Abstract
Aim:
The aim of our study was polymerase chain reaction (PCR) detection of the genes responsible for the multiple antibiotic resistance S. aureus isolated from food of animal origin in Egypt.
Materials and Methods:
A total of 125 samples were randomly collected from milk, meat, and their products from Giza and Beni-Suef Governorates markets. The S. aureus isolates were subjected to antimicrobial sensitivity tests using four antibacterial disks (Oxoid), and then the polymerase chain reaction (PCR) was performed for detection of antibiotic resistance genes.
Results:
Out of 125 samples, 19 S. aureus isolates were detected. All detected isolates were multiple drug resistance (MDR). The penicillin-, erythromycin-, kanamycin-, and tetracycline-resistant isolates were examined by PCR for resistance genes blaZ, (msrA, ermB, and ermC), aac(6’)aph (2”), and tetK. The isolates harbored these resistance genes with percentage of 100% (100%, 0%, and 100%), 62.5%, and 100%, respectively.
Conclusion:
Contaminated foods of animal origin may represent a source of MDR S. aureus that can be a major threat to public health.
Keywords: food of animal origin, multiple antibiotic resistance, polymerase chain reaction, resistance genes, Staphylococcus aureus
Introduction
The development of multiple antibiotic-resistant bacteria due to misuse of antibiotics in animals and poultry production is well authenticated for pathogenic bacteria [1,2]. Contaminated food of animal origins with antibiotic-resistant bacteria can be a great threat to public health, and the antibiotic resistance determinants can be transferred from antibiotic-resistant bacteria to other bacteria affecting human [3,4]. Identical elements of antibiotic-resistant genes found in bacteria that affect both animals and humans have shown the role of raw foods in the dissemination of these resistance genes through the food chains [5,6] or through occupational contact with livestock [7].
The multiple antibiotic-resistant bacteria were commonly isolated from food of animal origin such as raw milk and unpasteurized dairy products [2,8] and meat products [9], the resistance genes can be transferred from antibiotic-resistant bacteria to the intestinal flora of humans through food products, and the commensally flora can be a reservoir of resistant genes for pathogenic bacteria [10]. The high prevalence of multidrug-resistant Staphylococcus aureus was discovered from food of animal origin in Europe, Canada, and United States in multiple studies [11,12], which represents a huge problem in public health [13,14].
The aim of our study was polymerase chain reaction (PCR) detection of the genes responsible for the multiple antibiotic resistance S. aureus isolated from food of animal origin in Egypt.
Materials and Methods
Ethical approval
No animals were involved in the study at any stage.
Samples
A total of 125 samples were collected from meat, milk, and their products from Giza and Beni-Suef Governorates markets (Table-1), and all samples were aseptically collected and examined for detection of S. aureus.
Table-1.
Samples collected and their numbers from sale markets.
| Product | Milk | Yoghurt | Kareish | Minced meat | Burger | Luncheon | Total |
|---|---|---|---|---|---|---|---|
| Numbers | 28 | 18 | 19 | 20 | 20 | 20 | 125 |
Identification and characterization of S. aureus
One loopful from prepared incubated samples was plated onto nutrient agar (Difco) and mannitol salt agar (Difco), incubated for 18-24 h at 37°C and examined for bacterial growth. The suspected colonies were identified morphologically and biochemically [15].
Antimicrobial sensitivity test for identified strains
The antimicrobial sensitivity tests were done by disk diffusion technique [16] using 4 antibacterial disks (kanamycin, penicillin, erythromycin, and tetracycline) - Oxoid - the degree of sensitivity was interpreted according to Koneman et al., and CLSI [17,18].
PCR detection of the resistance genes
PCR was performed for detection of resistance genes in biotechnology center in the animal health institute according to Sambrook and Russel [19]. The primers were synthesized by Metabion Company, Germany, as mentioned in Table-2.
Table-2.
Oligonucleotide primers sequences of resistance genes.
| Antibiotic resistance | Target gene | Primer sequence (5’-3’) | Length of amplified product (bp) | References |
|---|---|---|---|---|
| Penicillin | blaZ | ACTTCAACACCTGCTGCTTTC | 173 | [20] |
| TGACCACTTTTATCAGCAACC | ||||
| Tetracycline | tet(K) | GTAGCGACAATAGGTAATAGT | 360 | |
| GTAGTGACAATAAACCTCCTA | ||||
| Aminoglycoside | aac(6’)aph (2”) | GAAGTACGCAGAAGAGA | 491 | |
| ACATGGCAAGCTCTAGGA | ||||
| Erythromycin | msr(A) | GCAAATGGTGTAGGTAAGACAACT | 400 | [21] |
| ATCATGTGATGTAAACAAAAT | ||||
| erm(C) | ATCTTTGAAATCGGCTCAGG | 295 | ||
| CAAACCCGTATTCCACGATT | ||||
| erm(B) | CATTTAACGACGAAACTGGC | 425 | ||
| GGAACATCTGTGGTATGGCG |
Results
Results of recovery rate of S. aureus isolates
Out of 125 collected samples, 19 S. aureus isolates were detected (Table-3) [20,21].
Table-3.
Prevalence of the isolated S. aureus.
| Source of the samples | Total number of samples examined | Recovered S. aureus examined/total number of original samples |
|---|---|---|
| n (%) | ||
| Milk | 28 | 8 (28.5) |
| Yogurt | 19 | 1 (5.2) |
| Kareish cheese | 18 | 0 (0) |
| Total milk and milk products | 65 | 9 (13.8) |
| Minced meat | 20 | 5 (25) |
| Burger | 20 | 2 (10) |
| Luncheon | 20 | 3 (15) |
| Total meat and meat products | 60 | 10 (16.6) |
| Total collected samples | 125 | 19 (15.2) |
S. aureus=Staphylococcus aureus
Results of antibacterial sensitivity
Results of antibiotic sensitivity test on 19 isolates of S. aureus recovered from raw milk, meat, and their products, 14 isolates exhibited resistance against penicillin (73.6%), 11 isolates were resistant against tetracycline (57.8%), while 5 isolates exhibited resistance to erythromycin (26.3%), and 8 isolates were resistant to kanamycin (42.1%) (Table-4).
Table-4.
Results of antibiotic sensitivity tests.
| Antibacterial agent | Milk and milk products (total n=9) | Meat and meat products (total n=10) | ||||
|---|---|---|---|---|---|---|
| Sensitive | Intermediate | Resistant | Sensitive | Intermediate | Resistant | |
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |
| Penicillin groups | ||||||
| Penicillin | 2 (22.2) | 0 (-) | 7 (77.7) | 3 (30) | 0 (-) | 7 (70) |
| Tetracycline group | ||||||
| Tetracycline | 2 (22.2) | 0 (-) | 7 (77.7) | 3 (30) | 3 (30) | 4 (40) |
| Aminoglycoside group | ||||||
| Kanamycin | 2 (22.2) | 2 (22.2) | 5 (55.5) | 4 (40) | 3 (30) | 3 (30) |
| Macrolide group | ||||||
| Erythromycin | 6 (66.6) | 1 (11) | 2 (22.2) | 5 (50) | 2 (20) | 3 (30) |
Results of PCR for amplification of blaz gene at 173 bp fragment
Fourteen isolates exhibited resistance against penicillin, out of them; eight isolates were randomly selected for genotypic detection of blaz gene. Amplification of blaz gene at amplicon size of 173 bp was detected in all the tested isolates (8) with a percentage of 100% (Figure-1).
Figure-1.
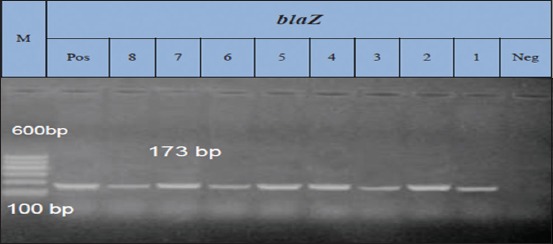
Lane: 1-8 positive amplification of blaz gene at 173 bp. Neg=Negative control, pos=Positive control, M=Marker.
Results of PCR for amplification of tet K gene at 360 bp fragment performed with its specific primer
Eleven isolates exhibited resistance against tetracycline, out of them; eight isolates were selected randomly for genotypic detection of tetK gene.
Amplification of tetK gene at amplicon size of 360 bp was detected in all the tested isolates with a percentage of 100% (Figure-2).
Figure-2.
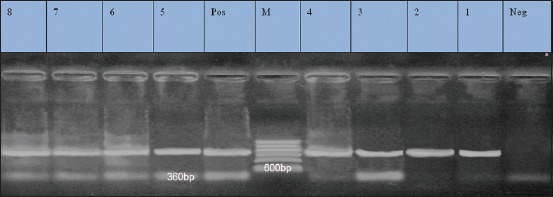
Lane 1-8: Positive amplification of tet K gene at 360 bp. Neg=Negative control, pos=Positive control, M=Marker.
Results of PCR for amplification of ermB (425 bp), msrA (400 bp), and ermC (295 bp) for erythromycin resistance isolates
Five isolates were resistant to erythromycin. They were examined by PCR for detection of ermB, msrA, and ermC genes. They exhibited prevalence of 0%, 100%, and 100% for ermB, msrA, and ermC genes, respectively (Figures-3-5).
Figure-3.
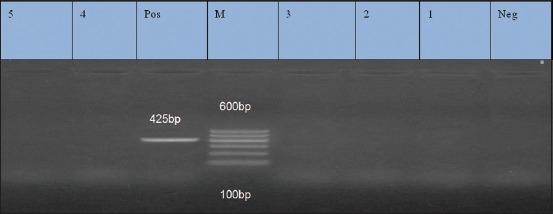
Lane 1, 2, 3, 4, 5: Negative amplification of ermB gene at 425 bp. Neg=Negative control, Pos=Positive control, M=Marker.
Figure-4.
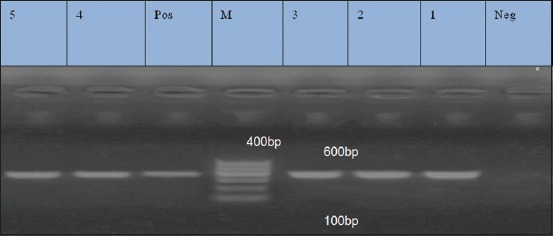
Lane 1-5: Positive amplification of msrA gene at 400 bp. Neg=Negative control, pos=Positive control, M=Marker.
Figure-5.

Lane 1-5: Positive amplification of ermC gene at 295 bp. Neg=Negative control, pos=Positive control, M=Marker.
Results of PCR for amplification of 491 bp fragment for aac(6’) aph (2”) gene (aminoglycoside)
Antibiotic susceptibility against aminoglycoside (kanamycin) using disk diffusion method revealed that eight isolates were resistant to kanamycin. They were examined by PCR for detection of aac(6’) aph(2”) gene, and the result revealed that the aac(6’) aph(2”) was detected in five (62.5%) out of eight isolates (Figure-6).
Figure-6.
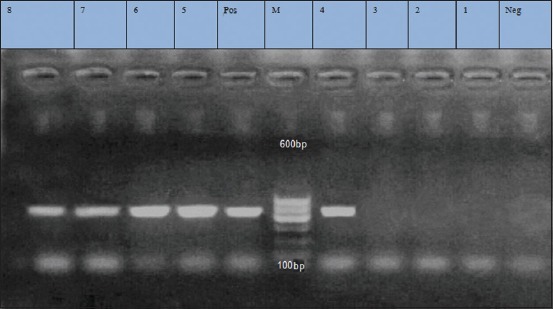
Lane 4, 5, 6, 7, 8 positive amplification of aac(6’) aph (2”) at 491 bp. Lane 1, 2, 3: Negative amplification of aac (6’) aph (2”). Neg=Negative control, pos=Positive control, M=Marker.
Discussion
The livestock products could be a source of exposure to multidrug-resistant S. aureus strains as a result of hazardous misuse of antibiotics in animal treatment and unhygienic livestock practices [2]. Food of animal origin is an ideal culture medium for growth of many organisms [22]. They are considered as a shelter of different types of microorganisms through processing, handling, preparation, and storage as well as distribution [23]. They are considered as major sources of foodborne diseases and have been linked to serious outbreaks of food poisoning worldwide.
The result showed in Table-3 reported that S. aureus isolated from raw milk and milk products (Kareish cheese and yoghurt), and from meat and meat products (burger and luncheon) were 13.8% and 16.6%, respectively. A higher recovery rate was obtained by El-Jakee et al. [24], they isolated S. aureus from cow milk (22.7%) and buffalo milk (16%), and on the other hand, El-Jakee et al. [25] revealed that raw milk contaminated with S. aureus with an incidence of 56%. While Jamali et al. [26] reported S. aureus with a percentage of 15.7% and 12.4% from dairy products and raw milk, respectively, Imani et al. [27] found 4% of milk and 32% of dairy products contaminated with S. aureus, similar results were obtained by Song et al. [28], who isolated S. aureus from raw milk with a percentage of 20.2%. Higher results of S. aureus contamination were reported in raw milk by Gwida and El-Gohary [29]. They recorded 56.66% of S. aureus present in market milk. The prevalence of S. aureus in Kareish cheese in our results is lower than the result reported by Hosny et al. [30], who isolated S. aureus with a percentage of 17% from milk shops and street vend but reported the same result from brand cheese. The presence of S. aureus in milk was variable in different regions, and these variations may be due to season, number of animals on the farm, farm size, hygiene status, variation in sampling, farm management practices, geographical location, and differences in detection methods and variation in types of samples evaluated. El-Sayed et al. [31] revealed that in Egypt the difference in white soft cheese due to acidity as Domiati or Kareish acid coagulation, enzyme coagulation, keeping temperatures, different salt concentrations, and ripening in brine solutions are factors affecting the microbiological quality of these varieties.
The incidence of S. aureus in meat products in our study agrees with the findings of the study by Fox et al. [32], who tested 124 raw meat samples for methicillin-resistant S. aureus (MRSA) including pig (n=63), poultry (n=50), and turkey (n=11) collected from England between March and July 2015. MRSA was isolated from 9 (73%) samples (4 poultry, 3 pig, and 2 turkeys) from different butchers and supermarkets. While Pesavento et al. [33] isolated S. aureus from raw meat 23.86%. Another study by Ge et al. [34] detected S. aureus from retail meats of turkey, pork, beef, and chickens. While Ali et al. [35] isolated S. aureus from meat samples with a percentage of 7%. Hassanin [36] isolated S. aureus from burger and luncheons agreed with our results with a percentage of 25% and 47.5%, respectively, but differ in minced meat (65%). S. aureus was isolated from meat products with a percentage of 30% by Abdaslam et al. [37]. While Li et al. [38] were recorded 27.9% of S. aureus isolates from food sample. Another study recovered S. aureus in 27 from 165 retail meat samples with a percentage of 16.4% [39]. On the other hand, Song et al. [28] isolated S. aureus 21.3% from frozen food and 28.1% from raw meat samples.
Transmission of antibiotic-resistant S. aureus strains can be done by contaminated foods with resistant bacteria [40]. Some researchers reported a primary relationship between the prevalence of antibiotic-resistant bacteria and the misusing of antibiotics for therapeutic purposes in animals [41].
The result of our S. aureus sensitivity test (Table-4) revealed that 14 out of 19 identified S. aureus isolates were resistant to penicillin (73.6%), while 11 isolates exhibited resistance against tetracycline (57.8%), 5 isolates were resistant to erythromycin (26.3%), and 8 isolates were resistant to kanamycin (42.1%). All the isolates were multidrug resistance (MDR) because they were resistant for more than one antibiotic class. The same results were found by Ammar et al. [42], and they observed MDR S. aureus (MDRSA) among 85% of isolates recovered from examined milk and meat product samples. Approximately 10.4% from S. aureus detected in retail meats in 1-year survey (2010-2011) collected from eight U.S. states were MDRSA Ge et al. [34]. While Jamali et al. [26] stated that 36.3%, 46.6%, and 12.8% of isolates were resistant to one, two, and more than two antimicrobial agents, respectively, and found that S. aureus resistant to tetracycline with a percentage of 56.1%, chloramphenicol (3.7%), and gentamicin (2.1%) but low incidence in case of erythromycin, kanamycin, streptomycin, penicillin G, and oxacillin. Our study agreed with many reports indicating a high percentage of multidrug-resistant S. aureus isolates from food of animal origin [32,43-45]. The same results obtained by Argudín et al. [46], and they found that S. aureus resistant to oxacillin (95%) and trimethoprim-sulfamethoxazole (4%) but differed in erythromycin (70%), tetracycline (100%), kanamycin (29%), and gentamicin (14%) and also reported that 4%, 30%, 21%, and 33% of S. aureus isolates were resistant to more than four, three, and five classes of antibiotics, respectively. Tan et al. [47] disagreed with our results, and they stated that 94.59% of the strains were resistant only to one of the antibiotics or did not resistant to all of the tested antibiotics, while only 5.41% of S. aureus strains were multidrug resistant, while Teramoto et al. [48] found that S. aureus isolates from conventional retail meat were resistant to both erythromycin 50.0% and tetracycline 58.3%.
PCR-based molecular methods are preferred for determination of antibiotic-resistant genes. Recently, many studies have demonstrated the extremely high capacity of molecular methods such as PCR and pulsed-field gel electrophoresis; these methods were increasingly used for their specific, rapid, reliable, and accurate detection of bacteria and genes of interest [49]. Nowadays, the detection of antibiotic-resistant genes was accomplished by PCR methods directed to the linA, tetK, msrA, msrB, ermA, ermC, aacA-D, and tetM gens [50]. In this work, PCR primers that can be used to survey clinically relevant antibiotic resistance genes frequently encountered in S. aureus.
The results revealed that all tested isolates of S. aureus which were resistant to penicillin carried blaZ genes, the erythromycin-resistant isolates carried ermB, msrA, and ermC with a percentage of 0%, 100%, and 100%, respectively, and aminoglycoside gene aac(6’) aph (2”) (kanamycin resistance gene) present in a percentage of 62.5% in the resistant isolates while the tetracycline-resistant isolates carried tetK gene with a percentage of 100%. Our findings agreed with McCallum et al. and Argudín et al. [46,51], they reported that tetK gene was present in a percentage of 91% of tetracycline resistant, 70% of erythromycin-resistant isolates carried resistance genes (encoded by ermC, ermA, and ermB, alone or in combination), and aac(6’)aph (2”) gene found in kanamycin-resistant gene, while penicillin-resistant isolates carried blaZ gene 94%. While Jamali et al. [26] found that blaZ (97.4%) and tetK (41.8%) present in penicillin- and tetracycline-resistant isolates, respectively, and msrA and ermC genes (erythromycin resistance gene) present in high prevalence as our results but with different prevalence of ermB gene. The high prevalence of the blaZ and tet M resistance genes in this study is in agreement with the results reported by Gao et al. [52]. The same findings were obtained by Li et al. [38], on msrA, ermC, tetK, and blaZ genes with different results for ermB. Duran et al. [20] reported erythromycin resistance genes (ermA, ermB, ermC, and msrA), one of them at least was present in erythromycin-resistant isolates, tetM or tetK or both resistance genes isolates were found in tetracycline-resistant isolates, aac(6’)aph(2”) presents in gentamicin susceptible S. aureus isolates, and major of staphylococci tested possessed the blaZ gene (89.9%). Argudín et al. [46] reported that all strains resistant to ampicillin–penicillin carried the blaZ gene, and they detected the genes responsible for erythromycin resistance together with inducible resistance to clindamycin were ermA and ermC while resistance to erythromycin only was associated with the presence of either msrB or msrB msrA. A high prevalence of ermB gene than ermC in food of animal origin was detected by Martineau et al. [53].
Conclusion
Foods of animal origin may represent a source of MDR S. aureus that can be a major threat to public health infection for humans.
Authors’ Contributions
FRE and AAS have planned the research work; also they participate laboratory work with HSHS, EAK, and AAK. All authors.
Acknowledgments
The authors acknowledge the National Research Center, Cairo, Egypt, for complete funding of our study. The authors are also thankful to Faculty of Veterinary Medicine, Beni-Suef University, Beni-Suef, Egypt.
Competing Interests
The authors declared that they have no competing interests.
References
- 1.Hawkey P.M, Jones A.M. The changing epidemiology of resistance. J. Antimicrob. Chemother. 2009;64(Suppl 1):i3–10. doi: 10.1093/jac/dkp256. [DOI] [PubMed] [Google Scholar]
- 2.Arenas N.E, Abril D.A, Valencia P, Khandige S, Soto C.Y, Moreno-Melo V. Screening food-borne and zoonotic pathogens associated with livestock practices in the Sumapaz region, Cundinamarca, Colombia. Trop. Anim. Health Prod. 2017;49(4):739–745. doi: 10.1007/s11250-017-1251-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Threlfall E.J, Ward L.R, Frost J.A, Willshaw G.A. The emergence and spread of antibiotic resistance in food-borne bacteria. Int. J. Food Microbiol. 2000;62:1–5. doi: 10.1016/s0168-1605(00)00351-2. [DOI] [PubMed] [Google Scholar]
- 4.Chiu C.H, Wu T.L, Su L.H, Chu C, Chia J.H, Kuo A.J, Chien M.S, Lin T.Y. The emergence in Taiwan of fluoroquinolone resistance in Salmonella enterica serotype choleraesuis. N. Engl. J. Med. 2002;346:413–419. doi: 10.1056/NEJMoa012261. [DOI] [PubMed] [Google Scholar]
- 5.O'Brien A.M, Hanson B.M, Farina S.A, Wu J.Y, Simmering J.E, Wardyn S.E. MRSA in conventional and alternative retail pork products. PLoS One. 2012;7(1):30092. doi: 10.1371/journal.pone.0030092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teuber M. Veterinary use and antibiotic resistance. Curr. Opin. Microbiol. 2001;4:493–499. doi: 10.1016/s1369-5274(00)00241-1. [DOI] [PubMed] [Google Scholar]
- 7.Leibler J.H, Jordan J.A, Brownstein K, Lander L, Price L.B, Perry M.J. Staphylococcus aureus nasal carriage among beef packing workers in a Midwestern United States slaughterhouse. PLoS One. 2016;11(2):e0148789. doi: 10.1371/journal.pone.0148789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munsch-Alatossava P, Alatossava T. Antibiotic resistance of raw-milk-associated psychrotrophic bacteria. Microbiol. Res. 2007;162(2):115–123. doi: 10.1016/j.micres.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 9.White D.G, Zhao S, Sudler R, Ayers S, Friedman S, Chen S, McDermott P.F, McDermott S, Wagner D.D, Meng J. The isolation of antibiotic-resistant Salmonella from retail ground meats. N. Engl. J. Med. 2001;345(16):1147–1154. doi: 10.1056/NEJMoa010315. [DOI] [PubMed] [Google Scholar]
- 10.Aarestrup F.M, Wegener H.C, Collignon P. Resistance in bacteria of the food chain:Epidemiology and control strategies. Expert Rev. Anti. Infect. Ther. 2008;6(5):733–750. doi: 10.1586/14787210.6.5.733. [DOI] [PubMed] [Google Scholar]
- 11.Khanna T, Friendship R, Dewey C, Weese J.S. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet. Microbiol. 2008;128:298–303. doi: 10.1016/j.vetmic.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Smith T.C, Pearson N. The emergence of Staphylococcus aureus ST398. Vector Borne Zoonotic Dis. 2011;11:327–339. doi: 10.1089/vbz.2010.0072. [DOI] [PubMed] [Google Scholar]
- 13.Morosini M.I, Garcia-Castillo M, Coque T.M, Valverde A, Novais A, Loza E, Canton R. Antibiotic coresistance in extended-spectrum-beta lactamase-producing Enterobacteriaceae and in vitro activity of tigecycline. Antimicrob. Agents Chemother. 2006;50:e2695–e2699. doi: 10.1128/AAC.00155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Jakee J.K, Nagwa S.A, Samy A.A, Bakry M.A, Elgabry E.A, Mai M.K, Gad El-Said W.A. Antimicrobial resistance in clinical isolates of Staphylococcus aureus from bovine and human sources in Egypt. Glob. Vet. 2011;7:581–586. [Google Scholar]
- 15.Quinn P.J, Markey B.K, Carter M.E, Donnelly W.J.C, Leonard F.C, Maguire D. Veterinary Microbiology and Microbial Diseases. 1st ed. Ames: Published Blackwell Science; 2002. [Google Scholar]
- 16.Finegold S.M, Martin W.J. Baileys and Scotts Diagnostic Microbiology. 6th ed. St. Louis, Toronto, London: The C.V. Mosby Company; 1982. [Google Scholar]
- 17.Koneman E, Allen S.D, Janda W.M, Schreckenberger P.C, Winn W.C. Color Atlas and Textbook of Diagnostic Microbiology. 5th ed. Philadelphia, PA, New York: Lippincott; 1997. [Google Scholar]
- 18.Clinical and Laboratory Standards Institute, CLSI. Approved Standards:Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard. 11th ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2013. [Google Scholar]
- 19.Sambrook X, Fritsch E.F, Maniatis T. Molecular Cloninga Laboratory Manual. 2nd ed. Cold Spring Harbor, New York: Laboratory Press; 1989. [Google Scholar]
- 20.Duran N, Ozer B, Duran G.G, Onlen Y, Demir C. Antibiotic resistance genes and susceptibility patterns in staphylococci. Indian J. Med. Res. 2012;135(3):389–396. [PMC free article] [PubMed] [Google Scholar]
- 21.Schlegelova J, Vlkova H, Babak V, Holasova M, Jaglic Z, Stosova T, Sauer P. Resistance to erythromycin of Staphylococcus spp. Isolates from the food chain. Vet. Med. 2008;53:307–314. [Google Scholar]
- 22.Hill W.E. The polymerase chain reaction:Applications for the detection of foodborne pathogens. Crit. Rev. Food Sci. Nutr. 1996;36(1-2):123–121. doi: 10.1080/10408399609527721. [DOI] [PubMed] [Google Scholar]
- 23.Hassanien F.S. Bacterial hazards associated with consumption of some meat products. Benha Vet. Med. J. 2004;15:41–54. [Google Scholar]
- 24.El-Jakee J, Ata-Nagwa S, Bakry M, Zouelfakar S.A, Elgabry E, Gad El-Said W.A. Characteristics of Staphylococcus aureus strains isolated from human and animal sources. Am. Eur. J. Agric. Environ. Sci. 2008;4(2):221–229. [Google Scholar]
- 25.El-Jakee J, Marouf S.A, Ata S.N, Abdel-Rahman H.E, Abd-Elmoez S, Samy A.A, El-Sayed E, Walaa E. Rapid method for detection of Staphylococcus aureus enterotoxins in food. Glob. Vet. 2013;11(3):335–341. [Google Scholar]
- 26.Jamali H, Paydar M, Radmehr B, Ismail S, Dadrasnia A. Prevalence and antimicrobial resistance of Staphylococcus aureus isolated from raw milk and dairy products. Food Control. J. 2015;54:383–388. [Google Scholar]
- 27.Imani F.A.A, Tavakoli H.R, Naderi A. Detection of enterotoxigenic S. aureus isolates in domestic dairy products. Iran. J. Microbiol. 2010;2(3):135–144. [PMC free article] [PubMed] [Google Scholar]
- 28.Song M, Bai Y, Xu J, Qiu C.M, Shi C, Shi X. Genetic diversity and virulence potential of Staphylococcus aureus isolates from raw and processed food commodities in Shanghai. Int. J. Food Microbiol. 2014;195:1–8. doi: 10.1016/j.ijfoodmicro.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 29.Gwida M.M, El-Gohary F.A. Zoonotic bacterial pathogens isolated from raw milk with special reference to Escherichia coli and Staphylococcus aureus in Dakahlia governorate, Egypt. Open Access Sci. Rep. 2013;2(4):705–708. [Google Scholar]
- 30.Hosny I.M, El-Kholy W.I, Murad H.A, El-Dairouty R.K. Antimicrobial activity of curcumin upon pathogenic microorganisms during manufacture and storage of a novel style cheese “Karishcum”. J. Am. Sci. 2011;7(5):611–618. [Google Scholar]
- 31.El-Sayed M.A, Hosny I.M, El-Kholy W.I, El-Dairouty R.K, Mohamed S.H.S. Microbiological evaluation of Egyptian white soft cheeses style. J. Am. Sci. 2011;7(5):517–526. [Google Scholar]
- 32.Fox A, Pichon B, Wilkinson H, Doumith M, Hill R.L, McLauchlin J, Kearns A.M. Detection and molecular characterization of livestock-associated MRSA in raw meat on retail sale in North West England. Lett. Appl. Microbiol. 2017;64(3):239–245. doi: 10.1111/lam.12709. [DOI] [PubMed] [Google Scholar]
- 33.Pesavento G, Ducci B, Comodo N, Lo Nostro A. Antimicrobial resistance profile of Staphylococcus aureus isolated from raw meat:A research for methicillin resistant Staphylococcus aureus(MRSA) Food Control. 2007;18:196–200. [Google Scholar]
- 34.Ge B, Mukherjee S, Hsu C.H, Davis J.A, Tran T.T, Yang Q, Abbott J.W, Ayers S.L, Young S.R, Crarey E.T, Womack N.A, Zhao S, McDermott P.F. MRSA and multidrug-resistant Staphylococcus aureus in U.S. Retail meats 2010-2011. Food Microbiol. 2017;62:289–297. doi: 10.1016/j.fm.2016.10.029. [DOI] [PubMed] [Google Scholar]
- 35.Ali N.H, Farooqui A, Khan A, Khan A.Y, Kazmi S.U. Microbial contamination of raw meat and its environment in retail shops in Karachi, Pakistan. J. Infect. Dev. Ctries. 2010;4:382–388. [PubMed] [Google Scholar]
- 36.Hassanin Z.H. Studies on Food Poisoning Microorganisms in Some Meat Products. M. V. Sc. Thesis, Faculty of Veterinary Medicine, Menoufia University, Sadat Branch. 2007 [Google Scholar]
- 37.Abdaslam S.A, Hassan M.A, Kaheel H.H, Abobaker T.M, Alnourain T.H, Hamdan H.A, Shankar S.G, Thambirajah J.J. Isolation of Escherichia coli O157 and other food borne pathogens from meat products and their susceptibility to different antimicrobial agents. Curr. Res. Microbiol. Biotechnol. 2014;2(3):391–397. [Google Scholar]
- 38.Li G, Wu C, Wang X, Meng J. Prevalence and characterization of methicillin susceptible Staphylococcus aureus ST398 isolates from retail foods. Int. J. Food Microbiol. 2015;196:94–97. doi: 10.1016/j.ijfoodmicro.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 39.Hanson B.M, Dressler A.E, Harper A.L, Scheibel R.P, Wardyn S.E, Roberts L.K, Kroeger J.S, Smith T.C. Prevalence of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus(MRSA) on retail meat in Iowa. J. Infect. Public Health. 2011;4:169–174. doi: 10.1016/j.jiph.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Gundogan N, Citak S, Turan E. Slime production, DNase activity and antibiotic resistance of Staphylococcus aureus isolated from raw milk, pasteurized milk and ice cream samples. Food Control. J. 2006;17(5):389–392. [Google Scholar]
- 41.Al-Zu'Bi E, Bdour S, Shehabi A.A. Antibiotic resistance patterns of mecA positive Staphylococcus aureus isolates from clinical specimens and nasal carriage. Microb. Drug Resist. Mech. Epidemiol. Dis. 2004;10(4):321–324. doi: 10.1089/mdr.2004.10.321. [DOI] [PubMed] [Google Scholar]
- 42.Ammar A.M, Attia A.M, Abd El-Hamid M.I, El-Shorbagy I.M, Abd El-Kader S.A. Genetic basis of resistance waves among methicillin resistant Staphylococcus aureus isolates recovered from milk and meat products in Egypt. Cell. Mol. Biol. Noisy-le-Grand. 2016;62(10):7–15. [PubMed] [Google Scholar]
- 43.Zhang L, Li Y, Bao H, Wei R, Zhou Y, Zhang H, Wang R. Population structure and antimicrobial profile of Staphylococcus aureus strains associated with bovine mastitis in China. Microb. Pathog. 2016;97:103–109. doi: 10.1016/j.micpath.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 44.Haran K, Godden S, Boxrud D, Jawahir S, Bender J, Sreevatsan S. Prevalence and characterization of Staphylococcus aureus including methicillin resistant Staphylococcus aureus isolated from bulk tank milk from Minnesota dairy farms. J. Clin. Microbiol. 2012;50(3):688–695. doi: 10.1128/JCM.05214-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waters A.E, Cuomo T.C, Buchhagen J, Liu C.M, Watson L, Pearce K, Foster J.T, Bowers J, Driebe E.M, Engelthaler D.M, Keim P.S, Price L.B. Multidrug-resistant Staphylococcus aureus in US meat and poultry. Clin. Infect. Dis. J. 2011;52:1227–1230. doi: 10.1093/cid/cir181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Argudín M.A, Mendoza M.C, González-Hevia M.A, Bances M, Guerra B, Rodicioa M.R. Genotypes, exotoxin gene content, and antimicrobial resistance of Staphylococcus aureus strains recovered from foods and food handlers. Appl. Environ. Microbiol. 2012;78(8):2930–2935. doi: 10.1128/AEM.07487-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan S.L, Lee H.Y, Mahyudin N.A. Antimicrobial resistance of Escherichia coli and Staphylococcus aureus isolated from food handler's hands. Food Control. J. 2014;44:203–207. [Google Scholar]
- 48.Teramoto H, Salaheen S, Biswas D. Contamination of post-harvest poultry products with multidrug resistant Staphylococcus aureus in Maryland-Washington DC metro area. Food Control. 2016;65:132–135. [Google Scholar]
- 49.Millan L.L, Gonicepero M.P, Ruiz J, Zolezzi P.C, Rubio C.M.C, Canales E.M. Molecular typing of Staphylococcus aureus clinical isolates by pulsed-field gel electrophoresis, staphylococcal cassette chromosome MEC type determination and dissemination of antibiotic resistance genes. Int. J. Antimicrob. Agents. 2007;30(6):505–513. doi: 10.1016/j.ijantimicag.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 50.Kumar R, Yadav B.R, Singh R.S. Genetic determinants of antibiotic resistance in Staphylococcus aureus isolates from milk of Mastitic crossbred cattle. Curr. Microbiol. 2010;60(5):379–386. doi: 10.1007/s00284-009-9553-1. [DOI] [PubMed] [Google Scholar]
- 51.McCallum N, Berger-Bachi B, Senn M.M. Regulation of antibiotic resistance in Staphylococcus aureus. Int. J. Med. Microbiol. 2010;300:118–129. doi: 10.1016/j.ijmm.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 52.Gao J, Ferreri M, Yu F, Liu Xi, Chen L, Su J, Han B. Molecular types and antibiotic resistance of Staphylococcus aureus isolates from bovine mastitis in a single herd in China. Vet. J. 2012;192:550–552. doi: 10.1016/j.tvjl.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 53.Martineau F, Picard F.J, Lansac N, MéNard C, Roy P.H, Ouellette M, Bergeron M.G. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2010;44(2):231–238. doi: 10.1128/aac.44.2.231-238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


