Abstract
Avoiding ingestion of excessively salty food is essential for cation homeostasis that underlies various physiological processes in organisms. The molecular and cellular basis of the aversive salt taste, however, remains elusive. Through a behavioral reverse genetic screening, we discover that feeding suppression by Na+-rich food requires Ionotropic Receptor 76b (Ir76b) in Drosophila labellar gustatory receptor neurons (GRNs). Concentrated sodium solutions with various anions caused feeding suppression dependent on Ir76b. Feeding aversion to caffeine and high concentrations of divalent cations and sorbitol was unimpaired in Ir76b-deficient animals, indicating sensory specificity of Ir76b-dependent Na+ detection and the irrelevance of hyperosmolarity-driven mechanosensation to Ir76b-mediated feeding aversion. Ir76b-dependent Na+-sensing GRNs in both L− and s-bristles are required for repulsion as opposed to the previous report where the L-bristle GRNs direct only low-Na+ attraction. Our work extends the physiological implications of Ir76b from low-Na+ attraction to high-Na+ aversion, prompting further investigation of the physiological mechanisms that modulate two competing components of Na+-evoked gustation coded in heterogeneous Ir76b-positive GRNs.
Keywords: capillary feeder assay, extracellular recordings, reverse genetic screening, salt taste aversion, variant ionotropic receptor
INTRODUCTION
Ingestion of salt is necessary for numerous physiological functions including neuronal excitability, transmembrane transport of organic compounds in various tissues, and osmotic homeostasis of body fluids. However, excessive ingestion of salt often leads to dysregulation of such functions adversely affecting the wellbeing of the organism. For example, elevated Na+ concentration in the body is suspected to cause hypertension (Frisoli et al., 2012), gastrointestinal cancer (Tsugane et al., 2004), osteoporosis (He and Mac-Gregor, 2008), and autoimmune diseases (Kleinewietfeld et al., 2013). For the control of optimal salt intake, salt taste can either promote or suppress food ingestion depending on the concentration of salt. Low concentrations of Na+ salt (<100 mM) are appetitive, and are mainly sensed by amiloride-sensitive epithelial sodium channel (ENaC) expressed on the apical surface of taste cells in mice (Chandrashekar et al., 2010). In contrast, the taste of concentrated salt (high-salt taste) is aversive to animals ranging from nematodes to rodents (Chatzigeorgiou et al., 2013; Niewalda et al., 2008; Oka et al., 2013). However, the molecular mechanism by which high-salt taste is sensed remains to be identified (Yarmolinsky et al., 2009). The transmembrane channel-like-1 (tmc-1) gene in Caenorhabditis elegans was originally proposed to encode a receptor of concentrated Na+ and mediate Na+ avoidance behavior (Chatzigeorgiou et al., 2013), but these results have recently become controversial (Wang et al., 2016).
Ionotropic receptors (IRs), which comprise a subgroup of the ionotropic glutamate receptor (iGluR) family, have been identified as critical sensory receptors in insects for environmental stimuli such as chemical compounds (Benton et al., 2009; Silbering et al., 2011), temperature changes (Ni et al., 2016) and moisture in the air (Knecht et al., 2016; 2017), rather than as regulators of synaptic transmission. In particular, IR76b expressed in L-bristle GRNs was proposed to serve as a functional Drosophila counterpart of mammalian ENaC, a receptor necessary for appetitive responses to Na+ at low concentrations (Zhang et al., 2013). The two receptors appear to have the common characteristic of constitutive cation channel activity. In the same study, GRNs in a few s-bristles were suggested to act as receptors of high-NaCl taste in an Ir76b-independent manner, while cells in L− and i-bristles had been reported to respond to high NaCl concentrations in an earlier study (Hiroi et al., 2004). However, little is known about the molecular mechanism of high-salt taste in Drosophila as in most other animals. Here, to unravel the gustatory mechanisms limiting excessive salt intake, we conducted a reverse genetic screening with salt receptor candidates related to ENaC/TMC or associated with constitutive cation conductance using the capillary feeder (CAFE) assay (Du et al., 2015; Ja et al., 2007). Unexpectedly, we find that feeding suppression by high Na+ concentrations relies on the Ir76b gene which is necessary for neuronal Na+ responses in the s-bristle taste cells besides the previously reported L-bristle GRNs. Our results indicate that IR76b plays a central role in gustation of both attractive and aversive Na+ concentrations possibly in combination with partner IRs instructing respective functions.
MATERIALS AND METHODS
Fly stocks
Ir76b1, Ir76b2 and Ir76b-Gal4 (on the third chromosome) lines generated in the previous study by Zhang et al. (2013), were provided by Dr. Seok Jun Moon at Yonsei University College of Dentistry, and subdued ko11is a gift from Dr. Yuh Nung Jan at the University of San Francisco. The wild-type line, wcs, is w1118 backcrossed with D. mel Canton S. TrpA1ins and UAS-TrpA1(A)10b was previously described (Du et al., 2016a; Rosenzweig et al., 2008). The following lines (stock number) were obtained from the Bloomington Stock Center: UAS-Ir76b (52610), Ir76b-Gal4RB (41730), Ir76b5 (9824), tmcf01875 (18483), axsD (1637), CG10353kg03540 (13793), CG6938MB01354 (23072), CG15270MB02602 (23582), nae04385 (18255), ppk5EY00388 (15022), ppk9EY10905 (20643), ppk12MB11059 (29179), ppk16MB11536 (27869), ppk17Mi03566 (36994), ppk18Mi04547 (37708), ppk18MB07822 (25571), ppk21Mi02964 (37590), ppk23G17320 (33300), ppk29f06838 (19016), ppk31Mi06637 (41105). The UAS-dsRNA stocks for the RNAi screen were obtained from the Bloomington Stock Center: ppk (29571), ppk6 (25880, 53010), ppk7 (25922), ppk8 (25814), ppk9 (25892), ppk13 (25817), ppk14 (27091), ppk15 (28012), ppk16 (25890), ppk21 (25849), ppk22 (28706), ppk24 (26006), ppk25 (27088), ppk26 (25825), ppk30 (25810), ripped pocket (ppk2; 25847, 39053), and NaCh (ppk4; 27262).
Capillary feeder (CAFÉ) assay
Capillary feeder assay was performed as described previously (Du et al., 2015). Briefly, twenty to forty flies (2–3 days old) were fasted for 16 h in a polypropylene vial containing a wet Kimwipe, and then transferred to a new vial for the feeding assay. Capillaries were filled with two types of liquid food by capillary action and inserted between the plug and vial wall with the depth of insertion consistent across inserted capillaries. In general, two sucrose capillary tubes and two sucrose-plus-chemical capillary tubes were offered to suppress noise of data between experiments. The flies were allowed to feed on the provided food sources under ambient lighting conditions for 30 min. Control vials without flies were used to determine the effects of evaporation in each experiment. After 30 min, the length of meniscus descent in capillaries was taken as food consumption, after the change due to evaporation was deducted. The number of live flies present in the vial was also recorded to determine the volume of consumption per fly. Avoidance index was obtained by deducting the fraction of consumed salty sucrose solution from the fraction of consumed sucrose solution with respect to total consumption in volume. Each allele of candidate genes in the reverse genetic screening was tested in four to five independent experiments.
Extracellular recording from taste sensilla in the labellum
Extracellular single-unit recordings were performed using the tip-recording method (Hodgson et al., 1955). Two to five-day-old flies were anesthetized by brief ice exposure. A reference electrode containing Drosophila Ringer solution was inserted through the thorax and head to the tip of the labellum, immobilizing the flies. Individual sensilla were exposed to tastants via a glass recording electrode (10–20 μm diameter) filled with tastant solution dissolved in 30 mM tricholine citrate. The recording electrode was connected to a TastePROBE (Syntech, The Netherlands), and electrical signals derived from taste sensilla were recorded using an acquisition controller (Syntech). These signals were amplified (10x), band-pass-filtered (100–3000 Hz), and sampled at 12 kHZ. Autospike 3.1 software (Syntech) was used to analyze neuronal firing rates, by counting the number of spikes generated over a 500 ms period beginning 200 ms after contact.
Statistical analysis
All data were analyzed with Sigmaplot 12. Two group comparison was conducted with Student’s t-test or the Mann-Whitney U test. Multiple-group comparisons were performed using one-way analysis of variance (ANOVA) followed by Tukey’s or Dunn’s multiple comparison tests depending on the result of normality test. Data were presented as the mean ± SEM.
RESULTS
A reverse genetic screening reveals the potential role of the Ir76b gene in tasting highly concentrated Na+
To identify genes critical for aversive gustation in response to concentrated Na+ in food, the capillary feeder (CAFE) assay (Deshpande et al., 2014; Ja et al., 2007) was employed for a candidate-based genetic screening (Fig. 1A). The CAFE assay is regarded as a consistent, sensitive and quantitative feeding assay compared to proboscis extension reflex and two color dye-based choice assays (Deshpande et al., 2014). In particular, the two color dye-based choice assay often used in gustation studies such as Zhang et al. (2013) is inferior to the CAFE assay in two aspects. First, the two color choice assay depends on the experimenter’s subjective judgement of whether the color of the fly abdomen is red, blue or purple. Second, it does not take into account of the volume of food consumption unlike CAFE; if one fly consumes a small amount of one substance and another fly ingests a large amount of another substance, the two choices are scored as even. In our CAFE assays, one of two pairs of calibrated glass capillaries contains sucrose only, while the other pair contains NaCl/sucrose solution. The glass tubes were offered to starved flies in an empty vial. Avoidance indices were calculated (see “Materials and Methods”) and compared between genotypes. Indicative of strong avoidance to high NaCl concentrations, wild-type flies (wcs) robustly avoided drinking 0.5 or 1 M NaCl in sucrose solution, mostly consuming sucrose solution without NaCl (Fig. 1B). Two categories of genes were targeted in our study: genes homologous to those encoding known receptors for Na+ detection and ion channel genes associated with various degrees of constitutive activity at rest. Constitutive ion channel opening has been attributed to the capability of detecting extracellular monovalent cations (Zhang et al., 2013). Transmembrane channel-like-1 (tmc-1) was found to code for a receptor of concentrated NaCl solution which allows C. elegans to avoid salty environments (Chatzigeorgiou et al., 2013). Drosophila has a tmc-1 orthologue and five distant tmc paralogues, anoctamins (axs, subdued, CG10353, CG6938 and CG15270) (Hahn et al., 2009). The basal level of channel opening has been linked to narrow abdomen (na) and TrpA1, which are the Drosophila orthologues of the NALCN (Na+ leak channel, nonselective) and TrpA1 (transient receptor potential ankyrin 1) genes encoding the nonselective Na+ leak channel and the reactive chemical receptor, TRPA1, respectively. The TRPA1(A) isoform, expressed from the Drosophila TrpA1 locus in GRNs, exhibits a low level of channel opening upon heterologous expression in Xenopus oocytes (Du et al., 2016b; Kang et al., 2012). Nonetheless, the alleles available for these candidates did not show impaired feeding aversion to concentrated NaCl (Fig. 1B). Note that transposons were inserted in the introns of CG10353, CG6938 and CG15270, while the tmc and na alleles contain transposons in exons (Gramates et al., 2017). The axsD allele accompanies a dominant negative point mutation (Zitron and Hawley, 1989), and the alleles of subdued (Wong et al., 2013) and TrpA1 (Rosenzweig et al., 2008) were produced by disrupting the loci by homologous recombination. Next, the pickpocket (ppk) gene family, which are Drosophila ENaC homologs, were examined for their potential roles in high NaCl taste. Among 31 members (Zelle et al., 2013), 27 ppk genes were tested by RNAi knockdown and exonal transposon insertion. Three ppk genes, ppk21, ppk27 and ppk29 appear to contribute to high-NaCl taste. RNAi knockdown (Fig. 1C) and exonal transposon insertion (Fig. 1D) of ppk21 caused defective salt aversion to 0.5 M NaCl but not to 1.0 M NaCl, suggesting its ancillary role in high-NaCl gustation. Alleles of ppk27 and ppk29 carrying exonal transposons showed statistically significant but very minute impairment in gustatory aversion to NaCl (Fig. 1D). Thus, high NaCl concentrations generally drive strong feeding avoidance in many different genetic backgrounds of Drosophila melanogaster upon behavioral evaluation by the CAFE assay.
Fig. 1. Various genotypes of Drosophila melanogaster strongly avoid drinking high-Na+ sucrose solution offered in a CAFE assay.
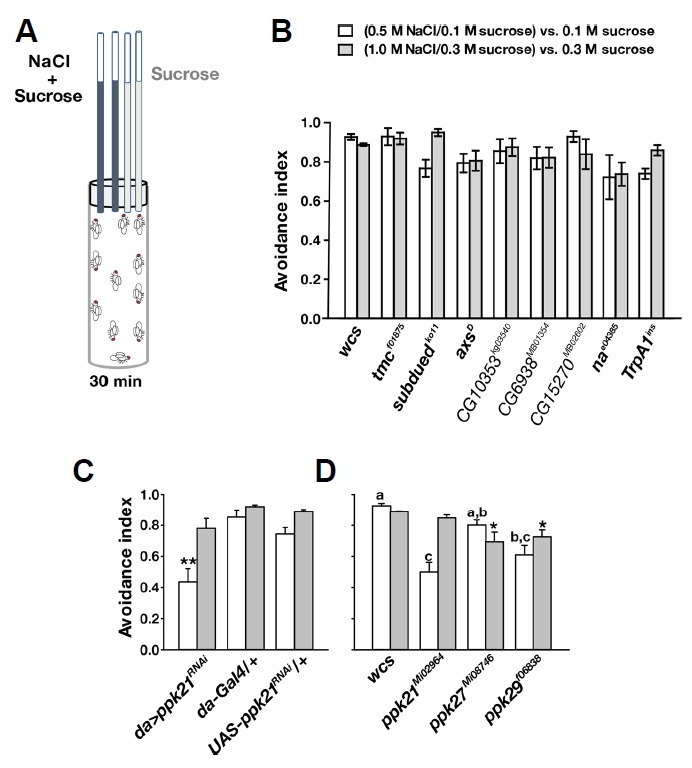
(A) Schematic illustration of the capillary feeder (CAFE) assay used in the study. (B) Wild type (wcs), and mutants of tmc, anoctamins, na, or TrpA1 genes show strong feeding avoidance to high concentrations of NaCl in sucrose solution as indicated. There is no statistically significant difference between the groups (n = 4–5). (C, D) Genetic disturbances of ppk21, ppk27 and pkk29 cause minor defects in high-NaCl feeding suppression (n = 4–5). **p < 0.01, Tukey’s test (C). Letters indicate statistically distinct groups: Tukey’s, p < 0.01. *p < 0.05, Dunn’s test (D).
In Drosophila, Ir76b was recently proposed to be critical for attraction to low-NaCl food, and its ability to detect low NaCl concentration was ascribed to its standing activity observed upon heterologous expression in HEK293 cells (Zhang et al., 2013). To our surprise, five different allelic combinations deficient for the functional Ir76b gene showed dramatic reduction in high-NaCl feeding avoidance (Fig. 2A). RNAi knockdown of Ir76b with pan-neuronal elavC155-Gal4 (Koushika et al., 1996) and dll-Gal4 covering chemosensory receptor cells (Calleja et al., 1996; Kang et al., 2010) also resulted in impaired NaCl avoidance (Fig. 2B). To examine if the Ir76b-dependent behavioral response concerns Na+, the chloride anion was replaced with others such as acetate or bicarbonate (Fig. 2C). Ir76b1 and Ir76b2 showed decreased avoidance indices in response to both anion-replaced Na+ salts, compared to wild-type flies that robustly avoided them. Sodium acetate (CH3COONa) at 1 M did not yield statistically significant difference between wild types and Ir76b mutants, probably because acetate stimulates an aversive acid tasting mechanism or olfactory aversion at high concentrations due to its volatility (Ko et al., 2015), independent of Ir76b. Decreased avoidance to sodium acetate and sodium bicarbonate in Ir76b1 and Ir76b2 strongly suggests that Na+ is the main component detected by the Ir76b-dependent gustatory mechanism rather than Cl−. In support of the sensory specificity of Ir76b-mediated Na+ detection, the mutant alleles showed robust feeding avoidance in response to other aversive tastants such as high concentrations of CaCl2 and BaCl2, the bitter chemical caffeine, and hyperosmolar solution of 2 M sorbitol (Fig. 2D). These results that the high osmolarity of ionic or nonionic compounds is avoided by Ir76b mutants at a level similar to wild types, suggest that Ir76b-mediated high-NaCl taste is unlikely to occur through mechanosensation.
Fig. 2. Ir76b is necessary for high-Na+ feeding avoidance.
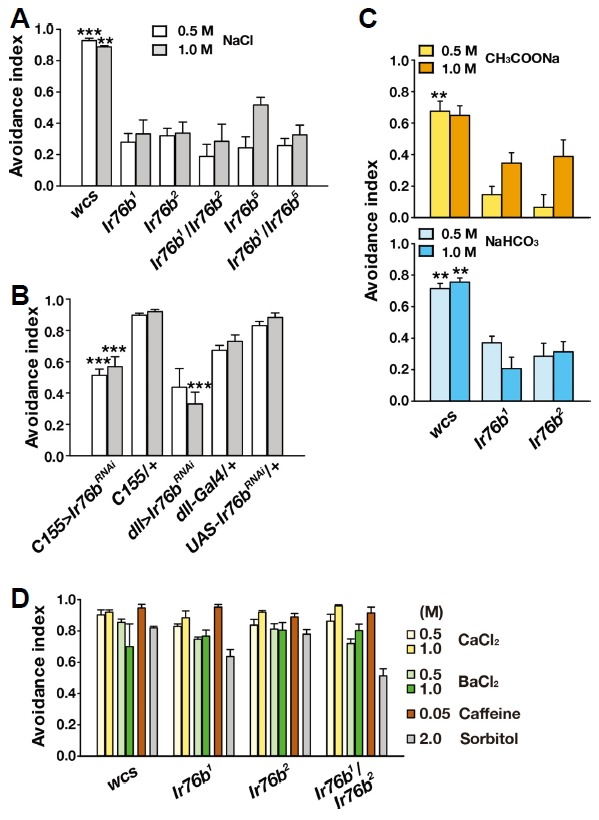
(A) Multiple Ir76b allelic combinations show defects in the CAFE assay testing high-Na+ feeding suppression (n = 4–9). (B) RNAi knockdown of Ir76b in neurons and the appendages by C155 and dll-Gal4, respectively leads to defective high-Na+ feeding avoidance (n = 4–8). (C). Ir76b is required for feeding deterrence to sodium solutions with acetate or bicarbonate (n = 4). (D) Ir76b alleles are not impaired in feeding avoidance to CaCl2, BaCl2, caffeine and sorbitol at the indicated concentrations (n = 4–6). **p < 0.01, ***p < 0.001, Tukey’s test.
Ir76b is required for the cellular response to concentrated NaCl in the s-bristle gustatory receptor neurons
GRNs in L-bristles were previously proposed to require Ir76b to signal for attractive low-Na+ taste, while Na+-responsive GRNs in s-bristles were suspected to code for aversive high-Na+ gustation by an unknown molecular mechanism (Zhang et al., 2013). A model for the decision of aversion over attraction between the inputs from the two classes of GRNs was put together mainly based on the observation that low-Na+-sensing GRNs in L-bristles show a bell-shaped spiking response curve to increasing Na+ doses, such that, at high Na+ concentrations, aversive input from the s-bristle GRNs prevails to direct feeding suppression. However, the bell shape relationship, where the responses of L-bristle cells supposedly decrease at high NaCl concentrations with a maximal spiking frequency at 0.1 M NaCl, was not recapitulated in our hands. Rather, the dose dependences of L− and s-bristle cells are similarly sigmoidal (Figs. 3A and 3B), while L-bristle GRNs showed higher Na+ sensitivities at 10 and 50 mM than the s-bristle cells as observed in the previous study (Zhang et al., 2013). The NaCl-evoked spiking frequency of the s6 GRN was low compared to other Na+-responding GRNs, suggesting a minor role in NaCl avoidance. For parallel comparison between cellular and behavioral responses, Na+ dose dependence of feeding avoidance was appraised (Fig. 3C). The avoidance index was dramatically increased at 0.3 M Na+, which is higher than the concentration of 0.1 M that yielded maximal spiking responses in L4, L6, s4 and s8 GRNs. Given that the CAFE assay offers sucrose with NaCl unlike extracellular recording of taste neurons, it may be that sweet taste raises the threshold of feeding avoidance to Na+-enriched food.
Fig. 3. Cellular response to high NaCl concentrations in Land s-bristle GRNs requires Ir76b.
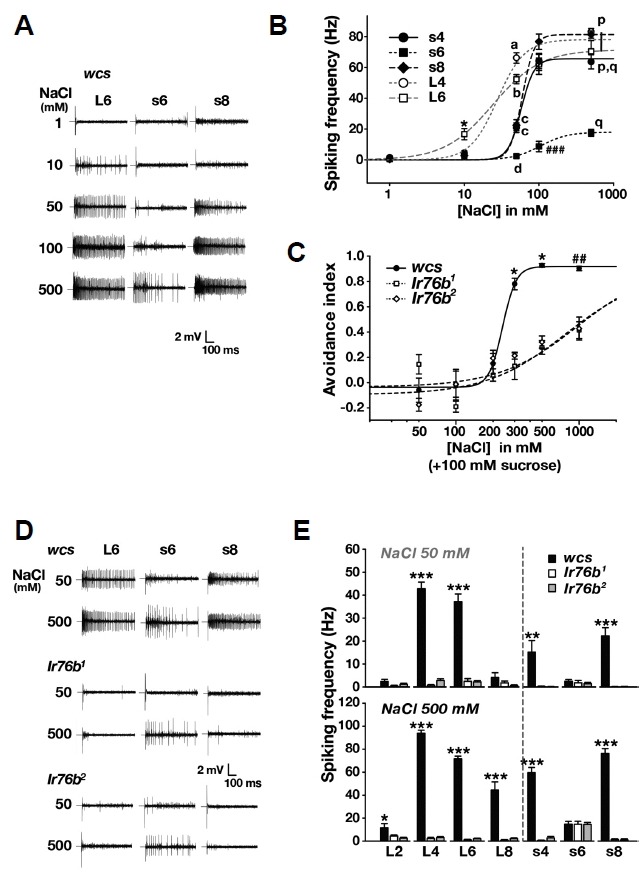
(A) Typical NaCl-induced spiking recordings registered from L− and s-bristles at increasing NaCl concentrations as indicated. (B) Averaged NaCl dose-dependent neuronal spiking frequencies from the indicated s− and L-bristles at incremental NaCl doses (n = 8–17). (C) NaCl dose-dependent behavioral responses of WT and Ir76b mutants (n = 4–9). Letters indicate statistically different groups: a–d, p < 0.001, Tukey’s test, and p and q, p < 0.05, Dunn’s test (B). *p < 0.05, Dunn’s Test. ##p < 0.01, ### p < 0.001, Tukey’s test (B, C). (D) Typical GRN recordings obtained from the L− and s-bristles of Ir76b1 and Ir76b2 at 50 and 500 mM NaCl. (E) Averaged GRN spiking frequencies from s− and L-bristles in Ir76b mutants in response to low and high NaCl doses as indicated (n = 8–21). *p < 0.05, **p < 0.01, ***p < 0.001, Tukey’s test (E).
Next, we examined if NaCl-provoked spikings in s-bristle GRNs are Ir76b-dependent (Figs. 3D and 3E). The robust responders in s4 and s8 showed dramatic decreases in spiking frequencies upon contact with both low and high NaCl concentrations at 50 and 500 mM, respectively. The NaCl spiking frequencies of the weakly responding s6 GRN persisted in Ir76b1 and Ir76b2, indicating that the mild response of s6 is independent of Ir76b. NaCl-induced spikings in L2, L4, L6 and L8 bristles relied on Ir76b as previously reported (Zhang et al., 2013). Interestingly, wild-type L2 and L8 GRNs were significantly activated by high NaCl concentration at 500 mM, but not at 50 mM, compared to Ir76b-deficient cells, suggesting that these cells may contribute to high-NaCl taste rather than low-NaCl taste. These results show that not only L-bristle GRNs but also s-bristle cells rely on Ir76b to respond to low and/or high concentrations of NaCl. Interestingly, the s4 and s8 bristles were previously categorized to be the s-c class that lacked responses to various bitter compounds (Weiss et al., 2011), implying that the s-bristle sensil la may be specialized for salt aversion.
Ir76b-Gal4RB transgene restores NaCl responsiveness in Ir76b-deficient s-bristle GRNs but not L-bristle GRNs
Using two different Ir76b-Gal4 lines, we attempted to restore the physiological defects of Ir76b-lacking L− and s-bristle GRNs. The Ir76b-Gal4 transgenic line was adopted from the study by Zhang et al. (2013). The Ir76b-Gal4RB line was reported to be weakly expressed in the labellum in comparison to the other Ir76b-Gal4 line, while expression of the two Gal4 transgenes was reported to be similar in the tarsi, the pharyngeal taste organ, and the taste pegs lining the oral surface of the labial palps (Ganguly et al., 2017). When the Ir76b cDNA was expressed with the use of Ir76b-Gal4 in the Ir76b mutant, the cellular responses were restored to levels comparable to wild-type in both L− and s-bristles, consistent with Ir76b requirement for NaCl detection (Figs. 4A and 4B). Interestingly, upon reintroduction of Ir76b cDNA with Ir76b-Gal4RB, only the s-bristle and not the L-bristle GRNs showed NaCl-evoked action potentials in Ir76b mutants. Therefore, the two Ir76b-Gal4 transgenic lines provide an intriguing neuroethological window to the physiological significance of NaCl-responding s− and L-bristle cells in NaCl-induced feeding behavior.
Fig. 4. Ir76b cDNA rescue by Ir76b-Gal4RB restores NaCl sensitivity in s-bristle but not L-bristle GRNs, while expression by Ir76b-Gal4 (CM) restores GRN NaCl responsiveness in both bristle types.
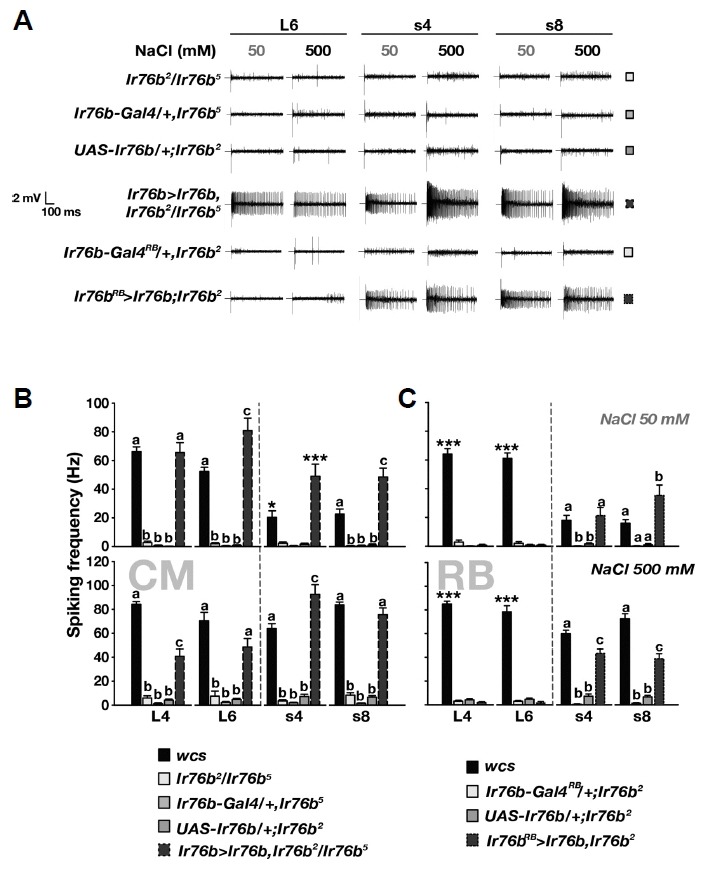
(A) Representative GRN action potential recordings in Ir76b cDNA rescue experiments. (B) Ir76b-Gal4 (CM) restores NaCl responses of L− and s-bristle GRNs in Ir76b-deficient animals (n = 8–18). (C) Ir76b-Gal4RB rescues only s-bristle GRNs in NaCl sensitivity (n = 16–27). Letters mark statistically distinct groups. *p < 0.05, **p < 0.01, ***p < 0.001. Tukey’s test.
Comparative behavioral analyses with the two Ir76b-Gal4 transgenes reveal that L-bristle Ir76b GRNs are dispensable for feeding attraction, and rather contribute to NaCl feeding avoidance
Reintroduction of Ir76b cDNA with Ir76b-Gal4 fully restored feeding avoidance to NaCl in Ir76b mutants, when assessed with the CAFE feeding assay. In contrast, expression of Ir76b cDNA in Ir76b-Gal4RB cells failed to significantly recue the Na+ avoidance deficit (Fig. 5A), suggesting that the s-bristle Ir76b GRNs are insufficient for full Na+ feeding avoidance. On the other hand, RNAi knockdown of Ir76b by either Ir76b-Gal4 or Ir76b-Gal4RB similarly attenuated the feeding avoidance (Fig. 5B), which indicates that s-bristle cells are insufficient but required for full Na+ feeding suppression. The Drosophila TRPA1 isoform, TRPA1(A), was recently identified as a receptor for nucleophilicity-bearing H2O2 that mediates feeding avoidance to H2O2-containing food in labellar bitter GRNs (Du et al., 2016b). To further characterize the Ir76b-Gal4 and Ir76b-Gal4RB cells, we attempted to ectopically rescue feeding aversion to H2O2 in the TrpA1-deficient mutant, TrpA1ins, using these Ir76b-Gal4 transgenes. As expected, Ir76b-Gal4 that physiologically covers both the s− and L-bristle Ir76b GRNs allowed TrpA1ins flies to avoid 100 mM H2O2-containing sucrose solution. However, TrpA1(A)10b expression (Du et al., 2016a) in TrpA1-deficient Ir76b-Gal4RB cells induced attraction rather than avoidance in response to 100 mM H2O2 (Fig. 5C). Given that Ir76b was previously implicated to be responsible for low-Na+ attraction (Zhang et al., 2013) as opposed to the high-Na+ aversion observed in this study, it is conceivable that the population of Ir76b-positive GRNs is heterogeneous with two groups relaying opposite messages to the brain upon firing. In this regard, the attraction to H2O2, achieved by Ir76b-Gal4RB, indicates that L-bristle Ir76b cells are expendable for Na+ attraction unlike the previous report by Zhang et al. and also suggests that the failure to fully restore Na+ avoidance by Ir76b-Gal4RB may be due to prevalent representation of attractive signals from unidentified attractive Ir76b cells. Conversely, L-bristle Ir76b cells not functionally encompassed by Ir76b-Gal4RB may pertain to high Na+ aversion, as lacking the cells shifts overall coding of Ir76b GRNs towards attraction.
Fig. 5. Comparative feeding analyses using Ir76b-Gal4RB and Ir76b-Gal4 suggests that L-bristle Ir76b GRNs are dispensable for feeding attraction.
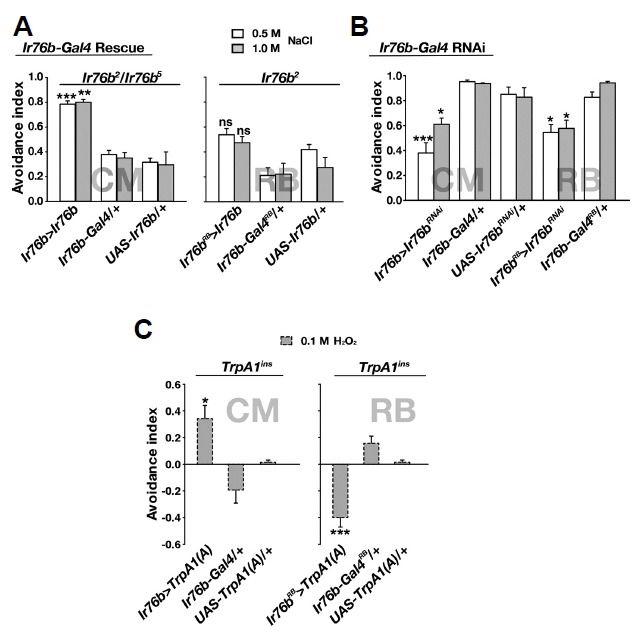
(A) Ir76b cDNA rescue of feeding avoidance to concentrated NaCl with either Ir76b-Gal4RB (Right, n = 4–5) or Ir76b-Gal4 (Left, n = 4) in Ir76b-lacking flies. (B) Ir76b RNAi knockdown effects on feeding avoidance to NaCl with either Ir76b-Gal4RB or Ir76b-Gal4 (n = 4–5). (C) TrpA1(A)10b cDNA rescue of feeding avoidance to hydrogen peroxide by the two Gal4 transgenes in TrpA1-deficient animals shows that overall output from Ir76b-Gal4RB cells codes for feeding attraction (n = 4–8). *p < 0.05, **p < 0.01, ***p < 0.001, and ns: not significant. Tukey’s test.
DISCUSSION
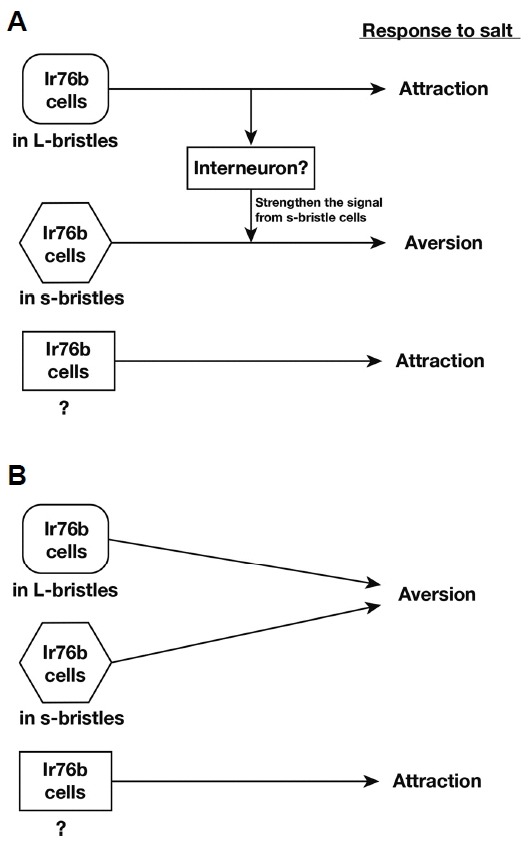
The molecular mechanism of high-salt taste has been enigmatic, while low-salt taste was identified to depend on ENaC in mammals. The only existing mechanistic model of high-Na+ taste is C. elegans TMC-1 that mediates negative chemotaxis to high concentrations of environmental Na+ (Chatzigeorgiou et al., 2013). However, the Na+ sensitivity of TMC-1 recently became debatable (Wang et al., 2016), and Drosophila tmc-1 mutants were found to be comparable to wild type regarding high-Na+ taste in our study (Fig. 1B). By conducting a reverse genetic screen, we discovered the Ir76b gene and s-bristle Ir76b-dependent GRNs as the genetic and cellular bases of Drosophila high-Na+ taste, respectively, extending the role of the Ir76b gene from low to high concentrations of Na+. Our study adopting the CAFE assay, a reliable quantitative feeding assay (Deshpande et al., 2014), however, failed to recapitulate Ir76b-dependent attraction to low Na+ concentrations (Fig. 3C), suggesting that a specific feeding context is necessary to attain Ir76b-dependent attraction. A subset of Ir76b-positive cells likely codes for feeding attraction, as expression of TrpA1(A)10b in Ir76b-Gal4 RB cells led to feeding attraction towards H2O2 in a TrpA1-deficient mutant (Fig. 5C). From these results, two models of feeding preference decision in response to Na+ may be drawn (Fig. 6). As proposed by Zhang et al., Ir76b GRNs in L-bristles may signal to the brain for feeding attraction. Through interneurons or gap junction, robust activation of L-bristle GRNs at high Na+ concentrations may then aid to warrant sufficient excitation of the aversive s-bristles GRNs (Fig. 6A). In this manner, Ir76b GRNs in L-bristles, which are active from low Na+ concentrations in comparison to s-bristle cells, are required for both attraction and aversion to increasing concentrations of Na+. Besides this view, the parsimonious model would be that the L-bristle GRNs code for repulsion, as opposed to the conclusion by Zhang et al. (Fig. 6B). In both cases, unidentified Ir76b cells ought to exist for feeding attraction, as lack of the L-bristle GRNs appears to move the balance of the whole Ir76b cell population towards attraction. Thus, our results expose the complexity of Ir76b-dependent Na+ gustation in Drosophila, and various Gal4 transgenes covering different subpopulations of the Ir76b cells would facilitate future studies further dissecting the cellular substrates for Ir76b-dependent Na+ feeding behavior.
Fig. 6. Schematic diagram of two taste-coding models for Ir76b-dependent high-Na+ avoidance.
Heterologous expression of Ir76b in HEK293 cells showed intrinsic Na+ sensitivity of the IR76b protein (Zhang et al., 2013). In addition, two recent studies demonstrate that Ir76b is critical for gustation to amino acids (Croset et al., 2016; Ganguly et al., 2017). Such dual responsiveness to salt and amino acids is reminiscent of extracellular Ca2+-sensing receptor (CaSR) (Mun et al., 2004) and mammalian kainate receptor iGluRs, GRIK1 and GRIK2 (Plested et al., 2008). CaSR, a GPCR, is activated by Ca2+ and other divalent cations as well as L-amino acids. In particular, iGluRs homologous to IR76b seem to be functionally conserved in salt sensitivity, as they bind Na+ ions to be activated by glutamate. In addition, iGluRs have been reported to be present in taste receptor cells (Brand et al., 1991; Chung et al., 2005). Thus, our study finds Ir76b to critically contribute to high-Na+ taste, and suggests the interesting possibility that iGluRs may in part compose the molecular mechanism of high Na+ taste in mammals which has yet to be identified.
ACKNOWLEDGMENTS
We thank Dr. Seok Jun Moon and the Bloomington Drosophila stock center for flies. This work was supported by Basic Science Research Program to KJK (NRF-2015R1D1A1 A01057288 and NRF-2017R1A4A1015534) and JYK (NRF-2016R1D1A1B03932743) through the National Research Foundation (NRF) of Korea funded by the Ministry of Education.
REFERENCES
- Benton R., Vannice K.S., Gomez-Diaz C., Vosshall L.B. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand J.G., Teeter J.H., Kumazawa T., Huque T., Bayley D.L. Transduction mechanisms for the taste of amino acids. Physiol Behav. 1991;49:899–904. doi: 10.1016/0031-9384(91)90201-x. [DOI] [PubMed] [Google Scholar]
- Calleja M., Moreno E., Pelaz S., Morata G. Visualization of gene expression in living adult Drosophila. Science. 1996;274:252–255. doi: 10.1126/science.274.5285.252. [DOI] [PubMed] [Google Scholar]
- Chandrashekar J., Kuhn C., Oka Y., Yarmolinsky D.A., Hummler E., Ryba N.J.P., Zuker C.S. The cells and peripheral representation of sodium taste in mice. Nature. 2010;464:297–301. doi: 10.1038/nature08783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzigeorgiou M., Bang S., Hwang S.W., Schafer W.R. tmc-1 encodes a sodium-sensitive channel required for salt chemosensation in C. elegans. Nature. 2013;494:95–99. doi: 10.1038/nature11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K.M., Lee S.B., Heur R., Cho Y.K., Lee C.H., Jung H.Y., Chung S.H., Lee S.P., Kim K.N. Glutamate-induced cobalt uptake elicited by kainate receptors in rat taste bud cells. Chem Senses. 2005;30:137–143. doi: 10.1093/chemse/bji009. [DOI] [PubMed] [Google Scholar]
- Croset V., Schleyer M., Arguello J.R., Gerber B., Benton R. A molecular and neuronal basis for amino acid sensing in the Drosophila larva. Sci Rep. 2016;6:34871. doi: 10.1038/srep34871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande S.A., Carvalho G.B., Amador A., Phillips A.M., Hoxha S., Lizotte K.J., Ja W.W. Quantifying Drosophila food intake: comparative analysis of current methodology. Nat Meth. 2014;11:535–540. doi: 10.1038/nmeth.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du E.J., Ahn T.J., Choi M.S., Kwon I., Kim H.-W., Kwon J.Y., Kang K. The mosquito repellent citronellal directly potentiates Drosophila TRPA1, facilitating feeding suppression. Mol Cells. 2015;38:911–917. doi: 10.14348/molcells.2015.0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du E.J., Ahn T.J., Kwon I., Lee J.H., Park J.-H., Park S.H., Kang T.M., Cho H., Kim T.J., Kim H.-W., et al. TrpA1 regulates defecation of food-borne pathogens under the control of the duox pathway. PLoS Genet. 2016a;12:e1005773. doi: 10.1371/journal.pgen.1005773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du E.J., Ahn T.J., Wen X., Seo D.-W., Na D.L., Kwon J.Y., Choi M., Kim H.-W., Cho H., Kang K. Nucleophile sensitivity of Drosophila TRPA1 underlies light-induced feeding deterrence. Elife. 2016b;5:e18425. doi: 10.7554/eLife.18425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisoli T.M., Schmieder R.E., Grodzicki T., Messerli F.H. Salt and hypertension: is salt dietary reduction worth the effort? Am. J Med. 2012;125:433–439. doi: 10.1016/j.amjmed.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Ganguly A., Pang L., Duong V.-K., Lee A., Schoniger H., Varady E., Dahanukar A. A Molecular and cellular context-dependent role for Ir76b in detection of amino acid taste. Cell Rep. 2017;18:737–750. doi: 10.1016/j.celrep.2016.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramates L.S., Marygold S.J., Santos Gdos Urbano J.-M., Antonazzo G., Matthews B.B., Rey A.J., Tabone C.J., Crosby M.A., Emmert D.B., et al. FlyBase at 25: looking to the future. Nucleic Acids Res. 2017;45:D663–D671. doi: 10.1093/nar/gkw1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn Y., Kim D.S., Pastan I.H., Lee B. Anoctamin and transmembrane channel-like proteins are evolutionarily related. Int J Mol Med. 2009;24:51–55. doi: 10.3892/ijmm_00000205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F.J., MacGregor G.A. A comprehensive review on salt and health and current experience of worldwide salt reduction programmes. J Hum Hypertens. 2008;23:363–384. doi: 10.1038/jhh.2008.144. [DOI] [PubMed] [Google Scholar]
- Hiroi M., Meunier N., Marion-Poll F., Tanimura T. Two antagonistic gustatory receptor neurons responding to sweet-salty and bitter taste in Drosophila. J Neurobiol. 2004;61:333–342. doi: 10.1002/neu.20063. [DOI] [PubMed] [Google Scholar]
- Hodgson E.S., Lettvin J.Y., Roedert K.D. Physiology of a primary chemoreceptor unit. Science. 1955;122:121–122. doi: 10.1126/science.122.3166.417-a. [DOI] [PubMed] [Google Scholar]
- Ja W.W., Carvalho G.B., Mak E.M., de la Rosa N.N., Fang A.Y., Liong J.C., Brummel T., Benzer S. Prandiology of Drosophila and the CAFE assay. Proc Natl Acad Sci USA. 2007;104:8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K., Pulver S.R., Panzano V.C., Chang E.C., Griffith L.C., Theobald D.L., Garrity P.A. Analysis of Drosophila TRPA1 reveals an ancient origin for human chemical nociception. Nature. 2010;464:597–600. doi: 10.1038/nature08848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K., Panzano V.C., Chang E.C., Ni L., Dainis A.M., Jenkins A.M., Regna K., Muskavitch M.A.T., Garrity P.A. Modulation of TRPA1 thermal sensitivity enables sensory discrimination in Drosophila. Nature. 2012;481:76–80. doi: 10.1038/nature10715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinewietfeld M., Manzel A., Titze J., Kvakan H., Yosef N., Linker R.A., Muller D.N., Hafler D.A. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature. 2013;496:518–522. doi: 10.1038/nature11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht Z.A., Silbering A.F., Ni L., Klein M., Budelli G., Bell R., Abuin L., Ferrer A.J., Samuel A.D., Benton R., et al. Distinct combinations of variant ionotropic glutamate receptors mediate thermosensation and hygrosensation in Drosophila. Elife. 2016;5:44–60. doi: 10.7554/eLife.17879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht Z.A., Silbering A.F., Cruz J., Yang L., Croset V., Benton R., Garrity P.A. Ionotropic Receptor-dependent moist and dry cells control hygrosensation in Drosophila. Elife. 2017;6 doi: 10.7554/eLife.26654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko K.I., Root C.M., Lindsay S.A., Zaninovich O.A., Shepherd A.K., Wasserman S.A., Kim S.M., Wang J.W., Pachter L., Lavista-Llanos S., et al. Starvation promotes concerted modulation of appetitive olfactory behavior via parallel neuromodulatory circuits. Elife. 2015;4:e50801. doi: 10.7554/eLife.08298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika S.P., Lisbin M.J., White K. ELAV, a Drosophila neuron-specific protein, mediates the generation of an alternatively spliced neural protein isoform. Curr Biol. 1996;6:1634–1641. doi: 10.1016/s0960-9822(02)70787-2. [DOI] [PubMed] [Google Scholar]
- Mun H.-C., Franks A.H., Culverston E.L., Krapcho K., Nemeth E.F., Conigrave A.D. The venus fly trap domain of the extracellular Ca2+-sensing receptor is required for l-amino acid sensing. J Biol Chem. 2004;279:51739–51744. doi: 10.1074/jbc.M406164/200. [DOI] [PubMed] [Google Scholar]
- Ni L., Klein M., Svec KV Budelli G., Chang E.C., Ferrer A.J., Benton R., Samuel A.D., Garrity P.A. The Ionotropic Receptors IR21a and IR25a mediate cool sensing in Drosophila. Elife. 2016;5:e13254. doi: 10.7554/eLife.13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewalda T., Singhal N., Fiala A., Saumweber T., Wegener S., Gerber B. Salt processing in larval Drosophila: choice, feeding, and learning shift from appetitive to aversive in a concentration-dependent way. Chem Senses. 2008;33:685–692. doi: 10.1093/chemse/bjn037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y., Butnaru M., von Buchholtz L., Ryba N.J.P., Zuker C.S. High salt recruits aversive taste pathways. Nature. 2013:1–5. doi: 10.1038/nature11905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plested A.J.R., Vijayan R., Biggin P.C., Mayer M.L. Molecular basis of kainate receptor modulation by sodium. Neuron. 2008;58:720–735. doi: 10.1016/j.neuron.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig M., Kang K., Garrity P.A.P.A. Distinct TRP channels are required for warm and cool avoidance in Drosophila melanogaster. Proc Natl Acad Sci USA. 2008;105:14668–73. doi: 10.1073/pnas.0805041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silbering A.F., Rytz R., Grosjean Y., Abuin L., Ramdya P., Jefferis G.S.X.E., Benton R. Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J Neurosci. 2011;31:13357–75. doi: 10.1523/JNEUROSCI.2360-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugane S., Sasazuki S., Kobayashi M., Sasaki S. Salt and salted food intake and subsequent risk of gastric cancer among middle-aged Japanese men and women. Br J Cancer. 2004;90:128–134. doi: 10.1038/sj.bjc.6601511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Li G., Liu J., Liu J., Xu X.Z.S., Wang X., Li G., Liu J., Liu J., Xu X.Z.S. TMC-1 mediates alkaline sensation in C. elegans through nociceptive neurons. Neuron. 2016;91:146–154. doi: 10.1016/j.neuron.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss L.A., Dahanukar A., Kwon J.Y., Banerjee D., Carlson J.R. The molecular and cellular basis of bitter taste in Drosophila. Neuron. 2011;69:258–272. doi: 10.1016/j.neuron.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong X.M., Younger S., Peters C.J., Jan Y.N., Jan L.Y., Shim W. Subdued, a TMEM16 family Ca2+ -activated Cl− channel in Drosophila melanogaster with an unexpected role in host defense. Elife. 2013;2:e00862. doi: 10.7554/eLife.00862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarmolinsky DA Zuker C.S., Ryba N.J.P. Common sense about taste: from mammals to insects. Cell. 2009;139:234–244. doi: 10.1016/j.cell.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelle K.M., Lu B., Pyfrom S.C., Ben-Shahar Y. The genetic architecture of degenerin/epithelial sodium channels in Drosophila. G3 (Bethesda) 2013;3:441–450. doi: 10.1534/g3.112.005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YV Ni J., Montell C. The molecular basis for attractive salt-taste coding in Drosophila. Science. 2013;340:1334–1338. doi: 10.1126/science.1234133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitron A.E., Hawley R.S. The genetic analysis of distributive segregation in Drosophila melanogaster. Genetics. 1989;122:801–821. doi: 10.1093/genetics/122.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]


