Abstract
The CRISPR/Cas system became a powerful genome editing tool for basic plant research and crop improvement. Thus far, CRISPR/Cas has been applied to many plants, including Arabidopsis, rice and other crop plants. It has been reported that CRISPR/Cas efficiency is generally high in many plants. In this study, we compared the genome editing efficiency of CRISPR/Cas in three different Arabidopsis accessions [Col-0, Ler, and C24RDLUC (C24 accession harboring the stress-responsive RD29A promoter-driven luciferase reporter)]. For the comparison, we chose to target the cold-responsive C-repeat/DRE-Binding Factor (CBF) genes. CBF1, CBF2, and CBF3 genes are tandemly located on Arabidopsis chromosome 4 with redundant functions as the key transcription factors functioning in cold stress signaling and tolerance. Due to the close proximity of these CBFs on the chromosome, it is impossible to generate cbf1, cbf2, cbf3 triple mutants (cbf123) by traditional genetic crosses. Therefore, using the CRISPR/Cas tool, we aimed to generate cbf123 mutants and compared the genome editing efficiency in different Arabidopsis accessions. Among the accessions, Ler was the most resilient to the CRISPR/Cas deletion with the lowest gene deletion ratio in both T1 and T2 generations. Interestingly, while C24RDLUC showed a high CBF123 deletion frequency in T2 only when the gene deletion was observed in T1 generation, Col-0 displayed high ratios of the CBF123 deletions in T2 regardless of the presence or absence of the CBF123 deletion in T1. Isolated cbf123 mutants in C24RDLUC background showed no expression of CBF1, CBF2, and CBF3 genes and proteins with reduction in the CBF target gene expression under cold stress.
Keywords: CRISPR, Cas, CBF, cold stress, cold signaling, accessions
Introduction
The clustered regularly interspaced short palindromic repeats/CRISPR-associated (CRISPR/Cas) system is a new technology for targeted genome editing. The CRISPR/Cas system was first studied as an adaptive immune system for prokaryotes to defend themselves from foreign nucleic acids invasion (Wiedenheft et al., 2012; Sampson and Weiss, 2014). Although other targeted genome editing methods, such as transcription activator-like effector nuclease (TALENs) and zinc finger nucleases (ZFN), can generate genome modifications, the CRISPR/Cas system is a more affordable, robust, and easy genome editing tool (Mali et al., 2013; Shan et al., 2013; Sampson and Weiss, 2014). As CRISPR/Cas uses a guide RNA to specify the editing target DNA sequence, CRIPSR/Cas system does not need the elaborate design and assembly of DNA-binding proteins and makes it possible to generate a construct ready for transformation with a synthesis of simple DNA oligomers. The synthesized DNA oligomers are transcribed into single guide RNA (sgRNA) that guides the Cas9 DNA endonuclease to the target sites by sgRNA hybridization. The endonuclease Cas9 makes a double strand break at 3 bp upstream of Palindromic Adjacent Motif (PAM) sequence. The DNA breakage is repaired by homologous recombination (HR) or the error-prone non-homologous end joining (NHEJ) mechanism (Schiml et al., 2014). The NHEJ mechanism is known to be the major double strand break repair pathway in plants (Britt, 1999). During the NHEJ DNA repair process, the errors can be introduced causing irreversible mutations at the target sites in plants. In addition, multiple DNA breakages are possible in plants by introducing multiple sgRNAs to the target plants. These multiple DNA breakages can cause multiple mutations or large deletions depending on distance among the target sites (Li et al., 2013; Mao et al., 2013).
The CRISPR/Cas system works in different rates depending on the target region and the sequence of the sgRNA (Mali et al., 2013; Mao et al., 2013). Thus, in the present study, we aimed to test the efficiency of CRISPR/Cas in various Arabidopsis accessions - Col-0, Ler, and C24RDLUC (C24 accession with the stress-responsive RD29A promoter-driven luciferase reporter). We chose these accessions because Col-0 and Ler are among the most commonly used ones in Arabidopsis and C24RDLUC would make it easy to examine CBF-target gene down-regulation by using luciferase imaging system (Ishitani et al., 1997). As genome editing target genes, we chose to delete C-repeat/DRE-Binding Factor (CBF) genes that are important in cold stress signaling in plants. Upon cold stress, plants increase the expression of Cold Regulated (COR) genes that molecularly adapt the plant to withstand cold stress (Chinnusamy et al., 2007; Park et al., 2015). The key signaling pathway for the expression of COR genes is the CBF signaling pathway. CBFs are transcription factors with a conserved DNA-binding domain found in the ethylene-responsive element-binding factors (ERF) and floral homeotic protein APETALA 2 (AP2) proteins. In Arabidopsis genome, there are four CBF genes. Among them, CBF1, CBF2, and CBF3 are early induced by cold, but not by drought and salt stresses (Yamaguchi-Shinozaki and Shinozaki, 2006). By contrast, CBF4 gene expression is up-regulated by drought stress, but not by low temperature (Haake et al., 2002). Accordingly, CBF1, CBF2, and CBF3 (CBF123 hereafter when all the three genes are mentioned) function as primary transcription factors for cold tolerance. CBF1, CBF2, and CBF3 are closely aligned within 7.1K base pairs on the chromosome 4 of Arabidopsis with a small intergenic distance (2–3 Kbp). Due to this tandem array of the three genes, it is almost impossible to generate cbf123 triple mutants by traditional crossings. However, with the emergence of the CRISPR/Cas system, generation of cbf123 triple mutants has become possible by targeting the tandemly located all three CBF genes. Indeed, very recently, four lines of cbf123 triple mutants were reported in Columbia-0 background (Jia et al., 2016; Zhao et al., 2016; Zhao and Zhu, 2016).
In this study, we found that Ler showed the lowest ratio of the CBF123 gene deletion by CRISPR/Cas in both T1 and T2 generations. In addition, Col-0 and C24RDLUC displayed generally high ratios of the CBF123 gene deletion in T1 and T2 generation. Interestingly, the high ratios of the CBF123 deletions in Col-0 were observed regardless of the presence or absence of the gene deletion tested in the leaves of each T2’s progenitor (T1) while C24RDLUC showed a high CBF123 deletion frequency in T2 generation when high gene deletion ratios were observed in T1 generation. Isolated cbf123 mutants in C24RDLUC background (cbf123LUC-2) showed no expression of CBF1, CBF2, and CBF3 genes and proteins were detected in these cbf123LUC-2 mutants after cold treatment, suggesting that cbf123LUC-2 is a null mutant. Accordingly, CBF target gene expression in cbf123LUC-2 was reduced under cold stress in comparison with its background wild type. cbf123LUC-2 displayed a smaller size than wild type at the early development stage.
Materials and Methods
Plant Growth
Arabidopsis seeds were surface-sterilized with bleach (∼4% sodium hypochlorite) and plated on MS plates. After plating, the plates were kept at 4°C for at least 2 days to obtain germination synchrony before being transferred to 22°C under constant illumination (80–100 μmol m-2S-1) and 70% relative humidity for germination and growth. Murashige and Skoog (MS) medium (pH 5.8) was made with full strength MS salts (Caisson Laboratories, United States), 2% sucrose, and 0.3% gelite (Duchefa, Netherlands). For selection plates, hygromycin B was added to a final 25 mg/L concentration to the MS media. For soil growth, the seeds planted on soil (Sungro mixture#5, Canada) were placed in a growth room operating at 22°C with the cycle of 16-h of light and 8-h of darkness (the light intensity of 80–100 μmol m-2S-1) after 2 days of cold stratification.
CRISPR/Cas9 Construct Generation
For the selection of multiple targeting sgRNA, multiple sequence alignment software Clustal W1 was used to align the coding sequence of CBF1, CBF2, and CBF3. Two 19-bp sequences (sgRNA12 and sgRNA23) immediately before a PAM sequence (5′-NGG-3′) were selected and used for DNA oligomer synthesis for sgRNA.
The forward and reverse DNA oligomers for sgRNA targeting CBF1 and CBF2 were CBF12-sgR1-F (5′-GATTGAGCTGCCATCTCAGCGGTT-3′), CBF12-sgR1-R (5′-AAACAACCGCTGAGATGGCAGCTC-3′) and for sgRNA targeting CBF2 and CBF3 were CBF23-sgR1-F (5′-GATTGGAGTCAGCGAAATTGAGAC-3′) and CBF23-sgR1-R (5′-AAACGTCTCAATTTCGCTGACTCC-3′).
Following the protocol suggested by Liu et al. (2015), psgR-Cas9-At was used to generate each single sgRNA-containing vector (i.e., sgRNA12-Cas9 vector and sgRNA23-Cas9 vector). To make a double sgRNA-containing sgRNA12-sgRNA23-Cas9 vector, sgRNA23 module from the sgRNA23-Cas9 plasmid was PCR-amplified using a following primer pair (sgR_U6_Kpn1-F, 5′-GCCGGTACCCATTCGGAGTTTTTGTAT-3′; sgR_end_EcoRI-R, 5′-TATGAATTCGCCATTTGTCTGCAGAATTG-3′). The resultant PCR product was inserted into the KpnI and EcoRI sites of sgRNA12–Cas9 vector. Finally, the whole cassette of sgRNA12-Cas9-sgRNA23 released by HindIII and EcoRI from the double sgRNA containing construct was subcloned to the HindIII-EcoRI sites of pCAMBIA1300, resulting in the pCAMBIA-sgRNA12-sgRNA23-Cas9 construct. A schematic drawing of the construct generation is shown in Supplementary Figure S1.
Construction of CRISPR/Cas Transgenic Arabidopsis and Detection of cbf123 Deletion
Arabidopsis thaliana plants (accession Col-0, Ler, and C24RDLUC) were transformed with pCAMBIA-sgRNA12-sgRNA23-Cas9 construct via Agrobacterium tumefaciens strain GV3101 by floral dipping (Clough and Bent, 1998). T1 seeds were collected from the floral dipped plants and then selected on MS plates with hygromycin B 25 μg/mL. The selected T1 plants were transferred to soil and genotyped with 3 different primer pairs (pCAM1300intC-R, 5′-GGCCTCTTCGCTATTACGC-3′ and CBF12-sgR1-R, 5′-AAACAACCGCTGAGATGGCAGCTC-3′ for sgRNA12; doublesgRNA-F, 5′-GATCGACCTGTCTCAGCTGG-3′ and pCAM1300intC-F, 5′-ATTAATGCAGCTGGCACGAC-3′ for sgRNA23; Cas9-F, 5′-CCCAACTTCAAGAGCAACTT-3′ and Cas9-R, 5′-TCACTTTGGTCAGCTCGTTA-3′ for Cas9). For the cbf123 deletion detection, nested PCR was employed with the following primer pairs were used (CBF1-PS-F, 5′-CGTGTGCTCCCCACATATC-3′ and CBF2-PS2-F, 5′-ATTTGTTGCTTATGGGGAGA-3′ for the first round PCR; CBF1_small_R, 5′-AATCCAAAAAGACTGAGAACTCTA-3′ and CBF2q-R, 5′-CTGCACTCAAAAACATTTGCA-3′ for the second round PCR). For the cbf123 deletion confirmation and homozygosity test, 4 different primers were used (CBF1_samll_R, CBF2q-R, CBF1-PS-R, 5′-CCGCTTTTTGGATATCCTTG-3′ and CBF2qRT-F(172), 5′-AACTCCGGTAAGTGGGTGTG-3′).
Luminescence Imaging
Luminescence intensities of cbf123LUC-2 were measured by the Lumazone luminescence imaging system (Roper Scientific, United States). Ten-day-old seedlings grown on MS agar plates were incubated at 0°C for the designated times and the luminescence images were taken after luciferin spray. The luminescence intensity of each seedlings was quantified with the WinView32 program.
Gene Expression Analysis
Total RNA was extracted from 11- to 13-day-old seedlings with or without cold (0°C) treatment using the RNAiso Plus reagent (Takara, Japan). After DNase I treatment (New England Biolabs, United States), cDNA was synthesized with 5 μg total RNA using the TOPScriptTM Reverse Transcriptase Kit (Enzynomics, Korea). cDNA was used as a template for quantitative real-time PCR (qRT-PCR) using SYBR® FAST (KAPA Biosystems, United States) on the LightCycler®96 (Roche, Switzerland). The following primer pairs were used for real-time PCR (CBF1q-F, 5′-GCATGTCTCAACTTCGCTGA-3′ and CBF1q-R, 5′-ATCGTCTCCTCCATGTCCAG-3′ for CBF1; CBF2q-F, 5′-TGACGTGTCCTTATGGAGCTA-3′ and CBF2q-R, 5′-CTGCACTCAAAAACATTTGCA-3′ for CBF2; CBF3q-F, 5′-GATGACGACGTATCGTTATGGA-3′ and CBF3q-R, 5′-TACACTCGTTTCTCAGTTTTACAAAC-3′ for CBF3; RD29A_RT-F, 5′-CTTGTCGACGAGAAGCAAAGAA-3′ and RD29A_RT-R, 5′-TCTTGATGGAGAATTCGTGTCC-3′ for RD29A; COR15A_RT-F, 5′-ACTCAGTTCGTCGTCGTTTCTC-3′ and COR15A_RT-R, 5′-TCTCACCATCTGCTAATGCCTC-3′ for COR15A; COR47_qRT-F, 5′-TGTCATCGAAAAGCTTCACCGA-3′ and COR47_qRT-R, 5′-ACCGGGATGGTAGTGGAAACTG-3′ for COR47; KIN1_qRT-F, 5′-ATGCCTTCCAAGCCGGTCAGAC-3′ and KIN1_qRT-R, 5′-CCGGTCTTGTCCTTCACGAAGT-3′ for KIN1; Clathrin-F, 5′-CTGACTGGCCCTGCTT-3′ and Clathrin-R, 5′-ATACGCGCTGAGTTCCC-3′ for Clathrin as the internal control).
Protein Blot Analysis
For protein blot analysis for CBF123 protein detection, 14-day-old seedlings of wild type and cbf123LUC-2 on MS agar plates were cold-treated at 0°C for 6 h, the seedlings were frozen-ground and suspended in protein sample buffer [130 mM Tris-Cl (pH 8.0), 4.6% (w/v) SDS, 0.02% Bromophenol blue, 2% DTT, 20% (w/v) Glycerol]. Samples were boiled for 3 min and centrifuged at 13,200 rpm for 10 min at 4°C. The supernatant was run on the sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel (Nguyen et al., 2016). The resultant gel was blotted on to the polyvinylidene fluoride (PVDF) membrane and the existence of CBF proteins were detected with rabbit anti-CBF123 antibody and anti-rabbit goat anti-rabbit IgG-HRP conjugate (1:10000, Abcam, United Kingdom). Anti-CBF123 antibody was raised with full length protein of CBF2. Further detail on the method of anti-CBF123 generation is provided in Supplementary Figure S3.
Statistical Analysis
Statistical analyses of the results from the experiments of three accessions were performed using two-way analysis of variance with Microsoft Excel program (2010 version), and significant differences between each index among accessions and among T2 ratios of the same accession were determined using Student’s T test.
Results
Small Guide RNA Sequence Selection for CBF1, CBF2 and CBF3
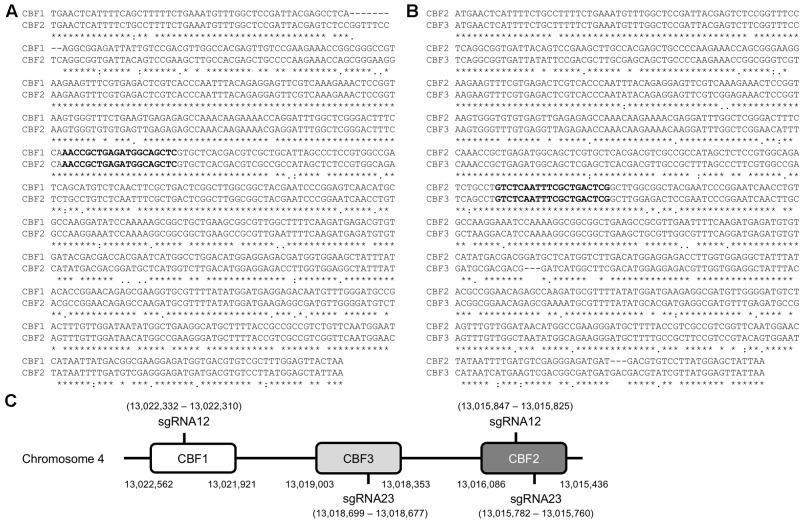
To generate a triple mutant of CBF1, CBF2, and CBF3 genes using the CRISPR/Cas system, two possible sgRNA target sites were selected by aligning CBF1 and CBF2, and CBF2 and CBF3 with a multiple sequence alignment program (Clustal W)2 (Figures 1A,B). The two sgRNA target sequences were named sgRNA12, which targets CBF1 and CBF2, and sgRNA23, which targets CBF2 and CBF3 (Figures 1A–C). There were no single sgRNA site that could target all three CBF genes in the coding sequence (CDS) region at the same time. By using these two sgRNAs, the CRISPR/Cas system can target the three CBFs in the four sites (Figure 1C). Since the sgRNAs will be used in three different accessions, Col-0, C24RDLUC and Ler, the CBF regions of these accessions were sequenced. We found that the DNA sequences for sgRNA12 and sgRNA23 were identical in all three accessions (data not shown). Then, the possible off-target sites of sgRNA12 and sgRNA23 were analyzed by the online program CRISPR-P3 (Lei et al., 2014). The analysis revealed that the off-target possibility of both sgRNA12 and sgRNA23 was extremely low (scores of 0.0–5.7 for off-target sites vs. 100 for CBF1, CBF2, and CBF3 genes), suggesting that each would very specifically target CBF1/CBF2 and CBF2/CBF3, respectively (Table 1).
FIGURE 1.
Design of DNA oligomer for sgRNA targeting CBF1, CBF2 and CBF3. (A,B) Alignments of coding sequences of CBF1/CBF2 (A) and CBF2/CBF3 (B). The bold sequences denote the selected sgRNA sequence and ∗indicates the same sequence between CBFs. (C) Schematic drawing of the location of DNA oligomer for sgRNAs targeting CBF1, CBF3, and CBF2 as in the order on the chromosome 4 of Arabidopsis thaliana. Numbers in parenthesis indicate the nucleotide numbers of chromosome 4.
Table 1.
Selected DNA sequence for sgRNA and possible off-target sequence analyzed by CRISPR-P program.
| Sequence | Score | AGI ID | Gene name | |
|---|---|---|---|---|
| sgRNA12 | GAGCTGCCATCTCAGCGGTTTGG | 100 | AT4G25480 | CBF2 |
| GAGCTGCCATCTCAGCGGTTTGG | 100 | AT4G25470 | CBF1 | |
| GAGCAGCCATGTCAGGGGCTTGG | 0 | AT3G17410 | Unknown protein | |
| GCGCTGCCATCTCCGCCGTGGGG | 0 | AT2G25820 | ESE2 | |
| sgRNA23 | CGAGTCAGCGAAATTGAGACAGG | 100 | AT4G25480 | CBF2 |
| CGAGTCAGCGAAATTGAGACAGG | 100 | AT4G25470 | CBF3 | |
| AGAATCAGCGAAATTGAGACAAG | 5.7 | AT5G51990 | CBF4 | |
| CGTTTCAGCGAAATTGATAAGGG | 0.1 | AT2G47790 | GTS1 | |
| CGAGACTGAGAAATTTAGACGGG | 0.1 | Intergenic region | ||
Identification of T1 Seedlings with a Complete Set of Transgenes
In order to target CBF1, CBF2, and CBF3 genes, DNA oligomers for sgRNA12 and sgRNA23 were inserted into psgR-Cas9-At vector containing the Cas9 sequence with a chimeric RNA backbone for the sgRNA (Feng et al., 2013; Mao et al., 2013; Liu et al., 2015). The whole cassette with Cas9 was then subsequently subcloned into pCAMBIA1300 to generate the CBF targeting CRISPR/Cas binary vector construct. The construct was used to transform three Arabidopsis accessions through Agrobacterium mediated plant transformation. The seeds from the floral-dipped plants (T1 seeds) were harvested and plated on hygromycin plates for the positive transformant selection. The presence of transgenes including sgRNA12, sgRNA23, and Cas9 in the positive T1 seedlings was then tested by PCR with the primers used for construct component confirmation. While generally high (75–100%), the detection ratios of each transgene in each accession varied, implying various occurrences of the whole transgene insertion in each accession (Table 2). The ratios of plants with a complete set of three transgenes were higher than 60% in all accessions with Columbia-0 being the highest (80%) (Table 2). These transgenic plants with a whole transgene set were further analyzed for CRISPR/Cas efficiency on the CBF genes.
Table 2.
Detection ratio of the sgRNA12-sgRNA23-Cas9 transgenes in T1 plants.
| Accession | sgRNA12 | Cas9 | sgRNA23 | sgRNA12 and Cas9 | Cas9 and sgRNA23 | sgRNA12 and sgRNA23 | sgRNA12, Cas9 and sgRNA23 |
|---|---|---|---|---|---|---|---|
| Col-0 | 95.00 | 100.00 | 95.00 | 95.00 | 95.00 | 90.00 | 16/20 ∗ 80.00% |
| C24RDLUC | 84.00 | 84.00 | 94.00 | 72.00 | 80.00 | 82.00 | 33/50 ∗ 66.00% |
| Ler | 87.50 | 89.58 | 87.50 | 81.25 | 81.25 | 75.00 | 35/48 ∗ 72.92% |
Detection ratio (%). ∗Number of plant with all transgenes of sgRNA12, Cas9 and sgRNA23/total number of plants examined.
Accession-Dependent CBF123 Deletion Efficiency in T1 Plants
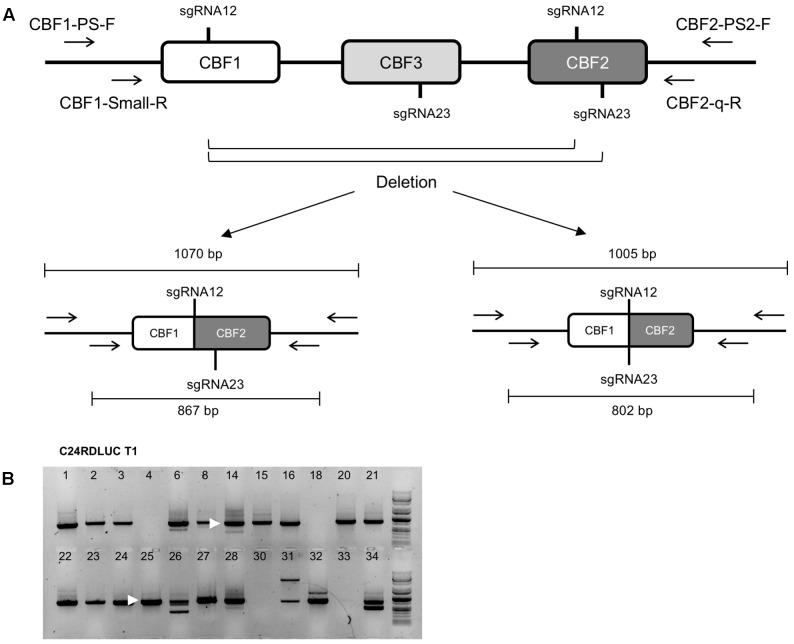
We then asked in T1 generation if our sgRNAs were functional in CRISPR/Cas-mediated genome editing and in what ratio the CRISPR/Cas-caused CBF123 deletions would occur in each accession. Our sgRNA design intended to target four sites (two sites per sgRNA) in CBF1, CBF2, and CBF3 genes (Figure 1C) to maximize the gene editing efficiency. Among the deletions that would be produced by our CRISPR/Cas system, the deletions for the cbf123 triple mutation should occur between the sgRNA12 site on CBF1 and sgRNA12 or sgRNA23 site on CBF2 (Figure 1C). In order to detect these deletions of the CBF gene region, we performed PCR using a primer set of one primer aligning to the 5′ UTR region of CBF1 and the other aligning at the 3’ UTR region of CBF2. Ca. 1070 bp and/or 1005 bp PCR products were expected if the deletion for cbf123 mutation took place (Figure 2A). Our initial PCR produced only several faint bands. It is highly likely because of the low numbers of cells with cbf123 deletion mutation in these T1 plants. It should be noted that T1 plants would contain heterogeneous cells with various CRISPR/Cas effects. Therefore, nested PCR was conducted with the 1000 times diluted initial PCR products. Nested PCR would also ensure that the faint bands are not non-specific and that the bands are also sufficiently intensified to be visible. As expected, in the nested PCR results, even the samples without initial visible bands displayed clear bands (Figure 2B). Among the T1 plants with all transgenes, the large deletion ratios for cbf123 triple mutations varied between 45.71 and 84.85% among the accessions with Ler being the lowest and C24RDLUC the highest (Table 3). These results suggest that the CRISPR/Cas system has accession-dependent efficiency in the generation of CBF123 deletion.
FIGURE 2.
Detection of large deletion for cbf123 triple mutation in T1 plants. (A) Schematic drawing of CBF123 deletion detection. Possible cut sites of sgRNA between sgRNA12 sites of CBF1 and CBF2 will generate a 1070 bp band and those between sgRNA12 of CBF1 and sgRNA23 of CBF2 will generate a 1005 base pair band. (B) Nested PCR results to detect a large deletion for cbf123 triple mutation. Arrow heads indicate the band sizes about 800 bp.
Table 3.
CBF123 deletion ratio among T1 plants harboring all transgenes of sgRNA12, Cas9 and sgRNA23.
| Accession | sgRNA12, Cas9 and sgRNA23∗ | Large deletion mutation∗∗ |
|---|---|---|
| Col-0 | 16/20 (80.00%) | 12/16 (75.00%) |
| C24RDLUC | 33/50 (66.00%) | 28/33 (84.85%) |
| Ler | 35/48 (72.92%) | 16/35 (45.71%) |
∗Number of plants with all transgenes of sgRNA12, Cas9 and sgRNA23/total number of plants examined. ∗∗Number of plants with CBF123 deletion/number of plants with all transgenes of sgRNA12, Cas9 and sgRNA23.
Various CBF123 Deletion in T1 Plants
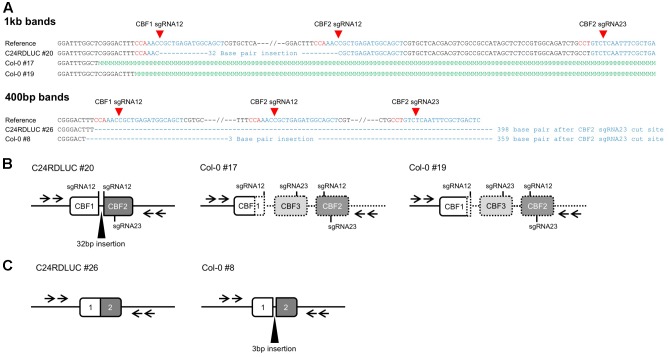
To determine whether the deletion occurred in the sgRNA cut sites, the nested PCR products from T1 plants were sequenced. Although the sizes of the PCR bands varied considerably, most bands showed a size of ca. 800 base pairs, which was in agreement with the assumption that the CBF123 deletion mutations would occur at the sgRNA cut sites present on CBF1 and CBF2 (Figure 2). CRISPR/Cas action can result in mutations with a few nucleotide additions or deletions at the sgRNA target site due to the error-prone non-homologous end joining pathway (Belhaj et al., 2013; Hsu et al., 2014; Lozano-Juste and Cutler, 2014; Liu et al., 2015). Because of this, T1 plants become genetically mosaic with cells containing different mutations at the sgRNA target sites. Thus, the PCR products similar in size could be heterogeneous and the sequencing of the PCR products could produce multiple peaks at the nucleotide after the sgRNA cut sites. Our sequencing results showed that the deletion started at the cut site of sgRNA or in close proximity to it, implying a specific double strand break on the target sites (Figures 3A,B). Our results also reveal that addition or deletion of nucleotides at the sgRNA target also occurred in our CBF123 deletion mutations (Figures 3A,B). Occasionally, we observed the sequencing results from some unexpected size products (Figures 3A,C). These could be due to the prolonged DNA repair which might lead to extra base pair deletion after the sgRNA cut site (Gorbunova and Levy, 1999).
FIGURE 3.
Various mutations found in T1 plants. (A) Alignments of sequences of the PCR product from T1 plants and the reference sequence. Red sequences indicate the PAM sequence, blue sequences denote the sgRNA sequences and the green “M” sequences mean multiple peaks shown in sequence electropherogram. (B) Drawing of the CBF123 deletion that will give rise to about 1 kb product from PCR with primers indicated by arrows. The drawings in dotted line indicate DNA regions with multiple sequencing peaks due to genetically mosaic T1 leaf tissues. (C) Drawing of the CBF123 deletion that will give rise to about 400 bp product from PCR with primers indicated by arrows.
Accession-Dependent CBF123 Deletion Inheritance to T2 Plants
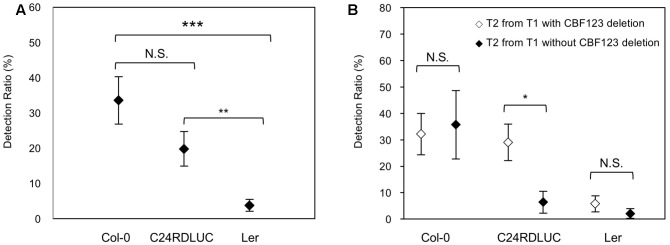
The T2 seeds from the T1 plants were harvested and used for the analysis of genetic inheritance of the CBF123 deletion and the cbf123 triple mutant screening. We examined T2 progenies from all T1 plants with a complete set of sgRNAs and Cas9 transgenes; T2 progenies from 16 T1 Col-0 plants, 33 T1 C24RDLUC plants, and 35 T1 Ler plants were analyzed (Table 3 and Supplementary Tables S1–S3). In most cases, we analyzed 24 T2 seedlings per T1 line. We found that CBF123 gene deletions could be observed in T2 seedlings not only from the T1 lines with the CBF123 deletion, but also from the T1 lines without the deletions. Also, there were some cases when the CBF123 gene deletion was not inherited to T2 seedlings from the T1 plants that contained the CBF123 deletions (Supplementary Tables S1–S3). This indicates that the CRISPR/Cas9-induced mutations in germ cells do not always occur even when the CBF123 deletion in somatic cells (leaf cells) exists at T1. Interestingly, the transmission ratios of CBF123 deletion from T1 to T2 generation were different among the Arabidopsis accessions. Although not significantly different, the ratios of the CBF123 deletion at the T2 generations in Col-0 was the highest (33.56%) followed by C24RDLUC (19.80%). Ler showed the lowest CBF123 gene deletion ratio (3.74%) (Figure 4A). We also noticed the different accession-dependent inheritance ratios of the CBF123 deletion among the T2 progenies from T1 lines with and without the CBF123 deletions (Figure 4B). In Col-0, the CBF123 deletions were detected at very similar ratios in the T2 progenies, regardless of the presence of CBF123 deletions at the T1 generation (32.20% in T2 from CBF123-deleted T1 vs. 35.71% in T2 from CBF123 not-deleted T1) (Figure 4B). By contrast, C24RDLUC showed a higher transmission ratio of the CBF123 deletions in T2 lines from T1 lines with the deletion rather than in those from T1 lines without the deletions (29.0% vs. 6.40%) (Figure 4B). Though similar in the CBF123 deletion ratios in either T2 lines of Ler (5.77% vs. 2.08%), Ler showed very low CBF123 deletion ratios in T2 (Figure 4B).
FIGURE 4.
Accession-dependent CBF123 deletion inheritance to T2. (A) Averages of each accession’s CBF123 deletion ratio in T2 plants. (B) Averages of each accession’s CBF123 deletion ratio in T2 plants from T1 with CBF123 deletion or without CBF123 deletion. Asterisks indicate a significant difference (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, N.S., non-significant). The error bars indicate the standard deviation.
Isolation of Homozygous cbf123 Triple Mutants in the T2 Generation
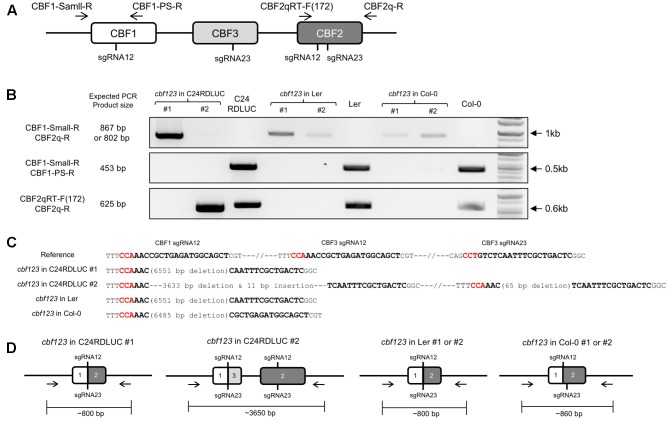
In order to isolate the homozygote mutants, we analyzed homozygosity of the candidate cbf123 triple mutants in each accession by PCR with three pairs of primers that were designed to detect the combination of CBF123 deletions (Figure 5A). Through the intensive PCR screening, we identified several homozygote mutants for the CBF123 deletion in each accession (Figure 5B).
FIGURE 5.
Homozygous cbf123 triple mutant generated by CRISPR/Cas system. (A) Alignment of the four primers used for homozygous cbf123 mutant detection. (B) CBF123 deletion confirmation. (C) Alignment of sequences of the PCR product from the homozygote mutants in C24RDLUC, Ler and Col-0 backgrounds and the reference sequence. Red sequences indicate the PAM sequence, bold sequences indicate the sgRNA sequences. (D) Schematic diagram of the homozygous cbf123 mutants in each accession.
In the case of the C24RDLUC background cbf123 mutant, we found two different homozygous mutants. cbf123#1 in C24RDLUC (hereafter cbf123LUC-1) had a 6,551 bp deletion between the cut sites of sgRNA 12 and sgRNA 23 in CBF2 while cbf123#2 in C24RDLUC (hereafter cbf123LUC-2) contained two deletions; one deletion was a 3,633 bp deletion between the cut sites of CBF1 sgRNA 12 and CBF3 sgRNA23 with an 11 bp insertion, and the other was a 65 bp deletion between the cut sites of sgRNA12 and sgRNA23 of the CBF2 region (Figures 5C,D and Supplementary Figure S2). Ler background cbf123 mutant had a 6,551 bp large deletion between the cut sites of CBF1 sgRNA 12 and CBF2 sgRNA 23 and Col-0 background cbf123 mutant showed a 6485 bp large deletion between the cut sites of CBF1 sgRNA 12 and CBF2 sgRNA 12 (Figures 5C,D and Supplementary Figure S2). In addition, no mutation was found in the potential off-target region of a closely related CBF paralog, the CBF4 gene (data not shown).
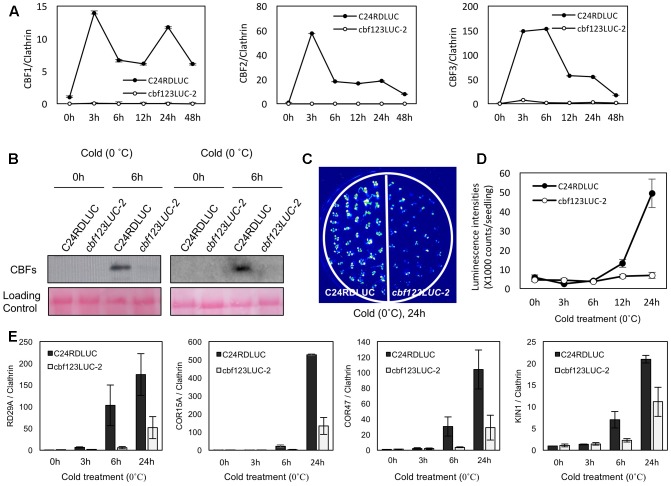
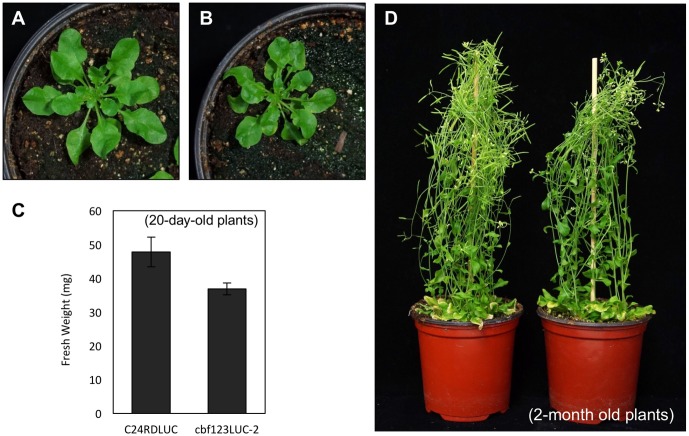
Among these cbf123 triple mutants, we decided to further characterize the cbf123LUC-2 because this mutant’s background line, C24RDLUC, contains a stress-inducible RD29A promoter-driven luciferase, which will be beneficial in monitoring the CBF target gene expression in vivo. In addition, the Cas9 transgene was segregated out of cbf123LUC-2, but not in other cbf123 triple mutants in other accession backgrounds. RNA transcripts and protein blot analysis confirmed the lack of the CBF proteins in cbf123LUC-2, indicating that cbf123LUC-2 is a null mutant (Figures 6A,B). Luminescence imaging after cold treatment showed clear reductions of the RD29A-LUC luminescence in cbf123LUC-2 in comparison to C24RDLUC (Figures 6C,D). This RD29A-LUC expression patterns were well correlated with those of the endogenous RD29A expression in cbf123LUC-2 (Figure 6E). Additionally, expressions of other CBF123 target genes were also reduced in cbf123LUC-2 (Figure 6E). In growth and development, cbf123LUC-2 appeared slightly smaller than C24RDLUC (Figures 7A–C) in early development stages. At the fully grown stage, there appeared to be no big differences in growth and development between C24RDLUC and cbf123LUC-2 (Figure 7D).
FIGURE 6.
Gene expression in cbf123LUC-2 mutant. (A) Expression of endogenous CBF1, CBF2 and CBF3 after cold treatment (0°C). 10-day-old seedlings were cold-treated for designated time before qRT-PCR analysis (B) Protein blots analysis to detect CBF proteins in 14-day-old seedlings using anti-CBF123 antibody with or without cold treatment (0°C, 6 h). Two biological repeats were shown. (C) Luminescence imaging of C24RDLUC and cbf123LUC-2 after cold treatment (0°C, 24 h). (D) Luminescence intensities of C24RDLUC and cbf123LUC-2 during cold treatment. (E) Expression of RD29A, COR15A, COR47, and KIN1 after cold treatment (0°C). 10-day-old seedlings were cold-treated for designated time before qRT-PCR analysis. For qRT-PCR quantification, clathrin was used as the internal control. Error bars indicate standard errors of the three biological replicates.
FIGURE 7.
Growth of C24RDLUC and cbf123LUC-2 plants. (A,B) Rosette leaf development of C24RDLUC (A) and cbf123LUC-2 (B) plants (20-day-old plants). (C) Fresh weights of C24RDLUC and cbf123LUC-2 (20-day-old plants). (D) Mature plants of C24RDLUC (left) and cbf123LUC-2 (right) plants (2-month old plants).
Discussion
In the present study, we found that CRISPR/Cas efficiency and inheritance could vary depending on Arabidopsis accessions. In the T1 generation, CRISPR/Cas efficiency was high in Col-0 and C24RDLUC with respective ratios of 75.00 and 84.85%, while it was only 45.71% in Ler (Table 3). CBF123 deletions were also observed differently in the T2 generation depending on Arabidopsis accessions. In T2, both Col-0 and C24RDLUC showed non-significantly different ratios of the CBF123 deletions, each displaying 33.56 and 19.80%, respectively (Figure 4A). However, the CBF123 deletion frequency was only 3.74% in Ler (Figure 4A). These results suggest that CRISPR/Cas efficiency is very low in Ler at least for CBF123 deletion. Interestingly, when T2 progenies from T1 with and without CBF123 deletions were separately analyzed, only C24RDLUC showed a correlation between T1 and T2 generations in CBF123 deletion frequencies (Figure 4B); T2 lines from T1 with CBF123 deletions showed a higher deletion ratio than T2 lines from T1 without CBF123 deletions. By contrast, Col-0 showed similar CBF123 deletions in T2 regardless of the frequency of CBF123 deletions at T1 (Figure 4B). Again, Ler T2 lines from T1 without CBF123 deletions showed an extremely low deletion ratio (2.08%). Thus, practically, if one uses Col-0, the most commonly used Arabidopsis accession, one should consider our observation that both T1 lines with and without the CRISPR/Cas target gene mutation could produce the target gene mutation in the T2 generation. Accession-dependent CRISPR/Cas efficiency and inheritance of its targeted mutation suggest that susceptibility to CRISPR/Cas system could vary among the naturally variant Arabidopsis accessions. Ler was the most resilient to CRISPR/Cas-mediated CBF123 deletion among the accessions tested. It showed the lowest CBF123 deletion ratio of 45.71% at T1 and a 3.74% deletion ratio at T2 (Figure 4A and Table 3).
CRISPR/Cas-induced gene modification can only be transmitted through the germ cells. Indeed, it has been reported that germ cell or egg cell-specific expression of CRISPR/Cas resulted in less or no-mosaic T1 plants with enhanced ratios of transmittable mutation (Wang et al., 2015; Mao et al., 2016). Thus, it is feasible to assume that Ler is in general not susceptible to CRISPR/Cas editing and that the activities of CRISPR/Cas in germ cells of Ler are very low.
It was also interesting that Col-0 displayed a high CBF123 deletion ratio at T2 regardless of the presence of CBF123 deletions in the parental lines (T1) of the T2 progenies, while C24RDLUC showed a strong correlation of CBF123 deletion ratio at T2 with CBF123 deletion ratio at the parental T1 (Figure 4B). CBF123 deletion at T1 are somatic mutations as genotyping was analyzed with the genomic DNA from young leaves of 15-day-old seedlings. Therefore, it might be possible that the CRISPR/Cas system is relatively highly active in the Col-0 germ cells independently of functional activation in somatic cells. By contrast, it might also be possible that there are not many differences in the CRISPR/Cas activity between somatic cells and germ line cells of C24RDLUC.
The differences among these accessions imply the presence of accession-specific genetic modifiers. The presence of such accession-specific modifiers is not uncommon. In particular, the mutation in ZWILLE/PINHEAD/AGO10 (ZLL) shows defects in maintenance of embryonic shoot apical meristem. However, these defects were apparent in Ler, but not in Col-0, because the defects were masked by the modifiers in Col-0 (Tucker et al., 2013). One of the modifiers in the ZLL function appeared to be the Arabidopsis Cyclophilin-40 orthologue SQUINT (SQN, AT2G15790) (Tucker et al., 2013). In addition, the altered meristem program (amp1) mutation differently affected the flowering time in Col-0 and Ler (Lee, 2009), presumably because of the different genetic modifiers in these accessions. Similarly, accession-dependent CRISPR/Cas efficiency and inheritance could be due to the different modifiers in these accessions. The candidate genetic modifiers could include the genes involved in DNA repair as CRISPR/Cas-induced mutations rely on error-prone DNA repair. The natural variations in these gene structures and regulation might contribute to these differences in CRISPR/Cas action in Arabidopsis accessions.
Another possibility for these differences is that Cas9 access to locus of CBF123 might be different in these accessions. It was shown that, in human cells, Cas9 access to the target sites was hindered by closed chromatin and restored upon induction of open chromatin status (Daer et al., 2017). In Arabidopsis, it has been known that the patterns of DNA methylation (hence the chromatin structure) is strongly correlated with the climate of accessions’ origin (Kawakatsu et al., 2016). Given that CBF1, 2, and 3 genes are cold-inducible, these Col-0, C24RDLUC, and Ler might have dynamic and different chromatin status affecting the efficiency of the CRISPR/Cas system. Therefore, the identification of such modifiers and epigenetic adjusters among these accessions will help improve the efficiency of CRISPR/Cas-mediated genome editing possibly in CRISPR/Cas-resilient crops for trait improvement.
The isolated cbf123 triple mutant in the C24RDLUC background (cbf123LUC-2) showed no transcripts of CBF1, 2, and 3 genes and their protein products, indicating that this cbf123LUC-2 mutant is a null mutant (Figures 6A,B). The previously reported cbf triple mutants showed different morphological phenotypes; Zhao et al found smaller sized cbf triple mutant (cbf123-1, cbf123-2) than Col-0, while Jia et al observed no difference in growth between Col-0 and their cbf triple mutant (cbfs-1) (Jia et al., 2016; Zhao et al., 2016). Our cbf123LUC-2 showed a smaller size than its background line (C24RDLUC) at the 20-day-old stage. It should be noted that Jia et al. (2016) generated the cbfs-1 mutant from cbf3 T-DNA mutant. Therefore, it is tempting to speculate that cbf3 T-DNA line might pose a different accession. Despite this uncertainty, the results from us and Zhao et al. suggest functions of CBF1, CBF2, and CBF3 genes in growth and development at normal temperature.
In summary, we found that CRISPR/Cas system has different efficiency in different Arabidopsis accessions. Our results imply the existence of genetic modifiers and/or chromatin access difference for CRISPR/Cas-mediated genome editing in different plant accessions.
Author Contributions
J-KZ, D-JY, and B-hL conceived and designed the research. SC, S-iY, JP, and YM performed the experiments. SC, S-iY, YM, J-KZ, D-JY, and B-hL discussed the results. SC, S-iY, and B-hL wrote the paper. SC and S-iY equally contributed to the work and should be regarded as joint First Authors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer SX declared a shared affiliation and past co-authorship with one of the authors J-KZ to the handling Editor.
Acknowledgments
This work was supported by Next-Generation BioGreen21 Program (PJ011006) of the Rural Development Administration, South Korea, and also by the Global Research Laboratory Program (2017K1A1A2013146) of the National Research Foundation (NRF) funded by the Ministry of Science and ICT (MSIT), South Korea.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01910/full#supplementary-material
FIGURE S1 | Schematic drawing of the CRISPR/Cas9 construct generation.
FIGURE S2 | Schematic drawing of the homozygous CBF123 deletion in each accession.
FIGURE S3 | Anti-CBF123 antibody generation and activity. (A) CBF2 protein detection by anti-CBF123 antibody. A serial dilutions of CBF2-GST extracts from E. coli (left) and plant protein extract from Arabidopsis Columbia (right) were loaded and proteins were detected by anti-CBF123 antibody. Red arrows indicate the expected protein size. For anti-CBF123 antibody generation, cDNA fragment corresponding to the coding region of CBF2 was amplified by PCR with primers containing attB1, attB2 sequence and cloned into donor vector (pDONR-zero) and subsequently moved into pDEST15 vector to produce recombinant protein with a GST protein The CBF2-GST recombinant vector was introduced into the BL21 strain of Escherichia coli. The E. coli transformants were grown at 37°C (OD600 = 0.8) and CBF2-GST was induced by 0.5 mM isoprophyl1-thio-β-D-galactopyranoside (IPTG). After 3 h at 30°C, cultures were harvested, resuspended in 1× PBS and incubated for 30min in the presence of 1% (v/v) Triton X-100 followed by disruption by sonication. The recombinant CBF2-GST fusion protein was purified by 12% SDS-PAGE and electro-eluted by use of Electro-Eluter (Model 422, BIO-RAD). The 500 mg of eluted CBF2 recombinant proteins were emulsified with the equal volume of Freund’s complete adjuvant and was injected subcutaneously at three sites in the rabbits’ backs. Two weeks later, the rabbits were injected with the 500 mg of CBF2 protein with Freund’s incomplete adjuvant as a booster immunization. A total of three booster injections were performed at 2-week intervals. One week following the final immunization, the antiserum was collected and stored at -20°C. (B) Cross-interaction of CBF123 proteins with anti-CBF123 antibody. Cross-interactions of the generated anti-CBF antibody were tested with Arabidopsis cbf2 or cbf3 single mutant (left) and transient expression of HA-tagged, CBF1-HA, CBF2-HA, and CBF3-HA proteins in tobacco leaves (right). The results suggest that the CBFs antibody could cross-interact with all CBF123 proteins. Red arrows indicate the expected protein size.
References
- Belhaj K., Chaparro-Garcia A., Kamoun S., Nekrasov V. (2013). Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9:39. 10.1186/1746-4811-9-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt A. B. (1999). Molecular genetics of DNA repair in higher plants. Trends Plant Sci. 4 20–25. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J., Zhu J. K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12 444–451. 10.1016/j.tplants.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Daer R. M., Cutts J. P., Brafman D. A., Haynes K. A. (2017). The impact of chromatin dynamics on Cas9-mediated genome editing in human cells. ACS Synth. Biol. 6 428–438. 10.1021/acssynbio.5b00299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Zhang B., Ding W., Liu X., Yang D. L., Wei P., et al. (2013). Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23 1229–1232. 10.1038/cr.2013.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V. V., Levy A. A. (1999). How plants make ends meet: DNA double-strand break repair. Trends Plant Sci. 4 263–269. [DOI] [PubMed] [Google Scholar]
- Haake V., Cook D., Riechmann J. L., Pineda O., Thomashow M. F., Zhang J. Z. (2002). Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 130 639–648. 10.1104/pp.006478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D., Lander E. S., Zhang F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157 1262–1278. 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M., Liming X., Stevenson B., Zhu J. K. (1997). Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-independent pathways. Plant Cell 9 1935–1949. 10.1105/tpc.9.11.1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Ding Y., Shi Y., Zhang X., Gong Z., Yang S. (2016). The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 212 345–353. 10.1111/nph.14088 [DOI] [PubMed] [Google Scholar]
- Kawakatsu T., Huang S. C., Jupe F., Sasaki E., Schmitz R. J., Urich M. A., et al. (2016). Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166 492–505. 10.1016/j.cell.2016.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. H. (2009). Ecotype-dependent genetic regulation of bolting time in the Arabidopsis mutants with increased number of leaves. J. Microbiol. Biotechnol. 19 542–546. 10.4014/jmb.0901.0082 [DOI] [PubMed] [Google Scholar]
- Lei Y., Lu L., Liu H. Y., Li S., Xing F., Chen L. L. (2014). CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 7 1494–1496. 10.1093/mp/ssu044 [DOI] [PubMed] [Google Scholar]
- Li J. F., Norville J. E., Aach J., Mccormack M., Zhang D., Bush J., et al. (2013). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31 688–691. 10.1038/nbt.2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. S., Zhu X. H., Lei M. G., Xia Q. Y., Botella J. R., Zhu J. K., et al. (2015). A detailed procedure for CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana. Sci. Bull. 60 1332–1347. 10.1007/s11434-015-0848-2 [DOI] [Google Scholar]
- Lozano-Juste J., Cutler S. R. (2014). Plant genome engineering in full bloom. Trends Plant Sci. 19 284–287. 10.1016/j.tplants.2014.02.014 [DOI] [PubMed] [Google Scholar]
- Mali P., Esvelt K. M., Church G. M. (2013). Cas9 as a versatile tool for engineering biology. Nat. Methods 10 957–963. 10.1038/nmeth.2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Zhang H., Xu N., Zhang B., Gou F., Zhu J. K. (2013). Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol. Plant 6 2008–2011. 10.1093/mp/sst121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Zhang Z., Feng Z., Wei P., Zhang H., Botella J. R., et al. (2016). Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol. J. 14 519–532. 10.1111/pbi.12468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen X. C., Kim S. H., Hussain S., An J., Yoo Y., Han H. J., et al. (2016). A positive transcription factor in osmotic stress tolerance, ZAT10, is regulated by MAP kinases in Arabidopsis. J. Plant Biol. 59 55–61. 10.1007/s12374-016-0442-4 [DOI] [Google Scholar]
- Park S., Lee C. M., Doherty C. J., Gilmour S. J., Kim Y., Thomashow M. F. (2015). Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 82 193–207. 10.1111/tpj.12796 [DOI] [PubMed] [Google Scholar]
- Sampson T. R., Weiss D. S. (2014). Exploiting CRISPR/Cas systems for biotechnology. Bioessays 36 34–38. 10.1002/bies.201300135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiml S., Fauser F., Puchta H. (2014). The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 80 1139–1150. 10.1111/tpj.12704 [DOI] [PubMed] [Google Scholar]
- Shan Q., Wang Y., Li J., Zhang Y., Chen K., Liang Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotech. 31 686–688. 10.1038/nbt.2650 [DOI] [PubMed] [Google Scholar]
- Tucker M. R., Roodbarkelari F., Truernit E., Adamski N. M., Hinze A., Lohmuller B., et al. (2013). Accession-specific modifiers act with ZWILLE/ARGONAUTE10 to maintain shoot meristem stem cells during embryogenesis in Arabidopsis. BMC Genomics 14:809. 10.1186/1471-2164-14-809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. P., Xing H. L., Dong L., Zhang H. Y., Han C. Y., Wang X. C., et al. (2015). Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16 144. 10.1186/s13059-015-0715-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B., Sternberg S. H., Doudna J. A. (2012). RNA-guided genetic silencing systems in bacteria and archaea. Nature 482 331–338. 10.1038/nature10886 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57 781–803. 10.1146/annurev.arplant.57.032905.105444 [DOI] [PubMed] [Google Scholar]
- Zhao C., Zhang Z., Xie S., Si T., Li Y., Zhu J. K. (2016). Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 171 2744–2759. 10.1104/pp.16.00533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Zhu J. K. (2016). The broad roles of CBF genes: from development to abiotic stress. Plant Signal. Behav. 11:e1215794. 10.1080/15592324.2016.1215794 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 | Schematic drawing of the CRISPR/Cas9 construct generation.
FIGURE S2 | Schematic drawing of the homozygous CBF123 deletion in each accession.
FIGURE S3 | Anti-CBF123 antibody generation and activity. (A) CBF2 protein detection by anti-CBF123 antibody. A serial dilutions of CBF2-GST extracts from E. coli (left) and plant protein extract from Arabidopsis Columbia (right) were loaded and proteins were detected by anti-CBF123 antibody. Red arrows indicate the expected protein size. For anti-CBF123 antibody generation, cDNA fragment corresponding to the coding region of CBF2 was amplified by PCR with primers containing attB1, attB2 sequence and cloned into donor vector (pDONR-zero) and subsequently moved into pDEST15 vector to produce recombinant protein with a GST protein The CBF2-GST recombinant vector was introduced into the BL21 strain of Escherichia coli. The E. coli transformants were grown at 37°C (OD600 = 0.8) and CBF2-GST was induced by 0.5 mM isoprophyl1-thio-β-D-galactopyranoside (IPTG). After 3 h at 30°C, cultures were harvested, resuspended in 1× PBS and incubated for 30min in the presence of 1% (v/v) Triton X-100 followed by disruption by sonication. The recombinant CBF2-GST fusion protein was purified by 12% SDS-PAGE and electro-eluted by use of Electro-Eluter (Model 422, BIO-RAD). The 500 mg of eluted CBF2 recombinant proteins were emulsified with the equal volume of Freund’s complete adjuvant and was injected subcutaneously at three sites in the rabbits’ backs. Two weeks later, the rabbits were injected with the 500 mg of CBF2 protein with Freund’s incomplete adjuvant as a booster immunization. A total of three booster injections were performed at 2-week intervals. One week following the final immunization, the antiserum was collected and stored at -20°C. (B) Cross-interaction of CBF123 proteins with anti-CBF123 antibody. Cross-interactions of the generated anti-CBF antibody were tested with Arabidopsis cbf2 or cbf3 single mutant (left) and transient expression of HA-tagged, CBF1-HA, CBF2-HA, and CBF3-HA proteins in tobacco leaves (right). The results suggest that the CBFs antibody could cross-interact with all CBF123 proteins. Red arrows indicate the expected protein size.