Abstract
Circular RNAs (circRNAs) own unique capabilities to communicate with nucleic acids and ribonucleoproteins and are emerging as indispensable compositions of the regulatory messages encoded in the genome. Due to lack of 3′ termini, circRNAs are more resistant to degradation by exonuclease RNase R and possess greater stability than linear RNAs. Moreover, circRNAs can act as microRNA (miRNA) sponge and affect messenger RNA (mRNA) splicing and transcription. By virtue of their great stability and elaborate regulatory mechanisms of gene expression, circRNAs play important roles in certain physiological activities. The development, homeostasis and stress response of the central nervous system (CNS) depend upon precise temporal and spatial regulation of gene networks. Moreover, emerging evidence has revealed that circRNAs are spatiotemporally regulated and dynamically expressed during brain development; therefore, they can exert significant influences on CNS development and diseases. In this review, we highlight the biogenesis of circRNAs and their central roles in regulation of CNS development and diseases.
Keywords: circular RNAs, back-splicing, microRNA sponge, central nervous system, biomarker
Introduction
Recent technical advances in next generation sequencing have greatly accelerated discoveries in the field of molecular genetics, among which the importance of non-protein coding transcripts have been increasingly highlighted (Sharma et al., 2016). In contrast to messenger RNAs (mRNAs), the term non-coding RNAs (ncRNAs) refers to a class of RNA transcripts that do not encode protein (Hirose et al., 2014). It was estimated that only 1–2% RNA in human genome could code for proteins and ncRNAs constitute a vast majority of the human transcriptome (Dunham et al., 2012). In addition to microRNAs (miRNAs), small interfering RNAs (siRNAs), long non-coding RNAs (lncRNAs) and small nuclear RNAs (snoRNAs), the recently recognized circular RNAs (circRNAs) represents a special type of ncRNAs that plays a central regulatory role in RNA metabolism (Hentze and Preiss, 2013).
As early as in the 1970s, several circRNAs were found in RNA viruses (Sanger et al., 1976; Hsu and Coca-Prados, 1979). However, they have long thought to be byproducts of mRNA splicing until recent technical advances of deep-sequencing revealed the existence of thousands of circRNAs in difference species (Liang et al., 2017; Reddy et al., 2017; Zhao W. et al., 2017). CircRNAs represent a novel class of widespread and diverse endogenous RNAs that, unlike linear RNAs, are characterized by a covalent bond linking the 5′ and 3′ ends (Guo et al., 2014). CircRNAs are generated from spliceosome-mediated precursor mRNA (pre-mRNA) splicing through a non-canonical splicing process in eukaryotes (Ashwal-Fluss et al., 2014; Chen, 2016). Many circRNAs originate from protein coding genes and contain exonic sequences; however, they have long been considered as RNAs that do not encode any proteins (Guo et al., 2014; You et al., 2015). Nevertheless, recent studies have shown that a subset of circRNAs can be translated endogenously (Pamudurti et al., 2017; Yang et al., 2017). Due to the absence of 3′ termini, circRNAs are resistant to degradation by exonuclease RNase R and thus being more stable than linear mRNAs (Chen, 2016). CircRNAs are evolutionarily conserved across multiple species (Jeck et al., 2013; Rybak-Wolf et al., 2015) indicating their functional importance. Given their abundance, long half-life and species conservation, circRNAs may effectively regulate a wide range of physiological activities and pathological changes (Zhang X.O. et al., 2016; Zheng et al., 2016). Although the function of circRNAs remains largely uncharacterized, recent studies have shown that some circRNAs can bind other ncRNAs or proteins and thus regulating the expression of other or even their parental genes (Abdelmohsen et al., 2017; Han et al., 2017). Moreover, recent discoveries of translated products from certain circRNA also suggest that they may have specific biological functions (Pamudurti et al., 2017; Yang et al., 2017).
Multiple recent studies have shown that circRNAs may play crucial roles in brain development and diseases. In a study testing the abundance of circRNAs in different organs, mammalian brain was found to have the highest amount of circRNAs among all tissues tested, including liver, lung, heart, and testis (You et al., 2015). In addition, circRNAs are especially highly enriched in synapses, and genes related to synapses can give rise to significantly abundant circRNAs (Earls et al., 2014; Rybak-Wolf et al., 2015; Szabo et al., 2015). Interestingly, the expression levels of certain circRNAs can be much higher than the canonical linear transcripts of their parent genes during the central nervous system (CNS) development (Rybak-Wolf et al., 2015), indicating that circRNAs may have different functions than their linear counterparts. It is known that other kinds of ncRNAs, such as miRNAs play an essential role in the process of CNS development and diseases (Hill and Lukiw, 2016; Persengiev et al., 2011; Yin et al., 2015). CircRNAs can serve as competing endogenous sponges to miRNAs and therefore by regulating their activities, circRNAs can have notable influence upon the development and diseases of the CNS (Taulli et al., 2013; Lu and Xu, 2016; Li et al., 2017).
In this review, we describe the biogenesis and characteristics of circRNAs. Their potential functions and possible roles in CNS development and diseases are also discussed.
Biogenesis of circRNAs
CircRNAs are derived from non-canonical splicing of precursor mRNAs (pre-mRNAs), which are regulated by the spliceosome and modulated by cis-regulatory elements and trans-acting factors (Zhang et al., 2013; Guo et al., 2014; Barrett and Salzman, 2016). Through RNA sequencing of non-polyadenylated transcriptomes, thousands of circRNAs were detected in different types of cells and tissues, and overall, they comprise over 10% of the transcripts encoded by their parent genes. Although the same gene contains sequences of both circRNAs and linear protein-coding mRNAs, the expression levels of circRNAs and their linear counterparts do not always correlate, indicating the presence of a distinct regulation mechanism of biosynthesis between the circRNA and linear mRNA (Jeck and Sharpless, 2014; Kelly et al., 2015; Thomas and Sætrom, 2014).
Canonical Spliceosomes Mediate Non-canonical circRNA Biogenesis
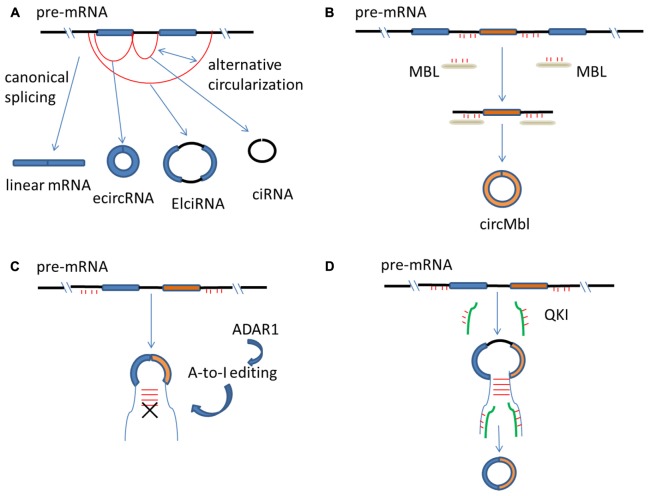
CircRNAs can arise from different regions of gene loci in eukaryotes, such as from exons (called exonic circRNA, or ecircRNA), from introns (called intronic circRNA, or ciRNA) or from both exons and introns (called exon–intron circRNA, or ElciRNA) (Li Z. et al., 2015; Zhang Y. et al., 2016) (Figure 1A). Nevertheless, most circRNAs are derived from exons of protein-coding genes through a non-canonical splicing process called ‘back-splicing,’ in which a downstream splice donor joins an upstream splice acceptor leading to exon circularization (Jeck et al., 2013; Salzman, 2016). The canonical splice machinery, however, is required for exon circularization as was shown by mutagenesis experiments (Chen and Yang, 2015; Starke et al., 2015). Since mRNAs that are generated from linear splicing is also mediated by the canonical splicesome, how circRNAs and mRNAs can both be generated from the same pre-mRNA remains to be an important question (Hamid and Makeyev, 2014). Similar to mRNA splicing, the presence of canonical splice sites of bracketing exons can determine, to a large degree, the circularization rates (Barrett et al., 2015; Wang and Wang, 2015; Zhang Y. et al., 2016). On the other hand, functional studies have elucidated that, during circular vs. linear splicing, different splice sites can be activated and the biogenesis of circRNAs and mRNAs may compete with each other (Tay et al., 2014; Barrett and Salzman, 2016). Moreover, there are other differences between canonical spliceosome mediated circRNA and mRNA biogenesis, which might largely determine which molecule will be predominantly generated from the pre-mRNA, leading to distinct expression patterns of circRNA and mRNA (Ashwal-Fluss et al., 2014; Starke et al., 2015). For example, the efficiency of back-splicing is lower than those of the corresponding linear counterparts, perhaps because spliceosomes are inefficiently aggregated at back-splicing sites at the steady-state of circRNA production (Ashwal-Fluss et al., 2014; Guo et al., 2014). Although the exact details on how the splicesome carry out exon circularization is not known, the canonical splice signals and canonical spliceosomal machinery are important in the processing of circRNAs biogenesis (Chen and Yang, 2015; Petkovic and Müller, 2015).
FIGURE 1.
Regulation of circRNA biogenesis. (A) mRNAs are generated from spliceosome-mediated canonical splicing, but circRNAs are generated from pre-mRNA splicing through a non-canonical splicing process. CircRNAs can arise from exons (called ecircRNA), from introns (called ciRNA) or from both exons and introns (called ElciRNA). (B) The flanking introns of circMbl have abundant MBL binding sites that are required for circMbl biogenesis. Therefore, MBL protein can control the production of the circRNA of its own. (C) ADAR1 converts Adenosine-to-Inosine (A-to-I), and then, inhibits circRNA biosynthesis by breaking RNA–RNA interactions that form during RNA base-pairing. (D) The presence of QKI binding sites in the intron of genes is able to trigger circRNA production. mRNA, messenger RNA; circRNA, circular RNA; pre-mRNA, precursor mRNA; ecircRNA, exonic circRNA; ciRNA, intronic circRNA; ElciRNA, exon–intron circRNA.
Cis- and Trans- Regulators of circRNA Biogenesis
In addition to the splicing machinery, both cis-regulatory elements and trans-acting factors are needed to bring the two splice sites into close proximity to facilitate back-splicing (Jeck and Sharpless, 2014; Kramer et al., 2015). For example, reverse and complementary sequences resided in flanking introns of circulated exons are able to form RNA duplexes, which enhance the efficiency of back-splicing (Ivanov et al., 2015; Zhang et al., 2014). RNA duplexes can form by repetitive sequences such as Alu elements (Kim et al., 2004; Jeck et al., 2013), as well as from non-repetitive but complementary sequences (Zhang et al., 2014). Longer complementary sequences in general have stronger RNA pairing strengths and enhanced circRNA biogenesis than shorter sequences; however, sequences as short as 30 nts also possess the ability to stimulate circRNA production (Liang and Wilusz, 2014). In addition, distinct ways of RNA pairing can lead to generation of different circRNAs. For example, repetitive sequences such as Alu elements can form different combinations of Alu pairs, which results in production of diverse circRNAs from a single gene – a process known as alternative circularization (Jeck et al., 2013) (Figure 1A). The types and expression levels of circRNAs are much higher in humans than in mice, and the increased circRNA complexity during species evolution is correlated with an increased number and RNA-pairing capability of those complementary sequences (Dong et al., 2016). Importantly, in addition to RNA base-pairing formed across flanking introns, RNA base-pairing can also form within an individual intron in linear RNA production (Hossain et al., 2011; Zhang et al., 2014). Therefore, there are competitions between non-canonical splicing and canonical splicing, based on choice of RNA base-pairing in flanking introns or within an individual intron (Jeck et al., 2013; Hamid and Makeyev, 2014).
Trans-acting factors such as RNA-binding proteins (RBPs) have been reported to be capable of regulating circRNA biogenesis (Castello et al., 2012; Conn et al., 2015; Dudekula et al., 2016). Three factors, muscleblind (MBL), Adenosine deaminae 1 acting on RNA (ADAR1) and quaking (QKI) have been shown so far to regulate circRNA production by different mechanisms (Ashwal-Fluss et al., 2014; Ivanov et al., 2015; Conn et al., 2015). MBL is a conserved regulator of RNA splicing. The second exon of MBL can be circularized to produce a circular transcript (called circMbl) and circMbl and its flanking introns have abundant MBL binding sites (Ashwal-Fluss et al., 2014). These sites are required for circMbl biogenesis and therefore, MBL protein can control the production of the circRNA of its own (Ashwal-Fluss et al., 2014) (Figure 1B). ADAR1 expression is negatively correlated with production of certain circRNAs (Szabo et al., 2015). ADAR1 converts Adenosine-to-Inosine (A-to-I), which predominantly occurs in double-stranded Alu repeats (Athanasiadis et al., 2004). Therefore, it is thought that ADAR1 inhibits circRNA biosynthesis by breaking the RNA stem of RNA-RNA interactions that form during RNA base-pairing (Ivanov et al., 2015) (Figure 1C). QKI regulates the production of many circRNAs and the presence of QKI binding sites in the intron of genes that are normally spliced in a linear fashion is sufficient to trigger circRNA production (Conn et al., 2015) (Figure 1D).
It was reported that circRNA production occurs post-transcriptionally, since a 3′ end processing signal is necessary for back-splicing and the 3′ polyadenylation is required for splicing of some introns (Liang and Wilusz, 2014). Nevertheless, other studies point out that circRNA biogenesis can occur both co-transcriptionally and post-transcriptionally, and the strength of base-pairing of complementary sequences in the flanking intronic repeats determines whether back-splicing occurs co- or post-transcriptionally (Wilusz and Sharp, 2013; Ashwal-Fluss et al., 2014; Kramer et al., 2015). While back-splicing mediated by minimal intronic repeats [<40 nucleotides (nt)] requires 3′ end processing and occur post-transcriptionally, long flanking repeats (∼400 nt) are able to produce circRNAs co-transcriptionally (Kramer et al., 2015).
CircRNAs in CNS Development
Central nervous system development is a complex and orderly process that requires precise regulation of proliferation and differentiation of progenitor cells and is intricately controlled by gene networks (Gailite et al., 2015; Villaescusa et al., 2016). Expression of circRNAs during CNS development is tissue-specific and stage-associated (Venø et al., 2015; Dou et al., 2016; van Rossum et al., 2016). In addition, circRNAs were reported to be preferentially back-spliced from neural genes and were widely and dynamically expressed in the brain. Therefore, circRNAs might play important roles in various processes of CNS development (Venø et al., 2015; You et al., 2015; Hanan et al., 2016; Filippenkov et al., 2017).
CircRNAs in Progenitor Cell Proliferation and Neural Differentiation
Proliferation and differentiation of neurons from stem cell precursors and progenitors are landmarks of neural development. NcRNAs such as miRNAs and lncRNAs have been shown to be important in these processes (Rani et al., 2016). Similarly, circRNAs have also become increasingly recognized to play crucial roles. During neural development, the expression pattern of neuronal circRNAs show conservation between mouse, pig, and human, and the highly expressed circRNAs is more conserved than average, indicating that these highly expressed circRNAs are potentially more important (Westholm et al., 2014; Rybak-Wolf et al., 2015) than others. In cell culture models of neural differentiation, the expression of the majority of circRNAs was shown to be significantly up-regulated (Rybak-Wolf et al., 2015). The host genes of these upregulated circRNAs are known to play essential roles in neuronal activities such as dendritic mRNA transport and synaptic membrane exocytosis (Ng et al., 2013; Heraud-Farlow and Kiebler, 2014; Jung et al., 2015). In addition, during porcine brain development, different anatomic regions were found to have different circRNA expression levels, with the minimum amount in the brain stem and the highest amount in the cortex and cerebellum (Venø et al., 2015). Moreover, at different stages of neural differentiation, there are distinct levels and types of up-regulated circRNAs (Rybak-Wolf et al., 2015). Taken together, circRNA expression is spatio-temporally regulated during brain development, suggesting that they may play an essential role in the process of neural proliferation and differentiation (Rybak-Wolf et al., 2015; Dang et al., 2016).
How do circRNAs control progenitor cell proliferation and neural differentiation? The origin of circRNAs may reflect their functions to some extent, assuming circRNAs have similar biological function to their host genes (Rybak-Wolf et al., 2015; Venø et al., 2015). In a study investigating spatio-temporal expression patterns of circRNAs during porcine embryonic brain development, three host gene associated pathways, Wnt signaling, axon guidance and the TGFβ signaling pathway, were found to be overrepresented at a developmental stage corresponding to major neurogenesis (Venø et al., 2015). These over-represented signaling pathways play a major role in neural stem cell proliferation, migration and differentiation (Rosso and Inestrosa, 2013; Mattar and Cayouette, 2014). How circRNAs impact these pathways remains to be an important question in understanding their contributions to neural proliferation and differentiation.
CircRNAs in Synaptogenesis
The expression change, localization and origin of circRNAs signify the importance of circRNAs in synaptogenesis to a large degree. During porcine and mouse brain maturation, the amount of circRNAs produced was found to be significantly increased, especially when synaptic connections and neural network forms (Venø et al., 2015; You et al., 2015). For example, in mice, the expression pattern of most circRNAs changes abruptly at postnatal day (P)10 when synaptogenesis begins (You et al., 2015). Moreover, circRNAs originated from synapse-related genes are consistently up-regulated during the establishment of mature neural networks (You et al., 2015). In addition, consistent with a fourfold increase of synaptic density in the human cortex compared to mice (Herculano-Houzel, 2009), higher levels of circRNAs were detected in human brains compared to mouse brains. Moreover, instead of being equally distributed in all neuronal compartments, circRNAs are highly enriched in synaptoneurosomes (You et al., 2015). Although both circRNAs and mRNAs can be detected in the cell body and dendrites, they are not co-localized. For example, circStau2a is mainly presented in synapses; however, the linear Stau2a mRNA transcripts primarily locate in the cytoplasm (Venø et al., 2015). Furthermore, neural circRNAs are disproportionally originated from host genes encoding proteins with synaptic functions. Genes related to synapse, synapse part, presynaptic active zone, presynaptic membrane and postsynaptic density can give rise to abundant circRNAs (Venø et al., 2015; You et al., 2015). For example, circHomer1_a, a circRNA originated from Homer1 that encodes a key protein in postsynaptic density regulation (Meyer et al., 2014), has the highest expression level during synaptic plasticity (You et al., 2015).
The function of circRNAs in mammalian brain remains elusive. Because of their primary localization to synapses, it has been hypothesized that circRNAs may function as topologically complex platforms which recruit ribonucleoprotein (RNP) granules and transport required RNAs and proteins (Rybak-Wolf et al., 2015) that are needed for synaptogenesis when synaptic connections forms (You et al., 2015). Moreover, due to their great stability and specificity, circRNAs might be used as synaptic tags to keep a molecular memory (Rybak-Wolf et al., 2015). Another possible function for circRNAs is to serve as miRNA sponges (Hansen et al., 2013). Certain circRNAs have been shown to contain multiple binding sites for core regions of miRNAs (Hansen et al., 2013; Zheng et al., 2016). It is possible that circRNAs can serve as competitive inhibitors for miRNA activities by inhibiting binding of miRNAs to their target transcripts. Multiple studies have shown that miRNAs play an important role in synapse formation, stabilization and plasticity (Rajasethupathy et al., 2009; McNeill and Van Vactor, 2012). Therefore, circRNAs may indirectly control synaptogenesis through regulating activities of synapse-related miRNAs and the expression of miRNA targeted mRNAs (Hansen et al., 2013; Zheng et al., 2016).
CircRNAs in CNS Diseases
CircRNAs play essential role in various processes of CNS development and might impact the function of aging brain (Westholm et al., 2014; Gruner et al., 2016), Recently, circRNAs have been investigated in neurological diseases (Chen and Schuman, 2016; Shao and Chen, 2016; Li et al., 2017). In addition, circRNAs have shown great potential to serve as prognostic biomarkers of many diseases due to their stability, specificity, sensitivity, and conservation (Memczak et al., 2015; Rybak-Wolf et al., 2015; Lyu and Huang, 2016).
CircRNAs in the Pathogenesis of CNS Diseases
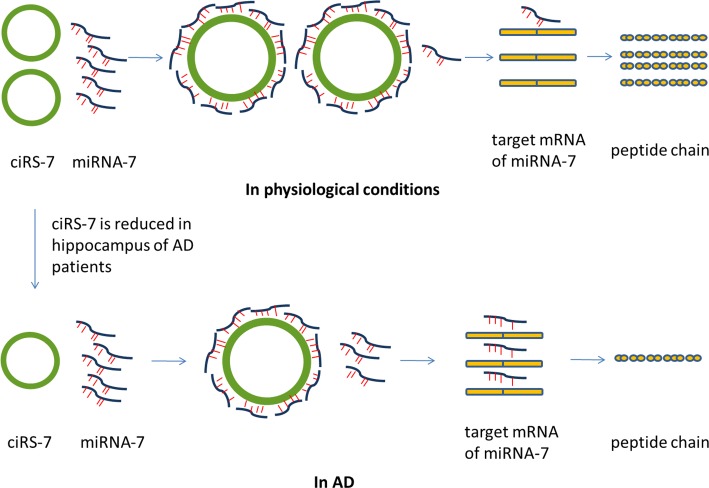
Several circRNAs were shown to function as miRNA sponges that sequester miRNAs and affect their interaction with downstream mRNAs (Floris et al., 2017; Piwecka et al., 2017) and an abnormal circRNA-miRNA-mRNA system has been indicated in the pathogenesis of CNS diseases (Li et al., 2017; Zhou et al., 2017). For example, Alzheimer’s disease (AD), a progressive neurodegenerative disorder, is caused by damage to synapses, neuronal cell bodies and axons due to the accumulation of pathological amyloid-β or hyper-phosphorylated Tau in the brain (Cohen et al., 2015). miRNA-7 is abundant in the brain and can directly regulate the expression of AD-related genes such Ubiquitin protein ligase A, which mediates the clearance of amyloid peptides in AD (Zhao et al., 2016). A circRNA, ciRS-7 (circRNA for miRNA-7), also termed as cerebellar degeneration-related protein 1 antisense RNA (Cdr1as), contains more than 70 selectively conserved anti-miRNA-7 sequences, thus acting as competing endogenous sponges to efficiently quench normal miRNA-7 activities (Hansen et al., 2013). miRNA-7 and ciRS-7 were found to be co-expressed in cultured primary murine neurons and in mouse brain sections (Hansen et al., 2013). In addition, expression of ciRS-7 is significantly reduced in hippocampus of AD patients (Lukiw, 2013), suggesting an abnormal miRNA-circRNA system may underline the disease mechanism of AD (Figure 2). Because of the importance of miRNA-7 in other diseases such as cancer and Parkinson’s disease (Junn et al., 2009; Saydam et al., 2011; Liu et al., 2014), it is possible that ciRS-7 may exert a regulatory role in brain cancer and other neurodegenerative diseases as well. Deleting the ciRS-7 locus in mice leads to dysfunctional synaptic transmission and abnormal neuropsychiatric-like behavior in mice (Piwecka et al., 2017). Other than miR-7, ciRS-7 also has a binding site to miR-671. Interestingly, the expression of miR-7 and miR-671 were both deregulated in all brain regions in ciRS-7 deficient mice; however, the direction of changes was opposite. It was thought that the binding site on ciRS-7 is completely complimentary to miR-671, and the interaction of these two molecules could lead to AGO mediated ciRS-7 slicing and miR-671 degradation. In contrast, the binding sites for miR-7 are partially complimentary which cannot trigger ciRS-7 cleavage, leading to stabilized miR-7. Therefore, it is possible that circRNAs can serve as a platform to store and transport certain miRNAs. Similarly, another circRNA generated from the gene Sry, has 16 putative binding sites to miR-138 and can attenuate activity of miR-138 in culture (Hansen et al., 2013). Expression of miR-138 was increased in cell culture models of AD suggesting the role of circRNA Sry in the disease pathology (Wang et al., 2015).
FIGURE 2.
Possible role of ciRS-7 in AD. CiRS-7 contains more than 70 selectively conserved anti-miRNA-7 sequences, thus acting as competing endogenous sponges to efficiently quench normal miRNA-7 activities. However, the expression of ciRS-7 is significantly reduced in hippocampus of AD patients. The abnormal miRNA-circRNA system might result in the abnormal mRNA expression of the target genes of miRNA-7. Therefore, the abnormal circRNA-miRNA-mRNA system may underline the disease mechanism of AD. miRNA, microRNA; circRNA, circular RNA; ciRS-7, circRNA for miRNA-7; AD, Alzheimer’s disease.
Other functions of circRNA in the CNS may be indicated by their host genes. For example, expression and complexity of circRNAs peak at embryonic day (E)60 during porcine brain development when dramatic morphological changes occur in the cortex. The top three highly expressed circRNAs at E60 are circCSPP1, circHDAC2 and circRIMS2 (Venø et al., 2015). The host genes of these circRNAs are crucial for CNS development and synaptic formation (Akizu et al., 2014). For instance, CSPP1 plays a part in the normal physiological functions of primary cilia and mutations in this gene lead to a developmental brain disorder called Joubert syndrome (Tuz et al., 2014). HDAC2 is important for neuronal survival and forebrain-specific deletion of this gene can lead to excessive repetitive behaviors in mice (Mahgoub et al., 2016). RIMS2 are involved in synaptic vesicles release and presynaptic plasticity and deletion of this gene can result in decreased activation of the presynaptic Ca2+-channel (Kaeser et al., 2012). CircRNAs might also play a role in nerve damage. Biosynthesis of circHomer1 may compete with synthesis of Homer1b/c mRNA (You et al., 2015). In an in vitro model inducing synaptic plasticity using primary hippocampal neurons, circHomer1a expression was remarkably increased, which could inhibit potential overexpression of Homer1b/c that is detrimental to synaptic plasticity (You et al., 2015). Down-regulation of Homer1b/c was also shown to improve neuronal survival in an in vitro traumatic nerve injury model (Fei et al., 2014). Therefore, circHomer1a may have great potential as a therapeutic target on repair and regeneration of injured nerves.
CircRNAs as Potential Biomarkers in CNS Disorders
Due to lack of 3′ termini, circRNAs are generally endowed with a strong resistance to exonuclease RNase R, thus being more stable than linear RNAs (Memczak et al., 2013; Enuka et al., 2016). The great stability and abundance of circRNAs make them as great candidates for molecular biomarkers in neurological diseases. Recently, several studies have reported the use of circRNAs as potential diagnostic biomarkers for atherosclerotic vascular disease, pre-diabetes and varieties of tumors including gastric cancer, hepatocellular carcinoma, lung cancer, colon carcinoma, laryngeal cancer and leukemia, et al (Ren et al., 2016; Xie et al., 2016; Zhao Z. et al., 2017). For example, circPVT1 could act as independent prognostic marker in gastric cancer (Chen J. et al., 2017). The expression of circPVT1 was up-regulated in gastric cancer tissues and it could promote cell proliferation through serving as a sponge for miR-125 family members (Chen J. et al., 2017). Also, similarly, circular RNA hsa_circ_0000190 was down-regulated in both human gastric cancer tissues and plasma samples and had higher sensitivity and specificity compared with traditional gastric cancer biomarkers, carcinoembryonic antigen (CEA) and CA19-9 (Chen S. et al., 2017). So far, multiple circRNAs have been explored as a biomarker for a variety of diseases especially in cancer (Meng et al., 2017).
Neural circRNAs show high conservation during evolution and are abundantly expressed in the brain (Venø et al., 2015). In Drosophila, circRNA expression increased substantially during CNS aging and could be regarded as a class of aging biomarkers (Westholm et al., 2014). Accumulation of circRNAs in aging brains has also been described in mice (Gruner et al., 2016) suggesting it could be a general theme for other species including humans. Therefore, although it has not been reported in CNS diseases, because circRNAs are stable, specific, conserved and sensitive, distinct circRNA profiles may be used to evaluate the physiologic and pathological circumstances in the brain (Memczak et al., 2013; Rybak-Wolf et al., 2015; Song et al., 2016). On the other hand, biopsies for neuronal tissues may be difficult to access. However, the pathological information of CNS tissues could be acquired through detecting the levels of certain components in the plasma and cerebrospinal fluid, etc. that are easily accessible (Luo et al., 2013). Therefore, detecting circRNAs from these sources can be possible and using circRNAs as biomarkers for neurological diseases should be further investigated.
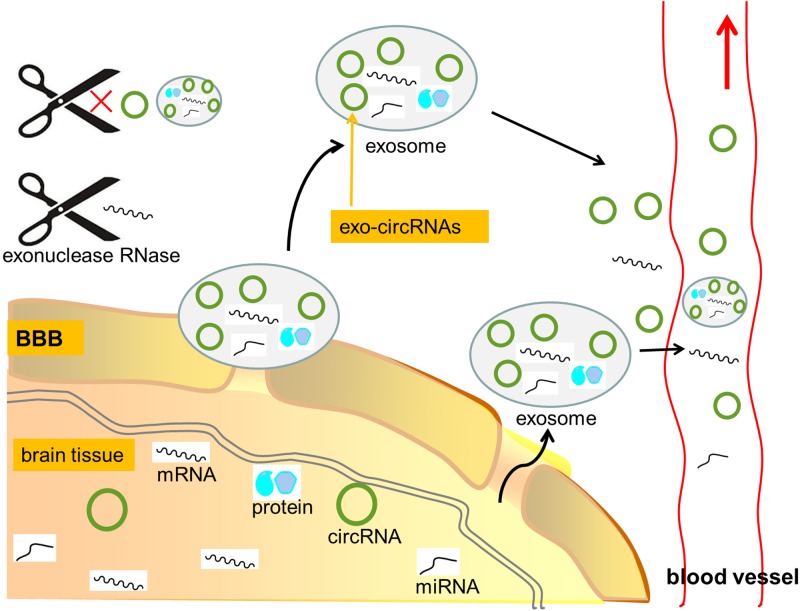
Furthermore, in CNS diseases, the blood–brain barrier (BBB) can be compromised and small molecules including circRNAs and exosomes can be transported out of BBB (Grapp et al., 2013). Exosomes are membranous small vesicles that can transfer circRNA, miRNA, mRNA and protein into extracellular fluid or cells (Cocucci and Meldolesi, 2015). Recent studies revealed that, circRNAs contained in exosomes (exo-circRNAs) were about twice more than those in their producer cells (Li Y. et al., 2015). CircRNAs tend to be more accessible to exosomes than linear RNAs, and the ratio of exo-circRNAs to their linear transcripts in exosomes is about six times higher than that in their producer cells (Li Y. et al., 2015) (Figure 3). In addition, exo-circRNAs showed high stability, perhaps because of their protein partners and the protection of exosomes (Li Y. et al., 2015). Taking together, the specific circRNAs profiles carried in exosomes might serve as potential biomarkers in CNS disorders (Lu and Xu, 2016).
FIGURE 3.
CircRNAs as potential biomarkers in CNS disorders. Due to lack of 3′ termini, circRNAs are generally endowed with a strong resistance to exonuclease RNase R, thus being more stable than linear RNAs. Furthermore, in CNS diseases, the blood–brain barrier (BBB) can be compromised and small molecules including circRNAs and exosomes can be transported out of BBB. CircRNAs contained in exosomes (exo-circRNAs) were about twice more than those in their producer cells. CircRNAs tend to be more accessible to exosomes than linear RNAs, and the ratio of exo-circRNAs to their linear transcripts in exosomes is about six times higher than that in their producer cells. In addition, exo-circRNAs showed high stability, perhaps because of their protein partners and the protection of exosomes. Thus, the great stability and abundance of circRNAs make them as great candidates for molecular biomarkers in neurological diseases. mRNA, messenger RNA; circRNA, circular RNA; miRNA, microRNA; BBB, blood–brain barrier; exo-circRNAs, circRNAs contained in exosome.
Future Exploration
Despite of recent discoveries that significantly expanded our knowledge on circRNAs (Danan et al., 2012; Kumar et al., 2016; Floris et al., 2017), there are still questions remain to be answered regarding circRNA biogenesis and their biological functions. These questions include: (i) how the spliceosome acts during the course of back-splicing reaction and the exact kinetics of back-splicing are currently not well known. (ii) In addition to acting as miRNA and RBP sponges, circRNAs could transport required RNAs and proteins and recruit ribonucleoproteins. However, we do not know enough details about the topological structure of circRNAs and how they interact with ribonucleoproteins and other RNAs. (iii) Neural circRNAs are abundant, conserved and spatio-temporally regulated. However, what controls the selectively high expression of certain neural circRNAs and the relative balance between the expression of circRNAs and their linear transcripts during CNS development is not known. (iv) In CNS development, how the highly expressed circRNAs act and how they are degraded are not clear. (v) Though miRNA sponge is the only known function for some circRNAs, other functions of most of circRNAs in the brain are not known. (vi) CircRNAs in certain tissues or in exosomes show high abundance and stability, but detection of their existence in exosomes can be costly. Thus, efficiency and convenient methods used for detecting circRNAs in exosomes are required for their widespread use as biomarkers. (vii) The repeatability, reliability and security of using circRNAs as biomarkers or therapeutic targets need to be further studied due to their complex roles in the body.
Author Contributions
LX prepared an initial draft and contributed to writing all sections and preparation of the final manuscript. MM contributed to overseeing revisions and preparation of the final manuscript. KX and BJ contributed to overseeing revisions and preparation of the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Natural Science Foundation of China (NSFC 81371012 to BJ and NSFC 81571939 to KX) and Nature Science Foundation of Hunan Province (2017jj2360 to BJ) and Development and Reform Commission of Hunan Province 2015-83.
References
- Abdelmohsen K., Panda A. C., Munk R., Grammatikakis I., Dudekula D. B., De S., et al. (2017). Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 14 361–369. 10.1080/15476286.2017.1279788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akizu N., Silhavy J. L., Rosti R. O., Scott E., Fenstermaker A. G., Schroth J., et al. (2014). Mutations in CSPP1 lead to classical Joubert syndrome. Am. J. Hum. Genet. 94 80–86. 10.1016/j.ajhg.2013.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwal-Fluss R., Meyer M., Pamudurti N. R., Ivanov A., Bartok O., Hanan M., et al. (2014). CircRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56 55–66. 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- Athanasiadis A., Rich A., Maas S. (2004). Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLOS Biol. 2:e391. 10.1371/journal.pbio.0020391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett S. P., Salzman J. (2016). Circular RNAs: analysis, expression and potential functions. Development 143 1838–1847. 10.1242/dev.128074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett S. P., Wang P. L., Salzman J. (2015). Circular RNA biogenesis can proceed through an exon-containing lariat precursor. eLife 4:e07540. 10.7554/eLife.07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B. M., Strein C., et al. (2012). Insight into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149 1393–1406. 10.1016/j.cell.2012.04.031 [DOI] [PubMed] [Google Scholar]
- Chen J., Li Y., Zheng Q., Bao C., He J., Chen B., et al. (2017). Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 388 208–219. 10.1016/j.canlet.2016.12.006 [DOI] [PubMed] [Google Scholar]
- Chen L. L. (2016). The biogenesis and emerging role of circular RNAs. Nat. Rev. Mol. Cell Biol. 17 205–211. [DOI] [PubMed] [Google Scholar]
- Chen L. L., Yang L. (2015). Regulation of circRNA biogenesis. RNA Biol. 12 381–388. 10.1080/15476286.2015.1020271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Li T., Zhao Q., Xiao B., Guo J. (2017). Using circular RNA has_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin. Chim. Acta 466 167–171. 10.1016/j.cca.2017.01.025 [DOI] [PubMed] [Google Scholar]
- Chen W., Schuman E. (2016). Circular RNAs in brain and other tissues: a functional enigma. Trends Neurosci. 39 597–604. 10.1016/j.tins.2016.06.006 [DOI] [PubMed] [Google Scholar]
- Cocucci E., Meldolesi J. (2015). Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 25 364–372. 10.1016/j.tcb.2015.01.004 [DOI] [PubMed] [Google Scholar]
- Cohen M. L., Kim C., Haldiman T., ElHag M., Mehndiratta P., Pichet T., et al. (2015). Rapidly progressive Alzheimer’s disease features distinct structures of amyloid-β. Brain 138 1009–1022. 10.1093/brain/awv006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn S. J., Pillman K. A., Toubia J., Conn V. M., Salmanidis M., Phillips C. A., et al. (2015). The RNA binding protein quaking regulates formation of circRNAs. Cell 160 1125–1134. 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- Danan M., Schwartz S., Edelheit S., Sorek R. (2012). Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 40 3131–3142. 10.1093/nar/gkr1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y., Yan L., Hu B., Fan X., Ren Y., Li R., et al. (2016). Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol. 17 130. 10.1186/s13059-016-0991-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R., Ma X. K., Chen L. L., Yang L. (2016). Increased complexity of circRNA expression during species evolution. RNA Biol. 10.1080/15476286.2016.1269999 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou C., Cao Z., Yang B., Ding N., Hou T., Luo F., et al. (2016). Changing expression profiles of lncRNAs, mRNAs, circRNAs and miRNAs during osteoclastogenesis. Sci. Rep. 6:21499. 10.1038/srep21499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudekula D. B., Panda A. C., Grammatikakis I., De S., Abdelmohsen K., Gorospe M. (2016). Circinteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 13 34–42. 10.1080/15476286.2015.1128065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I., Kundaje A., Aldred S. F., Collins P. J., Davis C. A., Doyle F., et al. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earls L. R., Westmoreland J. J., Zakharenko S. S. (2014). Non-coding RNA regulation of synaptic plasticity and memory: implication for aging. Ageing Res. Rev. 17 34–42. 10.1016/j.arr.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enuka Y., Lauriola M., Feldman M. E., Sas-Chen A., Ulitsky I., Yarden Y. (2016). Circular RNAs are long-lived and display only minimal early alterations in response to a growth factors. Nucleic Acids Res. 44 1370–1383. 10.1093/nar/gkv1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei F., Rao W., Zhang L., Chen B. G., Li J., Fei Z., et al. (2014). Downregulation of Homer1b/c improves neuronal survival after traumatic neuronal injury. Neuroscience 267 187–194. 10.1016/j.neuroscience.2014.02.037 [DOI] [PubMed] [Google Scholar]
- Filippenkov I. B., Kalinichenko E. O., Limborska S. A., Dergunova L. V. (2017). Circular RNAs-one of the enigmas of the brain. Neurogenetics 18 1–6. [DOI] [PubMed] [Google Scholar]
- Floris G., Zhang L., Follesa P., Sun T. (2017). Regulatory role of circular RNAs and neurological disorders. Mol. Neurobiol. 54 5156–5165. 10.1007/s12035-016-0055-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gailite I., Aerne B. L., Tapon N. (2015). Differential control of Yorkie activity by LKB1/AMPK and Hippo/Warts cascade in the central nervous system. Proc. Natl. Acad. Sci. U.S.A. 112 E5169–E5178. 10.1073/pnas.1505512112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grapp M., Wrede A., Schweizer M., Hüwel S., Galla H. J., Snaidero N., et al. (2013). Choroid plexus transcytosis and exosome shuttling deliver folate into brain parenchyma. Nat. Commun. 4:2123. 10.1038/ncomms3123 [DOI] [PubMed] [Google Scholar]
- Gruner H., Cortés-López M., Cooper D. A., Bauer M., Miura P. (2016). CircRNA accumulation in the aging mouse brain. Sci. Rep. 6:38907. 10.1038/srep38907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. U., Agarwal V., Guo H., Bartel D. P. (2014). Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 15 409. 10.1186/s13059-014-0409-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid F. M., Makeyev E. V. (2014). Regulation of mRNA abundance by polypyrimidine tract-binding protein-controlled alternate 5’ splice site choice. PLOS Genet. 10:e1004771. 10.1371/journal.pgen.1004771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Li J., Wang H., Su X., Hou J., Gu Y., et al. (2017). Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma. Hepatology 66 1151–1164. 10.1002/hep.29270 [DOI] [PubMed] [Google Scholar]
- Hanan M., Soreq H., Kadener S. (2016). CircRNAs in the brain. RNA Biol. 10.1080/15476286.2016.1255398 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T. B., Jensen T. I., Clausen B. H., Bramsen J. B., Finsen B., Damgaard C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495 384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Hentze M. W., Preiss T. (2013). Circular RNAs: splicing’s enigma variations. EMBO J. 32 923–925. 10.1038/emboj.2013.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heraud-Farlow J. E., Kiebler M. A. (2014). The multifunctional Staufen proteins: conserved roles from neurogenesis to synaptic plasticity. Trends Neurosci. 37 470–479. 10.1016/j.tins.2014.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S. (2009). The human brain in numbers: a linearly scaled-up primate brain. Front. Hum. Neurosci. 3:31. 10.3389/neuro.09.031.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. M., Lukiw W. J. (2016). MicroRNA (miRNA)-mediated pathogenetic signaling in Alzheimer’s disease (AD). Neurochem. Res. 41 96–100. 10.1007/s11064-015-1734-7 [DOI] [PubMed] [Google Scholar]
- Hirose T., Mishima Y., Tomari Y. (2014). Elements and machinery of non-coding RNAs: toward their taxonomy. EMBO Rep. 15 489–507. 10.1002/embr.201338390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M. A., Rodriguez C. M., Johnson T. L. (2011). Key features of the two-intron Saccharomyces cerevisiae gene SUS1 contribute to its alternative splicing. Nucleic Acids Res. 39 8612–8627. 10.1093/nar/gkr497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M. T., Coca-Prados M. (1979). Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280 339–340. 10.1038/280339a0 [DOI] [PubMed] [Google Scholar]
- Ivanov A., Memczak S., Wyler E., Torti F., Porath H. T., Orejuela M. R., et al. (2015). Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 10 170–177. 10.1016/j.celrep.2014.12.019 [DOI] [PubMed] [Google Scholar]
- Jeck W. R., Sharpless N. E. (2014). Detecting and characterizing circular RNAs. Nat. Biotechnol. 32 453–461. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W. R., Sorrentino J. A., Wang K., Slevin M. K., Burd C. E., Liu J., et al. (2013). Circular RNAs are abundant, conserved and associated with ALU repeats. RNA 19 141–157. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., Oshima-Takago T., Chakrabarti R., Wong A. B., Jing Z., Yamanbaeva G., et al. (2015). Rab3-interacting molecules 2αand 2β promote the abundance of voltage-gated CaV1.3 Ca2+ channels at hair cell active zones. Proc. Natl. Acad. Sci. U.S.A. 112 E3141–E3149. 10.1073/pnas.1417207112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junn E., Lee K. W., Jeong B. S., Chan T. W., Im J. Y., Mouradian M. M. (2009). Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc. Natl. Acad. Sci. U.S.A. 106 13052–13057. 10.1073/pnas.0906277106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser P. S., Deng L., Fan M., Südhof T. C. (2012). RIM genes differentially contribute to organizing presynaptic release sites. Proc. Natl. Acad. Sci. U.S.A. 109 11830–11835. 10.1073/pnas.1209318109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S., Greenman C., Cook P. R., Papantonis A. (2015). Exon skipping is correlated with exon circularization. J. Mol. Biol. 427 2414–2417. 10.1016/j.jmb.2015.02.018 [DOI] [PubMed] [Google Scholar]
- Kim D. D., Kim T. T., Walsh T., Kobayashi Y., Matise T. C., Buyske S., et al. (2004). Widespread RNA editing of embedded Alu elements in the human transcriptome. Genome Res. 14 1719–1725. 10.1101/gr.2855504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer M. C., Liang D., Tatomer D. C., Gold B., March Z. M., Cherry S., et al. (2015). Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 29 2168–2182. 10.1101/gad.270421.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L., Shamsuzzama, Haque R., Baghel T., Nazir A. (2016). Circular RNAs: the emerging class of non-coding RNAs and their potential role in human neurodegenerative diseases. Mol. Neurobiol. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Li T. R., Jia Y. J., Wang Q., Shao X. Q., Lv R. J. (2017). Circular RNA: a new star in neurological diseases. Int. J. Neurosci. 127 726–734. 10.1080/00207454.2016.1236382 [DOI] [PubMed] [Google Scholar]
- Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., et al. (2015). Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 25 981–984. 10.1038/cr.2015.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., et al. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22 256–264. 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- Liang D., Wilusz J. E. (2014). Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 28 2233–2247. 10.1101/gad.251926.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Yang Y., Niu G., Tang Z., Li K. (2017). Genome-wide profiling of Sus scrofa circular RNAs across nine organs and three developmental stages. DNA Res. 24 523–535. 10.1093/dnares/dsx022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Jiang Z., Huang J., Huang S., Li Y., Yu S., et al. (2014). MiR-7 inhibits glioblastoma growth by simultaneously interfering with the PI3K/AKT and Raf/MEK/ERK pathways. Int. J. Oncol. 44 1571–1580. 10.3892/ijo.2014.2322 [DOI] [PubMed] [Google Scholar]
- Lu D., Xu A. D. (2016). Mini review: circular RNAs as potential clinical biomarkers for disorders in the central nervous system. Front. Genet. 7:53. 10.3389/fgene.2016.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw W. J. (2013). Circular RNA (circRNA) in Alzheimer’s disease (AD). Front. Genet. 4:307. 10.3389/fgene.2013.00307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Hou L., Shi H., Zhong X., Zhang Y., Zheng D., et al. (2013). CSF levels of the neuronal injury biomarker visinin-like protein-1 in Alzheimer’s disease and dementia with Lewy bodies. J. Neurochem. 127 681–690. 10.1111/jnc.12331 [DOI] [PubMed] [Google Scholar]
- Lyu D., Huang S. (2016). The emerging role and clinical implication of human exonic circular RNA. RNA Biol. 10.1080/15476286.2016.1227904 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahgoub M., Adachi M., Suzuki K., Liu X., Kavalali E. T., Chahrour M. H., et al. (2016). MeCP2 and histone deacetylases 1 and 2 in dorsal striatum collectively suppress repetitive behaviors. Nat. Neurosci. 19 1506–1512. 10.1038/nn.4395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattar P., Cayouette M. (2014). Temporal control of neural progenitors: TGF-β switches the clock forward. Neuron 84 885–888. 10.1016/j.neuron.2014.11.013 [DOI] [PubMed] [Google Scholar]
- McNeill E., Van Vactor D. (2012). MicroRNAs shape the neuronal landscape. Neuron 75 363–379. 10.1016/j.neuron.2012.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495 333–338. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- Memczak S., Papavasileiou P., Peters O., Rajewsky N. (2015). Identification and characterization of circular RNA as a new class of putative biomarkers in human blood. PLOS ONE 10:e0141214. 10.1371/journal.pone.0141214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng S., Zhou H., Feng Z., Xu Z., Tang Y., Li P., et al. (2017). CircRNA: functions and properties of a novel potential biomarker for cancer. Mol. Cancer 16 94. 10.1186/s12943-017-0663-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D., Bonhoeffer T., Scheuss V. (2014). Balance and stability of synaptic structures during synaptic plasticity. Neuron 82 430–443. 10.1016/j.neuron.2014.02.031 [DOI] [PubMed] [Google Scholar]
- Ng S. Y., Bogu G. K., Soh B. S., Stanton L. W. (2013). The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 51 349–359. 10.1016/j.molcel.2013.07.017 [DOI] [PubMed] [Google Scholar]
- Pamudurti N. R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., et al. (2017). Translation of circRNAs. Mol. Cell 66 9–21. 10.1016/j.molcel.2017.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persengiev S., Kondova I., Otting N., Koeppen A. H., Bontrop R. E. (2011). Genome-wide analysis of miRNA expression reveals a potential role for miR-144 in brain aging and spinocerebellar ataxia pathogenesis. Neurobiol. Aging 32 e17–e27. 10.1016/j.neurobiolaging.2010.03.014 [DOI] [PubMed] [Google Scholar]
- Petkovic S., Müller S. (2015). RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 43 2454–2465. 10.1093/nar/gkv045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwecka M., Glažar P., Hernandez-Miranda L. R., Memczak S., Wolf S. A., Rybak-Wolf A., et al. (2017). Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357:eaam8526. 10.1126/science.aam8526 [DOI] [PubMed] [Google Scholar]
- Rajasethupathy P., Fiumara F., Sheridan R., Betel D., Puthanveettil S. V., Russo J. J., et al. (2009). Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron 63 803–817. 10.1016/j.neuron.2009.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani N., Nowakowski T. J., Zhou H., Godshalk S. E., Lisi V., Kriegstein A. R., et al. (2016). A primate lncRNA mediates Notch signaling during neuronal development by sequestering miRNA. Neuron 90 1174–1188. 10.1016/j.neuron.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. S., O’Brien D., Pisat N., Weichselbaum C. T., Sakers K., Lisci M., et al. (2017). A comprehensive analysis of cell type-specific nuclear RNA from neurons and glia of the brain. Biol. Psychiatry 81 252–264. 10.1016/j.biopsych.2016.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Xin S., Lan X., Lu G., Lin Y., Yang S., et al. (2016). Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget 7 26680–26691. 10.18632/oncotarget.8589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso S. B., Inestrosa N. C. (2013). WNT signaling in neuronal maturation and synaptogenesis. Front. Cell. Neurosci. 7:103 10.3389/fncel.2013.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A., Stottmeister C., Glažar P., Jens M., Pino N., Giusti S., et al. (2015). Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 58 870–885. 10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]
- Salzman J. (2016). Circular RNA expression: its potential regulation and function. Trends Genet. 32 309–316. 10.1016/j.tig.2016.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger H. L., Klotz G., Riesner D., Gross H. J., Kleinschmidt A. K. (1976). Viroids are single-stranded covalently closed circular RNA molecules existing are highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U.S.A. 73 3852–3856. 10.1073/pnas.73.11.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saydam O., Senol O., Würdinger T., Mizrak A., Ozdener G. B., Stemmer-Rachamimov A. O., et al. (2011). MiRNA-7 attenuation in Schwannoma tumors stimulates growth by upregulating three oncogenic signaling pathways. Cancer Res. 71 852–861. 10.1158/0008-5472.CAN-10-1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y., Chen Y. (2016). Roles of circular RNAs in neurologic disease. Front. Mol. Neurosci. 9:25. 10.3389/fnmol.2016.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma E., Sterne-Weiler T., O’Hanlon D., Blencowe B. J. (2016). Global mapping of human RNA-RNA interactions. Mol. Cell 62 618–626. 10.1016/j.molcel.2016.04.030 [DOI] [PubMed] [Google Scholar]
- Song X., Zhang N., Han P., Moon B. S., Lai R. K., Wang K., et al. (2016). Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 44 e87. 10.1093/nar/gkw075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starke S., Jost I., Rossbach O., Schneider T., Schreiner S., Hung L. H., et al. (2015). Exon circularization requires canonical splice signals. Cell Rep. 10 103–111. 10.1016/j.celrep.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Szabo L., Morey R., Palpant N. J., Wang P. L., Afari N., Jiang C., et al. (2015). Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 16 126. 10.1186/s13059-015-0690-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taulli R., Loretelli C., Pandolfi P. P. (2013). From pseudo-ceRNAs to circ-ceRNAs: a tale of cross-talk and competition. Nat. Struct. Mol. Biol. 20 541–543. 10.1038/nsmb.2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y., Rinn J., Pandolfi P. P. (2014). The multilayered complexity of ceRNA crosstalk and competition. Nature 505 344–352. 10.1038/nature12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L. F., Sætrom P. (2014). Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics 30 2243–2246. 10.1093/bioinformatics/btu257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuz K., Bachmann-Gagescu R., O’Day D. R., Hua K., Isabella C. R., Phelps I. G., et al. (2014). Mutations in CSPP1 cause primary cilia abnormalities and Joubert syndrome with or without Jeune asphyxiating thoracic dystrophy. Am. J. Hum. Genet. 94 62–72. 10.1016/j.ajhg.2013.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rossum D., Verheijen B. M., Pasterkamp R. J. (2016). Circular RNAs: novel regulators of neuronal development. Front. Mol. Neurosci. 9:74. 10.3389/fnmol.2016.00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venø M. T., Hansen T. B., Venø S. T., Clausen B. H., Grebing M., Finsen B., et al. (2015). Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 16 245. 10.1186/s13059-015-0801-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villaescusa J. C., Li B., Toledo E. M., Rivetti di Val Cervo P., Yang S., Stott S. R., et al. (2016). A PBX1 transcriptional network controls dopaminergic neuron development and is impaired in Parkinson’s disease. EMBO J. 35 1963–1978. 10.15252/embj.201593725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Tan L., Lu Y., Peng J., Zhu Y., Zhang Y., et al. (2015). MicroRNA-138 promotes tau phosphorylation by targeting retinoic acid receptor alpha. FEBS Lett. 589 726–729. 10.1016/j.febslet.2015.02.001 [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang Z. (2015). Efficient backsplicing produces translatable circular mRNAs. RNA 21 172–179. 10.1261/rna.048272.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm J. O., Miura P., Olson S., Shenker S., Joseph B., Sanfilippo P., et al. (2014). Genomewide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 9 1966–1980. 10.1016/j.celrep.2014.10.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J. E., Sharp P. A. (2013). Molecular biology. A circuitous route to noncoding RNA. Science 340 440–441. 10.1126/science.1238522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Xuan L., Qu L., Zhou H., Wang P., Yu H., et al. (2016). Circular RNA: a novel biomarker for progressive laryngeal cancer. Am. J. Transl. Res. 8 932–939. [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., et al. (2017). Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 27 626–641. 10.1038/cr.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Sun Y., Wu J., Yan S., Deng Z., Wang J., et al. (2015). Discovering novel microRNAs and age-related nonlinear changes in rat brains using deep sequencing. Neurobiol. Aging 36 1037–1044. 10.1016/j.neurobiolaging.2014.11.001 [DOI] [PubMed] [Google Scholar]
- You X., Vlatkovic I., Babic A., Will T., Epstein I., Tushev G., et al. (2015). Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 18 603–610. 10.1038/nn.3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. O., Dong R., Zhang Y., Zhang J. L., Luo Z., Zhang J., et al. (2016). Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 26 1277–1287. 10.1101/gr.202895.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. O., Wang H. B., Zhang Y., Lu X., Chen L. L., Yang L. (2014). Complementary sequence-mediated exon circularization. Cell 159 134–147. 10.1016/j.cell.2014.09.001 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xue W., Li X., Zhang J., Chen S., Zhang J. L., et al. (2016). The biogenesis of nascent circular RNAs. Cell Rep. 15 611–624. 10.1016/j.celrep.2016.03.058 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang X. O., Chen T., Xiang J. F., Yin Q. F., Xing Y. H., et al. (2013). Circular intronic long noncoding RNAs. Mol. Cell 51 792–806. 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- Zhao W., Cheng Y., Zhang C., You Q., Shen X., Guo W., et al. (2017). Genome-wide identification and characterization of circular RNAs by high throughput sequencing in soybean. Sci. Rep. 7:5636. 10.1038/s41598-017-05922-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Alexandrov P. N., Jaber V., Lukiw W. J. (2016). Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer’s disease (AD) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7). Genes 7:e116. 10.3390/genes7120116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Li X., Jian D., Hao P., Rao L., Li M. (2017). Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 54 237–245. 10.1007/s00592-016-0943-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., et al. (2016). Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 7:11215. 10.1038/ncomms11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Xiong Q., Chen H., Yang C., Fan Y. (2017). Identification of the spinal expression profile of non-coding RNAs involved in neuropathic pain following spared nerve injury by sequence analysis. Front. Mol. Neurosci. 10:91. 10.3389/fnmol.2017.00091 [DOI] [PMC free article] [PubMed] [Google Scholar]