Abstract
Milk spoilage is caused by the presence of proteolytic enzymes produced by Pseudomonas spp. during storage at low temperatures. The aim of this study was to identify Pseudomonas spp. in raw milk and investigate their associated proteolytic properties at low temperatures. Raw milk samples (n = 87) were collected from 87 bulk tanks in Shaanxi Province in China. Pseudomonas spp. were identified using Pseudomonas specific 16S, universal 16S rRNA sequencing, and rpoB gene sequencing. The proteolytic properties of Pseudomonas spp. were examined using milk agar, quantitative trinitrobenzenesulfonic acid assay, and by the presence of alkaline metallopeptidase gene (aprX). A total 143 isolates from all 87 samples were confirmed as Pseudomonas, and were identified as belonging to 14 Pseudomonas species. Of these, 40 (28.0%) isolates revealed proteolysis on milk agar at 2°C, 74 (51.8%) at 4°C, 104 (72.7%) at 7°C, and 102 (71.3%) at 10°C. However, proteolytic activity of 45 (31.5%) isolates exceeded 2 μmol of glycine equivalents per mL at 7°C, followed by 43 (30.1%) at 10°C, 18 (12.6%) at 4°C, and 7 (4.9%) at 2°C. The results reveal proteolytic activity of Pseudomonas spp. present in milk and their spoilage potential at different temperatures.
Keywords: milk, Pseudomonas spp., proteolytic activities, aprX gene, spoilage
Introduction
Spoilage of milk resulting from the contamination of dairy products with psychrotrophic microorganisms results in significant losses for the food industry and is a particular concern of the dairy industry (Dogan and Boor, 2003). Milk is usually stored at low temperatures for 2 to 5 days prior to heat treatment (De Jonghe et al., 2011; Baur et al., 2015). During storage, the microbiota shifts toward psychrotrophic microorganisms, which can reduce the quality of raw milk (Lafarge et al., 2004; Xin et al., 2017).
Pseudomonas has been identified as predominant milk-associated psychrotrophic bacteria, making it one of the most important bacterial groups in the dairy industry (Wiedmann et al., 2000; Marchand et al., 2009a). The most commonly detected Pseudomonas species in milk and milk products are P. fluorescens, P. gessardii, P. fragi, and P. lundensis (Mallet et al., 2012). Pseudomonas spp. can grow over a temperature range of 4–42°C, with an optimal growth temperature above 20°C (Chakravarty and Gregory, 2015). They are present in different environments and are frequently linked to food spoilage, especially, that of raw milk (Quigley et al., 2013; Chakravarty and Gregory, 2015). Pseudomonas can outgrow other bacteria at low temperatures, accounting for at least 50% of all bacteria in milk (Munsch-Alatossava and Alatossava, 2006; Fricker et al., 2011; von Neubeck et al., 2015). The growth of the Pseudomonas is often associated with the production of extracellular enzymes (e.g., peptidases and lipases).
Peptidases secreted by Pseudomonas during cold storage are heat-stable extracellular peptidases that can retain their activity after pasteurization or ultrahigh temperature (UHT) treatment (Marchand et al., 2009b; Glück et al., 2016). The residual enzyme activities can cause coagulation and degradation of milk and dairy products over time. They mainly belong to the class of metallopeptidase (EC 3.4.24.) (Dufour et al., 2008; Scatamburlo et al., 2015; Caldera et al., 2016). The most important peptidase, a heat-resistant peptidase alkaline metallopeptidase (AprX), belongs to the serralysin family, and has been characterized in several strains of Pseudomonas spp. It is responsible for the spoilage of milk with activity on casein (Dufour et al., 2008). The spoilage ability of Pseudomonas spp. varies dramatically depending on the strain and growth conditions (Chabeaud et al., 2001; Nicodeme et al., 2005). Many studies have quantified the proteolytic activity of Pseudomonas spp. within the temperature range 4–7°C (Kumaresan et al., 2007; Baur et al., 2015; Caldera et al., 2016). However, the assay conditions used in these studies did not simulate conditions during transport and at dairy plants (Machado et al., 2017). Furthermore, a low temperature (1–4°C) is recommended for the long-term storage of raw milk before further processing (De Jonghe et al., 2011).
The aims of the present work were (i) to isolate and identify Pseudomonas spp. from the raw cows’ milk samples; and (ii) to investigate the associated proteolytic properties stored at low temperatures (2, 4, 7, and 10°C, respectively).
Materials and Methods
Sampling of Raw Milk and the Identification of Pseudomonas spp.
Raw milk samples (n = 87; 25 mL each) were collected directly from 87 bulk tanks of 87 farms in Shaanxi Province in China in spring (average daily temperature > 20°C), when Pseudomonas spp. have high prevalence. The 87 farms were randomly chosen and were mainly located in Xi’an, Xianyang, Weinan, and Tongchuan, where the main milk producing areas are found (herd size ≤ 300, milking frequency 2–3 times per day, no clinical mastitis cow). All samples were transferred to sterile plastic bottles (Corning Inc., Corning, NY, United States), stored at 4°C, and transported to the laboratory within 4 h.
For the isolation and detection of Pseudomonas spp., all raw milk samples were first processed as described by Scatamburlo et al. (2015). Briefly, all raw milk samples were diluted 10-fold in 0.85% NaCl (wt/vol) and homogenized. Aliquots (1 mL) of selected dilutions were placed onto Pseudomonas agar (Oxoid Ltd., Basingstoke, United Kingdom) to selectively isolate Pseudomonas spp. The plates were incubated at 25°C for 48 h. Colonies (5–8 per plate) were then streaked onto new Pseudomonas agar plates (Oxoid), and incubated at 25°C for 48 h.
Single colonies were chosen from Pseudomonas agar and incubated in LB broth (Beijing Land Bridge Technology Co., Ltd., Beijing, China) overnight at 25°C. DNA was extracted using the InstaGene Matrix DNA extraction kit (Bio-Rad Laboratories, Hercules, CA, United States) according the manufacturer’s instructions. Pseudomonas spp. were then identified by PCR (Bio-Rad Laboratories). The primers were synthesized by GeneCreate Biological Engineering Co., Ltd. (Wuhan, China). A negative control (a sample without genomic DNA) and a positive control (DNA of P. fluorescens CICC 21620; China Center of Industrial Culture Collection, Beijing, China) were included in all PCR assays.
Pseudomonas spp. were initially identified by PCR amplification of the 16S DNA region. The following primers were used: PA-GS-F (5′-GACGGGTGAGTAATGCCTA-3′) and PA-GS-R (5′-CACTGGTGTTCCTTCCTATA-3′) (Spilker et al., 2004). The PCR reaction conditions were as described in Scatamburlo et al. (2015). Amplicons (618 bp) were considered indicative of Pseudomonas spp.
Isolates identified as Pseudomonas spp. (n = 143) were further analyzed by amplification of the 16S rRNA fragment and rpoB sequences using universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-CTACGGCTACCTTGTTACGA-3′), and PSF (5′-AGTTCATGGACCAGAACAACC-3′) and PTR (5′-CCTTGACGGTGAACTCGTTTC-3′) (Lane et al., 1985; Sajben et al., 2011). The amplification programs were performed according to Sajben et al. (2011) and Caldera et al. (2016).
Finally, the obtained amplicons were sequenced at Beijing Genomics Institute (Beijing, China). Sequence data of the isolated Pseudomonas species were analyzed using the Basic Local Alignments Search Tool (BLAST) program available from the National Center for Biotechnology Information (NCBI)1. During universal 16S rRNA fragment and rpoB sequence analysis, the cut-off level for pairwise comparisons was 99%.
Plate Assays of Proteolytic Activity
Pseudomonas single colonies were streaked onto Pseudomonas agar (Oxoid Ltd.) supplemented with penicillin (100,000 IU/L, Dr. Ehrenstorfer GmbH, Augsburg, Germany), pimaricin (0.01 g/L, Dr. Ehrenstorfer GmbH), and UHT milk [10%, vol/vol, Modern Farming (Group) Co., Ltd., Hebei, China] (henceforth referred to as “milk agar”) (Scatamburlo et al., 2015). The plates were incubated at 2, 4, 7, and 10°C for 5 days. The temperature of 25°C was set as a control. The plates were monitored daily. Proteolytic halos in the inoculated areas were indicative of proteolytic activity (Scatamburlo et al., 2015). The plate assays were performed twice.
Proteolytic Activity Quantification
To induce peptidases production at 2, 4, 7, and 10°C, Pseudomonas spp. isolates were first incubated in 5 mL of UHT milk [Modern Farming (Group) Co., Ltd.] for 5 days. Then the proteolytic activity was determined according to the protocol described by Caldera et al. (2016) to quantify the native proteolytic activity at different temperatures.
The trinitrobenzenesulfonic acid (TNBS) method was used to monitor the presence of free α-amino groups, indicators of protein hydrolysis (Polychroniadou, 1988; Marchand et al., 2009a). The TNBS reagent (Sigma–Aldrich, Taufkirchen, Germany) was reacted with the released α-amino groups at pH 9.2 in the dark for 100 min. The intensity of the yellow-orange color of the reaction products was measured by absorption values at 420 nm (VarioskanTM Flash Multimode Reader, Thermo Fisher Scientific, Waltham, MA, United States). Native proteolytic enzyme levels in raw milk were determined from the absorption ratios. Three independent replicate measurements were performed. Pseudomonas spp. isolates were considered peptidase active if the measured absorption exceeded 2 μmol glycine equivalents per mL. Standard curve was generated using glycine (Sigma–Aldrich).
Identification of aprX Gene
The amplification of the aprX gene from Pseudomonas spp. isolates was performed using SM2F (5′-AAATCGATAGCTTCAGCCAT-3′) and SM3R (5′-TTGAGGTTGATCTTCTGGTT-3′) primers according to Caldera et al. (2016). Amplicons of ca. 850 bp were considered typical for aprX.
Results
Molecular Classification and Identification of Pseudomonas spp.
Genus-specific PCR of 16S DNA fragments (618 bp) was used for preliminary characterization of their assignment at the Pseudomonas genus level. In total 143 Pseudomonas isolates were confirmed. In Supplementary Table S1 identifications are given for each isolate based on universal 16S rRNA and rpoB gene sequencing analyses. The 14 different Pseudomonas species (Table 1) were mainly based on the rpoB gene sequence data, since universal 16S rRNA gene sequences are less discriminative (Caldera et al., 2016).
Table 1.
Identification of Pseudomonas spp. in raw milk sampled from 87 farms.
| Group | Species | No. of isolates |
|---|---|---|
| Pseudomonas fluorescens group | Pseudomonas azotoformans | 6 |
| Pseudomonas brenneri | 2 | |
| Pseudomonas cedrina | 5 | |
| Pseudomonas fluorescens | 61 | |
| Pseudomonas gessardii | 5 | |
| Pseudomonas poae | 4 | |
| Pseudomonas proteolytica | 1 | |
| Pseudomonas chlororaphis group | Pseudomonas fragi | 37 |
| Pseudomonas lundensis | 2 | |
| Pseudomonas putida group | Pseudomonas putida | 2 |
| Unknown group | Pseudomonas baetica | 2 |
| Pseudomonas deceptionensis | 1 | |
| Pseudomonas lurida | 2 | |
| Pseudomonas psychrophila | 13 | |
| Total | 143 |
In the current study, the 14 Pseudomonas species are divided into 4 different groups, according to Anzai et al. (2000), namely, P. fluorescens group, P. chlororaphis group, P. putida group, and unknown group. The amount of P. fluorescens group (n = 84) was dominant, followed by P. chlororaphis group (n = 39) (Table 1).
Proteolytic Activity of the Identified Pseudomonas spp. Isolates
The detailed phenotypic results of Pseudomonas isolates were shown in Table 2 and Supplementary Table S1. We evaluated the proteolytic activity of the isolates at all tested temperatures. Extracellular peptidase activity on milk agar was determined by the appearance of halo in the inoculated area. Following a 5 days incubation on milk agar, extracellular peptidase activity was detected in 74.1% (106/143) of the isolates at 25°C, around 72% of the isolates at 7°C (104/143) and 10°C (102/143), 51.8% (74/143) at 4°C and 28.0% (40/143) at 2°C. No directly correlations were observed between Pseudomonas groups and the degree of proteolysis (Supplementary Table S1).
Table 2.
Pseudomonas spp. isolates1 with observable proteolytic activity2 at different temperatures.
| Day | Incubation temperature (°C) |
||||
|---|---|---|---|---|---|
| 2 | 4 | 7 | 10 | 25 | |
| 1 | 2 | 5 | 8 | 16 | 54 |
| 2 | 6 | 12 | 26 | 33 | 15 |
| 3 | 12 | 27 | 39 | 31 | 12 |
| 4 | 4 | 11 | 14 | 13 | 17 |
| 5 | 16 | 19 | 17 | 9 | 8 |
| Total | 40 | 74 | 104 | 102 | 106 |
1The total number of Pseudomonas spp. isolates obtained from raw milk was 143.
2As determined during growth on milk agar. The number of new isolates exhibiting extracellular proteolytic activity on a given day of incubation on milk agar is shown.
Quantification of the Proteolytic Activity of Pseudomonas spp. Isolates
The peptidase activity was quantified using UHT milk, and the results are shown in Figure 1 and Supplementary Table S1. In the standard curve, relating glycine concentration to absorbance in the TNBS assay, the R2 value was greater than 0.99 (data not shown). Strains were considered proteolytic active when the measured proteolytic activity exceeded 2 μmol of glycine equivalents per mL.
FIGURE 1.
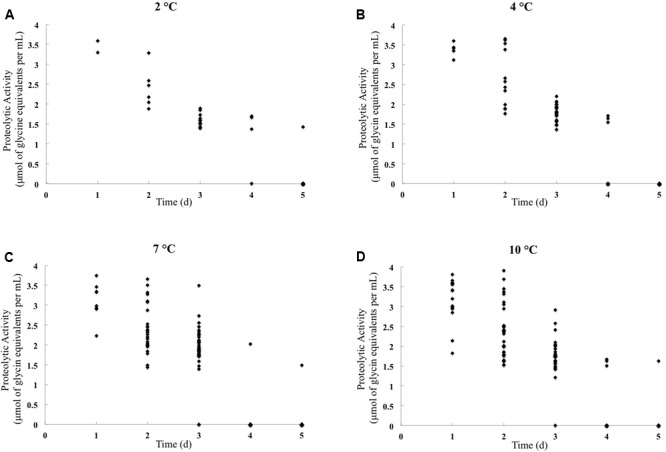
Proteolytic activity quantification of Pseudomonas isolates obtained from raw cow milk samples stored for 5 days at 2°C (A), 4°C (B), 7°C (C), and 10°C (D). The X-axis represents the storage day when the isolates displayed extracellular peptidase activity on milk agar, and the Y-axis represents proteolytic activity (μmol of glycine equivalents per mL). Species names and isolate numbers are not indicated.
The highest percentage of proteolytic activity isolates was obtained at 7°C (31.5%, n = 45) and 10°C (30.1%, n = 43). And 4.9% (7/143) isolates were proteolytic active at 2°C, and 12.6% (18/143) at 4°C. Using the TNBS method, peptidase activity was detected in 17.5% (7/40) of the isolates exhibiting extracellular peptidase activity on milk agar at 2°C and in 24.3% (18/74) of the isolates that were active at 4°C. At 7 and 10°C, the number of peptidase activity isolates was much larger than that at 2 and 4°C, reached 43.3% (45/104) and 42.2% (43/102), respectively (Table 2).
Analysis of the aprX Gene
We amplified the aprX gene from all isolates using PCR. An amplicon of the expected size (±850 bp) was obtained from reactions with genomic DNA of 133/143 isolates. Moreover, all the isolates, which displayed extracellular peptidase activity on milk agar, could detect the presence of aprX gene.
Discussion
After molecular confirmation at the genus level, the 143 isolates were assigned at species level. In general, we found a large diversity of Pseudomonas species. The frequencies of the Pseudomonas spp. confirm the findings of Jayarao and Wang (1999), who reported 98 isolates belonging to nine Pseudomonas species. Moreover, the Pseudomonas isolates were found to be randomly distributed among the different farms. These results demonstrate the ubiquitousness of Pseudomonas spp. in dairy farms, highlighting their relevance as natural contaminants (Scatamburlo et al., 2015).
PCR sequencing confirmed that 42.7% (n = 61) of the isolates were P. fluorescens, the dominant Pseudomonas specie. Our results are in agreement with those of Chambers (2003), who reported that P. fluorescens was the dominant Pseudomonas spp. in milk and those of Dogan and Boor (2003), who identified 51% of Pseudomonas isolates as P. fluorescens in fluid milk products and dairy processing plants using API20 NE. P. putida is another species of Pseudomonas found in milk (Senel and Gürsoy, 2014); however, we found only 2 P. putida isolates in the P. putida group in the current study (Table 1). P. psychrophila was the third most common strain identified here. By contrast, Vithanage et al. (2014) found only a few P. psychrophila isolates among psychrotrophic bacteria in raw milk samples from a commercial UHT milk processor. Although Pseudomonas spp. are ubiquitous in raw milk, our findings indicate that Pseudomonas spp. in milk differ considerably across studies. These differences could stem from regional and environmental differences.
All the isolates showed growth over a wide range of temperatures (2–25°C), although many studies have demonstrated that the growth of Pseudomonas isolates is temperature-dependent (Munsch-Alatossava and Alatossava, 2006; Caldera et al., 2016). By contrast, Munsch-Alatossava and Alatossava (2006) reported that Pseudomonas strains isolated from farms do not grow at 4 or 7°C. This could be because of differences in the growth characterization of the isolates.
The results showed that most isolates produced peptidase at 25°C, followed by 7, 10, 4, and 2°C. Similarly, peptidase production by P. fluorescens was reported previously to be highest at 22°C, followed by 7 and 32°C (Wang and Jayarao, 2001). Therefore, our results confirm those of other studies and demonstrate the spoilage capability of Pseudomonas spp. during storage and transport at low temperatures. In addition, we demonstrated that a number of Pseudomonas spp. isolates exhibit extracellular peptidase activity at low temperature after a short period of time, stressing the importance of controlling contamination during the early steps of milk storage.
Proteolytic activity was then confirmed by TNBS quantitative analysis and expressed as glycine equivalents per mL. The number of peptidase active isolates increased with temperature, most likely reflecting the fact that proteolytic activity is temperature regulated (Woods et al., 2001; McCarthy et al., 2004). Taken together, the extracellular peptidase activity and proteolytic activity data showed that the isolates could produce peptidases at different temperatures. This is a pertinent observation as it confirms the ability of Pseudomonas species to produce proteolytic enzymes under psychrotrophic conditions. More isolates produced peptidases with an increase in temperature; however, different peptidases were active at different temperatures (Supplementary Table S1).
According to current laws and standards in many countries, raw milk must be stored below 7°C (FDA, 2013; Scatamburlo et al., 2015; Zheng, 2015). Many studies have found that Pseudomonas spp. can produce peptidases under storage temperatures. Caldera et al. (2016) reported that 46.8% (15/32) of Pseudomonas isolates from milk and dairy products after incubation at 5°C for 5 days were peptidase active using a quantitative assay. Moreover, when Kumaresan et al. (2007) assessed the native proteolytic activity of psychrotrophs at 2, 4, and 7°C using the Kjeldahl method, they found that the storage of raw milk at these temperatures resulted in peptidase production by Pseudomonas spp. In this study, we confirmed these findings by showing that a number of Pseudomonas isolates could produce peptidases and exhibit proteolytic activity under different storage temperatures. Proteolytic activity was lower at 2°C than at 4°C, 7°C, or 10°C, as assessed by the TNBS method. However, we also identified many Pseudomonas isolates that acquired higher proteolytic activity after storage at 2°C than at 4°C, 7°C, and 10°C, indicating that spoilage ability is strain-dependent.
In some countries, the majority of raw milk is not processed after milking until delivery to a dairy, which might last up to 3 or 4 days. Additional storage before processing may also take place in the dairy (Fricker et al., 2011; von Neubeck et al., 2015). Therefore, 5 days was chosen to study the peptidase activities of the isolates in this study. Kumaresan et al. (2007) demonstrated that proteolytic activity increased with storage time. Stoeckel et al. (2016) also found a linear correlation between onset of product defects and enzyme activity in UHT milk samples. Our results showed that peptidases could be produced by a number of Pseudomonas isolates between 2°C and 10°C and that proteolytic activity increased with storage time. The peptidases secreted by Pseudomonas spp. in raw milk stored at low temperature are stable during heat treatment, which might explain the persistent proteolytic activity observed during storage of processed milk (Marchand et al., 2009a; Glück et al., 2016). Therefore, in order to prolong the storage time of milk products, raw milk should be preferentially stored at low temperature and processed within 48 h after milking to prevent the release of thermostable spoilage peptidases by Pseudomonas spp.
There was a good correlation between extracellular peptidase activity on milk agar and the presence of the aprX as determined by PCR, confirming the role of AprX in milk degradation as reported previously (Dufour et al., 2008; Caldera et al., 2016). However, some aprX-positive isolates do not show proteolytic activity at any storage temperature, indicating that AprX might be inhibited during milk protein degradation. In the genus Pseudomonas, the aprX gene is usually involved in nutrient utilization; the product of this gene degrades extracellular proteins, and therefore Pseudomonas spp. are usually associated with the spoilage of milk and dairy products (Dufour et al., 2008; Marchand et al., 2009b; Zhang et al., 2009). The production of this enzyme highlights the controlling of Pseudomonas spp. contamination in raw milk and dairy products.
Conclusion
This study revealed a diverse Pseudomonas spp. population in raw milk. In the dairy, milk is usually stored at temperatures below 7°C before processing. This temperature does not prevent the growth and proteolytic activity of Pseudomonas spp. in raw milk. Although all the Pseudomonas isolates could grow between 2°C and 10°C, proteolytic activity decreased when the milk storage temperature was reduced from 10°C to 2°C. There was no relation between specific Pseudomonas spp. and proteolytic activity. Low storage temperature and short periods before processing (within 48 h) could reduce peptidase production of Pseudomonas spp., but milking hygiene should also be properly controlled. It is necessary to acquire more information on Pseudomonas spp. with proteolytic activity and to develop sensitive and efficient tools to monitor for the presence of peptidases in raw milk.
Author Contributions
LM performed the major experiments and wrote the manuscript. YZ did many work in experiments and helped in writing. HL helped in the research and writing. SZ and JW gave the help in research plans. NZ is the corresponding author.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. The research was supported by Project of Risk Assessment on Raw Milk [grant no. GJFP2017008], Fundamental Research Funds for the Central Non-profit Research Institution [grant no. 2015ywf-zd-3], Special Fund for Agro-scientific Research in the Public Interest [grant no. 201403071], the Agricultural Science and Technology Innovation Program [grant no. ASTIP-IAS12], China, and the Modern Agro-Industry Technology Research System of the China [CARS-37].
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02158/full#supplementary-material
References
- Anzai Y., Kim H., Park J. Y., Wakabayashi H., Oyaizu H. (2000). Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 50 1563–1589. 10.1099/00207713-50-4-1563 [DOI] [PubMed] [Google Scholar]
- Baur C., Krewinkel M., Kranz B., von Neubeck M., Wenning M., Scherer S., et al. (2015). Quantification of the proteolytic and lipolytic activity of microorganisms isolated from raw milk. Int. Dairy J. 49 23–29. 10.1016/j.idairyj.2015.04.005 [DOI] [Google Scholar]
- Caldera L., Franzetti L., Van Coillie E., De Vos P., Stragier P., De Block J., et al. (2016). Identification, enzymatic spoilage characterization and proteolytic activity quantification of Pseudomonas spp. isolated from different foods. Food Microbiol. 54 142–153. 10.1016/j.fm.2015.10.004 [DOI] [Google Scholar]
- Chabeaud P., de Groot A., Bitter W., Tommassen J., Heulin T., Achouak W. (2001). Phase-variable expression of an operon encoding extracellular alkaline protease, a serine protease homolog, and lipase in Pseudomonas brassicacearum. J. Bacteriol. 183 2117–2120. 10.1128/JB.183.6.2117-2120.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty S., Gregory G. (2015). “The genus Pseudomonas,” in Practical Handbook of Microbiology eds Goldman E., Green L. H. (New York, NY: CRC Press; ) 321–344. [Google Scholar]
- Chambers J. V. (2003). “The microbiology of raw milk,” in Dairy Microbiology Handbook: The Microbiology of Milk and Milk Products 3rd Edn ed. Robinson R. K. (New York, NY: John Wiley and Sons, Inc.) 39–90. [Google Scholar]
- De Jonghe V., Coorevits A., Van Hoorde K., Messens W., Van Landschoot A., De Vos P., et al. (2011). Influence of storage conditions on the growth of Pseudomonas species in refrigerated raw milk. Appl. Environ. Microbiol. 77 460–470. 10.1128/AEM.00521-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan B., Boor K. J. (2003). Genetic diversity and spoilage potentials among Pseudomonas spp. isolated from fluid milk products and dairy processing plants. Appl. Environ. Microbiol. 69 130–138. 10.1128/AEM.69.1.130-138.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour D., Nicodeme M., Perrin C., Driou A., Brusseaux E., Humbert G., et al. (2008). Molecular typing of industrial strains of Pseudomonas spp. isolated from milk and genetical and biochemical characterization of an extracellular protease produced by one of them. Int. J. Food Microbiol. 125 188–196. 10.1016/j.ijfoodmicro.2008.04.004 [DOI] [PubMed] [Google Scholar]
- FDA (2013). Grade “A” Pasteurized Milk Ordinance. Silver Spring, MD: U.S. Food and Drug Administration. [Google Scholar]
- Fricker M., Skanseng B., Rudi K., Stessl B., Ehling-Schulz M. (2011). Shift from farm to dairy tank milk microbiota revealed by a polyphasic approach is independent from geographical origin. Int. J. Food Microbiol. 145(Suppl. 1) S24–S30. 10.1016/j.ijfoodmicro.2010.08.025 [DOI] [PubMed] [Google Scholar]
- Glück C., Rentschler E., Krewinkel M., Merz M., von Neubeck M., Wenning M., et al. (2016). Thermostability of peptidases secreted by microorganisms associated with raw milk. Int. Dairy J. 56 186–197. 10.1016/j.idairyj.2016.01.025 [DOI] [Google Scholar]
- Jayarao B. M., Wang L. (1999). A study on the prevalence of gram-negative bacteria in bulk tank milk. J. Dairy Sci. 82 2620–2624. 10.3168/jds.S0022-0302(99)75518-9 [DOI] [PubMed] [Google Scholar]
- Kumaresan G., Annalvilli R., Sivakumar K. (2007). Psychrotrophic spoilage of raw milk at different temperatures of storage. J. Appl. Sci. Res. 3 1383–1387. [Google Scholar]
- Lafarge V., Ogier J. C., Girard V., Maladen V., Leveau J. Y., Gruss A., et al. (2004). Raw cow milk bacterial population shifts attributable to refrigeration. Appl. Environ. Microbiol. 70 5644–5650. 10.1128/AEM.70.9.5644-5650.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. (1985). Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. U.S.A. 82 6955–6959. 10.1073/pnas.82.20.6955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado S. G., Baglinière F., Marchand S., Van Coillie E., Vanetti M. C. D., De Block J., et al. (2017). The biodiversity of the microbiota producing heat-resistant enzymes responsible for spoilage in processed bovine milk and dairy products. Front. Microbiol. 8:302. 10.3389/fmicb.2017.00302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet A., Guéguen M., Kauffmann F., Chesneau C., Sesboué A., Desmasures N. (2012). Quantitative and qualitative microbial analysis of raw milk reveals substantial diversity influenced by herd management practices. Int. Dairy J. 27 13–21. 10.1016/j.idairyj.2012.07.009 [DOI] [Google Scholar]
- Marchand S., Heylen K., Messens W., Coudijzer K., De Vos P., Dewettinck K., et al. (2009a). Seasonal influence on heat-resistant proteolytic capacity of Pseudomonas lundensis and Pseudomonas fragi, predominant milk spoilers isolated from Belgian raw milk samples. Environ. Microbiol. 11 467–482. 10.1111/j.1462-2920.2008.01785.x [DOI] [PubMed] [Google Scholar]
- Marchand S., Vandriesche G., Coorevits A., Coudijzer K., De Jonghe V., Dewettinck K., et al. (2009b). Heterogeneity of heat-resistant proteases from milk Pseudomonas species. Int. J. Food Microbiol. 133 68–77. 10.1016/j.ijfoodmicro.2009.04.027 [DOI] [PubMed] [Google Scholar]
- McCarthy C. N., Woods R. G., Beacham I. R. (2004). Regulation of the aprX-lipA operon of Pseudomonas fluorescens B52: differential regulation of the proximal and distal genes, encoding protease and lipase, by ompR-envZ. FEMS Microbiol. Lett. 241 243–248. 10.1016/j.femsle.2004.10.027 [DOI] [PubMed] [Google Scholar]
- Munsch-Alatossava P., Alatossava T. (2006). Phenotypic characterization of raw milk-associated psychrotrophic bacteria. Microbiol. Res. 161 334–346. 10.1016/j.micres.2005.12.004 [DOI] [PubMed] [Google Scholar]
- Nicodeme M., Grill J. P., Humbert G., Gaillard J. L. (2005). Extracellular protease activity of different Pseudomonas strains: dependence of proteolytic activity on culture conditions. J. Appl. Microbiol. 99 641–648. 10.1111/j.1365-2672.2005.02634.x [DOI] [PubMed] [Google Scholar]
- Polychroniadou A. (1988). A simple procedure using trinitrobenzenesulphonic acid for monitoring proteolysis in cheese. J. Dairy Res. 55 585–596. 10.1017/S0022029900033379 [DOI] [Google Scholar]
- Quigley L., O’Sullivan O., Stanton C., Beresford T. P., Ross R. P., Fitzgerald G. F., et al. (2013). The complex microbiota of raw milk. FEMS Microbiol. Rev. 37 664–698. 10.1111/1574-6976.12030 [DOI] [PubMed] [Google Scholar]
- Sajben E., Manczinger L., Nagy A., Kredics L., Vagvolgyi C. (2011). Characterization of pseudomonads isolated from decaying sporocarps of oyster mushroom. Microbiol. Res. 166 255–267. 10.1016/j.micres.2010.05.002 [DOI] [PubMed] [Google Scholar]
- Scatamburlo T. M., Yamazi A. K., Cavicchioli V. Q., Pieri F. A., Nero L. A. (2015). Spoilage potential of Pseudomonas species isolated from goat milk. J. Dairy Sci. 98 759–764. 10.3168/jds.2014-8747 [DOI] [PubMed] [Google Scholar]
- Senel E., Gürsoy A. (2014). “Microbiology of processed liquid milk,” in Dairy Microbiology and Biochemistry: Recent Developments eds Ozer B., Akdemir-Evrendilek G. (Boca Raton, FL: CRC Press; ) 95–112. [Google Scholar]
- Spilker T., Coenye T., Vandamme P., LiPuma J. J. (2004). PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J. Clin. Microbiol. 42 2074–2079. 10.1128/JCM.42.5.2074-2079.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckel M., Lidolt M., Achberger V., Glück C., Krewinkel M., Stressler T., et al. (2016). Growth of Pseudomonas weihenstephanensis, Pseudomonas proteolytica and Pseudomonas sp. in raw milk: Impact of residual heat-stable enzyme activity on stability of UHT milk during shelf-life. Int. Dairy J. 59 20–28. 10.1016/j.idairyj.2016.02.045 [DOI] [Google Scholar]
- Vithanage N. R., Yeager T. R., Jadhav S. R., Palombo E. A., Datta N. (2014). Comparison of identification systems for psychrotrophic bacteria isolated from raw bovine milk. Int. J. Food Microbiol. 189 26–38. 10.1016/j.ijfoodmicro.2014.07.023 [DOI] [PubMed] [Google Scholar]
- von Neubeck M., Baur C., Krewinkel M., Stoeckel M., Kranz B., Stressler T., et al. (2015). Biodiversity of refrigerated raw milk microbiota and their enzymatic spoilage potential. Int. J. Food Microbiol. 211 57–65. 10.1016/j.ijfoodmicro.2015.07.001 [DOI] [PubMed] [Google Scholar]
- Wang L., Jayarao B. M. (2001). Phenotypic and genotypic characterization of Pseudomonas fluorescens isolated from bulk tank milk. J. Dairy Sci. 84 1421–1429. 10.3168/jds.S0022-0302(01)70174-9 [DOI] [PubMed] [Google Scholar]
- Wiedmann M., Weilmeier D., Dineen S. S., Ralyea R., Boor K. J. (2000). Molecular and phenotypic characterization of Pseudomonas spp. isolated from milk. Appl. Environ. Microbiol. 66 2085–2095. 10.1128/AEM.66.5.2085-2095.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods R. G., Burger M., Beven C. A., Beacham I. R. (2001). The aprX-lipA operon of Pseudomonas fluorescens B52: a molecular analysis of metalloprotease and lipase production. Microbiology 147(Pt 2) 345–354. 10.1099/00221287-147-2-345 [DOI] [PubMed] [Google Scholar]
- Xin L., Meng Z. X., Zhang L. W., Cui Y. H., Han X., Yi H. X. (2017). The diversity and proteolytic properties of psychrotrophic bacteria in raw cows’ milk from North China. Int. Dairy J. 66 34–41. 10.1016/j.idairyj.2016.10.014 [DOI] [Google Scholar]
- Zhang W. W., Hu Y. H., Wang H. L., Sun L. (2009). Identification and characterization of a virulence-associated protease from a pathogenic Pseudomonas fluorescens strain. Vet. Microbiol. 139 183–188. 10.1016/j.vetmic.2009.04.026 [DOI] [PubMed] [Google Scholar]
- Zheng W. (2015). Translated English of Chinese Standard, GB12693-2010.:National Food Safety Standard Good Manufacturing Practice for Milk Products. Beijing: National Standards of the People’s Republic of China. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


