Abstract
An increasing number of variants of unknown significance (VUS) are being identified in leukemia patients with the application of deep sequencing and these include CSF3R cytoplasmic mutations. Previous studies have demonstrated oncogenic potential of certain CSF3R truncation mutations prior to internalization motifs. However, the oncogenic potential of truncating the more distal region of CSF3R cytoplasmic domain as well as cytoplasmic missense mutations remains uncharacterized. Here we identified that CSF3R distal cytoplasmic truncation mutations (Q793–Q823) also harbored leukemogenic potential. Mechanistically, these distal cytoplasmic truncation mutations demonstrated markedly decreased receptor degradation, probably due to loss of the de-phosphorylation domain (residues N818–F836). Furthermore, all truncations prior to Q823 demonstrated increased expression of the higher molecular weight CSF3R band, which is shown to be essential for the receptor surface expression and the oncogenic potential. We further demonstrated that sufficient STAT5 activation is essential for oncogenic potential. In addition, CSF3R K704A demonstrated transforming capacity due to interruption of receptor ubiquitination and degradation. In summary, we have expanded the region of the CSF3R cytoplasmic domain in which truncation or missense mutations exhibit leukemogenic capacity, which will be useful for evaluating the relevance of CSF3R mutations in patients and helpful in defining targeted therapy strategies.
Introduction
G-CSF and its receptor CSF3R play important roles in the regulation of hematopoiesis.(1) Wild type human CSF3R consists of an extracellular domain (amino acid positions 25–627), a transmembrane domain (amino acid positions 628–650) and a cytoplasmic domain (amino acid positions 651–836). Typical of the class I cytokine receptor family, CSF3R lacks intrinsic kinase activity and is coupled with cytoplasmic tyrosine kinases, including JAK/STAT, MEK/MAPK and PI3K3/AKT.(2),(3) The 4 conserved tyrosine residues Y727, Y752, Y767 and Y787 within the cytoplasmic domain provide docking sites for these signalling molecules. The signal termination of CSF3R is associated with membrane receptor endocytosis and degradation.(4) Briefly, CSF3R is internalized slowly and spontaneously or rapidly with G-CSF stimulation via the internalization motifs (amino acids 772–778 and 779–792, in particular the dileucine residues L776/777 and S772 inside the internalization motifs) followed by endosomal sorting and lysosomal degradation.(5) Ubiquitination of cytoplasmic lysine (K) residues, in particular K655 in the juxtamembrane domain is crucial for lysosomal routing and signal attenuation.(6)
Recently, an increasing number of studies have uncovered pathogenic roles of CSF3R variants in a variety of hematologic malignancies. These CSF3R variants are located in different regions of CSF3R. CSF3R cytoplasmic mutations include truncation (frameshift and nonsense) and missense mutations. CSF3R cytoplasmic truncation mutations (e.g. CSF3R T738X, previously known as d715) were first identified in severe congenital neutropenia (SCN) patients with high frequency.(7),(8) These mutations truncate 82–98 amino acids from the CSF3R c-terminal domain, resulting in the lack of 2 or 3 tyrosine residues and internalization motifs, and in this capacity the truncations contribute to leukemic transformation.(3,9) CSF3R isoform IV, represents a naturally occurring splice variant of CSF3R. It resembles the truncation mutant versions of CSF3R in that isoform IV replaces the carboxyl terminal 87 amino acids with a novel 34 amino acids peptide and lacks 3 of the 4 C-terminal tyrosine residues as well as the internalization motifs. CSF3R isoform IV has been shown to induce sustained STAT5 activation and was implicated in contributing to disease relapse in childhood acute myeloid leukemia (AML) patients treated with G-CSF.(10–12) Recently, a group of CSF3R truncation mutations were shown to be prevalent in chronic neutrophilic leukemia (CNL) and atypical chronic myeloid leukemia (aCML) patients.(13,14) Similar to CSF3R T738X, some of these truncating mutations (e.g. CSF3R D771fs and S783fs) were shown to cause receptor overexpression and hypersensitivity to G-CSF.(15) More recently, several CSF3R truncation mutations in the more distal region (e.g. F819fs) of the cytoplasmic domain are identified in AML patients.(16) However, the leukemogenic potential of these mutations has not yet been studied, therefore is one aim of this study.
In addition, CSF3R cytoplasmic missense mutations have been identified in pediatric AML patients.(14) However, the frequencies of CSF3R missense mutations in other hematologic malignancies and their pathogenic potential have not yet been characterized and are, therefore, another aim of the current study. Moreover, K704 and K705 have been predicted to be ubiquitination sites (http://www.ubpred.org/) for CSF3R in addition to K655 and K785, which have been characterized by previous studies. (4),(6) We, therefore, also investigated the leukemogenic potential of K704A and K705A.
Materials and Methods
Patient information and study approval
The study was approved by the Institutional Review Boards (IRB) from University of Texas Southwestern Medical Center, University of Colorado, Stanford University, Washington University in St. Louis, and Oregon health & Science University. Written informed consents were obtained from all the patients with IRB-approved protocols.
Deep Sequencing, TOPO TA cloning and Sanger confirmation
Exome sequencing was performed on a HiSeq 2500 using Illumina Nextera capture probes and paired end 100 cycle protocols. For RNA-Seq, libraries were constructed using the SureSelect stranded RNA-seq protocol (Agilent) on the Bravo robot (Agilent) and sequenced on the HiSeq 2500 using a 100 cycle paired end protocol. More information is provided in Supplementary data. TOPO TA cloning was performed with TOPO TA cloning kit (Thermo Scientific) according to the manufacture’s protocol. Sanger sequencing was performed as previously described.(13)
Cell lines and cell culture
HEK293T/17 and NIH/3T3 cells were maintained in DMEM (Invitrogen) supplemented with 10% FBS (Atlanta Biologicals), L-glutamine, penicillin/streptomycin (Invitrogen), and fungizone (Thermo Scientific). Ba/F3 cells were maintained in RPMI 1640 (Invitrogen) supplemented with 10% FBS, 15% WEHI-conditioned media, L-glutamine, penicillin/streptomycin and fungizone. Mycoplasma contamination was tested every other month. Only mycoplasma free cells were used in the experiments.
Transfection and transduction
CSF3R mutations were generated using the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies) as previously described(13,15). Briefly, retrovirus was produced by transfecting HEK293T/17 cells with FuGENE (Promega). After 2 days, the viral vector-containing supernatants were filtered, and infected to Ba/F3 cells followed by flow cytometry (FACS, AriaII, BD Biosciences) sorting of GFP positive cells. Of note, cells with equal intensity of GFP expression were sorted across all samples. For the co-immunoprecipitation (co-IP) assay, an HA-tagged ubiquitin plasmid (#17608, Addgene) was co-transfected with a V5 tagged pcDNACSF3R vector (#12290010, Thermo Scientific).
IL-3 withdrawal assay
Stably transduced Ba/F3 cells were washed 3 times and grown in cytokine-free media. Viable cell number was determined on a Guava Personal Cell Analysis System (Millipore).
Colony forming unit (CFU) assay
Bone marrow (BM) cells were infected with retrovirus expressing CSF3R variants, followed by FACS sorting. Seven thousand cells per well were seeded into a 6-well plate with 1.1mL of Methocult M3534 methylcellulose medium (StemCell Technologies) in the absence or presence of 0.5ng/ml G-CSF for 7–10 days. Colonies (>50 cells) were counted and images were taken via STEMvision™ colony counting software (StemCell Technologies).
FACS and phosphoflow
Cells were analyzed for surface expression of CSF3R via FACS with an anti-CD114 antibody (#346108, BioLegend). Intracellular signaling pathways were evaluated using phosphoflow technology. More information is provided in the supplementary data.
Immunoblotting
Immunoblotting was performed as previously described.(15) Detailed information is provided in supplementary data.
Receptor internalization
Membrane receptor internalization were quantified using FACS as previously described.(5) Briefly, for spontaneous internalization, cells were treated with cycloheximide (CHX, 100mg/ml) for 6 hour (h), washed with PBS+1%FBS+0.1%NaN3 and analyzed by FACS. To quantify G-CSF induced internalization, cells were stimulated with 10ng/ml G-CSF for 60 minutes (min), followed by FACS analysis of surface CSF3R expression. Total receptor internalization was quantified with immunoblotting after CHX or G-CSF treatment.
Glycosylation analysis
Cell lysates were treated with O-glycosidase and neuraminidase to remove O-linked oligosaccharides or PNGase F to remove N-linked oligosaccharides according to the manufacturer’s protocols (New England Biolabs) and quantified with immunoblotting as previously described(15).
Immunofluorescence Staining
Cells were grown on glass slides, fixed with 4% paraformaldehyde for 5min, rinsed with PBS and immunolabeled with anti-CD114 antibody (1:100) for 1h at room temperature. Slides were than washed with PBS, air dried, and mounted. All immunofluorescence staining images were captured with a confocal microscope (ApoTome.2, Zeiss).
Proliferation assay
Cells were labeled with 5 μM cell proliferation dye (CPD) eFluor 670 (#65-0840, eBiosciences) and cultured with or without brefeldin A (BFA, 100ng/ml, B7651, Sigma) for 2 days and analyzed via FACS.
Cell viability assay
Transformed Ba/F3 cells were seeded in 96-well plates (5000 cells per well) and exposed to increasing concentrations of ruxolitinib (Jakafi, Incyte), BFA or AKT Inhibitor X (CAS 925681-41-0) for 3 days. Cell relative number was assessed using a methanethiosulfonate (MTS)-based assay (CellTiter96 Aqueous One Solution; Promega). Cell viability was determined by comparing the absorbance of drug-treated cells to that of untreated controls set at 100%. IC50 values were calculated by regression curve fit analysis using GraphPad Prism software.
Statistical analysis
Statistical analyses were performed on GraphPad Prism software. The data were expressed as the mean ± standard error of mean (MED). Statistical significance was determined using Student’s two-tailed t tests comparing each condition to the respective CSF3R WT and expressed as p-values (*p<0.05, **p<0.01, and ***p<0.001). For BFA inhibition assay, one way ANOVA was used.
Result
CSF3R cytoplasmic mutations are identified in CNL/aCML/chronic myelomonocytic leukemia (CMML)/unclassified myeloproliferative neoplasms (MPN-U) and AML patients
CSF3R cytoplasmic mutations were determined by whole exome sequencing in a cohort of 843 patients with a variety of hematological malignances (Table 1 and Supplementary table 1). Detected mutations were validated with RNA-seq analysis and/or with Sanger sequencing from available samples (Supplementary figure 1).(13) We also referenced CSF3R mutation data from The Cancer Genome Atlas (TCGA) and a recent study of pediatric AML patients by Sano et al (Supplementary table 2).(14) Among 212 CNL/aCML/CMML/MPN-U patients in our cohort, 16(7.5%) patients harbor CSF3R cytoplasmic mutations, 14 (87.5 %) of which co-occur with CSF3R T618I, mostly in the same allele (Table 1). Five (1.3%) out of 378 adult AML patients in our cohort and 8 out of 534 (1.5%) pediatric AML patients in Sano’s study harbor CSF3R cytoplasmic mutations. We did not observe CSF3R cytoplasmic mutations among 183 CML, 70 lymphoid leukemia and 20 CMML patients in our cohort, nor in the 200 AML, 205 multiple myeloma and 48 diffuse large B-cell lymphoma patients in the TCGA database. In addition, we did not find CEBPa mutations in any of the cases with CSF3R cytoplasmic mutations.
Table 1.
Summary of CSF3R cytoplasmic mutations in hematologic malignances
| Disease group | Specimen | ID | subgroup | Cytoplasmic mutation | Membrane proximal mutation | Truncation/missense mutation | ||
|---|---|---|---|---|---|---|---|---|
| change | VAF | change | VAF | |||||
|
| ||||||||
| CNL/aCML/CMML/MPN-U (16/212) | 1 | 08-00423 | CNL | S783fs | 40% | T615A T640N |
12% 31% |
Same allele NA |
|
| ||||||||
| 2 | 09-00497 | aCML^ | S783fs | 9% | T615A | 7% | NA | |
|
| ||||||||
| 3 | 10-00694 | CNL | D771fs | NA | T618I | 40.4% | Same allele | |
|
| ||||||||
| 4 | 12-00121 | CMML | W791X | 12% | T618I | 0.41 | NA | |
|
| ||||||||
| 5 | 13-00068 | CMML | Q749X | 24% | ND | ND | ND | |
|
| ||||||||
| 6 | 13-00369 | aCML^ | S783fs | 13% | ND | ND | ND | |
|
| ||||||||
| 7 | 13-00437 | aCML | Y752X | 35% | T615A | 0.45 | NA | |
|
| ||||||||
| 8 | 13-00440 | aCML^ | W791X | 57% | T618I | 0.87 | NA | |
|
| ||||||||
| 9 | 13-00502 | CMML | Q754X | 52% | T618I | 53% | NA | |
|
| ||||||||
| 10 | 14-00201 | MPN-U^ | S783fs | 37% | T618I | 37% | Same allele | |
|
| ||||||||
| 11 | 13-00649* | CNL^ | Q754X | NA | T618I | NA | Same allele | |
|
| ||||||||
| 12 | 15-00430* | aCML^ | L780fs | 38% | T618I | 18% | Same allele | |
|
| ||||||||
| 13 | 15-00467* | CNL^ | W791X | 56% | T618I | 42% | Same allele | |
|
| ||||||||
| 14 | 13-00472* | MPN-U^ | YE787-788X | 27% | T618I | 53% | NA | |
|
| ||||||||
| 15 | 15-00257* | CNL^ | W791X | 54% | T618I | 52% | Same allele | |
|
| ||||||||
| 16 | 16-00105 | CNL^ | Q774X | 12% | T618I | 47% | NA | |
|
| ||||||||
| FAB group | Other potential pathogenic mutations~ | Karyotype | ||||||
|
| ||||||||
| AML (5/378) | 17 | 13-00625 | M1 | Q749X | 28% | BCORL1, CTCF, EZH2, NRAS | 45,XY,-7[6]; 47,XY,+13[5]; 46,XY[12] | |
|
| ||||||||
| 18 | 14-00608 | M2 | W791X | 55% | BCOR, CSF3R | 46,XX | ||
|
| ||||||||
| 19 | 14-00643 | M2 | Q823H | 6% | EZH2, KIT | 45,X,-Y, t(8;21)(q22;q22)[20] | ||
|
| ||||||||
| 20 | 15-00572 | M2 | Q752X | 16% | ASXL1, IDH2, PHF6, RUNX1, SMC1A, U2AF1 | 46,XY[20] | ||
|
| ||||||||
| 21 | 07-00008 | NA | Q741X | NA | NA | NA | ||
represent that CSF3R mutations were also identified in RNA-seq analysis.
represent unverified diagnosis due to the challenge of distinguishing CNL, aCML, and MPN-U.
represent that detailed mutation information is provided in supplementary table 1.
NA: not available. NT: not detected. [n] represent observed total cell number.
CSF3R distal cytoplasmic truncation mutations between amino acid Q793 and Q823 harbor oncogenic potential
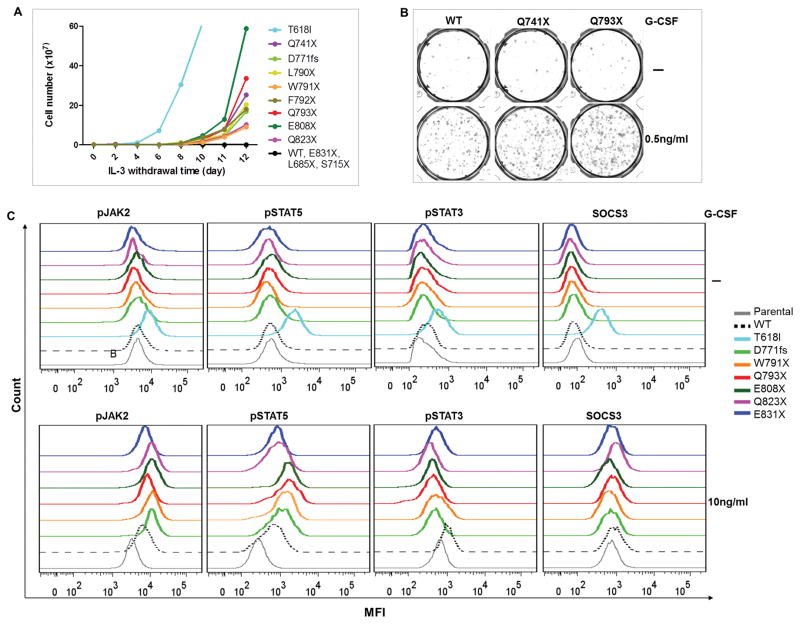
Previous studies have demonstrated G-CSF hypersensitivity of CSF3R truncations before the internalization domain (T738X, D771fs, etc.). To determine whether truncating the more distal region of CSF3R cytoplasmic domain enable CSF3R leukemogenic potential, we generated a series of CSF3R C-terminal truncation mutations (Supplementary figure 2A). As expected, we observed transformative potential, G-CSF hypersensitivity and sustained STAT5 activation in Q741X and D771fs (Figure 1 and supplementary figure 2B–C). Notably, L790X, F791X and W792X although only truncated one to three amino acids of the internalization domain, demonstrated oncogenic potential. Interestingly, even though Q793X, E808X and Q823X all retain the full internalization domain, these truncations also transformed Ba/F3 cells, demonstrated G-CSF hypersensitivity as shown in the CFU assay and induced marked sustained STAT5 activation compared to WT (Figure 1 and supplementary figure 2B–D). Truncation at E831 did not result in a phenotype different from WT. Consistent with previous studies, L685X and S715X did not transform Ba/F3 cells.
Figure 1. Truncating CSF3R at Q793–Q823 demonstrates leukemogenic potential.
(A) Representative transforming assay of Ba/F3 cells expressing CSF3R variants. T618I cells were used as a positive control. (B) Representative images of CFU assay demonstrate increased colony numbers in CSF3R truncation mutations in presence of 0.5ng/ml G-CSF as compared to CSF3R WT. (C) Representative histograms showing (upper panel) no ligand independent signaling activation in unstimulated and (lower panel) sustained pSTAT5/pJAK2 and reduced pSTAT3/SOCS3 activation of CSF3R truncation mutations when stimulated with G-CSF. Images shown are representative images of at least three independent experiments.
To better distinguish transformative truncation mutations, we hereafter refer to truncation mutations between T738 and F792 as early cytoplasmic truncation mutations, between Q793 and Q823 as distal cytoplasmic truncation mutations, and prior to T738 as juxtamembrane truncation mutations (Supplementary figure 2E).
CSF3R distal cytoplasmic truncation mutations interrupt receptor degradation
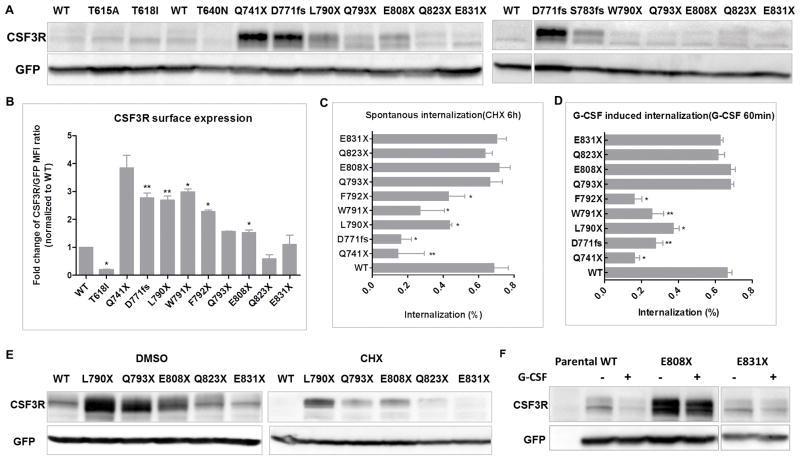
To understand the pathogenic mechanisms of CSF3R distal cytoplasmic truncation mutations, we first checked the receptor expression levels and the receptor internalization kinetics. Consistent with previous studies, we observed significantly higher (2.4–3.8 fold) receptor expression levels (surface and total protein) and delayed receptor internalization (spontaneous and ligand-induced) for early cytoplasmic truncation mutations (Figure 2A–D and supplementary Figure 3). Notably, F791X and W792X although only truncated one and two amino acids of the internalization domain, demonstrated significant reduced receptor internalization. Interestingly, Q793X and E808X also demonstrated 1.6 and 1.8 fold increase of surface receptor expression, whereas showed similar receptor internalization kinetics as WT (Figure 2A–D and supplementary Figure 3). To further understand the oncogenic mechanisms of the distal cytoplasmic truncation mutations, we quantified the receptor degradation by immunoblot. Interestingly, we observed that distal truncation mutations had marked more protein retained than WT after CHX and G-CSF treatment (Figure 2E–F), indicating a delayed spontaneous and ligand-induced receptor degradation despite intact internalization.
Figure 2. CSF3R truncation mutations between Q793 and Q823 interrupt receptor degradation.
(A) Representative immunoblots show CSF3R expression in (left panel) HEK293T/17 and (right panel) NIH3T3 cells. (B) CSF3R truncation mutations were transduced into Ba/F3 cells and cell surface expression of CSF3R was determined by FACS. Graph shows fold changes of CSF3R/GFP mean fluorescence intensity (MFI) ratio normalized to CSF3R WT. Graph depicts percentages of CSF3R internalization after (C) 6h CHX treatment normalized to DMSO control or (D) 60min G-CSF treatment normalized to no stimulation control. Representative immunoblot images of CSF3R expression in transduced HEK293T/17 cells treated with (E) 100mg/ml CHX for 6h or (F) 10ng/ml G-CSF for 60min. Data shown are mean ± SEM of biological replicates from at least 3 independent experiments. Images shown are representative of least three independent experiments. Statistical significance was assessed using a two tailed Student’s t-test: * p<.05; ** p<.01.
Increased higher molecular weight CSF3R bands were observed in CSF3R truncation mutations, which may be attributed to interrupted de-phosphorylation process
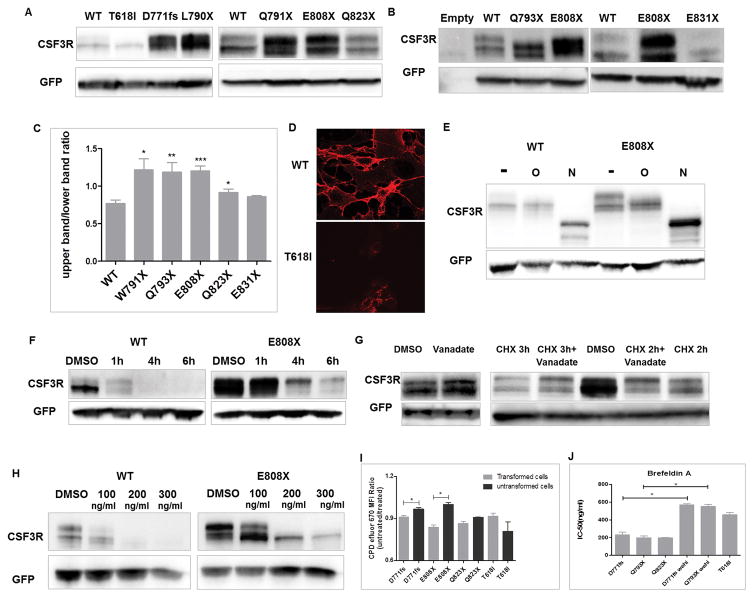
In addition to delayed kinetics of receptor internalization and/or degradation, we also observed altered CSF3R banding patterns in CSF3R truncation mutations both in HEK293T/17 and NIH3T3 cells (Figure 3A–B). CSF3R WT showed a stronger lower molecular weight band at around 105 KD, and a weaker higher molecular weight band at around 115 KD with an average higher/lower molecular weight band ratio of 0.77. In contrast, F791X, Q793X, E808X and Q823X, but not E831X demonstrated a reversed banding pattern: a stronger higher molecular weight band and a weaker or similar lower molecular weight band (Figure 3C). In addition, T618I demonstrated decreased higher molecular weight band and showed a low level of surface expression detected by FACS and fluorescence microscopy (Figure 2A–B and 3D), suggesting that the higher molecular weight band is the major form of CSF3R that is expressed on the cell surface. To further characterize the two bands of CSF3R, we performed glycosylation studies which showed that the higher molecular weight band is both N- and O-glycosylated (Figure 3E). Protein glycosylation can protect proteins from degradation. To test the stability of the two bands of CSF3R, we performed a time course CHX treatment. As shown in Figure 3F, the higher molecular weight band of CSF3R WT and E808X was significantly more retained after CHX treatment compared to the lower molecular weight band, indicating that the higher molecular weight band is more stable in the context of spontaneous degradation.
Figure 3. CSF3R truncation mutations demonstrate increased higher molecular weight band which is N and O-glycosylated.
Representative immunoblot images demonstrate CSF3R expression in (A) HEK293T/17 and (B) NIH3T3 cells. (C) Graph depicts CSF3R higher to lower molecular weight band ratios (mean ± SEM) shown in the immunoblot. (D) Detection of surface CSF3R expression in HEK293T/17 cells expressing CSF3R WT (upper panel), however showing a significantly low level in cells expressing CSF3R T618I construct (lower panel) at 63× magnification. Red represents CSF3R expression. (E) Representative immunoblot images of CSF3R WT and E808X expression before or after treatment to remove O-glycosylation or N-glycosylation. (F) Representative immunoblot images showing CSF3R expression in HEK293T/17 cells transient transfected with WT or E808X after a time course CHX treatment. (G) HEK293T/17 cells were transient transfected with CSF3R WT. Left panel: representative immunoblot image showing CSF3R expression in presence or absence of vanadate (1mM) for 1h. Right panel: immunoblot image showing CSF3R expression of transfected cells treated with CHX in presence or absence of vanadate (1mM) for 2 or 3h. For vanadate treated conditions, cells were pre-treated with vanadate for 1h before CHX treatment. The higher molecular weight bands of CSF3R truncation mutations are essential for the oncogenic potential. (H) Representative CSF3R expression of CSF3R WT and E808X transduced NIH3T3 cells before or after treatment with BFA for 48h. (I) Graph depicts CPD efluor 670 MFI ratios of transduced Ba/F3 cells expressing CSF3R variants grown with or without 100ng/ml BFA. (J) Transduced Ba/F3 cells were treated with a gradient concentration of BFA for 72h and the cell viabilities were determined as described in materials and methods. Graph depicts mean IC-50 of BFA. Images shown are representative of at least three independent experiments. Data shown are mean ± SEM of biological replicates of at least 3 independent experiments. Statistical significance was assessed using a two tailed Student’s t-test: * p<.05; ** p<.01: *** p<.001.
Previous studies have shown that the residues N818–F836 (here we call it the de-phosphorylation domain) at CSF3R c-terminal is important for peroxiredoxin mediated CSF3R de-phosphorylation, which subsequently mediates the receptor degradation process.(17) Hence, we tested whether a phosphatase inhibitor could protect CSF3R from degradation. As shown in Figure 3G, retention of CSF3R expression, especially the higher molecular weight band, could be observed after vanadate treatment for 1h. Moreover, vanadate protected CHX mediated CSF3R degradation (Figure 3G), indicating that the de-phosphorylation process contributes to the receptor expression level. This also explains why distal CSF3R truncation mutations, with deletion of only the de-phosphorylation domain, exhibited intact receptor internalization but delayed receptor degradation.
The higher molecular weight band is essential for the oncogenic potential of CSF3R truncation mutations
To further investigate the functional importance of the higher molecular weight band of CSF3R truncation mutations, we treated transformed cells with the glycosylation inhibitors. We observed that the glycosylation status of CSF3R was completely blocked by tunicamycin or BFA (at high concentration), indicated by the disappearance of CSF3R double bands and reduced surface CSF3R expression (Figure 3H and supplementary figure 4A). Importantly, we observed a disruption of only the higher molecular weight band at lower concentration of BFA (100ng/ml), indicating that ER to Golgi transport plays an important role in the formation of this higher molecular weight band (Figure 3H). In line with the reduced receptor levels, this low concentration of BFA also inhibited the proliferation of truncation mutation transformed cells, but had no effect on T618I transformed cells which demonstrated mostly the lower molecular weight band (Figure 3I). This low concentration of BFA also had no impact on growth of pre-transformed Ba/F3 cells expressing CSF3R truncation mutations growing in media containing IL-3 (Figure 3I). Accordingly, CSF3R truncation mutations demonstrated a lower IC50 for BFA compared to T618I and the respective untransformed cells (Figure 3J and supplementary figure 4B). These results indicate that the overexpressed CSF3R higher molecular weight band is indispensable to the leukemogenic potential of CSF3R truncation mutations.
Intact STAT5 activation is important for the oncogenic potential of CSF3R cytoplasmic truncation mutations
Previous studies have shown that the cytoplasmic membrane proximal 55 amino acids are important for STAT5 activation and G-CSF induced cell proliferation, whereas the following additional 30 amino acids (also called mitogenic enhancing domain (MED)) are required for the maximal STAT5 activation and cell proliferation.(18) Since the majority of CSF3R truncation mutations co-occur with CSF3R membrane proximal mutations, we therefore investigated whether non-transforming juxtamembrane region (amino acid positions 670–738) mutations could transform cells when combined with a T618I mutation. As shown in Supplementary figure 5A: T618I E700X, T618I T738X but not E700X and T618I L670X transformed Ba/F3 cells. To better characterize different compound mutations, we first checked their receptor expression and internalization profiles. L670X, E700X and V728X all demonstrated markedly increased receptor surface expression, altered CSF3R banding pattern and delayed internalization (Supplementary figure 5B–C). However, consistence with previous study, significantly reduced STAT3/5 activation was observed (Supplementary figure 5D–E). In agreement, decreased G-CSF responses at low concentration were observed (Supplementary figure 5F–G). These data indicate that, intact STAT5 activation is essential for the oncogenic potential of the truncation mutations alone; and the constitutive dimerization of T618I could compensate for decreased STAT5 activation in the context of E700X but not the absence of STAT5 activation of L670X. Furthermore, T618I with early or distal truncation mutation compound mutations demonstrated enhanced oncogenic potential (15), which may be attributed to delayed receptor degradation (Supplementary figure 5H).
Identification of oncogenic CSF3R cytoplasmic missense mutations
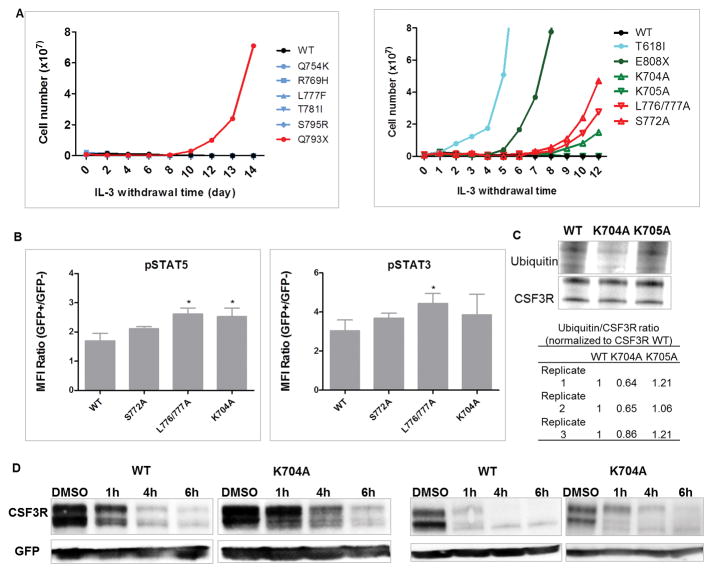
We next determined the oncogenic potential of CSF3R cytoplasmic missense mutations (Table 1, Supplementary table 2, and Supplementary figure 6A). None of these tested mutations (Q754A, R769H, L777F, T781I and S795R) seen in patients transformed Ba/F3 cells (Figure 4A) or demonstrated altered receptor expression or internalization pattern (Supplementary figure 6B–D). Consistent with previous studies, substitution of L776/777 and S772, which are important for receptor internalization demonstrated delayed receptor internalization, sustained STAT5 activation and transformative potential (Figure 4A–B and Supplementary figure 6C–D). Interestingly, K704A, but not K705A transformed Ba/F3 cells and induced sustained STAT5 activation (Figure 4A–B). In agreement, compared to WT, decreased receptor ubiquitination and delayed receptor degradation were observed in K704A, but not K705A in CSF3R and ubiquitin co-IP assay (Figure 4C–D), indicating that K704 plays a role in CSF3R ubiquitination and degradation.
Figure 4. CSF3R missense mutations seen in AML patients do not demonstrate leukemogenic potential; K704A is leukemogenic.
(A) Representative transforming assays of Ba/F3 cells expressing CSF3R missense mutations (left) seen in patients or (right) at potential functional residues. (B) Transduced Ba/F3 cells were serum starved for 90min and stimulated with 10ng/ml G-CSF for 90min. Graph depicts pSTAT5 and pSTAT3 MFI ratio of GFP+ transduced cells/GFP- non-transduced cells determined by phosphoflow. (C) Representative co-IP image demonstrates reduced ubiquitin binding in K704A cells. Table (lower panel) shows three replicates of K704A ubiquitin/CSF3R ratios normalized to CSF3R WT determined by integrated density quantification. K705A was used as a negative control. (D) Representative immunoblot images showing CSF3R expression in (left panel) HEK293T/17 and (right panel) NIH3T3 cells transient transfected with WT or K704A after a time course CHX treatment. Data and images shown are representative of three or more independent experiments. Statistical significance was assessed using a two tailed Student’s t-test: * p<.05.
Hypothesis and Validation
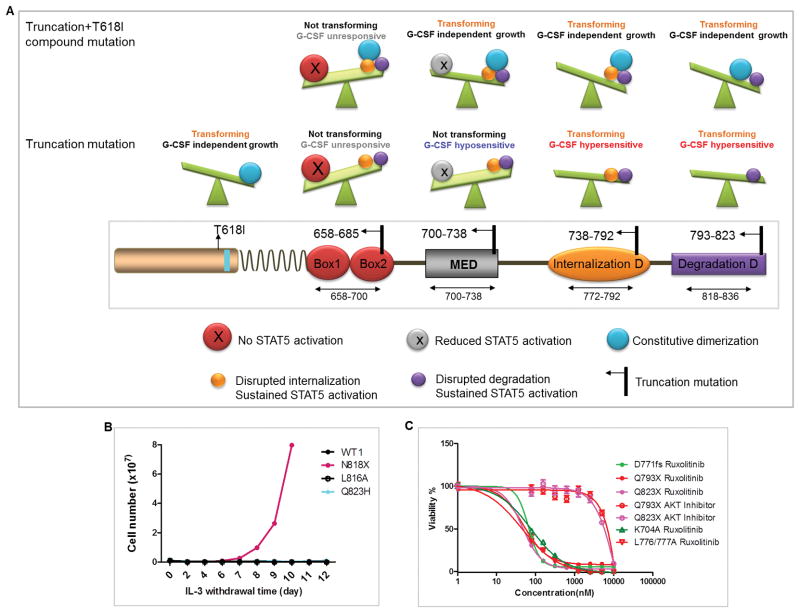
Based on the above observations, we hypothesized that CSF3R cytoplasmic truncation mutations disrupting receptor trafficking but preserving STAT5 activation are transforming (Figure 5A). Briefly, truncation mutations between T738 and Q823 are all leukemogenic and demonstrate reversed molecular weight banding patterns, whereas among missense mutations only L776/777A, S772A, and ubiquitination associated lysine mutations are leukemogenic. To validate this theory, we further generated N818X, L816A and Q823H mutations. As expected, N818X demonstrated reversed banding pattern and transformed Ba/F3 cells. L816A and Q823H showed similar banding pattern as CSF3R WT and did not transform Ba/F3 cells (Figure 5B and Supplementary figure 7). Notably, consistent with previous studies, transformed cells demonstrated sensitivity to ruxolitinib (13), but not an AKT inhibitor (Figure 5C).
Figure 5. Schematic illustration of the position and functional consequences of CSF3R truncation mutations and hypothesis validation.
(A) Briefly, truncation mutations at amino acid positions 658–685 which abrogate STAT5 activation do not transform BaF3 cells and do not respond to G-CSF, even when combined with T618I mutation. Truncation mutations at 700–738 do not transform Ba/F3 cells alone however transform Ba/F3 when combined with T618I and are hyposensitive to G-CSF due to the disruption of the MED. Truncation mutations at 738–792 and 793–823 transform Ba/F3 and induce G-CSF hypersensitivity and sustained STAT5 activation due to the interruption of the internalization and the de-phosphorylation domain respectively. (B) Representative IL-3 withdrawal assay showing leukemogenic potential of CSF3R N818X, but not L816A or Q823H. (C) Graph depicts mean ± SEM of cell viabilities of Ba/F3 cells expressing CSF3R variants after various doses of ruxolitinib treatment for 72h determined by MTS assay normalized to non-drug treatment controls.
Discussion
In the current study, we aim to understand the full spectrum of CSF3R cytoplasmic mutations in term of mutation disease distribution, oncogenic potential and the pathogenic mechanisms. Consistent with previous studies, we identified CSF3R cytoplasmic domain mutations in CNL/aCML/MPN-U and to a lesser extent in AML patients, but not in other hematological malignances. We further determined the driver mutation potential of these mutations by Ba/F3 transforming assays, G-CSF response assay, BM CFU assay and STAT5 activation assay. Moreover, we characterized the pathogenic mechanisms of those leukemogenic mutations.
We showed for the first time that CSF3R distal cytoplasmic truncation mutations (Q793–Q823) also harbour oncogenic potential which is associated with decreased receptor degradation, probably due to the disruption of the de-phosphorylation motif. Moreover, we identified another potential ubiquitination associated lysine residue K704, substitution of which reduced receptor ubiquitination and transformed Ba/F3 cells. With these data, together with previous studies, we summarize that CSF3R cytoplasmic mutations perturbing protein trafficking (internalization, degradation and ubiquitination), namely truncation mutations between T738 and Q823, and missense mutations including L776/777A, S772A, K655A, K785A and K704A are oncogenic. These data highlight the oncogenic role of defective receptor trafficking, which has been well recognized for other cytokine/growth factor receptors (e.g. EGFR)(19). Furthermore, this information will be helpful in predicting whether novel CSF3R cytoplasmic mutations observed in patients are potential drivers.
In addition, we further demonstrated that sustained STAT5 activation is the common altered downstream signaling pathway of oncogenic cytoplasmic mutations and sufficient STAT5 activation (box1-2 and MED for truncation mutations alone, box1-2 for compound mutations) is imperative for the oncogenic potential. Notably, all oncogenic cytoplasmic mutations showed sensitivity to ruxolitinib. These data will help define treatment strategies.
In addition, phenotypically, we observed that all oncogenic CSF3R truncation mutations demonstrate increased higher/lower molecular band ratios.. Our previous study showed that constitutively activated CSF3R membrane proximal and transmembrane mutations demonstrated decreased O-glycosylation.(15) At first glance, these two conclusions seem contradictory with one another; however, the pathogenic mechanisms of these two group mutations are different. Membrane proximal and transmembrane mutations transform cells in a ligand independent manner,(20) whereas truncation mutations are ligand dependent (Figure 1). The reduced higher molecular weight band of CSF3R in membrane proximal and transmembrane domain mutations suggests that the oncogenic capacity of these mutations does not rely on the higher molecular weight band and also may not rely on receptor surface expression. Similar phenomena were observed for other constitutively activated receptor mutations such as FLT3 ITD/D835Y and KIT V569G/D816V.(21,22) In contrast, CSF3R truncation mutations demonstrate increased higher/lower molecular weight band ratios. The increased higher molecular weight band may contribute to the decreased signalling termination phenotype seen in CSF3R truncation mutations, however still in a G-CSF dependent way, since no ligand independent signal activation is observed. Vanadate was shown previously to extend STAT5 activation. Interestingly, in the current study, we demonstrated that vanadate protected the higher molecular weight band from degradation, indicating a correlation of the higher molecular weight band with STAT5 activation.
In summary, increases of the higher molecular weight band of CSF3R and preservation of STAT5 activation capacity are important for the transforming potential of CSF3R truncation mutations. Our study provides comprehensive understanding of the connection between CSF3R structural domain, receptor expression pattern, receptor trafficking, signaling activation and biological function (G-CSF response and transforming potential). This information may provide insights to understanding oncogenic mechanisms contributing to mutations in other growth factor receptors. For clinical applications, the JAK inhibitor ruxolitinib was shown in case reports to be effective against CSF3R T618I and a clinical trial is undergoing to evaluate ruxolitinib in a larger cohort of CNL/aCML/MPN-U patients. Similar trials would be useful for AML patients with CSF3R mutations. Given the poor prognosis of CSF3R active mutations seen in CNL patient, the information from our study may eventually benefit patients with CSF3R oncogenic truncation mutations.
Supplementary Material
Acknowledgments
The authors thank Christopher Eide and Samantha Savage for sharing the CML sequence data. The authors also acknowledge Larry David and John Klimek for mass spectrometry analysis; Aurelie Snyder and Stefanie Kaech Petrie for ApoTome microscope assistance and Brianna Garcia for her help in the FACS sorting.
Footnotes
Conflict-of-interest statements: JWT receives research support from Agios Pharmaceuticals, Array Biopharma, Aptose Biosciences, AstraZeneca, Constellation Pharmaceuticals, Genentech, Gilead, Incyte Corporation, Janssen Pharmaceutica, Seattle Genetics, Syros, Takeda Pharmaceutical Company and is a consultant for Leap Oncology. The other authors do not have any conflict of interests.
Authorship
HZ designed and performed the experiment, analyzed the data and prepared the manuscript; ARS performed the experiment, analyzed the data and proof-read the manuscript. SL helped perform the mouse CFU assay and proof-read the manuscript; AR prepared DNA and RNA sequence samples. YS participated in the FACS experiments. SM helped maintain the cell culture and performed western blot experiment for the signaling pathway analysis. DB, BW and SKM performed and analyzed exome and RNA sequencing data. JWT designed the experiment, interpreted the data, wrote and revised the manuscript.
References
- 1.Panopoulos AD, Watowich SS. Granulocyte colony-stimulating factor: Molecular mechanisms of action during steady state and “emergency” hematopoiesis. Cytokine. 2008;42:277–88. doi: 10.1016/j.cyto.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hermans MHA, Van de Geijn GJ, Antonissen C, Gits J, Van Leeuwen D, Ward AC, et al. Signaling mechanisms coupled to tyrosines in the granulocyte colony-stimulating factor receptor orchestrate G-CSF-induced expansion of myeloid progenitor cells. Blood. 2003;101(7):2584–90. doi: 10.1182/blood-2002-07-2062. [DOI] [PubMed] [Google Scholar]
- 3.Touw IP, Palande K, Beekman R. Granulocyte Colony-Stimulating Factor Receptor Signaling. Implications for G-CSF Responses and Leukemic Progression in Severe Congenital Neutropenia. Hematology/Oncology Clinics of North America. 2013;27:61–73. doi: 10.1016/j.hoc.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Ai J, Druhan LJ, Loveland MJ, Avalos BR. G-CSFR ubiquitination critically regulates myeloid cell survival and proliferation. PLoS One. 2008;3(10):e3422. doi: 10.1371/journal.pone.0003422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aarts LHJ, Roovers O, Ward AC, Touw IP. Receptor activation and 2 distinct COOH-terminal motifs control G-CSF receptor distribution and internalization kinetics. Blood. 2004;103(2):571–9. doi: 10.1182/blood-2003-07-2250. [DOI] [PubMed] [Google Scholar]
- 6.Wölfler A, Irandoust M, Meenhuis A, Gits J, Roovers O, Touw IP. Site-specific ubiquitination determines lysosomal sorting and signal attenuation of the granulocyte colony-stimulating factor receptor. Traffic. 2009;10(8):1168–79. doi: 10.1111/j.1600-0854.2009.00928.x. [DOI] [PubMed] [Google Scholar]
- 7.Dong F, Brynes RK, Tidow N, Welte K, Löwenberg B, Touw IP. Mutations in the gene for the granulocyte colony-stimulating-factor receptor in patients with acute myeloid leukemia preceded by severe congenital neutropenia. N Engl J Med [Internet] 1995;333(8):487–93. doi: 10.1056/NEJM199508243330804. Available from: http://www.nejm.org/doi/full/10.1056/NEJM199508243330804#t=abstract%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/7542747. [DOI] [PubMed] [Google Scholar]
- 8.Germeshausen M, Ballmaier M, Welte K. Incidence of CSF3R mutations in severe congenital neutropenia and relevance for leukemogenesis: Results of a long-term survey. Blood. 2007;109(1):93–9. doi: 10.1182/blood-2006-02-004275. [DOI] [PubMed] [Google Scholar]
- 9.Hermans MH, Antonissen C, Ward AC, Mayen AE, Ploemacher RE, Touw IP. Sustained receptor activation and hyperproliferation in response to granulocyte colony-stimulating factor (G-CSF) in mice with a severe congenital neutropenia/acute myeloid leukemia-derived mutation in the G-CSF receptor gene. J Exp Med [Internet] 1999;189(4):683–92. doi: 10.1084/jem.189.4.683. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2192935&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehta HM, Futami M, Glaubach T, Lee DW, Andolina JR, Yang Q, et al. Alternatively spliced, truncated GCSF receptor promotes leukemogenic properties and sensitivity to JAK inhibition. Leukemia [Internet] 2014;28(5):1041–51. doi: 10.1038/leu.2013.321. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24170028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, Goudeva L, Immenschuh S, Schambach A, Skokowa J, Eiz-Vesper B, et al. miR-155 is associated with the leukemogenic potential of the class IV granulocyte colony-stimulating factor receptor in CD34+ progenitor cells. Mol Med [Internet] 2014;20(4):736–46. doi: 10.2119/molmed.2014.00146. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4398671&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehlers S, Herbst C, Zimmermann M, Scharn N, Germeshausen M, von Neuhoff N, et al. Granulocyte Colony-Stimulating Factor (G-CSF) Treatment of Childhood Acute Myeloid Leukemias That Overexpress the Differentiation-Defective G-CSF Receptor Isoform IV Is Associated With a Higher Incidence of Relapse. J Clin Oncol [Internet] 2010;28(15):2591–7. doi: 10.1200/JCO.2009.25.9010. Available from: http://jco.ascopubs.org/cgi/doi/10.1200/JCO.2009.25.9010%5Cnpapers2://publication/doi/10.1200/JCO.2009.25.9010. [DOI] [PubMed] [Google Scholar]
- 13.Maxson JE, Gotlib J, Pollyea Da, Fleischman AG, Agarwal A, Eide Ca, et al. Oncogenic CSF3R mutations in chronic neutrophilic leukemia and atypical CML. N Engl J Med [Internet] 2013;368(19):1781–90. doi: 10.1056/NEJMoa1214514. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3730275&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sano H, Ohki K, Park MJ, Shiba N, Hara Y, Sotomatsu M, et al. CSF3R and CALR mutations in paediatric myeloid disorders and the association of CSF3R mutations with translocations, including t(8; 21) Br J Haematol. 2015;170(3):391–7. doi: 10.1111/bjh.13439. [DOI] [PubMed] [Google Scholar]
- 15.Maxson JE, Luty SB, MacManiman JD, Abel ML, Druker BJ, Tyner JW. Ligand independence of the T618I mutation in the colony-stimulating factor 3 receptor (CSF3R) protein results from loss of O-linked glycosylation and increased receptor dimerization. J Biol Chem. 2014;289(9):5820–7. doi: 10.1074/jbc.M113.508440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maxson JE, Ries RE, Wang Y-C, Gerbing RB, Kolb EA, Thompson SL, et al. CSF3R mutations have a high degree of overlap with CEBPA mutations in pediatric AML. Blood [Internet] 2016 Jun 16;127(24):3094 LP–3098. doi: 10.1182/blood-2016-04-709899. Available from: http://www.bloodjournal.org/content/127/24/3094.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palande K, Roovers O, Gits J, Verwijmeren C, Iuchi Y, Fujii J, et al. Peroxiredoxin-controlled G-CSF signalling at the endoplasmic reticulum-early endosome interface. J Cell Sci. 2011;124(November):3695–705. doi: 10.1242/jcs.089656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong F, Liu X, de Koning JP, Touw IP, Hennighausen L, Larner A, et al. Stimulation of Stat5 by granulocyte colony-stimulating factor (G-CSF) is modulated by two distinct cytoplasmic regions of the G-CSF receptor. J Immunol UNITED STATES. 1998 Dec;161(12):6503–9. [PubMed] [Google Scholar]
- 19.Tomas A, Futter CE, Eden ER. EGF receptor trafficking: Consequences for signaling and cancer. Trends in Cell Biology. 2014;24:26–34. doi: 10.1016/j.tcb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehta HM, Glaubach T, Long A, Lu H, Przychodzen B, Makishima H, et al. Granulocyte colony-stimulating factor receptor T595I (T618I) mutation confers ligand independence and enhanced signaling. Leukemia. 2013;27:2407–10. doi: 10.1038/leu.2013.164. [DOI] [PubMed] [Google Scholar]
- 21.Bougherara H, Subra F, Crepin R, Tauc P, Auclair C, Poul M-A. The aberrant localization of oncogenic kit tyrosine kinase receptor mutants is reversed on specific inhibitory treatment. Mol Cancer Res. United States. 2009 Sep;7(9):1525–33. doi: 10.1158/1541-7786.MCR-09-0138. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto Y, Kiyoi H, Nakano Y, Suzuki R, Kodera Y, Miyawaki S, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood. 2001;97(8):2434–9. doi: 10.1182/blood.v97.8.2434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.