Abstract
Germline mutations in the novel tumor suppressor gene FLCN are responsible for the autosomal dominant inherited disorder Birt-Hogg-Dubé (BHD) syndrome that predisposes to fibrofolliculomas, lung cysts and spontaneous pneumothorax, and an increased risk for developing kidney tumors. Although the encoded protein, folliculin (FLCN), has no sequence homology to known functional domains, x-ray crystallographic studies have shown that the C-terminus of FLCN has structural similarity to DENN (differentially expressed in normal cells and neoplasia) domain proteins that act as guanine nucleotide exchange factors (GEFs) for small Rab GTPases. FLCN forms a complex with folliculin interacting proteins 1 and 2 (FNIP1, FNIP2) and with 5’ AMP-activated protein kinase (AMPK). This review summarizes FLCN functional studies which support a role for FLCN in diverse metabolic pathways and cellular processes that include modulation of the mTOR pathway, regulation of PGC1α and mitochondrial biogenesis, cell-cell adhesion and Rho A signaling, control of TFE3/TFEB transcriptional activity, amino acid-dependent activation of mTORC1 on lysosomes through Rag GTPases, and regulation of autophagy. Ongoing research efforts are focused on clarifying the primary FLCN-associated pathway(s) that drives the development of fibrofolliculomas, lung cysts and kidney tumors in BHD patients carrying germline FLCN mutations.
Keywords: FLCN, folliculin, FNIP1, FNIP2, Birt-Hogg-Dubé syndrome, Kidney tumor, DENN domain, mTOR, AMPK, PGC1α
1. Introduction
In 2002, germline mutations in a novel gene, folliculin (FLCN), were identified as causative for Birt-Hogg-Dubé (BHD) syndrome (Nickerson et al., 2002), an autosomal dominant hamartoma syndrome characterized by benign cutaneous papules (Birt et al., 1977), pulmonary cysts, spontaneous pneumothorax and kidney tumors (Zbar et al., 2002).The encoded FLCN protein was unique with no known functional domains and no homology to known proteins. This review will discuss events leading to the discovery of the FLCN gene, clinical manifestations of BHD syndrome and FLCN mutational spectrum, functional clues gained from resolving the crystal structure of the FLCN protein, and identification of interacting protein partners FNIP1 and FNIP2. Here we summarize research from multiple laboratories proposing functional roles for tumor suppressor FLCN in a number of metabolic pathways and biological processes mainly through study of FLCN deficient in vitro and in vivo models (Table 1).
Table 1.
In vivo models with Flcn inactivation
| Organism | Type of model | Target tissue | Phenotype | Pathways affected | Reference |
|---|---|---|---|---|---|
| Mouse | GEM, Cre-lox | Kidney, distal nephron | Large cystic kidneys, renal failure | ↑AKT-mTOR activation ↑Raf/MEK/Erk1/2 axis |
(Baba et al., 2008) |
| Mouse | GEM, Cre-lox | Kidney, distal nephron | Large cystic kidneys, cystic RCC, renal failure | ↑mTOR activation | (Chen et al., 2008) |
| Mouse | GEM, Cre-lox | Heterozygous deletion in whole animal | Late onset kidney tumors w/ LOH | ↑AKT, mTORC1 & mTORC2 activation | (Hasumi et al., 2009) |
| Mouse | GEM, gene trap KO plus ENU treatment | Heterozygous deletion in whole animal | Cysts and low frequency of oncocytic tumors | ↓mTORC1 activation | (Hartman et al., 2009) |
| Mouse | GEM, gene trap KO | Heterozygous deletion in whole animal | Cysts, adenomas & multicystic kidneys | Both ↑ & ↓ mTORC1 activation; ↑ERK1/2 | (Hudon et al., 2010) |
| Mouse | GEM, Cre-lox | Skeletal muscle | Red colored muscle & ↑mitochondrial number in muscle | ↑PGC1α expression & mitochondrial biogenesis | (Hasumi et al., 2012) |
| Mouse | GEM, Cre-lox | Epidermal layer of skin | Delayed eye opening, wavy fur, hair loss, epidermal hyperplasia | ↑mTORC1 activation | (Medvetz et al., 2012) |
| Mouse | GEM, Cre-lox | Heart | Cardiac hypertrophy & cardiac dysfunction | ↓pAMPK, ↑mTOR, ↑PGC1α expression & mitochondrial biogenesis | (Hasumi et al., 2014) |
| Mouse | GEM, Cre-lox | Lung, alveolar epithelial type II cell | Alveolar enlargement, impaired lung function | ↓E-cadherin-LKB1-AMPK axis | (Goncharova et al., 2014) |
| Mouse | GEM, Cre-lox | Kidney, proximal tubule | Kidney cysts & tumors | ↑AKT-mTOR activation ↑TGF-β pathway activation |
(Chen et al., 2015) |
| Mouse | GEM, Cre-lox | Adipose tissue | ↑beige adipocytes in WAT; high energy expenditure; resist cold exposure | ↑pAMPK-PGC1α-ERRα axis & mitochondrial biogenesis | (Yan et al., 2016) |
| Mouse | GEM, Cre-lox | Adipose tissue | ↑beige adipocytes in WAT; resist cold exposure | ↑TFE3 activity & targets PGC1α, PGC1β,GPNMB; ↑mitochondrial biogenesis |
(Wada et al., 2016) |
| Mouse | GEM, transgenic | Flcn H255Y transgene in kidney-targeted Flcn KO background | Cannot rescue kidney-targeted Flcn KO phenotype | Acts as Flcn KO allele; strong mutation | (Hasumi et al., 2017) |
| Mouse | GEM, transgenic | Flcn K508R transgene in kidney-targeted Flcn KO background | Partially, not entirely rescues kidney-targeted Flcn KO phenotype | Acts to partially reverse Flcn KO phenotype; weak mutation & potential DN function | (Hasumi et al., 2017) |
| Yeast (S. pombe) | Replacement of bhd ORF with kanamycin cassette | Deletion in whole organism | None evident | ↑amino acid permeases & amino acids suggesting ↓Tor2 activity | (van Slegtenhorst et al., 2007) |
| Fruit fly (D. melanogaster) | DBHD dsRNA-mediated RNAi | Knockdown in whole organism | Defects in male germline stem cell maintenance in testis | DBHD functions in parallel or downstream of JAK-STAT & Dpp pathways | (Singh et al., 2006) |
| Fruit fly (D. melanogaster) | DBHD null allele | Homozygous knockout in whole organism | Growth retardation, small body, larval lethality | ↑autophagy; rescue by leucine feeding suggesting dTOR inhibition in DBHD−/− | (Liu et al., 2013) |
| Round worm (C. elegans) | flcn-1 (ok975) null allele | Knockout in whole organism | Increased resistance to oxidative stress | ↑AAK-2, ↑autophagy, ↑ATP levels, ↓apoptosis | (Possik et al., 2014) |
| Zebrafish (D. rerio) | flcn knockout allele using morpholino anti-sense oligonucleotide | Knockout in whole organism | 18 somite stage developmental arrest; hydrocephalus ; tail circulatory defects | Disruption of the cell cycle and loss of proliferation in embryonic brain | (Kenyon et al., 2016) |
Abbreviations: GEM, genetically engineered model; RCC, renal cell carcinoma; Cre, cre recombinase; lox, flanked by loxP sites; LOH, loss of heterozygosity; KO, knockout; ORF, open reading frame; WAT, white adipose tissue; DN, dominant negative; AAK-2, AMPK ortholog
1.1 FLCN gene structure and encoded FLCN protein
The human folliculin (FLCN) gene encodes at least two major transcript variants on chromosome 17p11.2. Transcript variant 1 (NCBI RefSeq NM_144997.5) encodes the longer isoform that is 3723 bp in length containing 14 exons of which 11 exons are coding. Folliculin (FLCN) isoform 1 (NCBI RefSeq NP_659434.2; UniProtKB Q8NFG4) encoded by transcript variant 1 is 579 amino acids in length with a mass of 64kDa and no classic functional domains or sequence homology to other known proteins. The FLCN protein is conserved across species from man to Caenorhabditis elegans (C. elegans). Transcript variant 2 (NCBI RefSeq NM_144606.5) is a shorter isoform (1760bp) containing 8 exons (5 coding exons) and uses an alternate splice site in the 3’ coding region to produce a distinctly different carboxy (C)-terminus compared to transcript variant 1. FLCN isoform 2 (NCBI RefSeq NP_653207.1) comprised of 342 amino acids is encoded by transcript variant 2. To date all reported studies are based on transcript variant 1 and FLCN isoform 1. The function of transcript variant 2/FLCN isoform 2 has not been investigated. This information was obtained from NCBI Gene (http://www.ncbi.nlm.nih.gov/gene) and UCSC Genome Browser (http://genome.ucsc.edu/) sources.
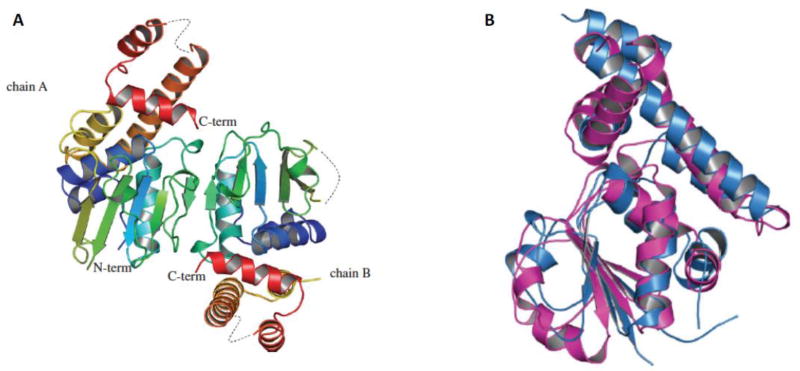
The C-terminal region of folliculin (FLCN) was crystallized following the introduction of mutations in three cysteine residues after efforts to crystallize the full length FLCN protein were unsuccessful. The three-dimensional structure of the FLCN C-terminal domain was determined to 2 Å resolution (Figure 1A) (Protein Data Bank accession code 3V42) (Nookala et al., 2012). The C-terminal domain of FLCN shares significant structural similarity with the differentially expressed in normal cells and neoplasia (DENN) domain of the DENN1B protein (Wu et al., 2011) (Figure 1B) showing the same order and orientation of strands. The DENN domain family of proteins share common structural features and have been shown to be guanine nucleotide exchange factors (GEF) for Rab GTPases (Yoshimura et al., 2010), facilitating GDP-GTP exchange to activate the vesicular transport function of these small GTPases. Further support for a similarity between FLCN and DENN1B came from the prediction by protein structure software of an N-terminal Longin domain in FLCN, which is a feature present in the N-terminus of DENN1B (Nookala et al., 2012). The structural similarity of C-terminal FLCN to the DENN domain protein DENN1B and predicted FLCN N-terminal Longin domain, which is also present in DENN1B, suggest that FLCN may possess GEF activity towards Rab GTPases and function in membrane trafficking (see section 5.6).
Figure 1.
CT-FLCN has structural similarity to DENN domain proteins. A) Crystal structure of carboxy-terminal (CT) domain of FLCN at 2 Å resolution. Two FLCN molecules are in the asymmetric unit. The N-terminus is blue and the C-terminus is red. B) CT-FLCN is structurally similar to the DENN domain of DENN1B protein. CT-FLCN is in blue and DENN domain of DENN1B is in magenta. Adapted from (Nookala et al., 2012).
1.2 FLCN expression
Northern blot analysis revealed a 3.8 kb FLCN transcript that was detected in a wide variety of adult tissues including brain, heart, placenta, testis, skin, lung and kidney, and in fetal lung, liver, brain and kidney (Nickerson et al., 2002). Fluorescent in situ hybridization studies have demonstrated FLCN mRNA expression in skin, distal nephron of the kidney, stromal cells and pneumocytes of the lung, acinar cells of pancreas and parotid gland, epithelial ducts of the breast and prostate, neurons of the cerebrum and Purkinje cells of the cerebellum. FLCN mRNA was expressed in macrophages and lymphocytes of the spleen but had reduced expression in heart, muscle and liver (Warren et al., 2004).
2. The folliculin (FLCN) gene and Birt-Hogg-Dubé (BHD) syndrome
2.1 The discovery of the folliculin (FLCN) gene as responsible for Birt-Hogg-Dubé syndrome
Birt-Hogg-Dubé (BHD) syndrome is an autosomal dominant inherited disorder in which patients develop benign tumors of the hair follicle called fibrofolliculomas, lung cysts, spontaneous pneumothorax, and an increased risk for kidney tumors (Birt et al., 1977; Nickerson et al., 2002; Zbar et al., 2002). The discovery that the FLCN gene was responsible for BHD syndrome followed a report describing another rare genetic disorder called familial renal oncocytoma (FRO), in which multiple individuals within a family presented with bilateral multifocal renal oncocytomas (Weirich et al., 1998). When these FRO patients were subjected to routine evaluation by a dermatologist, cutaneous facial papules consistent with fibrofolliculomas were identified in the affected members of one FRO kindred. Since fibrofolliculomas were the hallmark cutaneous feature of BHD syndrome, additional patients with familial renal tumors were evaluated for cutaneous lesions to determine if renal neoplasia were associated with BHD. Cosegregation of renal tumors with fibrofolliculomas was identified in three of these kindreds, thereby expanding the phenotypic manifestations of BHD syndrome to include renal cancer (Toro et al., 1999). Genetic linkage analysis of additional families who were recruited based on BHD cutaneous lesions narrowed the BHD disease locus to the short arm of chromosome 17 (Khoo et al., 2001; Schmidt et al., 2001). Fine mapping of recombinants in additional BHD families and positional cloning identified a novel gene, FLCN, located at chromosome 17p11.2, which was mutated in the germline of BHD-affected individuals (Nickerson et al., 2002).
2.2 Clinical manifestations of BHD syndrome
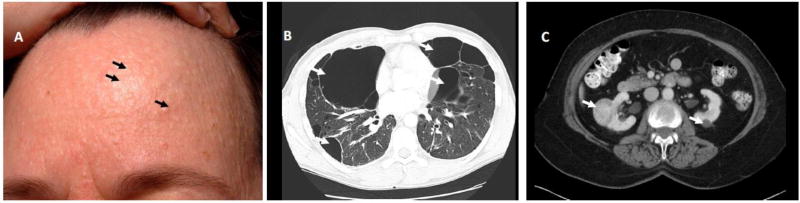
In the initial report of this disorder, Birt and colleagues defined a triad of dermatologic lesions including fibrofolliculomas, trichodiscomas and acrochordons as the classic features of BHD syndrome (Birt et al., 1977). Over a decade later, several reports were published that described an association of lung cysts, often leading to spontaneous pneumothoraces, with BHD skin manifestations in BHD kindreds (Binet et al., 1986; Chung et al., 1996; Toro et al., 1999). After an initial observation of bilateral multifocal chromophobe renal cancer in a BHD patient (Roth et al., 1993), cosegregation of renal tumors with fibrofolliculomas was described in three kindreds supporting renal neoplasia as part of the BHD phenotype (Toro et al., 1999). (Figure 2)
Figure 2.
Clinical manifestations of Birt-Hogg-Dubé syndrome. A) Fibrofolliculomas on the face of a BHD patient. B) Bilateral pulmonary cysts in a BHD patient with a small pneumothorax on the right. C) CT scan showing bilateral multifocal kidney tumors in a BHD patient. Adapted from (Schmidt and Linehan, 2015).
Fibrofolliculomas are the most common manifestation of BHD presenting on the face, neck and upper torso of greater than 85% of BHD-affected individuals over 25 years of age (Schmidt et al., 2005; Leter et al., 2008; Toro et al., 2008). Multiple bilateral lung cysts are also highly penetrant manifestations occurring in more than 75% of individuals with BHD. Approximately one-fourth of BHD patients report a history of spontaneous pneumothorax usually occurring before 40 years of age (Gupta et al., 2013). Renal cysts can present in BHD patients (Furuya et al., 2016), sometimes as the only renal manifestation (Kluger et al., 2010). Renal tumors develop in up to one-third of BHD-affected individuals with an average age of onset of 50 years (Zbar et al., 2002; Schmidt et al., 2005; Toro et al., 2008). BHD patients most frequently present with chromophobe renal carcinoma (34%) or hybrid oncocytic tumors (50%) comprised of features of chromophobe renal carcinoma and renal oncocytoma (Pavlovich et al., 2002). In a risk assessment evaluating family members of BHD kindreds, individuals with BHD syndrome had a 7-fold greater risk for developing kidney neoplasia and a 50-fold greater risk for developing spontaneous pneumothorax, when adjusted for age, than their unaffected siblings (Zbar et al., 2002). Although no association between BHD-affected status and colon polyps or cancer was found in the same risk assessment (Zbar et al., 2002), findings from a different study identified a significantly greater risk for developing colon neoplasia in BHD patients who carried the FLCN c.1285dupC mutation compared with the c.610delGCinsTA mutation (Nahorski et al., 2010). Other manifestations (i.e., parotid oncocytomas, lipomas, thyroid nodules/cancer, liver cysts) have been infrequently reported in BHD patients but none has been confirmed as part of the BHD clinical phenotype.
2.3 FLCN germline mutations in BHD syndrome
Over 150 unique mutations spanning the entire FLCN coding region have been identified in BHD families and in families with spontaneous pneumothorax as the only manifestation, and documented in the FLCN Leiden Open Variation Database (Lim et al., 2010) (http://www.lovd.nl/flcn). No clear association between mutation type or location within the FLCN gene and cutaneous or renal manifestations has been reported, although a statistically significant correlation between FLCN mutations in exon 9 and increased number of lung cysts and a trend toward more episodes of spontaneous pneumothorax in BHD patients with exons 9 and 12 mutations were observed (Toro et al., 2007). The majority of FLCN mutations identified in the germline of BHD patients are frameshift (insertion/deletion), nonsense or splice site mutations predicted to truncate and presumably inactivate the FLCN protein (Lim et al., 2010). An insertion/deletion of a cytosine in a tract of 8 cytosines (c.1285dupC/delC) in FLCN exon 11 was the most frequent mutation observed in several large BHD patient cohorts suggesting a mutational “hot spot” caused by slippage of the DNA polymerase during replication of the mononucleotide stretch (Nickerson et al., 2002; Toro et al., 2008; Nahorski et al., 2010)(Furuya et al., 2016).
Less frequently, large intragenic partial deletions or duplications of FLCN, including deletion of the putative promoter region in exon 1 have been reported (Kunogi et al., 2010; Benhammou et al., 2011; Houweling et al., 2011; Ding et al., 2015; Rossing et al., 2017). FLCN missense mutations that result in amino acid substitution (Toro et al., 2008; Lim et al., 2010) are also less common in BHD syndrome comprising only 11% of unique mutations in the FLCN Leiden Open Variation Database. Additional experimental evidence will be important for proving that they are disease causing variants. Two FLCN missense mutations, p.Arg239Cys and p.His255Pro, have been shown to reduce FLCN protein stability in vitro, thereby supporting their pathogenicity. On the other hand, the p.Lys508Arg mutant protein was expressed and stable comparable to wildtype FLCN (Nahorski et al., 2011). When another missense FLCN mutation, p.His255Tyr, was genetically introduced into kidney-targeted Flcn knockout mice, the Flcn-deficient polycystic kidney phenotype could not be rescued, thus confirming its pathogenicity. However, introduction of the p.Lys508Arg mutation prolonged survival and diminished the polycystic kidney phenotype in kidney-targeted Flcn knockout mice, but the mice eventually developed cystic kidneys and succumbed to renal failure. These findings suggest that p.Lys508Arg is, in fact, a pathogenic FLCN mutation, albeit a weak one (Hasumi et al., 2017). Interestingly, when the p.Lys508Arg mutation was introduced into Flcn heterozygous knockout mice, a portion of the mice developed a later onset cystic kidney phenotype with retention of the wild type Flcn allele in the cysts, suggesting that the p.Lys508Arg FLCN mutant protein might have a dominant negative effect on wild type FLCN (Hasumi et al., 2017).
2.3.1 FLCN mutations in sporadic kidney tumors
The identification of FLCN mutations in BHD syndrome, an inherited form of kidney cancer, led researchers to investigate whether FLCN mutations might be responsible for sporadic renal tumors that were histologically similar to BHD-associated tumors. However, the frequency of FLCN mutations in sporadic renal tumors was quite low. In one study, 5 of 46 chromophobe renal tumors, of which 3 were likely germline, and 1 of 18 renal oncocytomas had FLCN mutations (Gad et al., 2007). In other reports, FLCN mutations were detected in only 1 of 39 sporadic renal tumors (Khoo et al., 2003), 2 of 30 mixed sporadic tumors and cell lines (da Silva et al., 2003), and in none of 16 renal oncocytomas and chromophobe renal tumors (Nagy et al., 2004). In a recent report from the Cancer Genome Atlas project, whole exome sequencing of 66 sporadic chromophobe renal tumors did not detect any FLCN mutations (Davis et al., 2014). These studies suggest that somatic FLCN mutations are not major drivers of sporadic renal cancer.
3. FLCN as a tumor suppressor gene
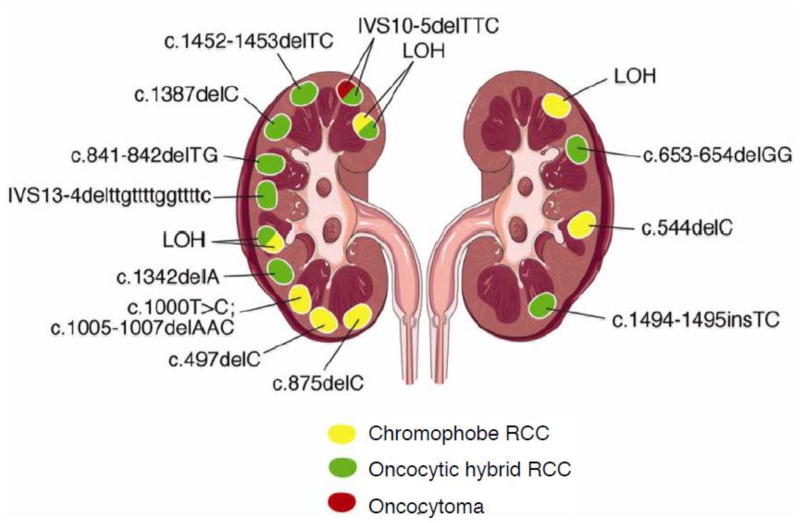
3.1 Second hit FLCN mutations in BHD renal tumors
FLCN is a classic tumor suppressor gene as defined by the Knudson two-hit model of tumor suppression (Knudson, 1971). In a study of 77 renal tumors from BHD patients with germline FLCN mutations, loss of heterozygosity (LOH) on the short arm of chromosome 17 was identified in 17% and somatic “second hit” mutations in 53% of the BHD-associated renal tumors (Vocke et al., 2005). (Figure 3) A somatic FLCN mutation was also found in a BHD-associated chromophobe renal tumor (Khoo et al., 2002). UOK257, a cell line established from a BHD patient renal tumor was determined to have only the mutant copy of FLCN (Yang et al., 2008), thereby demonstrating LOH in the original tumor. UOK257 was tumorigenic when injected into athymic nude mice, but lost its tumorigenic properties upon restoration of wild type FLCN expression (Hong et al., 2010b). Further support for FLCN as a tumor suppressor was provided by a study in which FLCN knockdown in ACHN, a clear cell renal tumor cell line, led to the development of significantly larger tumors in immunocompromised mice than the parent ACHN cells expressing wild type FLCN (Hudon et al., 2010). These researchers also observed that overexpression of FLCN in the tumorigenic VHL deficient cell line 786-O reduced tumor growth in a xenograft model.
Figure 3.
Different somatic second hit FLCN mutations in multiple tumors that developed in the kidneys of a BHD patient with a germline FLCN mutation demonstrating that FLCN is a tumor suppressor gene. Histologic subtypes are color coded. Chromophobe renal carcinoma, yellow; oncocytic hybrid tumor, green; renal oncocytoma, red. LOH, loss of heterozygosity. From (Vocke et al., 2005).
3.2 Naturally-occurring animal models of FLCN inactivation
Two naturally-occurring animal models of BHD have been described that further confirm a tumor suppressor function for FLCN. Kidney tumors develop in the Nihon rat model of BHD, a Sprague-Dawley rat strain that spontaneously acquired a germline Flcn mutation (Okimoto et al., 2004). When a transgene expressing wild type Flcn was genetically introduced into the heterozygous Flcn+/− Nihon rat, tumorigenic potential was lost (Togashi et al., 2006). German Shepherd dogs with the inherited disorder renal cystadenocarcinoma and nodular dermatofibrosis (RCND) develop multiple kidney tumors, uterine fibroids and skin nodules with dense collagen fibers. This disorder results from the inheritance of a germline missense mutation in the canine Flcn gene changing a highly conserved histidine to an arginine (p.His255Arg)(Lingaas et al., 2003). Somatic second hit mutations in the remaining wild type Flcn allele or LOH was observed in the RCND-associated renal tumors (Bonsdorff et al., 2008). Both in vivo models support a tumor suppressor function for FLCN.
4. Interacting partners of FLCN
4.1 Folliculin-interacting protein 1 (FNIP1)
Initial efforts to determine FLCN function based on protein sequence did not identify any homology to known functional domains, so researchers searched for FLCN interacting partners to provide clues to FLCN function. The first folliculin protein partner identified through FLCN coimmunoprecipitation studies was folliculin interacting protein 1, designated FNIP1, a novel 130kDa protein also without recognizable functional domains (Baba et al., 2006). FNIP1 is widely expressed in a pattern similar to FLCN and conserved through Xenopus (Baba et al., 2006). Binding studies with FLCN structural mutants demonstrated that FNIP1 binds through the carboxy-terminus of FLCN (Baba et al., 2006). Notably, additional co-immunoprecipitation experiments demonstrated an interaction of FNIP1 with the γ-1 subunit of 5’-AMP activated protein kinase (AMPK), a heterotrimeric protein kinase that serves as a critical energy sensor in cells and negative regulator of mechanistic target of rapamycin (mTOR) (Baba et al., 2006) (Shackelford and Shaw, 2009). FLCN was confirmed to interact with the FNIP1-AMPK complex but was not essential for their interaction. FNIP1 preferentially bound the active phosphorylated form of AMPK and both FLCN and FNIP1 were shown to be phosphorylated by AMPK (Baba et al., 2006).
4.1.1 FLCN phosphorylation facilitated by FNIP1 expression may involve AMPK and mTOR
Western blot analysis of the UOK257 renal tumor cell line with restored FLCN expression demonstrated that FLCN exists in several phosphorylated forms that have different electrophoretic mobilities, and that FNIP1 immunoprecipitates are enriched for the phosphorylated forms of FLCN (Baba et al., 2006). The phosphorylated FLCN bands are diminished, to different degrees, by inhibitors of either AMPK or mTOR, suggesting that FLCN has multiple phosphorylation sites that are phosphorylated through mTOR and AMPK signaling (Baba et al., 2006). Mutation of candidate residues in the amino-terminal region of FLCN revealed that mutation of serine 62 to alanine completely eliminated the slower migrating bands (phospho-FLCN) on immunoblots, and mass spectrophotometric analysis of tryptic peptides from transiently expressed GST-FLCN identified a single phospho-peptide, which contained phospho-serine 62 (Wang et al., 2010). Mutation of serine 62 to alanine did not affect FLCN binding to FNIP1 but did slightly reduce its binding to the AMPKα1 subunit suggesting that FLCN phosphorylation at serine 62 may stabilize or enhance a complex that includes AMPK and FLCN (Wang et al., 2010).
4.2 Folliculin-interacting protein 2 (FNIP2)
A second folliculin-interacting protein, FNIP2 (also known as FNIPL, KIAA1450; NCBI RefSeq NM_020840), was identified by bioinformatics searching of sequence databases for FNIP1 homologs (Hasumi et al., 2008; Takagi et al., 2008). FNIP2 has 49% identity and 74% similarity to FNIP1 and is conserved across species through Xenopus. D. melanogaster and C. elegans each have a single transcript encoding a protein with homology to both FNIPs (Hasumi et al., 2008). Tissue expression of FNIP2 mRNA is generally similar to FNIP1 and FLCN suggesting they may cooperate in these organs, but some specificity for FNIP2 in fat, liver and pancreas compared to FNIP1 may imply a specific function in metabolic tissues (Hasumi et al., 2008). FNIP1, on the other hand, was more highly expressed in muscle, salivary gland, parathyroid and nasal mucosa. Binding studies demonstrated interaction of FNIP2 with the C-terminus of FLCN and with the subunits of AMPK in a manner identical to FNIP1 (Hasumi et al., 2008; Takagi et al., 2008). Interestingly, FNIP1 and FNIP2 were shown to form homodimers and heterodimers with each other, and in all cases, FLCN and AMPK subunits were present in the immunoprecipitates containing these FNIP1/FNIP2 multimeric complexes (Hasumi et al., 2008). The functional significance of the FNIP1/FNIP2 multimeric complexes awaits further investigation to clarify their impact on FLCN function.
Interestingly, a novel gene designated as Mapo1 (O6-methyguanine induced apoptosis 1) was identified as one of the components that function in apoptosis triggered by O6 methylguanine (O6-meG) produced by alkylating agents (Komori et al., 2009). O6-meG can mispair with thymine as well as cytosine during DNA replication and introduce mutations in the next round of replication. The encoded protein MAPO1 was determined to be identical to FNIP2/FNIPL, and coimmunoprecipitated with FLCN and AMPKα (Lim et al., 2012). Suppression of O6 methylguanine induction of apoptosis was observed with either FLCN or AMPK knockdown suggesting a potential role for FLCN in complex with MAPO1/FNIP2/FNIPL in DNA mismatch repair.
4.3 Functional studies in FNIP1/FNIP2-deficient in vivo models
Although FNIP1 and FNIP2 were identified as FLCN binding partners, they were novel proteins that had not previously been reported. Studies are in progress to determine how FNIP1 and FNIP2 function in association with FLCN. In three different mouse models, knockout of Fnip1 produced a dramatic B-cell deficiency resulting from a block in B-cell development at the pre-B cell stage. Results in two of the models suggest that this phenotype was caused by rapid caspase-induced cell death that was rescued by expression of the anti-apoptotic protein Bcl2 (Baba et al., 2012; Siggs et al., 2016). In the third model, Fnip1 deficiency resulted in dysregulation of pre-B cells with increased AMPK and PGC1α (PPARGC1A; peroxisome proliferator-activated receptor gamma coactivator 1 alpha) expression leading to increased mitochondrial biogenesis (Park et al., 2012). Fnip1-deficient mice were also reported to develop cardiomyopathy (Hasumi et al., 2015; Siggs et al., 2016) and a switch from type II “fast twitch” to type 1 “slow twitch” skeletal muscle fiber type as a result of increased AMPK activation and expression of its target PGC1α, transcriptional coactivator and master regulator of mitochondrial biogenesis (Reyes et al., 2015). The physiologic significance of the findings in Fnip1-deficient mice to BHD is unclear since BHD patients with germline FLCN mutations do not develop manifestations in B-cells or muscle tissues.
Of more relevance, however, to BHD syndrome is the observation that mice with kidney-targeted Fnip1 and Fnip2 inactivation (but not with inactivation of either Fnip1 or Fnip2 alone) developed highly cystic kidneys that expressed elevated levels of PGC1α and increased mitochondrial biogenesis (Hasumi et al., 2015) mimicking the phenotype of kidney-targeted Flcn knockout mice (Baba et al., 2008; Chen et al., 2008). Furthermore, mice with Fnip1 heterozygous and Fnip2 homozygous inactivation developed frank tumors at 24 months of age, suggesting that these proteins are somewhat redundant and function together with Flcn to regulate normal cell growth in the kidney, whereas loss of Fnip1/2 expression abrogates the tumor suppressive properties of FLCN. (Hasumi et al., 2015).
5. Potential functions of FLCN
Since the discovery 15 years ago that FLCN germline mutations are responsible for Birt-Hogg-Dubé syndrome, multiple research groups have focused their efforts on understanding FLCN function and how dysregulation of the pathway(s) in which FLCN interacts leads to the skin and lung manifestations and kidney cancer associated with BHD syndrome. Investigations have uncovered potential roles for FLCN in a number of diverse molecular pathways, which are summarized in the following sections.
5.1 AKT-mTOR pathway signaling
Studies in FLCN-deficient in vivo models provided the first clues to a potential role for FLCN in modulating the mTOR pathway. Mice with Flcn inactivation targeted to the distal nephron of the kidney developed polycystic kidneys and cystic renal disease, and died at 3 weeks of age due to renal failure (Baba et al., 2008; Chen et al., 2008). Flcn deletion targeted to kidney proximal tubules produced renal cysts and early onset (≥6 months) renal neoplasms with high tumor penetrance (Chen et al., 2015). Cysts and tumors from the Flcn-deficient mouse kidneys displayed elevated levels of phospho-AKT, phospho-mTOR and phosphorylated ribosomal protein S6 (phospho-S6) demonstrating activation of the AKT-mTOR pathway. Treatment with the mTOR inhibitor rapamycin suppressed cyst formation (Baba et al., 2008; Chen et al., 2008) and renal tumor growth (Chen et al., 2015). Flcn homozygous knockout mice die in utero, but Flcn heterozygous knockout mice develop kidney tumors with a long latency (>10 months) and show loss of the Flcn wild type allele (Hasumi et al., 2009). These Flcn-deficient tumors demonstrated activation of AKT, mTORC1 and mTORC2. Renal tumors from BHD patients with germline FLCN mutations also showed activated AKT-mTOR signaling (Baba et al., 2008; Hasumi et al., 2009), and increased phospho-S6 protein levels were seen in human lung-derived cells with FLCN knockdown (Khabibullin et al., 2014). Taken together these in vitro and in vivo data support a role for FLCN in suppressing mTOR activation.
On the other hand, results from other in vivo models support a role for FLCN in activating the mTOR pathway. Deletion of the FLCN homolog, bhd, in Schizosaccharomyces pombe (S. pombe) produced an expression profile of permease and transporter genes that was opposite to results obtained in S. pombe with deletion of TSC1 or TSC2 homologs, which are negative regulators of mTOR. S. pombe lacking bhd was hypersensitive to rapamycin suggesting that Bhd activates Tor (van Slegtenhorst et al., 2007) These researchers also generated a mouse model using a gene trap vector technique that expressed a truncated Flcn protein, and treated these mice with N-ethyl-N-nitrosourea to accelerate renal cyst and tumor development. These mice developed microcysts and rare tumors with late onset that displayed reduced phospho-S6 immunostaining (readout for mTOR) relative to adjacent normal kidney (Hartman et al., 2009b). Reduced phospho-S6 was also observed in several mammalian cell lines treated with FLCN siRNA. Another Flcn knockout mouse model generated in a similar manner with a gene-trap vector developed multiple renal cysts and adenomas over a wide age range. In this model, phospho-S6 immunostaining was variable depending on cyst size and number with elevated expression in large multilocular cysts and weak to no phospho-S6 staining in small single cysts (Hudon et al., 2010).
Based upon the conflicting data from multiple studies, it has been proposed that the role of Flcn loss in renal cysts may be more complex than simply activation of mTOR and may depend upon cell type and nutritional/energy status (Hudon et al., 2010). FLCN loss may activate other signaling pathways (i.e., Raf-MEK-ERK signaling)(Baba et al., 2008) that in turn can affect mTOR signaling.
5.2 PPARGC1A/PGC1α regulation and mitochondrial biogenesis
The first indication that a physiological relationship might exist between FLCN loss and regulation of PPARGC1A/PGC1α came from gene expression profiling studies comparing BHD-associated renal tumors with sporadic chromophobe renal tumors and oncocytomas in an effort to determine if the inherited and sporadic tumors with the same histologies had similar or distinct gene profiles (Klomp et al., 2010). Although there were clear differences in expression of some genes in BHD tumors compared with sporadic tumors, both sporadic counterpart tumors and BHD renal tumors with reduced FLCN expression were enriched for mitochondria- and oxidative phosphorylation-associated genes reflecting the abundance of mitochondria in both tumor types. BHD tumors were distinct from their sporadic counterparts in demonstrating upregulation of two transcription factors PPARGC1A(PGC1α) and transcription factor A, mitochondrial (TFAM) along with a subset of PGC1α regulated genes, all of which are important drivers of mitochondrial biogenesis and mitochondria replication, and suggest that tumor development in BHD is driven by deregulation of the PGC1α-TFAM axis (Klomp et al., 2010).
Studies in Flcn-deficient in vitro and in vivo models provide further support for regulation of PPARGC1A by FLCN. Mice with muscle-targeted Flcn inactivation underwent a marked metabolic shift toward oxidative phosphorylation as evidenced by increased mRNA expression of mitochondrial genes, elevated mitochondrial number and enhanced flux through the tricarboxylic acid cycle in Flcn-deficient muscle (Hasumi et al., 2012). Mitochondrial respiratory capacity was enhanced, ATP production was high, and membrane potential was increased in Flcn-deficient muscle relative to controls demonstrating that the mitochondria were functioning normally. These observations were confirmed in FLCN-deficient UOK257 renal tumor cells and reversed by re-expression of FLCN. Subsequently, a screen of genes involved in mitochondrial biogenesis revealed increased PPARGC1A mRNA and protein in the Flcn-deficient muscle and in UOK257 cells. Genetic inactivation of PPARGC1A in the muscle-targeted Flcn-knockout mice or FLCN re-expression in UOK257 cells reversed the PPARGC1A–driven oxidative metabolism phenotypes (Hasumi et al., 2012). PPARGC1A mRNA expression was also high in human BHD-associated renal tumors relative to normal kidney from patients with BHD or sporadic counterpart renal tumors, and in Flcn-deficient mouse kidney tumors from Flcn heterozygous knockout mice. Flcn-deficient kidneys from kidney-targeted Flcn knockout mice demonstrated increased PPARGC1A expression and higher numbers of mitochondria relative to littermate controls, and the polycystic mouse kidney phenotype was partially reversed by genetic inactivation of PPARGC1A (Hasumi et al., 2012). In a mouse model in which Flcn inactivation was targeted to the heart, mice died after 3 months of age due to dilated cardiomyopathy and the Flcn-deficient hearts displayed elevated ATP levels, increased mitochondrial numbers and respiratory capacity, and elevated PPARGC1A protein levels (Hasumi et al., 2014). Targeting Flcn inactivation to mouse adipose tissues in two independent models led to a complete metabolic reprogramming of adipose tissues resulting in “browning” of white adipose tissue (WAT) through increased mitochondrial biogenesis and enhanced oxidative metabolism. In the first model, Flcn loss induced AMPK-dependent PGC1α/estrogen-related receptor α (ERRα) transcriptional control of energy metabolism resulting in increased mitochondrial biogenesis in both white and brown adipose tissues (Yan et al., 2016). Interestingly, in a second adipose tissue-targeted Flcn knockout model, mitochondrial biogenesis was driven by increased expression of both PGC1α and PGC1β in a TFE3- and mTOR-dependent manner (Wada et al., 2016) (see section 5.4). Elevated PGC1α-mediated mitochondrial biogenesis was also seen in Flcn−/− mouse embryonic fibroblasts (MEFs) (Yan et al., 2014). These results from FLCN-deficient renal tumor cell lines and MEFs, mouse tissues with targeted Flcn deletion, and BHD-associated renal tumors underscore a role for FLCN as a negative regulator of PPARGC1A/PGC1α and mitochondrial biogenesis.
5.2.1 Regulation of AMPK activation, a driver of PGC1α-regulated oxidative metabolism
FLCN interacts with 5’AMP activated protein kinase (AMPK) through its protein partners FNIP1 and FNIP2 (Baba et al., 2006; Hasumi et al., 2008; Lim et al., 2012) and both FLCN and FNIP1 could serve as substrates for AMP kinase activity in vitro (Baba et al., 2006). The association of FLCN, FNIP1/2, and AMPK in immunoprecipitates from multiple cell types suggests that they exist in a complex, which led investigators to consider whether FLCN and FNIP1/2 might regulate the activity of AMPK or conversely, whether AMPK might modulate the function or expression level of FLCN and FNIP1/2. Investigations into the functional relationship between FLCN and AMPK in in vitro and in vivo systems with FLCN inactivation have produced somewhat equivocal results. In Flcn-deficient (Flcn−/−) MEFs, AMPK was activated (elevated phospho-AMPKα) leading to increased PGC1α expression, mitochondrial biogenesis and ATP production, resulting in reactive oxygen species (ROS)-dependent activation of HIFα and a metabolic switch to aerobic glycolysis (Yan et al., 2014). However, when Flcn was knocked down in Ampk−/− MEFs, the expression levels of PGC1α and ROS remained unchanged similar to those in Flcn-expressing Ampk-deficient MEFs. These results support AMPK activation as the driving force for the cascade of events initiated by PGC1α activation and culminating in the switch to aerobic glycolysis in Flcn-deficient MEFs (Yan et al., 2014). In a mouse model with targeted loss of Flcn in adipose tissue, chronic hyperactivation of AMPK was observed resulting in PGC1α-driven mitochondrial biogenesis and activity (Yan et al., 2016). In a C elegans model, the flcn-1 loss of function mutant demonstrated increased phosphorylation of AAK-2, the ortholog of AMPKα (Possik et al., 2014). On the other hand, in a mouse model in which Flcn inactivation was targeted to heart, mice died early due to cardiac hypertrophy and Flcn-deficient hearts displayed reduced phospho-AMPKα (T172) but still demonstrated elevated PPARGC1A expression leading to increasing mitochondrial biogenesis, respiratory capacity and ATP production (Hasumi et al., 2014). Mice with heart-targeted double inactivation of Flcn and PPARGC1A displayed phospho-AMPK levels comparable to wildtype littermates. The investigators suggested that excess ATP production resulting from the PPARGC1A cascade caused the decrease in AMPK activity. The results obtained from most of these in vivo and in vitro models suggest that FLCN acts as a negative regulator of AMPK; however, further work will be necessary to understand the disparate findings in the Flcn-deficient hearts.
5.3 Regulation of cell-cell adhesion, cell polarity, and Rho A activity
In an effort to identify FLCN protein partners, a yeast two hybrid screen was performed independently by two groups using full length FLCN as bait, and p0071 (PKP4/ plakophilin) was identified, and validated in vitro by coimmunoprecipitation with FLCN (Medvetz et al., 2012; Nahorski et al., 2012). p0071 is member of the armadillo repeat-containing protein family, a component of the adherens junctions, and regulates the activity of the small GTPase Rho A. FLCN colocalized with p0071 at cell junctions during interphase and at the midbody during cytokinesis (Nahorski et al., 2012). Studies from one of the groups demonstrated deregulation of Rho A signaling (increased GTP-bound Rho A) and multinucleation in FLCN-deficient FTC-133 thyroid cancer cells reflecting defects in cytokinesis, which were reversed by restoration of FLCN expression (Nahorski et al., 2012). In a wound healing assay, FLCN-deficient FTC-133 cells demonstrated a more migratory phenotype (invasion/metastasis) than FLCN-restored counterpart cells, and inhibition of downstream Rho A signaling with an inhibitor of Rho-associated kinase (ROCK) significantly reduced cell migration. In the ACHN kidney cancer cell line, levels of FLCN knockdown by shRNA were inversely correlated with Rho A activity in a dose- dependent manner (Nahorski et al., 2012).
On the other hand, a report from the second group found that loss of FLCN led to decreased Rho A activity as measured by reduced activity of ROCK (Medvetz et al., 2012). Rho A signaling and cell polarity are required for correct closure in a wound healing assay, which was delayed in FLCN-deficient UOK257 cells compared to FLCN-restored isogenic cells. FLCN was shown to negatively regulate cell-cell adhesions in three-dimensional cell cluster assays. Both normal and aberrant desmosomes necessary for cell-cell adhesions were present in FLCN- deficient UOK257 cells but not in FLCN-expressing UOK257 cells (Medvetz et al., 2012). Although data are conflicting as to whether loss of FLCN upregulates or down regulates Rho A signaling, the phenotypes are consistent in supporting a role for the interaction of FLCN and p0071 in regulating cell-cell adhesion through Rho A signaling.
Loss of cell polarity and cell-cell adhesion is frequently observed in epithelial cancers such as BHD-associated kidney cancer, and may also be the cause for the development of BHD lung cysts. In polarized IMCD-3 (mouse inner medullary collecting duct) cells treated with FLCN siRNA, immunostaining of claudin-1, a tight junction component, and E-cadherin, an adherens junction component, were significantly reduced and disordered. Additionally FLCN knockdown was associated with reduced trans-epithelial electrical resistance in IMCD-3 cells suggesting a delay in tight junction formation leading to an overall disruptive effect on cell polarity and cell-cell adhesion (Nahorski et al., 2012). In contrast, loss of FLCN in kidney-derived UOK257 and HEK293 cells (Medvetz et al., 2012) and human lung-derived cells (Khabibullin et al., 2014) was shown to enhance cell-cell adhesion in cell aggregate assays. p0071 loss phenocopied FLCN deficiency in this assay (Medvetz et al., 2012) supporting the concept that either FLCN or p0071 deficiency leads to increased cell-cell adhesion and defects in cell polarity mediated by loss of the FLCN-p0071 interaction. Since these groups used different FLCN-deficient cells in their studies, the effect of FLCN deficiency on cell polarity and cell-cell adhesion may be cell context-dependent. Clarification of these discordant results awaits further investigation of cell-cell adhesions and cell polarity in additional FLCN-deficient cell types.
One potential mechanism by which FLCN loss may lead to imbalance of cell-cell adhesions and cell polarity is suggested by studies in a mouse model in which Flcn inactivation was targeted to the lung, specifically the alveolar epithelial type II cells (AEC). Flcn-deficient lungs from this model displayed impaired lung function, alveolar enlargement and AEC apoptosis with elevated cleaved caspase-3 (Goncharova et al., 2014). Flcn inactivation in cultured mouse AECs and NMuMG cells resulted in loss of AMPK activation, and inactivation of both membrane and cytosolic LKB1, an upstream kinase that phosphorylates and activates AMPK (Goncharova et al., 2014). These observations were recapitulated in UOK257 cells. E-cadherin, which has been shown to regulate the localization of LKB1, was significantly reduced and mislocalized from adherens junctions to cytoplasm in Flcn-deficient mouse AECs. Given that LKB1 is a well-known regulator of cell polarity (Martin-Belmonte and Perez-Moreno, 2011) and E-cadherin is an important component of adherens junctions, a pathway in which loss of Flcn leads to decreased AMPK activation through loss of LKB1, and dysregulation of E-cadherin at adherens junctions may contribute to increased cell-cell adhesions and loss of cell polarity under Flcn deficiency.
5.4 TFE3/TFEB transcriptional activation
Another experimental approach to elucidating FLCN function involved comparing the gene expression profile of FLCN-deficient UOK257 cells with FLCN-restored counterpart cells (Hong et al., 2010b). Twelve of the top 15 differentially expressed gene candidates were upregulated and 3 were down regulated by restoration of FLCN expression. Bioinformatics searching of promoter regions in these genes revealed that one of the candidates, glycoprotein non-metastatic B (GPNMB), was transcriptionally regulated through a highly conserved M box sequence in its promoter by the microphthalmia transcription factor (MiTF), a member of the MiTF family of transcription factors that includes transcription factors E3 and B (TFE3, TFEB) (Hong et al., 2010a). Although UOK257 expressed a high level of GPNMB, only a low level of MiTF was detected, but a moderate level of another MiTF family member, TFE3, was observed in these FLCN-deficient cells. Confirmation that GPNMB expression was transcriptionally upregulated by TFE3 in response to FLCN knockdown was made in several cell lines, and electrophoretic mobility shift assays confirmed that TFE3 bound to the GPNMB promoter M box sequence. FLCN inactivation was correlated with a decrease in TFE3 phosphorylation and its nuclear localization in UOK257 cells and Flcn-deficient MEFs. On the other hand, re-expression of FLCN in UOK257 or MEFs resulted in increased TFE3 phosphorylation and its movement to the cytoplasm where TFE3 is no longer active (Hong et al., 2010a). Expression of GPNMB, as a surrogate for TFE3 activity, was found to be high in BHD-associated kidney tumors (Hong et al., 2010a; Furuya et al., 2015) and kidney tumors from Flcn heterozygous knockout mice (Hong et al., 2010a), and may be a useful biomarker for BHD-associated kidney cancer. These findings confirmed a role for FLCN as a negative regulator of TFE3 and its target genes including GPNMB.
Although the evidence that FLCN functions as a negative regulator of TFE3 was convincing, the mechanistic details were lacking and it was not known whether FLCN could also regulate other MiTF family members such as TFEB. TFEB has been described as the master regulator of lysosomal biogenesis (Raben and Puertollano, 2016). Previous work had established that nuclear localization of TFEB was controlled by mTORC1-dependent phosphorylation of TFEB on serine 211 that facilitates its interaction with 14-3-3 proteins and inhibits its nuclear localization when lysosome function is adequate. Conversely, inhibition of lysosome function reduced mTORC1-dependent phosphorylation, released 14-3-3 proteins, and enabled nuclear translocation of TFEB (Martina et al., 2012) (Roczniak-Ferguson et al., 2012). To evaluate a role for FLCN in TFEB regulation, researchers knocked down FLCN in HeLa cells expressing TFEB and observed increased nuclear localization of TFEB and loss of TFEB-positive puncta (indicating lysosomal localization) in the cytoplasm (Petit et al., 2013). Importantly, TFEB phosphorylation on serine 211 was reduced with release of 14-3-3 and an increase in lysosomal proteins, reflecting transcriptionally active TFEB in HeLa cells with FLCN knockdown (Petit et al., 2013). Pharmacologic inhibition of mTOR produced a similar increase in lysosome biogenesis as was seen under FLCN knockdown, supporting a role for FLCN in mTOR-dependent regulation of TFEB.
Results from studies in an adipose tissue-targeted Flcn knockout mouse model support a similar mechanism for regulation of TFE3 activity by FLCN. These researchers showed in immortalized mouse preadipocytes that FLCN was necessary for mTORC1-dependent phosphorylation of TFE3 on mouse serine 320 (homologous to serine 321 in human TFE3 and serine 211 in TFEB) leading to interaction with 14-3-3 proteins and sequestration of TFE3 in the cytoplasm where it is inactive. In Flcn-deficient cells, mTORC1-dependent phosphorylation of TFE3 on serine 320 was inhibited, thereby enabling TFE3 nuclear localization and transcriptional activation of its target genes (Wada et al., 2016).
5.4.1 Embryonic stem cell exit from pluripotency via Tfe3 regulation
Factors that maintain mouse embryonic stem cell (ESC) self renewal are well studied, but the mechanisms by which ESCs exit from pluripotency are not well understood. A recent report presents data to support a rather intriguing role for Flcn in driving exit from pluripotency in ESCs through regulation of Tfe3 activity. In a large scale siRNA screen, Flcn was identified as one of the genes required for progression from pluripotency to a state primed for differentiation (Betschinger et al., 2013). Flcn with its binding partners Fnip1 and Fnip2 was able to drive differentiation by sequestering Tfe3 in the cytoplasm, where it remains inactive. Conversely, in ESCs with Flcn or Fnip1/2 knockdown, nuclear localization of Tfe3 was sustained, which constrained exit from pluripotency. Moreover, Tfe3 was shown to transcriptionally upregulate the orphan nuclear receptor Esrrb, a core pluripotency factor, thereby maintaining the ESC pluripotent state. These findings provide further evidence that Flcn in complex with Fnip1/Fnip2 negatively regulates Tfe3 and present support for a previously undescribed role of Flcn in developmental progression from the pluripotent state in ESCs.
5.5 Interaction with Rag GTPases for amino acid-dependent mTORC1 activation on lysosomes
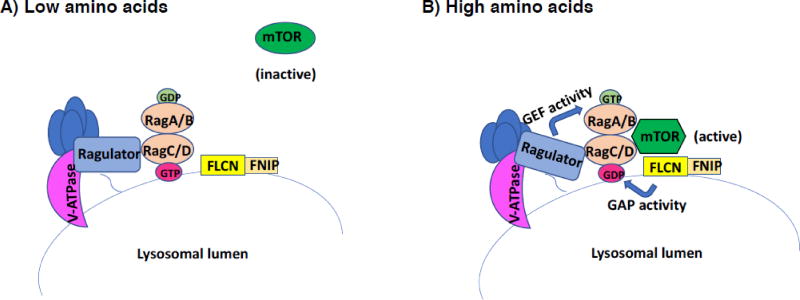
mTOR activation by amino acids in mammalian cells involves a protein complex on the lysosome consisting of vacuolar (v)-ATPase, the Ragulator complex and Rag GTPases. The Rag proteins function as obligate heterodimers of Rag A or B with Rag C or D. Amino acids signal from within the lysosomal lumen to Ragulator in a v-ATPase- dependent manner. Ragulator then activates Rag A/B through its guanine exchange factor (GEF) activity, and the Rag heterodimer recruits mTORC1 to the lysosome surface for activation by the small GTPase Rheb (Bar-Peled and Sabatini, 2014). Rag A/B has been most intensely studied, whereas less was known about Rag C/D, although it was shown to be essential for amino acid sensing by mTORC1. Using proteomic approaches to identify other mTORC1 pathway components, investigators identified peptides derived from FLCN and its interacting partners FNIP1 and FNIP2 in HEK293 immunoprecipitates with Rag proteins (Tsun et al., 2013). Endogenous FLCN localized to the lysosomal surface in response to amino acid starvation and dispersed upon amino acid stimulation. FLCN was shown to be necessary for mTORC1 activation by amino acids in cell based assays and for localization of mTORC1 to the lysosomal surface upon amino acid stimulation. Binding of FLCN to Rag proteins required FNIP2, and purified FLCN- FNIP2 stimulated GTP hydrolysis by Rag C and Rag D, but not by Rag A or Rag B (Tsun et al., 2013). These results indicate that FLCN-FNIP2 acts as a GTPase activating protein (GAP) for Rag C (and to a lesser extent Rag D) and promotes mTORC1 binding to the Rag heterodimer through its GAP activity towards Rag C/D (Figure 4).
Figure 4.
FLCN acts as a GAP for RagC/D to activate mTOR on the lysosome surface in response to amino acids. mTORC1 activation by amino acids requires a lysosome-associated complex of proteins that include the vacuolar (v)-ATPase, the Ragulator complex, and Rag GTPases, which function as obligate heterodimers RagA or B and RagC or D. Amino acids signal from within the lysosomal lumen to Ragulator, which exerts its guanine nucleotide exchange factor (GEF) activity toward RagA/B. GTP-loaded RagA/B then recruits mTORC1 to the lysosomal surface for activation. Additionally, when amino acids are low, FLCN-FNIP associates with the lysosomal surface. When amino acid levels are sufficient, FLCN-FNIP acts as a GTPase activating protein (GAP) towards RagC/D. The GDP-loaded RagC/D can then facilitate mTORC1 recruitment to the lysosome for activation (Petit et al., 2013; Tsun et al., 2013).
Independently, researchers who identified a potential role for FLCN in mTOR-dependent regulation of TFEB (see section 5.4) also reported that loss of FLCN resulted in reduced mTORC1 activity and showed that FLCN was required for enrichment of mTORC1 on lysosomes and its activation by amino acids. However, they found that FNIP1, not FNIP2, facilitated this process (Petit et al., 2013). FLCN was selectively recruited to the lysosome after amino acid depletion and interacted with Rag GTPase heterodimers, but in this study FLCN preferentially bound the inactive or GDP loaded Rag A through its GTPase domain (and presumably Rag B given their homology) and FNIP1 was required for the Rag A interaction (Petit et al., 2013). The results of this study add support for a functional role of FLCN in recruitment of mTORC1 to lysosomes through amino acid- dependent activation of Rag heterodimers.
The signaling cues that send FLCN to the lysosome under nutrient depleted conditions remain unclear but recent work has provided a potential mechanism. Using HeLa cells and FLCN-deficient and FLCN-restored UOK257 renal tumor cells, investigators demonstrated that FNIP1 and FNIP2 were unstable under nutrient replete conditions when mTORC1 is activated. FNIP2 was shown to be phosphorylated within a consensus binding motif for the F-box protein β-TRCP by casein kinase 1, and subsequently targeted by the SCFPβ–TRCP (Skp1-Cullin 1-F box protein) E3 ubiquitin ligase complex for ubiquitin-mediated proteasomal degradation in a nutrient-dependent manner (Nagashima et al., 2017). FNIP1/FNIP2 were shown to be necessary for starvation-induced localization of FLCN on lysosomes and for dissociation of mTORC1 from the lysosomes. These data underscore the role of the FNIP proteins in facilitating the nutrient-dependent lysosome association of FLCN.
5.5.1 Species conservation of FLCN GAP activity towards Rag GTPases for mTORC1 activation on lysosomes
The discovery that a FLCN-FNIP complex activates Rag heterodimers to facilitate amino acid-dependent mTORC1 recruitment and activation on lysosomes has led to an appreciation for the evolutionarily conserved relationship between FLCN and Rag GTPases in response to amino acid availability. Drosophila melanogaster larvae with Flcn inactivation (DBHD−/−) displayed a nutrient starved phenotype including growth retardation and small body size that was rescued by dietary yeast or leucine (but not by other amino acids). The rescue effect was correlated with elevated mTOR signaling since rapamycin treatment reversed the amino acid feeding rescue (Liu et al., 2013). In earlier studies in yeast, the FLCN ortholog Lst7 was identified in a Saccharomyces cerevisiae synthetic lethal screen with sec13 mutants to characterize new factors involved in vesicle formation at the endoplasmic reticulum. Lst7, Lst4 (ortholog of FNIP1/2) and Lst8 (ortholog of mLST8, a subunit of mTOR complex) appeared to function in a manner similar to Sec13 in amino acid permease (Gap1p) transport from the Golgi to the plasma membrane (Roberg et al., 1997), and mutations in Lst4 or Lst7 blocked multivesicular endosome (MVE) to plasma trafficking of Gap1p (Rubio-Texeira and Kaiser, 2006). The recent identification of Lst4 and Lst7 as orthologs of FNIP 1/2 and FLCN was based in part on their classification as DENN domain proteins (Nookala et al., 2012; Zhang et al., 2012; Pacitto et al., 2015).
Evolutionary conservation was further verified by more recent studies of the FLCN and FNIP orthologs in the yeast system. The Lst4-Lst7 complex was found to localize to the vacuolar (lysosomal) membrane in response to amino acid starvation. Upon amino acid refeeding, Lst4-Lst7 bound to and acted as a GAP for the Rag family GTPase Gtr2, the ortholog of mammalian Rag C/D, within the context of a Gtr1-Gtr2 heterodimer, enabling recruitment of mTORC1 to the vacuolar membrane for activation, and release of Lst4-Lst7 from the membrane (Peli-Gulli et al., 2015). Although the regulatory mechanism of Rag GTPases for amino-acid dependent mTORC1 activation appears highly conserved in yeast, there were several distinct differences from the mammalian system. First, the Lst4-Lst7 complex, which associated with the vacuolar membrane under amino acid starvation, did not require Rag GTPases for the association and was antagonized by mTORC1, which was not the case in the mammalian cells. Second, the preference of Lst4 for Gtr2 in its GTP-loaded state was contrary to the preferred association of FLCN-FNIP with inactive or GDP-loaded Rag A in the mammalian system. Finally, refeeding with glutamine or methionine (or substrates that can be converted to these amino acids) provided strong signals to activate mTORC1 through Gtr2 and release Lst4-Lst7 from the vacuolar membrane (Peli-Gulli et al., 2015). The evolutionary conservation of the mechanism in S. cerevisiae by which the Rag GTPase orthologs Gtr1/2 recruit and activate mTORC1 on the vacuolar membrane through the FLCN-FNIP orthologs Lst4-Lst7 is remarkable, and makes this an excellent in vivo model for teasing out the mechanistic details of this complex amino acid sensing system.
5.6 Structural homology to DENN proteins and membrane trafficking
Recent structural studies using x-ray crystallography and fold recognition and structure prediction software have confirmed that FLCN and FNIP1/2 have both an N-terminal Longin domain and a C-terminal DENN (differentially expressed in normal and neoplastic cells) domain (Nookala et al., 2012; Zhang et al., 2012; Pacitto et al., 2015) placing them within the family of Rab guanine nucleotide exchange factors (GEF) that are regulators of most membrane trafficking events in eukaryotes (Zhang et al., 2012). In fact, C-terminal FLCN was shown to have GEF activity in vitro towards Rab35, a Rab GTPase implicated in endocytic trafficking (Nookala et al., 2012).
A novel interaction between another Rab GTPase, Rab7A, and FLCN was identified by mass spectrometric analysis of protein complexes from FLCN deficient and FLCN restored UOK257 cells (Laviolette et al., 2017). Rab7A plays an important role in endosomal recycling and lysosomal degradation of epidermal growth factor receptor (EGFR), processes that are important for regulation of EGFR signaling and stability. Rab7A was shown to bind to FLCN through its C-terminal DENN domain, and wild-type FLCN but not mutant forms stimulated GTP hydrolysis of Rab7A supporting a role for FLCN as a Rab7A GAP (Laviolette et al., 2017). FLCN-deficient UOK257 cells demonstrated increased EGFR signaling in response to ligand stimulation, which was reversed by exogenous Rab7A expression. FLCN loss in FLCN−/− FTC-133 cells resulted in slower endocytic trafficking and the accumulation of EGFR in early endosomes where the ligand-stimulated EGFR signaling cascade is still active. Decreased EGFR and downstream signaling was observed with Rab7A expression in the FLCN−/− cells. EGFR activation was also seen in kidney tumors from BHD patients and Flcn-deficient mouse tumors, and treatment of human FLCN−/− xenografts in mice with the EGFR inhibitor Afatinib slowed tumor growth (Laviolette et al., 2017). These data suggest a role for FLCN as a GAP for Rab7A and support a function for FLCN in endocytic trafficking of EGFR, and potentially other receptor tyrosine kinases, through its interaction with Rab7A.
In another recent study the potential role of FLCN in membrane trafficking was investigated by examining the involvement of FLCN in the spatial distribution of lysosomes in response to nutrient status. Perinuclear clustering of lysosomes occurs in response to nutrient/amino acid withdrawal, when mTORC1 activity is suppressed. When nutrients are sufficient and mTORC1 activity is high, lysosomes are dispersed near the cell periphery. During starvation conditions, peri-nuclear localization of lysosomes was inhibited by FLCN knockdown and by siRNA targeting of FNIP1 and FNIP2 in HeLa cells, suggesting that the FLCN/FNIP complex is required for nutrient–dependent distribution of lysosomes (Starling et al., 2016). FLCN/FNIP ectopic expression promoted the formation of dynamic lysosome-associated tubules. Rab34, a small GTPase known to play a role in lysosomal transport in complex with its effector RILP (Rab interacting lysosomal protein) promotes perinuclear clustering of lysosomes under starvation conditions, and this interaction was shown to require FLCN (Starling et al., 2016). FLCN was found to interact directly with RILP through its C-terminal DENN domain and this interaction promoted loading of active GTP- bound Rab34 onto RILP to form an active RILP-Rab34 complex. These observations were recapitulated in UOK257 kidney cancer cells with restored FLCN expression, which displayed perinuclear localization of lysosomes and Rab34-RILP interaction that were absent from FLCN-deficient UOK257 cells (Starling et al., 2016). These findings support a previously unknown function for FLCN connecting the lysosomal nutrient signaling network with the cellular machinery that controls the intracellular distribution of the lysosome itself.
5.7 Regulation of autophagy
Autophagy is an evolutionarily conserved process that carries out well controlled degradation and recycling of damaged organelles and macromolecules to replenish cellular energy and amino acid supply for maintenance of cellular homeostasis, and for survival as an adaptive response to stress. The cytoplasmic material is sequestered in double membrane vesicles called autophagosomes, which eventually become fused with lysosomes at maturation. Subsequently, lysosomal enzymes degrade the material allowing permeases to transport the resulting amino acids and lipids back into the cytoplasm for biosynthesis or energy production [for review see (Choi et al., 2013)]. The recent reports demonstrating that FLCN localizes to lysosomes and monitors nutrient sensing through Rag GTPases (see sections 5.5 and 5.6) led investigators to ask whether the FLCN- FNIP complex might be involved in autophagy. Knockdown of FLCN in HK2 cells resulted in the accumulation of the autophagic marker sequestosome1 (SQSTM1/p62), which signals impaired autophagy, and was reversed by FLCN re-expression. Similarly, elevated SQSTM1/p62 expression was seen in Flcn −/−MEFs and in kidney tumor tissue from a BHD patient with a germline FLCN mutation. Impaired maturation of autophagosomes (i.e., reduced fusion with lysosomes) and increased endogenous levels of GABA(A) receptor-associated protein (GABARAP), a component of mature autophagosomes, were observed in FLCN-deficient HEK 293 and HK2 cells (Dunlop et al., 2014). FLCN was shown to interact with GABARAP both in in vitro binding assays and endogenously, with a requirement for either FNIP1 or FNIP2, but a preference for FNIP2. Unc-51 like autophagy activating kinase 1 (ULK1) is an activator of the autophagic process and a GABARAP interactor. ULK1 was shown to interact with and phosphorylate FLCN, potentially at one of three serines (Ser406, Ser537, Ser542) identified by mass spectrometry and predicted by modeling studies on the FLCN C-terminal crystal structure to be exposed to solvent and accessible to the kinase (Dunlop et al., 2014). Coimmunoprecipitation experiments with FLCN-FNIP2, kinase dead ULK1 or wild type ULK1 demonstrated that the kinase activity of ULK1 was required for FLCN-FNIP2 dissociation from GABARAP. Mutant proteins expressing the truncating FLCN mutations seen in BHD patients interacted more strongly with ULK1 than wild type FLCN protein in vitro, showed impaired binding to GABARAP and were not able to repress SQSTM1/p62 levels as efficiently as wildtype FLCN (Dunlop et al., 2014). Taken together, these data suggest that FLCN is a positive modulator of autophagy and FLCN loss leads to impaired autophagy that may contribute to the BHD syndrome phenotype.
On the other hand, results from studies exploring the relationship between AMPK and FLCN in the flcn deficient Caenorhabditis elegans model, flcn-1(ok975), support an opposing relationship between FLCN and autophagy. In this in vivo model, loss of flcn-1 increased resistance to oxidative stress imparted by paraquat, a superoxide inducer, and this was dependent upon aak-2, the ortholog of AMPK (Possik et al., 2014). The increased resistance to oxidative stress was shown not to be from reactive oxygen species pathways but rather from increased autophagy. Increased numbers of autophagic vacuoles containing LGG-1 (LC3 ortholog) were observed in the flcn-1 mutants and this higher autophagic activity was shown to be aak-2 dependent and required for resistance to oxidative stress. Autophagy increased ATP levels in the flcn-1 mutants and protected against cell death by apoptosis. These findings were recapitulated in Flcn−/− MEFs and in FTC-133 cells lacking FLCN. Overall, loss of FLCN in worm, mouse and human in vivo models resulted in activation of AMPK, elevated autophagy and increased ATP levels conferring resistance to metabolic stress (Possik et al., 2014). In a report in which FLCN was shown to contribute to VHL tumor suppressing activity in renal cancer, FLCN was demonstrated to be necessary for the LC3B- and LC3C- mediated autophagic pathways (Bastola et al., 2013). More in-depth studies will be required to understand these divergent results regarding a role for FLCN in regulating autophagy.
5.8 Ciliogenesis and cilia-dependent flow sensory mechanisms
Patients with both von Hippel-Lindau and tuberous sclerosis complex diseases are predisposed to develop renal cysts and tumors, and these diseases have been linked to dysfunctional primary cilia (Esteban et al., 2006; Hartman et al., 2009a). Individuals affected with BHD syndrome also develop cysts in the lung, kidney and, in one report, liver (Kluger et al., 2010), and mouse models with kidney targeted Flcn inactivation produce multi-cystic kidneys (Baba et al., 2008; Chen et al., 2008), which has led to the hypothesis that FLCN may play a role in primary cilia formation and that dysfunctional cilia may lead to the manifestations associated with BHD syndrome. In one study designed to test this hypothesis, both endogenous FLCN protein and exogenously expressed FLCN were found to localize to primary cilia in multiple cell types (Luijten et al., 2013), and both wild type and mutant forms of FLCN were found on cilia, the basal body and the centrosome. Knockdown of FLCN in serum starved human kidney (HK-2) cells resulted in delayed development of cilia. Over expression of wild type FLCN in FLCN-restored UOK257 kidney cancer cells led to reduction in cilia numbers as well (Luijten et al., 2013). In kidney cells grown as spheroids in 3D culture, treatment with siRNA against Flcn resulted in severe impairment of spheroid formation with decreased luminal size, fewer cilia, and increased numbers of abnormally oriented cell divisions and, as was seen in monolayer culture, this phenotype also developed in spheroids exogenously expressing wild type FLCN, suggesting that tight regulation of FLCN levels is necessary for proper ciliogenesis. Primary cilia restrict canonical Wnt signaling by sequestration of β catenin in the basal body (Lancaster et al., 2011), and abnormal Wnt/β catenin signaling has been attributed to renal cyst formation. In this study, elevated levels of unphosphorylated (active) β catenin and its downstream targets were observed in cultured mouse IMCD3 cells treated with siRNA against Flcn. These findings suggest that Flcn deficiency may lead to kidney and lung cyst formation through defective ciliogenesis resulting in inappropriate activation of the canonical Wnt/β catenin pathway (Luijten et al., 2013).
Further evidence that FLCN may be a ciliary protein has come from a yeast two-hybrid screen that identified KIF3A as a FLCN-interacting protein. Primary cilium assembly and maintenance require intraflagellar transport (IFT) that is driven by kinesin-2 motor comprised of two motor subunits, KIF3A and KIF3B (Kim and Dynlacht, 2013). FLCN was shown to interact with both of the motor subunits in a cilium-dependent manner, and as was shown previously, FLCN localized to primary cilia in FLCN-expressing but not FLCN depleted cells (Zhong et al., 2016). Cilia function as flow sensors and down regulate mTOR signaling through flow-mediated activation of the serine/threonine kinase LKB1 in the cilia-basal body compartment of non-cycling cells. LKB1 in turn phosphorylates and activates AMPK, which negatively regulates mTORC1 via phosphorylation of TSC2 (Boehlke et al., 2010). In this study, flow stress reduced mTORC1 signaling (as measured by downstream readouts) in FLCN–expressing HKC-8 and UOK257-2 cells but not with knockdown of FLCN or its partner FNIP1, in a cilium-dependent manner (Zhong et al., 2016). In FLCN-replete but not FLCN-deficient HKC-8 and UOK257-2 cells grown under flow conditions, AMPK activity was elevated and Tsc2 was phosphorylated leading to mTORC1 inhibition. FLCN was shown to recruit LKB1 to cilia and induce its association with AMPK for activation in the basal body in response to flow stress (Zhong et al., 2016). These results support a role for FLCN as part of the mechanosensory signaling machinery of the cell that controls LKB1 levels and AMPK activation leading to mTORC1 pathway inhibition through primary cilia, and offer a potential mechanism by which FLCN loss leads to mTORC1 dysregulation in Flcn-deficient preclinical models and BHD-associated kidney cancer.
5.9 Other potential functions of FLCN
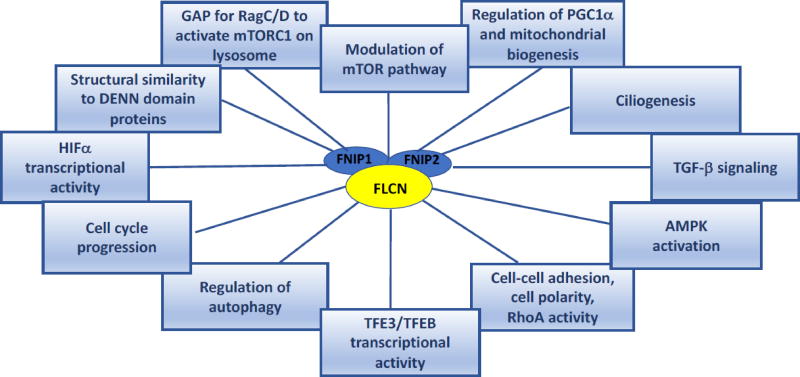
In addition to the potential functions of FLCN described in the above sections (Figure 5), a number of published reports support roles for FLCN in other signaling pathways and cellular processes including TGF-β signaling (Singh et al., 2006; Hong et al., 2010b; Cash et al., 2011; Khabibullin et al., 2014; Chen et al., 2015), regulation of HIF1α transcriptional activity (Preston et al., 2010; Yan et al., 2014), cell cycle progression (Laviolette et al., 2013; Kenyon et al., 2016), regulation of rRNA synthesis (Gaur et al., 2013), and interaction with HSP90 as client with co-chaperones FNIP1/2 (Woodford et al., 2016). Understanding FLCN function is a research area under intense investigation in multiple laboratories and it is expected that additional FLCN interactors and pathways affected by FLCN will continue to emerge.
Figure 5.
Molecular pathways and cellular processes in which FLCN may have a functional role. Research supporting potential functions of FLCN in these pathways and processes is summarized in section 5.
6. Conclusions and future directions
As summarized in the preceding sections, a number of potential roles for FLCN in regulating important metabolic pathways and cellular processes have been proposed. Research that supports a role for FLCN in mTORC1 activation raises questions as to the rationale for a tumor suppressor to positively regulate a pathway that drives protein synthesis and cell growth and is upregulated in many cancers. Previous work had established that nuclear localization and activation of TFEB was negatively regulated by mTORC1-dependent phosphorylation of TFEB on serine 211 facilitating its interaction with 14-3-3 proteins and inhibiting its movement to the nucleus (Hager et al., 2010; Martina et al., 2012; Roczniak-Ferguson et al., 2012). As discussed above, FLCN has been shown to negatively regulate another MiTF family member TFE3. Loss of FLCN was associated with decreased phosphorylation of TFE3 and its nuclear localization and transcriptional activation (Hong et al., 2010a). Additionally, FLCN was shown to positively regulate mTORC1 by recruiting mTORC1 to the lysosome for activation through Rag GTPases in response to amino acids (Petit et al., 2013; Tsun et al., 2013). If TFE3, like TFEB, is negatively regulated by mTORC1 through phosphorylation on a critical serine as suggested by findings in Flcn-deficient mouse preadipocytes (Wada et al., 2016), then it is possible that loss of FLCN could result in TFE3 activation through loss of its negative regulation by mTOR. Loss of mTORC1 activation in FLCN-deficient cells and tissues would enable TFE3 to escape phosphorylation by mTORC1 and permit its nuclear localization where it becomes transcriptionally active. Enhanced TFE3 transcriptional activity as a result of somatic chromosome Xp11.2 translocations between TFE3 and a variety of fusion partners causes an aggressive type of kidney cancer (Argani, 2015) and could potentially contribute to BHD-associated renal tumorigenesis. Additional experiments will be necessary to test this hypothesis.
The primary role for FLCN in modulating cellular processes that regulate cell growth and become dysregulated in BHD syndrome, remains to be determined. Understanding FLCN function and the cellular pathways with which it interacts will inform the development of targeted therapies to treat BHD-associated kidney cancer, fibrofolliculomas and pulmonary manifestations. The mTOR inhibitors sirolimus and rapamycin were effective in reducing the number and size of renal tumors and cysts in Flcn-deficient mouse models and inhibiting Flcn-deficient mouse allograft tumor growth (Baba et al., 2008; Chen et al., 2008; Wu et al., 2015). Although therapies that target the mTOR pathway have not been successful in treating BHD-associated fibrofolliculomas (Gijezen et al., 2014), a limited survival advantage was observed with everolimus as a sixth-line systemic treatment in a BHD patient, when compared to the response of the patient to other tyrosine kinase inhibitors (Nakamura et al., 2013). Several cases have been reported in which BHD patients with metastatic RCC have been treated with systemic small molecule tyrosine kinase and/or mTOR inhibitors and have responded with stable disease (Benusiglio et al., 2014). The encouraging result with mTOR inhibitors in Flcn-deficient preclinical models was the basis for a phase 2 study of everolimus therapy in kidney cancer patients with Birt-Hogg-Dubé syndrome and sporadic chromophobe renal cancer (clinicaltrials.gov; NCT02504892). Elucidating the primary function of FLCN that leads to cutaneous, lung and renal manifestations when FLCN is mutated in the germline of BHD patients awaits exciting discoveries to be uncovered in future research efforts and will inform the development of effective targeted therapies for BHD patients.
Highlights.
Birt-Hogg-Dubé (BHD) syndrome is caused by germline mutations in the FLCN gene
BHD patients are predisposed to fibrofolliculomas, lung cysts and kidney cancer
Somatic “second hit” FLCN mutations drive BHD-associated kidney tumorigenesis
FLCN interacts with AMPK through FNIP1/FNIP2 and modulates mTOR signaling
FLCN is a GAP for RagC/D for amino acid dependent mTORC1 activation on lysosomes
Recent work highlights the role of FLCN in multiple cellular pathways and processes
Acknowledgments
This review and the corresponding Gene Wiki article are written as part of the GeneWiki Review series—a series resulting from a collaboration between the journal GENE and the Gene Wiki Initiative. The Gene Wiki Initiative is supported by the National Institutes of Health (GM089820). Additional support for Gene Wiki Reviews is provided by Elsevier, the publisher of GENE. This work was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This project has been funded in part with federal funds from the Frederick National Laboratory for Cancer Research, NIH, under Contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. The corresponding Gene Wiki entry for this review can be found here: http://en.wikipedia.org/wiki/Folliculin.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Argani P. MiT family translocation renal cell carcinoma. Semin Diagn Pathol. 2015;32:103–13. doi: 10.1053/j.semdp.2015.02.003. [DOI] [PubMed] [Google Scholar]
- Baba M, Furihata M, Hong SB, Tessarollo L, Haines DC, Southon E, Patel V, Igarashi P, Alvord WG, Leighty R, Yao M, Bernardo M, Ileva L, Choyke P, Warren MB, Zbar B, Linehan WM, Schmidt LS. Kidney-targeted Birt-Hogg-Dubé, gene inactivation in a mouse model: Erk1/2 and Akt-mTOR activation, cell hyperproliferation, and polycystic kidneys. Journal of the National Cancer Institute. 2008;100:140–154. doi: 10.1093/jnci/djm288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Hong SB, Sharma N, Warren MB, Nickerson ML, Iwamatsu A, Esposito D, Gillette WK, Hopkins RF, III, Hartley JL, Furihata M, Oishi S, Zhen W, Burke TR, Jr, Linehan WM, Schmidt LS, Zbar B. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci U S A. 2006;103:15552–15557. doi: 10.1073/pnas.0603781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Keller JR, Sun HW, Resch W, Kuchen S, Suh HC, Hasumi H, Hasumi Y, Kieffer-Kwon KR, Gonzalez CG, Hughes RM, Klein ME, Oh HF, Bible P, Southon E, Tessarollo L, Schmidt LS, Linehan WM, Casellas R. The folliculin-FNIP1 pathway deleted in human Birt-Hogg-Dubé syndrome is required for murine B-cell development. Blood. 2012;120:1254–1261. doi: 10.1182/blood-2012-02-410407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014;24:400–6. doi: 10.1016/j.tcb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastola P, Stratton Y, Kellner E, Mikhaylova O, Yi Y, Sartor MA, Medvedovic M, Biesiada J, Meller J, Czyzyk-Krzeska MF. Folliculin contributes to VHL tumor suppressing activity in renal cancer through regulation of autophagy. PLoS One. 2013;8:e70030. doi: 10.1371/journal.pone.0070030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhammou JN, Vocke CD, Santani A, Schmidt LS, Baba M, Seyama K, Wu X, Korolevich S, Nathanson KL, Stolle CA, Linehan WM. Identification of intragenic deletions and duplication in the FLCN gene in Birt-Hogg-Dubé syndrome. Genes Chromosomes Cancer. 2011;50:466–477. doi: 10.1002/gcc.20872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benusiglio PR, Giraud S, Deveaux S, Mejean A, Correas JM, Joly D, Timsit MO, Ferlicot S, Verkarre V, Abadie C, Chauveau D, Leroux D, Avril MF, Cordier JF, Richard S and French National Cancer Institute Inherited Predisposition to Kidney Cancer, N. Renal cell tumour characteristics in patients with the Birt-Hogg-Dubé cancer susceptibility syndrome: a retrospective, multicentre study. Orphanet J Rare Dis. 2014;9:163. doi: 10.1186/s13023-014-0163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J, Nichols J, Dietmann S, Corrin PD, Paddison PJ, Smith A. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell. 2013;153:335–47. doi: 10.1016/j.cell.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binet O, Robin J, Vicart M, Ventura G, Beltzer-Garelly E. Fibromes Perifolliculaires Polypose Colique Familaile Pneumothorax Spontanes Familiaux. Annales de Dermotologie et de Venereologie. 1986;113:928–930. [Google Scholar]
- Birt AR, Hogg GR, Dube WJ. Hereditary multiple fibrofolliculomas with trichodiscomas and acrochordons. Archives Dermatology. 1977;113:1674–1677. [PubMed] [Google Scholar]
- Boehlke C, Kotsis F, Patel V, Braeg S, Voelker H, Bredt S, Beyer T, Janusch H, Hamann C, Godel M, Muller K, Herbst M, Hornung M, Doerken M, Kottgen M, Nitschke R, Igarashi P, Walz G, Kuehn EW. Primary cilia regulate mTORC1 activity and cell size through Lkb1. Nat Cell Biol. 2010;12:1115–22. doi: 10.1038/ncb2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsdorff TB, Jansen JH, Lingaas F. Second hits in the FLCN gene in a hereditary renal cancer syndrome in dogs. Mamm Genome. 2008;19:121–6. doi: 10.1007/s00335-007-9088-3. [DOI] [PubMed] [Google Scholar]
- Cash TP, Gruber JJ, Hartman TR, Henske EP, Simon MC. Loss of the Birt-Hogg-Dubé tumor suppressor results in apoptotic resistance due to aberrant TGFbeta-mediated transcription. Oncogene. 2011;30:2534–46. doi: 10.1038/onc.2010.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Futami K, Petillo D, Peng J, Wang P, Knol J, Li Y, Khoo SK, Huang D, Qian CN, Zhao P, Dykema K, Zhang R, Cao B, Yang XJ, Furge K, Williams BO, Teh BT. Deficiency of FLCN in mouse kidney led to development of polycystic kidneys and renal neoplasia. PLoS ONE. 2008;3:e3581. doi: 10.1371/journal.pone.0003581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Huang D, Rubera I, Futami K, Wang P, Zickert P, Khoo SK, Dykema K, Zhao P, Petillo D, Cao B, Zhang Z, Si S, Schoen SR, Yang XJ, Zhou M, Xiao GQ, Wu G, Nordenskjold M, Tauc M, Williams BO, Furge KA, Teh BT. Disruption of tubular Flcn expression as a mouse model for renal tumor induction. Kidney Int. 2015;88:1057–69. doi: 10.1038/ki.2015.177. [DOI] [PubMed] [Google Scholar]
- Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:1845–6. doi: 10.1056/NEJMc1303158. [DOI] [PubMed] [Google Scholar]
- Chung JY, Ramos-Caro FA, Beers B, Ford MJ, Flowers F. Multiple lipomas, angiolipomas, and parathyroid adenomas in a patient with Birt-Hogg-Dubé syndrome. International Journal of Dermatology. 1996;35:365–367. doi: 10.1111/j.1365-4362.1996.tb03642.x. [DOI] [PubMed] [Google Scholar]
- da Silva NF, Gentle D, Hesson LB, Morton DG, Latif F, Maher ER. Analysis of the Birt-Hogg-Dubé (BHD) tumour suppressor gene in sporadic renal cell carcinoma and colorectal cancer. J Med Genet. 2003;40:820–824. doi: 10.1136/jmg.40.11.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CF, Ricketts CJ, Wang M, Yang L, Cherniack AD, Shen H, Buhay C, Kang H, Kim SC, Fahey CC, Hacker KE, Bhanot G, Gordenin DA, Chu A, Gunaratne PH, Biehl M, Seth S, Kaipparettu BA, Bristow CA, Donehower LA, Wallen EM, Smith AB, Tickoo SK, Tamboli P, Reuter V, Schmidt LS, Hsieh JJ, Choueiri TK, Hakimi AA, Chin L, Meyerson M, Kucherlapati R, Park WY, Robertson AG, Laird PW, Henske EP, Kwiatkowski DJ, Park PJ, Morgan M, Shuch B, Muzny D, Wheeler DA, Linehan WM, Gibbs RA, Rathmell WK, Creighton CJ. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26:319–330. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Zhu C, Zou W, Ma D, Min H, Chen B, Ye M, Pan Y, Cao L, Wan Y, Zhang W, Meng L, Mei Y, Yang C, Chen S, Gao Q, Yi L. FLCN intragenic deletions in Chinese familial primary spontaneous pneumothorax. Am J Med Genet A. 2015;167A:1125–33. doi: 10.1002/ajmg.a.36979. [DOI] [PubMed] [Google Scholar]
- Dunlop EA, Seifan S, Claessens T, Behrends C, Kamps MA, Rozycka E, Kemp AJ, Nookala RK, Blenis J, Coull BJ, Murray JT, van Steensel MA, Wilkinson S, Tee AR. FLCN, a novel autophagy component, interacts with GABARAP and is regulated by ULK1 phosphorylation. Autophagy. 2014;10:1749–60. doi: 10.4161/auto.29640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban MA, Harten SK, Tran MG, Maxwell PH. Formation of primary cilia in the renal epithelium is regulated by the von Hippel-Lindau tumor suppressor protein. J Am Soc Nephrol. 2006;17:1801–6. doi: 10.1681/ASN.2006020181. [DOI] [PubMed] [Google Scholar]
- Furuya M, Hong SB, Tanaka R, Kuroda N, Nagashima Y, Nagahama K, Suyama T, Yao M, Nakatani Y. Distinctive expression patterns of glycoprotein non-metastatic B and folliculin in renal tumors in patients with Birt-Hogg-Dubé syndrome. Cancer Sci. 2015;106:315–23. doi: 10.1111/cas.12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya M, Yao M, Tanaka R, Nagashima Y, Kuroda N, Hasumi H, Baba M, Matsushima J, Nomura F, Nakatani Y. Genetic, epidemiologic and clinicopathologic studies of Japanese Asian patients with Birt-Hogg-Dubé syndrome. Clin Genet. 2016;90:403–412. doi: 10.1111/cge.12807. [DOI] [PubMed] [Google Scholar]
- Gad S, Lefevre SH, Khoo SK, Giraud S, Vieillefond A, Vasiliu V, Ferlicot S, Molinie V, Denoux Y, Thiounn N, Chretien Y, Mejean A, Zerbib M, Benoit G, Herve JM, Allegre G, Bressac-de PB, Teh BT, Richard S. Mutations in BHD and TP53 genes, but not in HNF1beta gene, in a large series of sporadic chromophobe renal cell carcinoma. Br J Cancer. 2007;96:336–340. doi: 10.1038/sj.bjc.6603492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur K, Li J, Wang D, Dutta P, Yan SJ, Tsurumi A, Land H, Wu G, Li WX. The Birt-Hogg-Dubé tumor suppressor Folliculin negatively regulates ribosomal RNA synthesis. Hum Mol Genet. 2013;22:284–99. doi: 10.1093/hmg/dds428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijezen LM, Vernooij M, Martens H, Oduber CE, Henquet CJ, Starink TM, Prins MH, Menko FH, Nelemans PJ, van Steensel MA. Topical rapamycin as a treatment for fibrofolliculomas in Birt-Hogg-Dubé syndrome: a double-blind placebo-controlled randomized split-face trial. PLoS One. 2014;9:e99071. doi: 10.1371/journal.pone.0099071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncharova EA, Goncharov DA, James ML, Atochina-Vasserman EN, Stepanova V, Hong SB, Li H, Gonzales L, Baba M, Linehan WM, Gow AJ, Margulies S, Guttentag S, Schmidt LS, Krymskaya VP. Folliculin controls lung alveolar enlargement and epithelial cell survival through E-cadherin, LKB1, and AMPK. Cell Rep. 2014;7:412–23. doi: 10.1016/j.celrep.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N, Seyama K, McCormack FX. Pulmonary manifestations of Birt-Hogg-Dubé syndrome. Fam Cancer. 2013;12:387–96. doi: 10.1007/s10689-013-9660-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager M, Haufe H, Lusuardi L, Schmeller N, Kolbitsch C. p-AKT overexpression in primary renal cell carcinomas and their metastases. Clin Exp Metastasis. 2010;27:611–7. doi: 10.1007/s10585-010-9351-y. [DOI] [PubMed] [Google Scholar]
- Hartman TR, Liu D, Zilfou JT, Robb V, Morrison T, Watnick T, Henske EP. The tuberous sclerosis proteins regulate formation of the primary cilium via a rapamycin-insensitive and polycystin 1-independent pathway. Hum Mol Genet. 2009a;18:151–63. doi: 10.1093/hmg/ddn325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman TR, Nicolas E, Klein-Szanto A, Al-Saleem T, Cash TP, Simon MC, Henske EP. The role of the Birt-Hogg-Dubé protein in mTOR activation and renal tumorigenesis. Oncogene. 2009b;28:1594–604. doi: 10.1038/onc.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasumi H, Baba M, Hasumi Y, Huang Y, Oh HF, Hughes RM, Klein ME, Takikita S, Nagashima K, Schmidt LS, Linehan WM. Regulation of Mitochondrial Oxidative Metabolism by Tumor Suppressor FLCN Journal of the National Cancer Institute. 2012;104:1750–64. doi: 10.1093/jnci/djs418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasumi H, Baba M, Hasumi Y, Lang M, Huang Y, Oh HF, Matsuo M, Merino MJ, Yao M, Ito Y, Furuya M, Iribe Y, Kodama T, Southon E, Tessarollo L, Nagashima K, Haines DC, Linehan WM, Schmidt LS. Folliculin-interacting proteins Fnip1 and Fnip2 play critical roles in kidney tumor suppression in cooperation with Flcn. Proc Natl Acad Sci U S A. 2015;112:E1624–31. doi: 10.1073/pnas.1419502112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasumi H, Baba M, Hong SB, Hasumi Y, Huang Y, Yao M, Valera VA, Linehan WM, Schmidt LS. Identification and characterization of a novel folliculin-interacting protein FNIP2. Gene. 2008;415:60–67. doi: 10.1016/j.gene.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasumi H, Hasumi Y, Baba M, Nishi H, Furuya M, Vocke CD, Lang M, Irie N, Esumi C, Merino MJ, Kawahara T, Isono Y, Makiyama K, Warner AC, Haines DC, Wei MH, Zbar B, Hagenau H, Feigenbaum L, Kondo K, Nakaigawa N, Yao M, Metwalli AR, Marston Linehan W, Schmidt LS. H255Y and K508R missense mutations in tumour suppressor folliculin (FLCN) promote kidney cell proliferation. Hum Mol Genet. 2017;26:354–366. doi: 10.1093/hmg/ddw392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasumi Y, Baba M, Ajima R, Hasumi H, Valera VA, Klein ME, Haines DC, Merino MJ, Hong SB, Yamaguchi TP, Schmidt LS, Linehan WM. Homozygous loss of BHD causes early embryonic lethality and kidney tumor development with activation of mTORC1 and mTORC2. Proc Natl Acad Sci U S A. 2009;106:18722–18727. doi: 10.1073/pnas.0908853106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasumi Y, Baba M, Hasumi H, Huang Y, Lang M, Reindorf R, Oh HB, Sciarretta S, Nagashima K, Haines DC, Schneider MD, Adelstein RS, Schmidt LS, Sadoshima J, Marston Linehan W. Folliculin (Flcn) inactivation leads to murine cardiac hypertrophy through mTORC1 deregulation. Hum Mol Genet. 2014;23:5706–19. doi: 10.1093/hmg/ddu286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SB, Oh H, Valera VA, Baba M, Schmidt LS, Linehan WM. Inactivation of the FLCN tumor suppressor gene induces TFE3 transcriptional activity by increasing its nuclear localization. PLoS ONE. 2010a;5:e15793. doi: 10.1371/journal.pone.0015793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SB, Oh H, Valera VA, Stull J, Ngo DT, Baba M, Merino MJ, Linehan WM, Schmidt LS. Tumor suppressor FLCN inhibits tumorigenesis of a FLCN-null renal cancer cell line and regulates expression of key molecules in TGF-beta signaling. Molecular Cancer. 2010b;9:160. doi: 10.1186/1476-4598-9-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houweling AC, Gijezen LM, Jonker MA, van Doorn MB, Oldenburg RA, van Spaendonck-Zwarts KY, Leter EM, van Os TA, van Grieken NC, Jaspars EH, de Jong MM, Bongers EM, Johannesma PC, Postmus PE, van Moorselaar RJ, van Waesberghe JH, Starink TM, van Steensel MA, Gille JJ, Menko FH. Renal cancer and pneumothorax risk in Birt-Hogg-Dubé syndrome; an analysis of 115 FLCN mutation carriers from 35 BHD families. Br J Cancer. 2011;105:1912–9. doi: 10.1038/bjc.2011.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudon V, Sabourin S, Dydensborg AB, Kottis V, Ghazi A, Paquet M, Crosby K, Pomerleau V, Uetani N, Pause A. Renal tumour suppressor function of the Birt-Hogg-Dubé syndrome gene product folliculin. J Med Genet. 2010;47:182–9. doi: 10.1136/jmg.2009.072009. [DOI] [PubMed] [Google Scholar]
- Kenyon EJ, Luijten MN, Gill H, Li N, Rawlings M, Bull JC, Hadzhiev Y, van Steensel MA, Maher E, Mueller F. Expression and knockdown of zebrafish folliculin suggests requirement for embryonic brain morphogenesis. BMC Dev Biol. 2016;16:23. doi: 10.1186/s12861-016-0119-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khabibullin D, Medvetz DA, Pinilla M, Hariharan V, Li C, Hergrueter A, Laucho Contreras M, Zhang E, Parkhitko A, Yu JJ, Owen CA, Huang H, Baron RM, Henske EP. Folliculin regulates cell-cell adhesion, AMPK, and mTORC1 in a cell-type-specific manner in lung-derived cells. Physiol Rep. 2014;2:e12107. doi: 10.14814/phy2.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo SK, Bradley M, Wong FK, Hedblad MA, Nordenskjold M, Teh BT. Birt-Hogg-Dubé syndrome: mapping of a novel hereditary neoplasia gene to chromosome 17p12-q11.2. Oncogene. 2001;20:5239–5242. doi: 10.1038/sj.onc.1204703. [DOI] [PubMed] [Google Scholar]
- Khoo SK, Giraud S, Kahnoski K, Chen J, Motorna O, Nickolov R, Binet O, Lambert D, Friedel J, Levy R, Ferlicot S, Wolkenstein P, Hammel P, Bergerheim U, Hedblad MA, Bradley M, Teh BT, Nordenskjold M, Richard S. Clinical and genetic studies of Birt-Hogg-Dubé syndrome. J Med Genet. 2002;39:906–912. doi: 10.1136/jmg.39.12.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo SK, Kahnoski K, Sugimura J, Petillo D, Chen J, Shockley K, Ludlow J, Knapp R, Giraud S, Richard S, Nordenskjold M, Teh BT. Inactivation of BHD in sporadic renal tumors. Cancer Research. 2003;63:4583–4587. [PubMed] [Google Scholar]
- Kim S, Dynlacht BD. Assembling a primary cilium. Curr Opin Cell Biol. 2013;25:506–11. doi: 10.1016/j.ceb.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klomp JA, Petillo D, Niemi NM, Dykema KJ, Chen J, Yang XJ, Saaf A, Zickert P, Aly M, Bergerheim U, Nordenskjold M, Gad S, Giraud S, Denoux Y, Yonneau L, Mejean A, Vasiliu V, Richard S, MacKeigan JP, Teh BT, Furge KA. Birt-Hogg-Dubé renal tumors are genetically distinct from other renal neoplasias and are associated with up-regulation of mitochondrial gene expression. BMC Med Genomics. 2010;3:59. doi: 10.1186/1755-8794-3-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluger N, Giraud S, Coupier I, Avril MF, Dereure O, Guillot B, Richard S, Bessis D. Birt-Hogg-Dubé syndrome: clinical and genetic studies of 10 French families. Br J Dermatol. 2010;162:527–37. doi: 10.1111/j.1365-2133.2009.09517.x. [DOI] [PubMed] [Google Scholar]
- Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori K, Takagi Y, Sanada M, Lim TH, Nakatsu Y, Tsuzuki T, Sekiguchi M, Hidaka M. A novel protein, MAPO1, that functions in apoptosis triggered by O6-methylguanine mispair in DNA. Oncogene. 2009;28:1142–1150. doi: 10.1038/onc.2008.462. [DOI] [PubMed] [Google Scholar]
- Kunogi M, Kurihara M, Ikegami TS, Kobayashi T, Shindo N, Kumasaka T, Gunji Y, Kikkawa M, Iwakami S, Hino O, Takahashi K, Seyama K. Clinical and genetic spectrum of Birt-Hogg-Dubé syndrome patients in whom pneumothorax and/or multiple lung cysts are the presenting feature. J Med Genet. 2010;47:281–287. doi: 10.1136/jmg.2009.070565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Schroth J, Gleeson JG. Subcellular spatial regulation of canonical Wnt signalling at the primary cilium. Nat Cell Biol. 2011;13:700–7. doi: 10.1038/ncb2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laviolette LA, Mermoud J, Calvo IA, Olson N, Boukhali M, Steinlein OK, Roider E, Sattler EC, Huang D, Teh BT, Motamedi M, Haas W, Iliopoulos O. Negative regulation of EGFR signalling by the human folliculin tumour suppressor protein. Nat Commun. 2017;8:15866. doi: 10.1038/ncomms15866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laviolette LA, Wilson J, Koller J, Neil C, Hulick P, Rejtar T, Karger B, Teh BT, Iliopoulos O. Human folliculin delays cell cycle progression through late S and G2/M-phases: effect of phosphorylation and tumor associated mutations. PLoS One. 2013;8:e66775. doi: 10.1371/journal.pone.0066775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leter EM, Koopmans AK, Gille JJ, van Os TA, Vittoz GG, David EF, Jaspars EH, Postmus PE, van Moorselaar RJ, Craanen ME, Starink TM, Menko FH. Birt-Hogg-Dubé syndrome: clinical and genetic studies of 20 families. J Invest Dermatol. 2008;128:45–9. doi: 10.1038/sj.jid.5700959. [DOI] [PubMed] [Google Scholar]
- Lim DH, Rehal PK, Nahorski MS, Macdonald F, Claessens T, Van Geel M, Gijezen L, Gille JJ, Giraud S, Richard S, van Steensel M, Menko FH, Maher ER. A new locus-specific database (LSDB) for mutations in the folliculin (FLCN) gene. Hum Mutat. 2010;31:E1043–51. doi: 10.1002/humu.21130. [DOI] [PubMed] [Google Scholar]
- Lim TH, Fujikane R, Sano S, Sakagami R, Nakatsu Y, Tsuzuki T, Sekiguchi M, Hidaka M. Activation of AMP-activated protein kinase by MAPO1 and FLCN induces apoptosis triggered by alkylated base mismatch in DNA. DNA Repair (Amst) 2012;11:259–266. doi: 10.1016/j.dnarep.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Lingaas F, Comstock KE, Kirkness EF, Sorensen A, Aarskaug T, Hitte C, Nickerson ML, Moe L, Schmidt LS, Thomas R, Breen M, Galibert F, Zbar B, Ostrander EA. A mutation in the canine BHD gene is associated with hereditary multifocal renal cystadenocarcinoma and nodular dermatofibrosis in the German Shepherd dog. Human Molecular Genetics. 2003;12:3043–3053. doi: 10.1093/hmg/ddg336. [DOI] [PubMed] [Google Scholar]
- Liu W, Chen Z, Ma Y, Wu X, Jin Y, Hou S. Genetic characterization of the Drosophila birt-hogg-dubé syndrome gene. PLoS One. 2013;8:e65869. doi: 10.1371/journal.pone.0065869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijten MN, Basten SG, Claessens T, Vernooij M, Scott CL, Janssen R, Easton JA, Kamps MA, Vreeburg M, Broers JL, van Geel M, Menko FH, Harbottle RP, Nookala RK, Tee AR, Land SC, Giles RH, Coull BJ, van Steensel MA. Birt-Hogg-Dubé syndrome is a novel ciliopathy. Hum Mol Genet. 2013;22:4383–97. doi: 10.1093/hmg/ddt288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Belmonte F, Perez-Moreno M. Epithelial cell polarity, stem cells and cancer. Nat Rev Cancer. 2011;12:23–38. doi: 10.1038/nrc3169. [DOI] [PubMed] [Google Scholar]
- Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–14. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvetz DA, Khabibullin D, Hariharan V, Ongusaha PP, Goncharova EA, Schlechter T, Darling TN, Hofmann I, Krymskaya VP, Liao JK, Huang H, Henske EP. Folliculin, the product of the Birt-Hogg-Dubé tumor suppressor gene, interacts with the adherens junction protein p0071 to regulate cell-cell adhesion. PLoS One. 2012;7:e47842. doi: 10.1371/journal.pone.0047842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima K, Fukushima H, Shimizu K, Yamada A, Hidaka M, Hasumi H, Ikebe T, Fukumoto S, Okabe K, Inuzuka H. Nutrient-induced FNIP degradation by SCFbeta-TRCP regulates FLCN complex localization and promotes renal cancer progression. Oncotarget. 2017;8:9947–9960. doi: 10.18632/oncotarget.14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Zoubakov D, Stupar Z, Kovacs G. Lack of mutation of the folliculin gene in sporadic chromophobe renal cell carcinoma and renal oncocytoma. Int J Cancer. 2004;109:472–475. doi: 10.1002/ijc.11694. [DOI] [PubMed] [Google Scholar]
- Nahorski MS, Lim DH, Martin L, Gille JJ, McKay K, Rehal PK, Ploeger HM, van SM, Tomlinson IP, Latif F, Menko FH, Maher ER. Investigation of the Birt-Hogg-Dubé tumour suppressor gene (FLCN) in familial and sporadic colorectal cancer. J Med Genet. 2010;47:385–390. doi: 10.1136/jmg.2009.073304. [DOI] [PubMed] [Google Scholar]
- Nahorski MS, Reiman A, Lim DH, Nookala RK, Seabra L, Lu X, Fenton J, Boora U, Nordenskjold M, Latif F, Hurst LD, Maher ER. Birt Hogg-Dube syndrome-associated FLCN mutations disrupt protein stability. Hum Mutat. 2011;32:921–9. doi: 10.1002/humu.21519. [DOI] [PubMed] [Google Scholar]
- Nahorski MS, Seabra L, Straatman-Iwanowska A, Wingenfeld A, Reiman A, Lu X, Klomp JA, Teh BT, Hatzfeld M, Gissen P, Maher ER. Folliculin interacts with p0071 (plakophilin-4) and deficiency is associated with disordered RhoA signalling, epithelial polarization and cytokinesis. Hum Mol Genet. 2012;21:5268–79. doi: 10.1093/hmg/dds378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Yao M, Sano F, Sakata R, Tatenuma T, Makiyama K, Nakaigawa N, Kubota Y. [A case of metastatic renal cell carcinoma associated with Birt-Hogg-Dubé syndrome treated with molecular-targeting agents] Hinyokika Kiyo. 2013;59:503–6. [PubMed] [Google Scholar]
- Nickerson ML, Warren MB, Toro JR, Matrosova V, Glenn GM, Turner ML, Duray P, Merino MJ, Choyke P, Pavlovich CP, Sharma N, Walther MM, Munroe D, Hill R, Maher E, Greenberg C, Lerman MI, Linehan WM, Zbar B, Schmidt LS. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dubé syndrome. Cancer Cell. 2002;2:157–164. doi: 10.1016/s1535-6108(02)00104-6. [DOI] [PubMed] [Google Scholar]
- Nookala RK, Langemeyer L, Pacitto A, Ochoa-Montano B, Donaldson JC, Blaszczyk BK, Chirgadze DY, Barr FA, Bazan JF, Blundell TL. Crystal structure of folliculin reveals a hidDENN function in genetically inherited renal cancer. Open Biol. 2012;2:120071. doi: 10.1098/rsob.120071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okimoto K, Sakurai J, Kobayashi T, Mitani H, Hirayama Y, Nickerson ML, Warren MB, Zbar B, Schmidt LS, Hino O. A germ-line insertion in the Birt-Hogg-Dubé (BHD) gene gives rise to the Nihon rat model of inherited renal cancer. Proc Natl Acad Sci U S A. 2004;101:2023–2027. doi: 10.1073/pnas.0308071100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacitto A, Ascher DB, Wong LH, Blaszczyk BK, Nookala RK, Zhang N, Dokudovskaya S, Levine TP, Blundell TL. Lst4, the yeast Fnip1/2 orthologue, is a DENN-family protein. Open Biol. 2015;5:150174. doi: 10.1098/rsob.150174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Staehling K, Tsang M, Appleby MW, Brunkow ME, Margineantu D, Hockenbery DM, Habib T, Liggitt HD, Carlson G, Iritani BM. Disruption of Fnip1 reveals a metabolic checkpoint controlling B lymphocyte development. Immunity. 2012;36:769–81. doi: 10.1016/j.immuni.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovich CP, Walther MM, Eyler RA, Hewitt SM, Zbar B, Linehan WM, Merino MJ. Renal tumors in the Birt-Hogg-Dubé, syndrome. Am J Surg Pathol. 2002;26:1542–1552. doi: 10.1097/00000478-200212000-00002. [DOI] [PubMed] [Google Scholar]
- Peli-Gulli MP, Sardu A, Panchaud N, Raucci S, De Virgilio C. Amino Acids Stimulate TORC1 through Lst4-Lst7, a GTPase-Activating Protein Complex for the Rag Family GTPase Gtr2. Cell Rep. 2015;13:1–7. doi: 10.1016/j.celrep.2015.08.059. [DOI] [PubMed] [Google Scholar]
- Petit CS, Roczniak-Ferguson A, Ferguson SM. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol. 2013;202:1107–22. doi: 10.1083/jcb.201307084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possik E, Jalali Z, Nouet Y, Yan M, Gingras MC, Schmeisser K, Panaite L, Dupuy F, Kharitidi D, Chotard L, Jones RG, Hall DH, Pause A. Folliculin regulates ampk-dependent autophagy and metabolic stress survival. PLoS Genet. 2014;10:e1004273. doi: 10.1371/journal.pgen.1004273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston RS, Philp A, Claessens T, Gijezen L, Dydensborg AB, Dunlop EA, Harper KT, Brinkhuizen T, Menko FH, Davies DM, Land SC, Pause A, Baar K, van Steensel MA, Tee AR. Absence of the Birt-Hogg-Dubé gene product is associated with increased hypoxia-inducible factor transcriptional activity and a loss of metabolic flexibility. Oncogene. 2010;30:1159–1173. doi: 10.1038/onc.2010.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raben N, Puertollano R. TFEB and TFE3: Linking Lysosomes to Cellular Adaptation to Stress. Annu Rev Cell Dev Biol. 2016;32:255–278. doi: 10.1146/annurev-cellbio-111315-125407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes NL, Banks GB, Tsang M, Margineantu D, Gu H, Djukovic D, Chan J, Torres M, Liggitt HD, Hirenallur SD, Hockenbery DM, Raftery D, Iritani BM. Fnip1 regulates skeletal muscle fiber type specification, fatigue resistance, and susceptibility to muscular dystrophy. Proc Natl Acad Sci U S A. 2015;112:424–9. doi: 10.1073/pnas.1413021112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberg KJ, Bickel S, Rowley N, Kaiser CA. Control of amino acid permease sorting in the late secretory pathway of Saccharomyces cerevisiae by SEC13, LST4, LST7 and LST8. Genetics. 1997;147:1569–1584. doi: 10.1093/genetics/147.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The Transcription Factor TFEB Links mTORC1 Signaling to Transcriptional Control of Lysosome Homeostasis. Sci Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossing M, Albrechtsen A, Skytte AB, Jensen UB, Ousager LB, Gerdes AM, Nielsen FC, Hansen TV. Genetic screening of the FLCN gene identify six novel variants and a Danish founder mutation. J Hum Genet. 2017;62:151–157. doi: 10.1038/jhg.2016.118. [DOI] [PubMed] [Google Scholar]
- Roth JS, Rabinowitz AD, Benson M, Grossman ME. Bilateral renal cell carcinoma in the Birt-Hogg-Dubé syndrome. J Am Acad Dermatol. 1993;29:1055–1056. doi: 10.1016/s0190-9622(08)82049-x. [DOI] [PubMed] [Google Scholar]
- Rubio-Texeira M, Kaiser CA. Amino acids regulate retrieval of the yeast general amino acid permease from the vacuolar targeting pathway. Mol Biol Cell. 2006;17:3031–50. doi: 10.1091/mbc.E05-07-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt LS, Linehan WM. Molecular genetics and clinical features of Birt-Hogg-Dubé syndrome. Nat Rev Urol. 2015;12:558–69. doi: 10.1038/nrurol.2015.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt LS, Nickerson ML, Warren MB, Glenn GM, Toro JR, Merino MJ, Turner ML, Choyke PL, Sharma N, Peterson J, Morrison P, Maher ER, Walther MM, Zbar B, Linehan WM. Germline BHD-mutation spectrum and phenotype analysis of a large cohort of families with Birt-Hogg-Dubé, syndrome. American Journal of Human Genetics. 2005;76:1023–1033. doi: 10.1086/430842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt LS, Warren MB, Nickerson ML, Weirich G, Matrosova V, Toro JR, Turner ML, Duray P, Merino MJ, Hewitt S, Pavlovich CP, Glenn GM, Greenberg CR, Linehan WM, Zbar B. Birt-Hogg-Dubé syndrome, a genodermatosis associated with spontaneous pneumothorax and kidney neoplasia, maps to chromosome 17p11.2. American Journal of Human Genetics. 2001;69:876–882. doi: 10.1086/323744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–75. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggs OM, Stockenhuber A, Deobagkar-Lele M, Bull KR, Crockford TL, Kingston BL, Crawford G, Anzilotti C, Steeples V, Ghaffari S, Czibik G, Bellahcene M, Watkins H, Ashrafian H, Davies B, Woods A, Carling D, Yavari A, Beutler B, Cornall RJ. Mutation of Fnip1 is associated with B-cell deficiency, cardiomyopathy, and elevated AMPK activity. Proc Natl Acad Sci U S A. 2016;113:E3706–15. doi: 10.1073/pnas.1607592113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SR, Zhen W, Zheng Z, Wang H, Oh SW, Liu W, Zbar B, Schmidt LS, Hou SX. The Drosophila homolog of the human tumor suppressor gene BHD interacts with the JAK-STAT and Dpp signaling pathways in regulating male germline stem cell maintenance. Oncogene. 2006;25:5933–5941. doi: 10.1038/sj.onc.1209593. [DOI] [PubMed] [Google Scholar]
- Starling GP, Yip YY, Sanger A, Morton PE, Eden ER, Dodding MP. Folliculin directs the formation of a Rab34-RILP complex to control the nutrient-dependent dynamic distribution of lysosomes. EMBO Rep. 2016;17:823–41. doi: 10.15252/embr.201541382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi Y, Kobayashi T, Shiono M, Wang L, Piao X, Sun G, Zhang D, Abe M, Hagiwara Y, Takahashi K, Hino O. Interaction of folliculin (Birt-Hogg-Dubé gene product) with a novel Fnip1-like (FnipL/Fnip2) protein. Oncogene. 2008 doi: 10.1038/onc.2008.261. [DOI] [PubMed] [Google Scholar]
- Togashi Y, Kobayashi T, Momose S, Ueda M, Okimoto K, Hino O. Transgenic rescue from embryonic lethality and renal carcinogenesis in the Nihon rat model by introduction of a wild-type Bhd gene. Oncogene. 2006;25:2885–9. doi: 10.1038/sj.onc.1209317. [DOI] [PubMed] [Google Scholar]
- Toro JR, Glenn G, Duray P, Darling T, Weirich G, Zbar B, Linehan M, Turner ML. Birt-Hogg-Dubé syndrome: a novel marker of kidney neoplasia. Archives Dermatology. 1999;135:1195–1202. doi: 10.1001/archderm.135.10.1195. [DOI] [PubMed] [Google Scholar]
- Toro JR, Pautler SE, Stewart L, Glenn GM, Weinreich M, Toure O, Wei MH, Schmidt LS, Davis L, Zbar B, Choyke P, Steinberg SM, Nguyen DM, Linehan WM. Lung Cysts, Spontaneous Pneumothrorax and Genetic Associations in 89 Families with Birt-Hogg-Dubé, Syndrome. American Journal of Respiratory and Critical Care Medicine. 2007;175:1044–1053. doi: 10.1164/rccm.200610-1483OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro JR, Wei MH, Glenn GM, Weinreich M, Toure O, Vocke CD, Turner M, Choyke P, Merino MJ, Pinto PA, Steinberg SM, Schmidt LS, Linehan WM. BHD mutations, clinical and molecular genetic investigations of Birt-Hogg-Dubé syndrome: a new series of 50 families and a review of published reports. J Med Genet. 2008;45:321–331. doi: 10.1136/jmg.2007.054304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Slegtenhorst M, Khabibullin D, Hartman TR, Nicolas E, Kruger WD, Henske EP. The Birt-Hogg-Dubé and tuberous sclerosis complex homologs have opposing roles in amino acid homeostasis in Schizosaccharomyces pombe. J Biol Chem. 2007;282:24583–24590. doi: 10.1074/jbc.M700857200. [DOI] [PubMed] [Google Scholar]
- Vocke CD, Yang Y, Pavlovich CP, Schmidt LS, Nickerson ML, Torres-Cabala CA, Merino MJ, Walther MM, Zbar B, Linehan WM. High Frequency of Somatic Frameshift BHD Gene Mutations in Birt-Hogg-Dubé-Associated Renal Tumors. Journal of the National Cancer Institute. 2005;97:931–935. doi: 10.1093/jnci/dji154. [DOI] [PubMed] [Google Scholar]
- Wada S, Neinast M, Jang C, Ibrahim YH, Lee G, Babu A, Li J, Hoshino A, Rowe GC, Rhee J, Martina JA, Puertollano R, Blenis J, Morley M, Baur JA, Seale P, Arany Z. The tumor suppressor FLCN mediates an alternate mTOR pathway to regulate browning of adipose tissue. Genes Dev. 2016;30:2551–2564. doi: 10.1101/gad.287953.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Kobayashi T, Piao X, Shiono M, Takagi Y, Mineki R, Taka H, Zhang D, Abe M, Sun G, Hagiwara Y, Okimoto K, Matsumoto I, Kouchi M, Hino O. Serine 62 is a phosphorylation site in folliculin, the Birt-Hogg-Dubé gene product. FEBS Lett. 2010;584:39–43. doi: 10.1016/j.febslet.2009.11.033. [DOI] [PubMed] [Google Scholar]
- Warren MB, Torres-Cabala CA, Turner ML, Merino MJ, Matrosova VY, Nickerson ML, Ma W, Linehan WM, Zbar B, Schmidt LS. Expression of Birt-Hogg-Dubé gene mRNA in normal and neoplastic human tissues. Mod Pathol. 2004;17:998–1011. doi: 10.1038/modpathol.3800152. [DOI] [PubMed] [Google Scholar]
- Weirich G, Glenn GM, Junker K, Merino MJ, Storkel S, Lubensky I, Choyke P, Pack S, Amin M, Walther MM, Linehan WM, Zbar B. Familial renal oncocytoma: clinicopathological study of 5 families. Journal of Urology. 1998;160:335–340. doi: 10.1016/s0022-5347(01)62888-x. [DOI] [PubMed] [Google Scholar]
- Woodford MR, Dunn DM, Blanden AR, Capriotti D, Loiselle D, Prodromou C, Panaretou B, Hughes PF, Smith A, Ackerman W, Haystead TA, Loh SN, Bourboulia D, Schmidt LS, Marston Linehan W, Bratslavsky G, Mollapour M. The FNIP co-chaperones decelerate the Hsp90 chaperone cycle and enhance drug binding. Nat Commun. 2016;7:12037. doi: 10.1038/ncomms12037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Si S, Li Y, Schoen S, Xiao GQ, Li X, Teh BT, Wu G, Chen J. Flcn-deficient renal cells are tumorigenic and sensitive to mTOR suppression. Oncotarget. 2015;6:32761–73. doi: 10.18632/oncotarget.5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Bradley MJ, Cai Y, Kummel D, De La Cruz EM, Barr FA, Reinisch KM. Insights regarding guanine nucleotide exchange from the structure of a DENN-domain protein complexed with its Rab GTPase substrate. Proc Natl Acad Sci U S A. 2011;108:18672–18677. doi: 10.1073/pnas.1110415108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Audet-Walsh E, Manteghi S, Dufour CR, Walker B, Baba M, St-Pierre J, Giguere V, Pause A. Chronic AMPK activation via loss of FLCN induces functional beige adipose tissue through PGC-1alpha/ERRalpha. Genes Dev. 2016;30:1034–46. doi: 10.1101/gad.281410.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Gingras MC, Dunlop EA, Nouet Y, Dupuy F, Jalali Z, Possik E, Coull BJ, Kharitidi D, Dydensborg AB, Faubert B, Kamps M, Sabourin S, Preston RS, Davies DM, Roughead T, Chotard L, van Steensel MA, Jones R, Tee AR, Pause A. The tumor suppressor folliculin regulates AMPK-dependent metabolic transformation. J Clin Invest. 2014;124:2640–50. doi: 10.1172/JCI71749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Padilla-Nash HM, Vira MA, bu-Asab MS, Val D, Worrell R, Tsokos M, Merino MJ, Pavlovich CP, Ried T, Linehan WM, Vocke CD. The UOK 257 cell line: a novel model for studies of the human Birt-Hogg-Dubé gene pathway. Cancer Genetics and Cytogenetics. 2008;180:100–109. doi: 10.1016/j.cancergencyto.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S, Gerondopoulos A, Linford A, Rigden DJ, Barr FA. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J Cell Biol. 2010;191:367–81. doi: 10.1083/jcb.201008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbar B, Alvord WG, Glenn GM, Turner M, Pavlovich CP, Schmidt LS, Walther MM, Choyke P, Weirich G, Hewitt SM, Duray P, Gabril F, Greenberg C, Merino MJ, Toro J, Linehan WM. Risk of renal and colonic neoplasms and spontaneous pneumothorax in the Birt-Hogg-Dubé syndrome. Cancer Epidemiology, Biomarkers and Prevention. 2002;11:393–400. [PubMed] [Google Scholar]
- Zhang D, Iyer LM, He F, Aravind L. Discovery of Novel DENN Proteins: Implications for the Evolution of Eukaryotic Intracellular Membrane Structures and Human Disease. Front Genet. 2012;3:283. doi: 10.3389/fgene.2012.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M, Zhao X, Li J, Yuan W, Yan G, Tong M, Guo S, Zhu Y, Jiang Y, Liu Y, Jiang Y. Tumor Suppressor Folliculin Regulates mTORC1 through Primary Cilia. J Biol Chem. 2016;291:11689–97. doi: 10.1074/jbc.M116.719997. [DOI] [PMC free article] [PubMed] [Google Scholar]