heterocomplex between IGF1R and the integrin β3 subunit (Tahimic et al., 2016). We therefore compared the transcription of ITGB3 and CHL1 in LCLs maintained in SF vs. serum-containing media. In addition, we examined the transcription of ITGAV, coding for the integrin alpha-v subunit, which together with integrin beta-3, forms the vitronectin receptor (αvβ3), in SF vs. serum-containing media. The expression levels of ITGAV, ITGB3, and CHL1 were all elevated following 21-day growth of human LCLs in SF media (Figure 7A) and a large inter-individual variation in the extent of up-regulated expression was observed for ITGB3 and CHL1 (Figure 7B). A correlation (R2 = 0.46; P = 0.031) was observed between the fold-change of the elevated expression levels of these two genes (Figure 7C). This finding is intriguing as ITGB3 interacts with CHL1 at the cell membrane (Katic et al., 2014), and both ITGB3 and CHL1 were implicated in our hypothesized model regarding the mode of action of SSRI drugs (Oved et al., 2013). This model depicts the cell membrane proteins encoded by CHL1 and SLC6A4 (coding for the serotonin transporter), competing on a limited cell membrane protein reservoir of integrin beta-3 (encoded by ITGB3) (Oved et al., 2013).
Das et al. reported that cultivation of cells in serum-free medium in the presence of fibronectin up-regulates the activity of MMP-2 and MMP-9, two matrix metaloproteinases implicated in cell migration and invasion (Das et al., 2008). Expression of αvβ3 correlates with activation of MT1-MMP and MMP-2 in human melanoma cells (Hofmann et al., 2000). Improved cell adhesion of non-adherent cells in serum-free media has been demonstrated, including for Jurkat lymphoblastoid cells, an immortalized cell line of human T-lymphocytes (Thirumala et al., 2007; Audiffred et al., 2010; Nakayama et al., 2014). These findings correspond to our observations of higher expression levels of the cell adhesion genes ITGAV, ITGB3, and CHL1 in human LCLs maintained in serum-free, compared with serum-supplemented media.
Study limitations
The present study has several limitations. Although we have previously observed inverse expression patterns between miRNAs and the target gene mRNAs levels in human LCLs and have now experimentally validated miRNA-targets using luciferase reporter assays, one has to keep in mind that our experiments were carried out in vitro in human cell lines. We used LCLs from healthy unrelated donors, while the expression of the following miRNAs and target genes may differ in brain tissues of MDD patients. In addition, in order to measure the effect of miR-151a-3p upon SSRI response we used an in vitro assay of LCL growth inhibition by the SSRI drug paroxetine as a surrogate for clinical SSRI drug response. Our findings should thus be considered as tentative miRNA/target gene biomarkers for SSRI response until studies with MDD patient blood samples and/or studies with brain tissues of animal MDD models validate our results.
Conclusions
Taken together, our current observations lend further support for the putative role of miR-151a-3p and miR-221/222 and their (here confirmed) respective target-genes, CHL1 and ITGB3, in SSRI responsiveness, and possibly in the pathology of MDD. We therefore propose that miR-151a-3p and miR-221/222 could tentatively serve as SSRI response biomarkers, as well as potential novel antidepressant therapeutic targets, following additional experimental validation in animal models as well as in clinical trials.
Author contributions
NS, DG, and KO were responsible for the study design and interpretation of the data. KO, LF, AG, II, and DH were responsible for acquisition of the data.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the donors of the NLGIP biobank at Tel Aviv University, Israel, whose altruism and trust in biomedical research have made this study possible. The authors also thank A. L. Polsky (Tel-Aviv University) for insightful discussion and help with writing the paper.
1WHO. Depression [Internet]. WHO. [cited 2017 Jul 24]. Available from: http://www.who.int/mediacentre/factsheets/fs369/en/.
2miRBase [Internet]. [cited 2017 Jan 19]. Available from: http://www.mirbase.org/
3TargetScanHuman 7.1 [Internet]. [cited 2016 Nov 20]. Available from: http://www.targetscan.org/vert_71/
4microRNAviewer-all [Internet]. [cited 2017 Jan 19]. Available from: http://people.csail.mit.edu/akiezun/microRNAviewer/index.html.
5UCSC Genome Browser Home [Internet]. [cited 2016 Nov 20]. Available from: https://genome-euro.ucsc.edu/index.html.
6Amiel. Homepage [Internet]. Natl. Lab. Genet. Isr. Popul. [cited 2017 Jan 2]. Available from: http://yoran.tau.ac.il/nlgip/.
Funding. This study was supported by the Chief Scientist office, Ministry of Health, Israel, in the frame of ERA-Net Neuron. The Shomron laboratory is supported by the Israel Cancer Research Fund (ICRF), Research Career Development Award (RCDA); Wolfson Family Charitable Fund; Claire and Amedee Maratier Institute for the Study of Blindness and Visual Disorders; I-CORE Program of the Planning and Budgeting Committee, The Israel Science Foundation (grant number 41/11); the Israeli Ministry of Defense, Office of Assistant Minister of Defense for Chemical, Biological, Radiological and Nuclear (CBRN) Defense; Foundation Fighting Blindness; Saban Family Foundation, Melanoma Research Alliance; Binational Science Foundation (BSF); Israel Cancer Research Fund (ICRF) Acceleration Grant; Israel Cancer Association (ICA); Donation from the Kateznik K. Association Holocaust; Margot Stoltz Foundation through the Faculty of Medicine grants of Tel-Aviv University; The Varda and Boaz Dotan Research Center in Hemato-Oncology, Idea Grant; “Lirot” Association and the Consortium for Mapping Retinal Degeneration Disorders in Israel; Interdisciplinary grant of the Israeli Ministry of Science, Technology and Space on the Science, Technology and Innovation for the Third Age; The Edmond J. Safra Center for Ethics at Tel Aviv University; Check Point Institute for Information Security; Joint Core Program of Research on the Molecular Basis of Human Disease, Shabbetai Donnolo Fellowships supported by the Italian Ministry of Foreign Affairs; Israel Science Foundation (ISF, 1852/16); The Edmond J. Safra Center for Bioinformatics at Tel Aviv University; Adelis Foundation. DG is supported by the Yoran Institute for Human Genome Research at Tel Aviv University. We thank the anonymous donors of the NLGIP biobank at Tel Aviv University, Israel, whose altruism and trust in biomedical research have made this study possible. KO was supported by a graduate student scholarship from the Sagol School of Neuroscience at Tel Aviv University and by Buchman Fellowship from the Faculty of Medicine at Tel Aviv University.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2017.00355/full#supplementary-material
References
- Ashhab M. U., Omran A., Gan N., Kong H., Peng J., Yin F. (2013). microRNA s (9, 138, 181A, 221, and 222) and mesial temporal lobe epilepsy in developing brains. Transl. Neurosci. 4, 357–362. 10.2478/s13380-013-0128-z [DOI] [Google Scholar]
- Audiffred J. F., De Leo S. E., Brown P. K., Hale-Donze H., Monroe W. T. (2010). Characterization and applications of serum-free induced adhesion in Jurkat suspension cells. Biotechnol. Bioeng. 106, 784–793. 10.1002/bit.22728 [DOI] [PubMed] [Google Scholar]
- Baldessarini R. J., Faedda G. L., Offidani E., Vázquez G. H., Marangoni C., Serra G., et al. (2013). Antidepressant-associated mood-switching and transition from unipolar major depression to bipolar disorder: a review. J. Affect. Disord. 148, 129–135. 10.1016/j.jad.2012.10.033 [DOI] [PubMed] [Google Scholar]
- Bambico F. R., Belzung C. (2013). Novel insights into depression and antidepressants: a synergy between synaptogenesis and neurogenesis? Curr. Top. Behav. Neurosci. 15, 243–291. 10.1007/7854_2012_234 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M., Adli M., Baethge C., Berghöfer A., Sasse J., Heinz A., et al. (2003). Lithium augmentation therapy in refractory depression: clinical evidence and neurobiological mechanisms. Can. J. Psychiatry Rev. Can. Psychiatr. 48, 440–448. 10.1177/070674370304800703 [DOI] [PubMed] [Google Scholar]
- Biernacka J. M., Sangkuhl K., Jenkins G., Whaley R. M., Barman P., Batzler A., et al. (2015). The International SSRI Pharmacogenomics Consortium (ISPC): a genome-wide association study of antidepressant treatment response. Transl. Psychiatry 5:e553. 10.1038/tp.2015.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocchio-Chiavetto L., Maffioletti E., Bettinsoli P., Giovannini C., Bignotti S., Tardito D., et al. (2013). Blood microRNA changes in depressed patients during antidepressant treatment. Eur. Neuropsychopharmacol. J. Eur. Coll. Neuropsychopharmacol. 23, 602–611. 10.1016/j.euroneuro.2012.06.013 [DOI] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R. B., Cohen S. M. (2005). Principles of microRNA-target recognition. PLoS Biol. 3:e85. 10.1371/journal.pbio.0030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhusi M., Midkiff B. R., Gates A. M., Richter M., Schachner M., Maness P. F. (2003). Close homolog of L1 is an enhancer of integrin-mediated cell migration. J. Biol. Chem. 278, 25024–25031. 10.1074/jbc.M303084200 [DOI] [PubMed] [Google Scholar]
- Carneiro A. M. D., Cook E. H., Murphy D. L., Blakely R. D. (2008). Interactions between integrin alphaIIbbeta3 and the serotonin transporter regulate serotonin transport and platelet aggregation in mice and humans. J. Clin. Invest 118, 1544–1552. 10.1172/JCI33374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M. D., Shah C. R., Muller C. L., Crawley J. N., Carneiro A. M. D., Veenstra-VanderWeele J. (2011). Absence of preference for social novelty and increased grooming in integrin β3 knockout mice: initial studies and future directions. Autism Res. Off. J. Int. Soc. Autism Res. 4, 57–67. 10.1002/aur.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A. F., McIntyre R. S., Dimelis D., Gonda X., Berk M., Nunes-Neto P. R., et al. (2014). Predominant polarity as a course specifier for bipolar disorder: a systematic review. J. Affect. Disord. 163, 56–64. 10.1016/j.jad.2014.03.035 [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) (2010). Current depression among adults—United States, 2006 and 2008. MMWR Morb. Mortal. Wkly. Rep. 59, 1229–1235. Available online at: https://www.cdc.gov/mmwr/pdf/other/su6203.pdf [PubMed] [Google Scholar]
- Chen Q.-Y., Chen Q., Feng G.-Y., Lindpaintner K., Chen Y., Sun X., et al. (2005). Case-control association study of the close homologue of L1 (CHL1) gene and schizophrenia in the Chinese population. Schizophr. Res. 73, 269–274. 10.1016/j.schres.2004.06.001 [DOI] [PubMed] [Google Scholar]
- Cheng H., Zhou L., Li B., Zhu M., Too H.-P., Choi W. K. (2014). Nano-topology guided neurite outgrowth in PC12 cells is mediated by miRNAs. Nanomed. Nanotechnol. Biol. Med. 10, 1871–1875. 10.1016/j.nano.2014.07.011 [DOI] [PubMed] [Google Scholar]
- Chu Y., Ouyang Y., Wang F., Zheng A., Bai L., Han L., et al. (2014). MicroRNA-590 promotes cervical cancer cell growth and invasion by targeting CHL1. J. Cell. Biochem. 115, 847–853. 10.1002/jcb.24726 [DOI] [PubMed] [Google Scholar]
- Cingolani L. A., Goda Y. (2008). Differential involvement of beta3 integrin in pre- and postsynaptic forms of adaptation to chronic activity deprivation. Neuron Glia Biol. 4, 179–187. 10.1017/S1740925X0999024X [DOI] [PubMed] [Google Scholar]
- Cingolani L. A., Thalhammer A., Yu L. M. Y., Catalano M., Ramos T., Colicos M. A., et al. (2008). Activity-dependent regulation of synaptic AMPA receptor composition and abundance by β3 integrins. Neuron 58, 749–762. 10.1016/j.neuron.2008.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czéh B., Fuchs E., Wiborg O., Simon M. (2016). Animal models of major depression and their clinical implications. Prog. Neuropsychopharmacol. Biol. Psychiatry 64, 293–310. 10.1016/j.pnpbp.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Dale E., Bang-Andersen B., Sánchez C. (2015). Emerging mechanisms and treatments for depression beyond SSRIs and SNRIs. Biochem. Pharmacol. 95, 81–97. 10.1016/j.bcp.2015.03.011 [DOI] [PubMed] [Google Scholar]
- Danzer S. C. (2012). Depression, stress, epilepsy and adult neurogenesis. Exp. Neurol 233, 22–32. 10.1016/j.expneurol.2011.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Banerji A., Frei E., Chatterjee A. (2008). Rapid expression and activation of MMP-2 and MMP-9 upon exposure of human breast cancer cells (MCF-7) to fibronectin in serum free medium. Life Sci. 82, 467–476. 10.1016/j.lfs.2007.12.013 [DOI] [PubMed] [Google Scholar]
- Demyanenko G. P., Halberstadt A. I., Rao R. S., Maness P. F. (2010). CHL1 cooperates with PAK1-3 to regulate morphological differentiation of embryonic cortical neurons. Neuroscience 165, 107–115. 10.1016/j.neuroscience.2009.09.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demyanenko G. P., Schachner M., Anton E., Schmid R., Feng G., Sanes J., et al. (2004). Close homolog of L1 modulates area-specific neuronal positioning and dendrite orientation in the cerebral cortex. Neuron 44, 423–437. 10.1016/j.neuron.2004.10.016 [DOI] [PubMed] [Google Scholar]
- Demyanenko G. P., Siesser P. F., Wright A. G., Brennaman L. H., Bartsch U., Schachner M., et al. (2011). L1 and CHL1 cooperate in thalamocortical axon targeting. Cereb. Cortex N. Y. N 1991 21, 401–412. 10.1093/cercor/bhq115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desarnaud F., Jakovcevski M., Morellini F., Schachner M. (2008). Stress downregulates hippocampal expression of the adhesion molecules NCAM and CHL1 in mice by mechanisms independent of DNA methylation of their promoters. Cell Adhes. Migr. 2, 38–44. 10.4161/cam.2.1.6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman C. H., Duman R. S. (2015). Spine synapse remodeling in the pathophysiology and treatment of depression. Neurosci. Lett. 601, 20–29. 10.1016/j.neulet.2015.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman R. S. (2014). Neurobiology of stress, depression, and rapid acting antidepressants: remodeling synaptic connections. Depress. Anxiety 31, 291–296. 10.1002/da.22227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y. (2011). Evidence demonstrating role of microRNAs in the etiopathology of major depression. J. Chem. Neuroanat. 42, 142–156. 10.1016/j.jchemneu.2011.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisch A. J., Petrik D. (2012). Depression and hippocampal neurogenesis: a road to remission? Science 338, 72–75. 10.1126/science.1222941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegood J., Henkelman R. M., Lerch J. P. (2012). Neuroanatomical assessment of the integrin β3 mouse model related to autism and the serotonin system using high resolution MRI. Front. Psychiatry 3:37. 10.3389/fpsyt.2012.00037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enatescu V. R., Papava I., Enatescu I., Antonescu M., Anghel A., Seclaman E., et al. (2016). Circulating plasma micro RNAs in patients with major depressive disorder treated with antidepressants: a pilot study. Psychiatry Investig. 13, 549–557. 10.4306/pi.2016.13.5.549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre H., Baune B. T. (2012). Neuroplastic changes in depression: a role for the immune system. Psychoneuroendocrinology 37, 1397–1416. 10.1016/j.psyneuen.2012.03.019 [DOI] [PubMed] [Google Scholar]
- Friedman R. C., Farh K. K.-H., Burge C. B., Bartel D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frints S. G. M., Marynen P., Hartmann D., Fryns J.-P., Steyaert J., Schachner M., et al. (2003). CALL interrupted in a patient with non-specific mental retardation: gene dosage-dependent alteration of murine brain development and behavior. Hum. Mol. Genet. 12, 1463–1474. 10.1093/hmg/ddg165 [DOI] [PubMed] [Google Scholar]
- Garbett K. A., Vereczkei A., Kálmán S., Brown J. A., Taylor W. D., Faludi G., et al. (2015). Coordinated messenger RNA/microRNA changes in fibroblasts of patients with major depression. Biol. Psychiatry 77, 256–265. 10.1016/j.biopsych.2014.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriock H. A., Kraft J. B., Shyn S. I., Peters E. J., Yokoyama J. S., Jenkins G. D., et al. (2010). A genomewide association study of citalopram response in major depressive disorder. Biol. Psychiatry 67, 133–138. 10.1016/j.biopsych.2009.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GENDEP Investigators MARS Investigators, STAR*D Investigators (2013). Common genetic variation and antidepressant efficacy in major depressive disorder: a meta-analysis of three genome-wide pharmacogenetic studies. Am. J. Psychiatry 170, 207–217. 10.1176/appi.ajp.2012.12020237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind D. H., Rakic P. (2013). Cortical evolution: judge the brain by its cover. Neuron 80, 633–647. 10.1016/j.neuron.2013.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi P., Pompili M., Innamorati M., Mancini M., Serafini G., Mazzarini L., et al. (2009). Duloxetine in acute major depression: review of comparisons to placebo and standard antidepressants using dissimilar methods. Hum. Psychopharmacol. 24, 177–190. 10.1002/hup.1005 [DOI] [PubMed] [Google Scholar]
- Greenberg P. E., Fournier A.-A., Sisitsky T., Pike C. T., Kessler R. C. (2015). The economic burden of adults with major depressive disorder in the United States (2005 and 2010). J. Clin. Psychiatry 76, 155–162. 10.4088/JCP.14m09298 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H. K., van Dongen S., Enright A. J. (2008). miRBase: tools for microRNA genomics. Nucleic Acids Res. 36, D154–D158. 10.1093/nar/gkm952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A., Farh K. K.-H., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. (2007). MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91–105. 10.1016/j.molcel.2007.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurwitz D. (2015). Exosomal microRNAs in tissue crosstalk. Drug Dev. Res. 76, 259–262. 10.1002/ddr.21264 [DOI] [PubMed] [Google Scholar]
- Hamada N., Fujita Y., Kojima T., Kitamoto A., Akao Y., Nozawa Y., et al. (2012). MicroRNA expression profiling of NGF-treated PC12 cells revealed a critical role for miR-221 in neuronal differentiation. Neurochem. Int. 60, 743–750. 10.1016/j.neuint.2012.03.010 [DOI] [PubMed] [Google Scholar]
- Hanson N. D., Owens M. J., Nemeroff C. B. (2011). Depression, antidepressants, and neurogenesis: a critical reappraisal. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 36, 2589–2602. 10.1038/npp.2011.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann U. B., Westphal J. R., Van Kraats A. A., Ruiter D. J., Van Muijen G. N. (2000). Expression of integrin alpha(v)beta(3) correlates with activation of membrane-type matrix metalloproteinase-1 (MT1-MMP) and matrix metalloproteinase-2 (MMP-2) in human melanoma cells in vitro and in vivo. Int. J. Cancer 87, 12–19. [DOI] [PubMed] [Google Scholar]
- Huang X., Zhu L., Zhao T., Wu L., Wu K., Schachner M., et al. (2011). CHL1 negatively regulates the proliferation and neuronal differentiation of neural progenitor cells through activation of the ERK1/2 MAPK pathway. Mol. Cell. Neurosci. 46, 296–307. 10.1016/j.mcn.2010.09.013 [DOI] [PubMed] [Google Scholar]
- Ising M., Lucae S., Binder E. B., Bettecken T., Uhr M., Ripke S., et al. (2009). A genomewide association study points to multiple loci that predict antidepressant drug treatment outcome in depression. Arch. Gen. Psychiatry 66, 966–975. 10.1001/archgenpsychiatry.2009.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issler O., Haramati S., Paul E. D., Maeno H., Navon I., Zwang R., et al. (2014). MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron 83, 344–360. 10.1016/j.neuron.2014.05.042 [DOI] [PubMed] [Google Scholar]
- Ji Y., Biernacka J. M., Hebbring S., Chai Y., Jenkins G. D., Batzler A., et al. (2013). Pharmacogenomics of selective serotonin reuptake inhibitor treatment for major depressive disorder: genome-wide associations and functional genomics. Pharmacogen. J. 13, 456–463. 10.1038/tpj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katic J., Loers G., Kleene R., Karl N., Schmidt C., Buck F., et al. (2014). Interaction of the cell adhesion molecule CHL1 with vitronectin, integrins, and the plasminogen activator inhibitor-2 promotes CHL1-induced neurite outgrowth and neuronal migration. J. Neurosci. Off. J. Soc. Neurosci. 34, 14606–14623. 10.1523/JNEUROSCI.3280-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Serretti A. (2010). Review and meta-analysis of antidepressant pharmacogenetic findings in major depressive disorder. Mol. Psychiatry 15, 473–500. 10.1038/mp.2008.116 [DOI] [PubMed] [Google Scholar]
- Kiezun A., Artzi S., Modai S., Volk N., Isakov O., Shomron N. (2012). miRviewer: a multispecies microRNA homologous viewer. BMC Res. Notes 5:92. 10.1186/1756-0500-5-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene R., Chaudhary H., Karl N., Katic J., Kotarska A., Guitart K., et al. (2015). Interaction between CHL1 and serotonin receptor 2c regulates signal transduction and behavior in mice. J. Cell Sci. 128, 4642–4652. 10.1242/jcs.176941 [DOI] [PubMed] [Google Scholar]
- Kopczak A., Stalla G. K., Uhr M., Lucae S., Hennings J., Ising M., et al. (2015). IGF-I in major depression and antidepressant treatment response. Eur. Neuropsychopharmacol. J. Eur. Coll. Neuropsychopharmacol. 25, 864–872. 10.1016/j.euroneuro.2014.12.013 [DOI] [PubMed] [Google Scholar]
- Labermaier C., Masana M., Müller M. B. (2013). Biomarkers predicting antidepressant treatment response: how can we advance the field? Dis. Markers 35, 23–31. 10.1155/2013/984845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage A. D., Boyer R., Grunberg F., Vanier C., Morissette R., Ménard-Buteau C., et al. (1994). Suicide and mental disorders: a case-control study of young men. Am. J. Psychiatry 151, 1063–1068. 10.1176/ajp.151.7.1063 [DOI] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B., Bartel D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20. 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Li Y., Shang Y. M., Wang Q. W. (2016). MicroRNA-21 promotes the proliferation and invasion of neuroblastoma cells through targeting CHL1. Minerva Med. 107, 287–293. [PubMed] [Google Scholar]
- Liang Y., Ridzon D., Wong L., Chen C. (2007). Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8:166. 10.1186/1471-2164-8-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., Castle J., et al. (2005). Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433, 769–773. 10.1038/nature03315 [DOI] [PubMed] [Google Scholar]
- Long M.-J., Wu F.-X., Li P., Liu M., Li X., Tang H. (2012). MicroRNA-10a targets CHL1 and promotes cell growth, migration and invasion in human cervical cancer cells. Cancer Lett. 324, 186–196. 10.1016/j.canlet.2012.05.022 [DOI] [PubMed] [Google Scholar]
- Lopez J. P., Lim R., Cruceanu C., Crapper L., Fasano C., Labonte B., et al. (2014). miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nat. Med. 20, 764–768. 10.1038/nm.3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maness P. F., Schachner M. (2007). Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat. Neurosci. 10, 19–26. 10.1038/nn1827 [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D., Maccarrone G., Ising M., Kloiber S., Lucae S., Holsboer F., et al. (2014). Blood mononuclear cell proteome suggests integrin and Ras signaling as critical pathways for antidepressant treatment response. Biol. Psychiatry 76, e15–e17. 10.1016/j.biopsych.2014.01.022 [DOI] [PubMed] [Google Scholar]
- Mateus-Pinheiro A., Pinto L., Bessa J. M., Morais M., Alves N. D., Monteiro S., et al. (2013). Sustained remission from depressive-like behavior depends on hippocampal neurogenesis. Transl. Psychiatry 3:e210. 10.1038/tp.2012.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazalouskas M., Jessen T., Varney S., Sutcliffe J. S., Veenstra-VanderWeele J., Cook E. H., et al. (2015). Integrin β3 haploinsufficiency modulates serotonin transport and antidepressant-sensitive behavior in mice. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 40, 2015–2024. 10.1038/npp.2015.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milanesi E., Hadar A., Maffioletti E., Werner H., Shomron N., Gennarelli M., et al. (2015). Insulin-like growth factor 1 differentially affects lithium sensitivity of lymphoblastoid cell lines from lithium responder and non-responder bipolar disorder patients. J. Mol. Neurosci. MN 56, 681–687. 10.1007/s12031-015-0523-8 [DOI] [PubMed] [Google Scholar]
- Modai S., Shomron N. (2016). Molecular risk factors for schizophrenia. Trends Mol. Med 22, 242–253. 10.1016/j.molmed.2016.01.006 [DOI] [PubMed] [Google Scholar]
- Montag-Sallaz M., Baarke A., Montag D. (2003). Aberrant neuronal connectivity in CHL1-deficient mice is associated with altered information processing-related immediate early gene expression. J. Neurobiol 57, 67–80. 10.1002/neu.10254 [DOI] [PubMed] [Google Scholar]
- Montag-Sallaz M., Schachner M., Montag D. (2002). Misguided axonal projections, neural cell adhesion molecule 180 mRNA upregulation, and altered behavior in mice deficient for the close homolog of L1. Mol. Cell. Biol. 22, 7967–7981. 10.1128/MCB.22.22.7967-7981.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor E., Kano S.-I., Colantuoni C., Sawa A., Navon R., Shomron N. (2013). MicroRNA-382 expression is elevated in the olfactory neuroepithelium of schizophrenia patients. Neurobiol. Dis. 55, 1–10. 10.1016/j.nbd.2013.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morag A., Kirchheiner J., Rehavi M., Gurwitz D. (2010). Human lymphoblastoid cell line panels: novel tools for assessing shared drug pathways. Pharmacogenomics 11, 327–340. 10.2217/pgs.10.27 [DOI] [PubMed] [Google Scholar]
- Morag A., Pasmanik-Chor M., Oron-Karni V., Rehavi M., Stingl J. C., Gurwitz D. (2011). Genome-wide expression profiling of human lymphoblastoid cell lines identifies CHL1 as a putative SSRI antidepressant response biomarker. Pharmacogenomics 12, 171–184. 10.2217/pgs.10.185 [DOI] [PubMed] [Google Scholar]
- Mouillet-Richard S., Baudry A., Launay J.-M., Kellermann O. (2012). MicroRNAs and depression. Neurobiol. Dis. 46, 272–278. 10.1016/j.nbd.2011.12.035 [DOI] [PubMed] [Google Scholar]
- Müller D. W., Bosserhoff A.-K. (2008). Integrin beta 3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene 27, 6698–6706. 10.1038/onc.2008.282 [DOI] [PubMed] [Google Scholar]
- Nakayama T., Mihara K., Kawata J., Kimura H., Saitoh H. (2014). Adhesion of suspension cells on a coverslip in serum-free conditions. Anal. Biochem. 466, 1–3. 10.1016/j.ab.2014.07.023 [DOI] [PubMed] [Google Scholar]
- Ni R., Huang Y., Wang J. (2015). miR-98 targets ITGB3 to inhibit proliferation, migration, and invasion of non-small-cell lung cancer. OncoTargets Ther. 8, 2689–2697. 10.2147/OTT.S90998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor R. M., Grenham S., Dinan T. G., Cryan J. F. (2013). microRNAs as novel antidepressant targets: converging effects of ketamine and electroconvulsive shock therapy in the rat hippocampus. Int. J. Neuropsychopharmacol. Off. Sci. J. Coll. Int. Neuropsychopharmacol. CINP 16, 1885–1892. 10.1017/S1461145713000448 [DOI] [PubMed] [Google Scholar]
- Oved K., Morag A., Pasmanik-Chor M., Oron-Karni V., Shomron N., Rehavi M., et al. (2012). Genome-wide miRNA expression profiling of human lymphoblastoid cell lines identifies tentative SSRI antidepressant response biomarkers. Pharmacogenomics 13, 1129–1139. 10.2217/pgs.12.93 [DOI] [PubMed] [Google Scholar]
- Oved K., Morag A., Pasmanik-Chor M., Rehavi M., Shomron N., Gurwitz D. (2013). Genome-wide expression profiling of human lymphoblastoid cell lines implicates integrin beta-3 in the mode of action of antidepressants. Transl. Psychiatry 3:e313. 10.1038/tp.2013.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo K., Cingolani L. A., Bassani S., Laurent F., Passafaro M., Goda Y. (2012). β3 integrin interacts directly with GluA2 AMPA receptor subunit and regulates AMPA receptor expression in hippocampal neurons. Proc. Natl. Acad. Sci. U.S.A 109, 1323–1328. 10.1073/pnas.1113736109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price L. H., Charney D. S., Delgado P. L., Heninger G. R. (1990). Lithium and serotonin function: implications for the serotonin hypothesis of depression. Psychopharmacology (Berl.) 100, 3–12. 10.1007/BF02245781 [DOI] [PubMed] [Google Scholar]
- Ruhé H. G., Huyser J., Swinkels J. A., Schene A. H. (2006). Switching antidepressants after a first selective serotonin reuptake inhibitor in major depressive disorder: a systematic review. J. Clin. Psychiatry 67, 1836–1855. 10.4088/JCP.v67n1203 [DOI] [PubMed] [Google Scholar]
- Rukov J. L., Shomron N. (2011). MicroRNA pharmacogenomics: post-transcriptional regulation of drug response. Trends Mol. Med. 17, 412–423. 10.1016/j.molmed.2011.04.003 [DOI] [PubMed] [Google Scholar]
- Rukov J. L., Vinther J., Shomron N. (2011). Pharmacogenomics genes show varying perceptibility to microRNA regulation. Pharmacogenet. Genomics 21, 251–262. 10.1097/FPC.0b013e3283438865 [DOI] [PubMed] [Google Scholar]
- Rukov J. L., Wilentzik R., Jaffe I., Vinther J., Shomron N. (2014). Pharmaco-miR: linking microRNAs and drug effects. Brief. Bioinform 15, 648–659. 10.1093/bib/bbs082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzezniczek S., Obuchowicz M., Datka W., Siwek M., Dudek D., Kmiotek K., et al. (2016). Decreased sensitivity to paroxetine-induced inhibition of peripheral blood mononuclear cell growth in depressed and antidepressant treatment-resistant patients. Transl. Psychiatry 6:e827. 10.1038/tp.2016.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saegusa J., Yamaji S., Ieguchi K., Wu C.-Y., Lam K. S., Liu F.-T., et al. (2009). The direct binding of insulin-like growth factor-1 (IGF-1) to integrin alphavbeta3 is involved in IGF-1 signaling. J. Biol. Chem 284, 24106–24114. 10.1074/jbc.M109.013201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangkuhl K., Klein T. E., Altman R. B. (2009). Selective serotonin reuptake inhibitors pathway. Pharmacogenet. Genomics 19, 907–909. 10.1097/FPC.0b013e32833132cb [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlatter M. C., Buhusi M., Wright A. G., Maness P. F. (2008). CHL1 promotes Sema3A-induced growth cone collapse and neurite elaboration through a motif required for recruitment of ERM proteins to the plasma membrane. J. Neurochem. 104, 731–744. 10.1111/j.1471-4159.2007.05013.x [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc 3, 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Serafini G., Pompili M., Hansen K. F., Obrietan K., Dwivedi Y., Shomron N., et al. (2014). The involvement of microRNAs in major depression, suicidal behavior, and related disorders: a focus on miR-185 and miR-491-3p. Cell. Mol. Neurobiol. 34, 17–30. 10.1007/s10571-013-9997-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao N.-Y., Hu H. Y., Yan Z., Xu Y., Hu H., Menzel C., et al. (2010). Comprehensive survey of human brain microRNA by deep sequencing. BMC Genomics 11:409. 10.1186/1471-2164-11-409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomron N. (2009). MicroRNAs and their antagonists as novel therapeutics. Eur. J. Cancer Oxf. Engl. 1990 45(Suppl. 1), 388–390. 10.1016/S0959-8049(09)70060-7 [DOI] [PubMed] [Google Scholar]
- Shomron N. (2010). MicroRNAs and pharmacogenomics. Pharmacogenomics 11, 629–632. 10.2217/pgs.10.26 [DOI] [PubMed] [Google Scholar]
- Souery D., Oswald P., Massat I., Bailer U., Bollen J., Demyttenaere K., et al. (2007). Clinical factors associated with treatment resistance in major depressive disorder: results from a European multicenter study. J. Clin. Psychiatry 68, 1062–1070. 10.4088/JCP.v68n0713 [DOI] [PubMed] [Google Scholar]
- Sun L., Liu B., Lin Z., Yao Y., Chen Y., Li Y., et al. (2015). MiR-320a acts as a prognostic factor and Inhibits metastasis of salivary adenoid cystic carcinoma by targeting ITGB3. Mol. Cancer 14:96. 10.1186/s12943-015-0344-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahimic C. G. T., Long R. K., Kubota T., Sun M. Y., Elalieh H., Fong C., et al. (2016). Regulation of Ligand and Shear Stress-induced Insulin-like Growth Factor 1 (IGF1) Signaling by the Integrin Pathway. J. Biol. Chem. 291, 8140–8149. 10.1074/jbc.M115.693598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansey K. E., Guipponi M., Perroud N., Bondolfi G., Domenici E., Evans D., et al. (2012). Genetic predictors of response to serotonergic and noradrenergic antidepressants in major depressive disorder: a genome-wide analysis of individual-level data and a meta-analysis. PLoS Med. 9:e1001326. 10.1371/journal.pmed.1001326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasawa K., Ichimura A., Sato F., Shimizu K., Tsujimoto G. (2009). Sustained activation of ERK1/2 by NGF induces microRNA-221 and 222 in PC12 cells. FEBS J. 276, 3269–3276. 10.1111/j.1742-4658.2009.07041.x [DOI] [PubMed] [Google Scholar]
- Thaler K. J., Morgan L. C., Van Noord M., Gaynes B. N., Hansen R. A., Lux L. J., et al. (2012). Comparative effectiveness of second-generation antidepressants for accompanying anxiety, insomnia, and pain in depressed patients: a systematic review. Depress. Anxiety 29, 495–505. 10.1002/da.21951 [DOI] [PubMed] [Google Scholar]
- Thirumala S., Forman J. M., Monroe W. T., Devireddy R. V. (2007). Freezing and post-thaw apoptotic behaviour of cells in the presence of palmitoyl nanogold particles. Nanotechnology 18:195104 10.1088/0957-4484/18/19/195104 [DOI] [Google Scholar]
- Thomas R. M., Peterson D. A. (2008). Even neural stem cells get the blues: evidence for a molecular link between modulation of adult neurogenesis and depression. Gene Expr 14, 183–193. [PMC free article] [PubMed] [Google Scholar]
- Tsankova N., Renthal W., Kumar A., Nestler E. J. (2007). Epigenetic regulation in psychiatric disorders. Nat. Rev. Neurosci. 8, 355–367. 10.1038/nrn2132 [DOI] [PubMed] [Google Scholar]
- Uher R., Perroud N., Ng M. Y. M., Hauser J., Henigsberg N., Maier W., et al. (2010). Genome-wide pharmacogenetics of antidepressant response in the GENDEP project. Am. J. Psychiatry 167, 555–564. 10.1176/appi.ajp.2009.09070932 [DOI] [PubMed] [Google Scholar]
- Voorhoeve P. M., le Sage C., Schrier M., Gillis A. J. M., Stoop H., Nagel R., et al. (2006). A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124, 1169–1181. 10.1016/j.cell.2006.02.037 [DOI] [PubMed] [Google Scholar]
- Wan Y., Liu Y., Wang X., Wu J., Liu K., Zhou J., et al. (2015). Identification of differential microRNAs in cerebrospinal fluid and serum of patients with major depressive disorder. PLoS ONE 10:e0121975. 10.1371/journal.pone.0121975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J. A., Baxter D. H., Zhang S., Huang D. Y., Huang K. H., Lee M. J., et al. (2010). The microRNA spectrum in 12 body fluids. Clin. Chem. 56, 1733–1741. 10.1373/clinchem.2010.147405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte A., Jessen T., Varney S., Carneiro A. M. D. (2014). Serotonin transporter and integrin beta 3 genes interact to modulate serotonin uptake in mouse brain. Neurochem. Int. 73, 122–126. 10.1016/j.neuint.2013.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Han H., Chen J., Zhang Z., Li S., Fang F., et al. (2014). MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 342, 43–51. 10.1016/j.canlet.2013.08.030 [DOI] [PubMed] [Google Scholar]
- Zhou R., Yuan P., Wang Y., Hunsberger J. G., Elkahloun A., Wei Y., et al. (2009). Evidence for selective microRNAs and their effectors as common long-term targets for the actions of mood stabilizers. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 34, 1395–1405. 10.1038/npp.2008.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Fang J., Zhang J., Zhao Z., Liu L., Wang J., et al. (2014). miR-182 targets CHL1 and controls tumor growth and invasion in papillary thyroid carcinoma. Biochem. Biophys. Res. Commun. 450, 857–862. 10.1016/j.bbrc.2014.06.073 [DOI] [PubMed] [Google Scholar]
- Zweig R. A., Hinrichsen G. A. (1993). Factors associated with suicide attempts by depressed older adults: a prospective study. Am. J. Psychiatry 150, 1687–1692. 10.1176/ajp.150.11.1687 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.