Abstract
Macrophorins are representative members of isoprenoid epoxycyclohexenones, containing cyclized drimane moieties. We located and characterized the biosynthetic gene cluster of macrophorin from Penicillium terrestris. MacJ encoded by this cluster was characterized to be the first example of membrane-bound type II terpene cyclases catalyzing the cyclization of meroterpenoids via direct protonation of terminal olefinic bond in the acyclic yanuthone. The late-stage functionalization and substrate promiscuity of MacJ make it a potential biocatalyst for synthesizing macrophorin analogs.
Graphical Abstract
Isoprenoid epoxycyclohexenones constitute a large family of natural products isolated from a variety of microorganisms, including fungi.1 These molecules feature an epoxydon core substituted with isoprenoid units, frequently a C15 farnesyl chain. The biological activities of members of this family are diverse, and differ depending on whether the isoprenoid chain is linear or cyclized. Linear members include yanuthone A (antimicrobial)2, oligosporons (nematicidal)3 and tricholomenyns (antimitotics)4. The farnesyl chain can be cyclized into the bicyclic drimane scaffold, as found in macrophorins (immunosuppressive)5 and epoxy-phomadins (nanomolar cytotoxic).6 Because of the structural diversity and biological activities, the sesquiterpene epoxycyclohexenones have received significant interest from the synthetic community.7 The biosynthesis of the linear compound yanuthone was recently reported, confirming the origin of the cyclohexenone ring is derived from 6-methyl-salicylic acid (6-MSA) synthesized by the corresponding polyketide synthase.8 Given the epoxycyclohexenone ring is shared between the linear and cyclized members of this family, it is proposed that a terpene cyclase must be present in the biosynthetic gene clusters of the cyclized compounds.
Cyclization of the linear chain in yanuthone to the drimane unit in macrophorin is expected to be catalyzed by a type II terpene cyclase (TC) that uses an active site general acid to initiate the carbocation reaction cascade.9 The best characterized class II TCs are squalene-hopene cyclase (SHC) and oxidosqualene cyclase (OSC). SHC directly protonates the terminal olefinic bond to initiate the cation mediated cyclization of squalene into hopene.10 In contrast, OSC cannot directly protonate the terminal olefinic bond, but requires squalene to first undergo epoxidation to form 2,3-oxidosqualene with the epoxide serving as the electrophilic functional group in subsequent cyclization steps to afford lanosterol (Figure 1B).11 During cyclization of the terpene portion of most hybrid terpene natural products such as in indole diterpenes and meroterpenoids (polyketide-terpene), a family of TCs, known as membrane-bound meroterpenoid TCs, use the epoxide-opening mechanism as in OSC to initiate the reaction.12 In these pathways, TC-catalyzed cyclization typically occurs early in the biosynthetic pathway as a scaffold building step, and is following by post-cyclization modifications to yield the final products. In contrast, we expect cyclization of macrophorin to occur as a late-stage biosynthetic transformation, with the linear yanuthone as a possible precursor. In addition, absence of oxygen substitution in the decalin ring suggests the responsible TC should follow the mechanism of SHC. This mode of cyclization in natural product biosynthesis is most notably observed in the cyclization of geranylgeranyl-diphosphate into ent-copalyl-diphosphate, catalyzed by ent-copalyl-PP synthase.13 However, a TC that catalyzes cyclization of meroterpenoid through direct olefinic bond protonation has not been reported. Recently, soluble type II TC MstE that catalyzes the analogous reaction in the merosterol biosynthetic pathway was discovered.14
Figure 1.
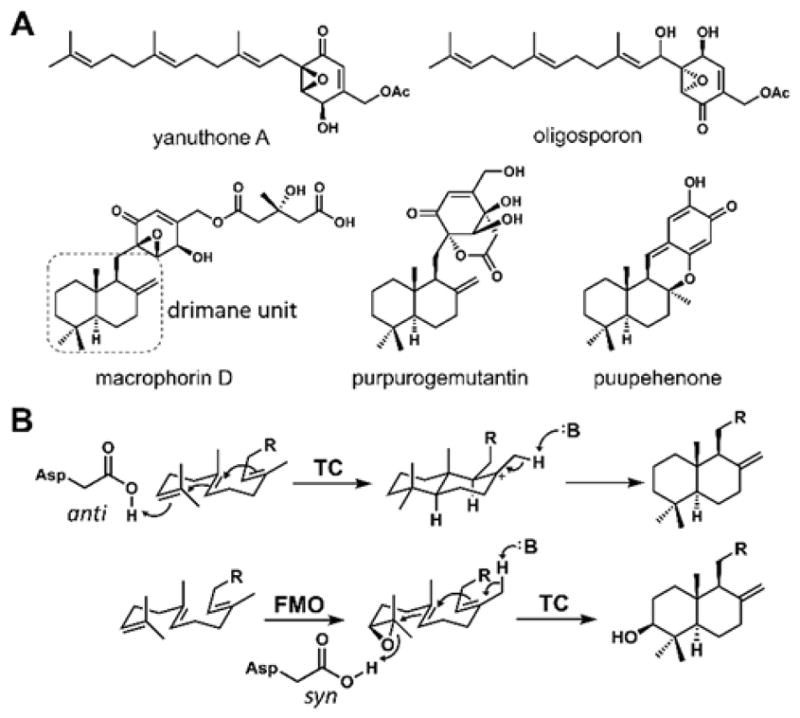
A) The structures of the representative sesquiterpene epoxycyclohexenones. B) Two different cyclization mechanisms of class II terpene cyclases: protonation on the terminal olefinic bond or the epoxide to trigger the cyclization cascade. TC, terpene cyclase; FMO, flavin-dependent monooxygenase.
To identify gene clusters that may produce cyclized isoprenoid epoxycyclohexenes and a responsible TC for the cyclization step, we searched sequenced fungal genomes using the entire yanuthone (yan) cluster8 identified from Aspergillus niger (Figure 2A) as a query. We found several highly homologous clusters in the genomes database, including from the genomes of Penicillium terrestris LM2 (named mac) and Penicillium chrysogenum. All of the genes from the mac cluster from P. terrestris LM2 are in synteny and highly identical to those in the yan cluster, except for a single gene macJ located at one terminus of the cluster that was initially assigned to encode a hypothetical protein. Further bioinformatic analysis revealed MacJ is predicted to contain seven transmembrane helices and is weakly homologous to TCs that promote indole diterpene and meroterpenoid cyclization via acid catalyzed epoxide-opening, including AndB15, Pyr416, Trt117, AtmB18 and PaxB19. We previously classified these TCs into clades based on sequence and function (Figure S2).20 MacJ does not clade with the known members above, and instead belongs to a large sub-clade of unknown function, indicating its catalytic mechanism may be distinct. Overall, of the 12 of clusters that are highly identical to the yan cluster, 10 contain a macJ homolog (Figure S1).
Figure 2.
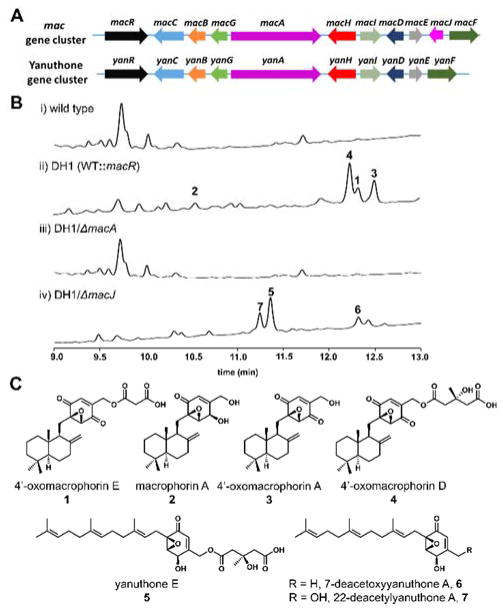
Identification of a macroporin gene cluster. A) Comparison of the mac gene cluster identified from Penicillium terrestris LM2 with the yan cluster of yanuthones from Aspergillus niger. B) HPLC analysis (λ=220 nm) of the extracts of P. terrestris LM2: i) wild type; ii) macR overexpression strain DH1; iii) DH1/ΔmacA; iv) DH1/ΔmacJ. C) Structures of compounds purified from the extracts of strains.
To identify the natural product encoded in the mac cluster, we cultured P. terrestris LM2 under different culture conditions in attempt to isolate epoxycyclohexenone-containing compounds. However, no related compound can be isolated (Figure 2B, i), indicating this cluster may be silent under these conditions. We then overexpressed a putative fungal transcriptional regulator found in the gene cluster, macR, using the constitutive gpdA promoter.21 After confirming the ecotopic integration of the overexpression cassette, the strain DH1 was cultured on TGA plates and extracted for metabolite analysis. As shown in Figure 2, several new metabolites (compounds 1–4) related to epoxycyclohexenone were produced (Figure 2B, ii). These compounds were isolated and characterized by 1D and 2D NMR spectroscopy to be related to macrophorin. Together with the known compounds macrophorin A (2), 4′-oxomacrophorin A (3), and 4′-oxomacrophorin D (4), a new compound named 4′-oxomacrophorin E (1) that is the malonate ester of 3 was also identified (Figure 2C). Optical rotation measurement confirmed the absolute stereochemistry of 1–4 (1, +17; 2, +31; 3, +15; 4, +44) to be the same as that reported for known macrophorins (2, +29; 3, +10; 4, +35),5 of which the drimane portion is cyclized from a “normal” chair/chair conformation of the linear terpene.
To establish the link between the production of 1–4 and the mac gene cluster, the 6-MSA synthase (6-MSAS) gene macA was deleted from the macR-overexpression strain by gene replacement to yield the mutant strain DH1/ΔmacA. HPLC analysis showed that deletion of macA completely abolished the production of compound 1–4 (Figure 2B, iii). Moreover, heterologous expression of macA in Saccharomyces cerevisiae strain BJ5464-NpgA indeed resulted in the production of 6-MSA (Figure S3), confirming the function of MacA is the same as the YanA from the yanuthone biosynthetic pathway.8 To examine if the putative membrane-bound MacJ is the enzyme responsible for terpene cyclization, we deleted macJ in DH1 to yield the strain DH1/ΔmacJ. Metabolite analysis from DH1/ΔmacJ showed that while production of 1–4 were completely abolished, three new metabolites 5–7 were instead accumulated (Figure 2B, iv). The three compounds were structurally elucidated to be yanuthone E (5), 7-deacetoxyyanuthone A (6) and 22-deacetylyanuthone A (7) (Figure 2C) based on NMR data, all of which contain the linear farnesyl chain. Hence, MacJ is directly involved in the formation of the drimane unit.
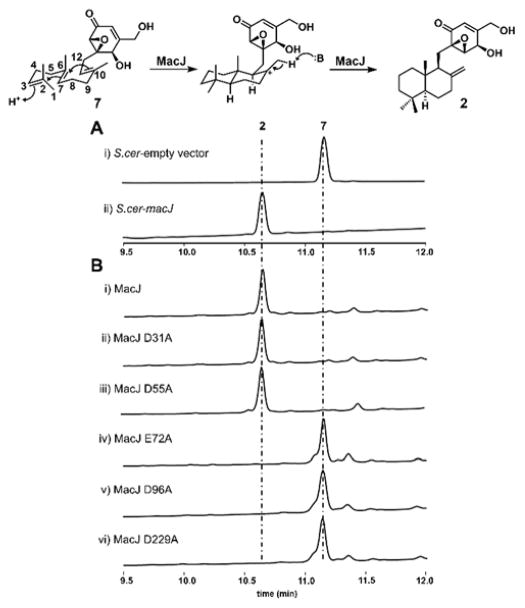
To further verify the function of MacJ, the enzyme was cloned from cDNA and expressed in BJ5464-NpgA. The function of MacJ was first examined by in vivo bio-transformation using the purified acyclic compound 7 as the substrate. Compared to the yeast strain harboring the empty vector, LC-MS analysis results indicated that 7 can be completely converted to the cyclized product 2 (Figure 3A, i and ii). The function of MacJ was further assayed by using the yeast microsomal fraction (Figure 3B, i), during which 0.2 mg/mL of 7 can be completely converted to 2 in 12 hrs using 2 mg/mL of microsomal protein. These results therefore confirm the direct role of MacJ as the TC that directly cyclizes the linear farnesyl unit into the drimane unit in 7. The cyclization activity of MacJ towards other acyclic substrates 5 and 6 were similarly assayed. Both compounds can be efficiently converted to the corresponding cyclized macrophorin analogs, indicating MacJ has significant tolerance towards substitutions in the epoxyhexenone ring as substitution as large as the mevalonate ester can be tolerated (Figure S4). However, when farnesylhydroquinone was tested as substrate, only trace amount of the cyclized product can be detected by LC-MS (Figure S4). Hence the epoxide moiety appears to be crucial for recognition by MacJ. These assays support the assignment of MacJ as a late-stage terpene cyclase that can transform the fully decorated linear epoxyhexenones into the cyclized versions. Its substrate promiscuity towards functionalized epoxydon therefore makes it a potential biocatalyst for generating analogs.
Figure 3.
Biochemical characterization of MacJ and its mutants: A) HPLC analysis (λ=220 nm) of MacJ catalyzed biotransformation of 7 to 2. B) HPLC analysis (λ=220 nm) of in vitro biochemical assays of MacJ and its mutants by using yeast microsomal fractions.
The mechanism of MacJ is likely analogous to that of SHC and ent-copalyl-diphosphate synthase. In both of these soluble enzymes, an active site aspartic acid in the anti configuration serves as the strong Brønsted acid to generate the carbocation at C2 of the isoprenoid chain. MacJ bears no sequence homology to either of these enzymes, and is predicted to be an integral membrane protein. To search for potential catalytic residues of MacJ, mutagenesis of conserved acidic residues identified from sequence alignment of homologous enzymes (Figure S6) was performed. These included Glu72 that is strictly conserved among MacJ homologs, as well as Asp31, Asp55, Asp96, and Asp229 that are conserved in most members. The mutants were expressed in yeast and assayed using microsomal fractions. Mutations to Glu72, Asp96 and Asp229 completely abolished the cyclase activity (Figure 3B, iv), suggesting that one of the three may serve as the corresponding Brønsted acid. In contrast, mutations to Asp31 and Asp55 had no effect (Figure 3B, ii and iii). Previous mutagenesis studies with Pyr4 showed the acidic residues corresponding to Glu72 and Asp229 are also crucial for the cyclization activity.16 However, Pyr4 catalyzes cyclization of meroterpenoid via protonation mediated epoxide-opening.
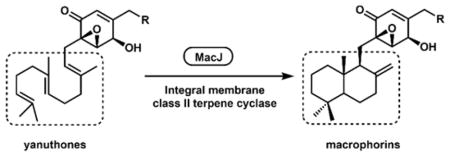
MacJ adds to the growing list of fungal terpene cyclases that are found to be integral membrane proteins compared to the soluble counterparts found in bacteria. This includes both type I and type II terpene cyclases. The fumagillin TC that synthesizes β-trans-bergamotene from farnesyl-diphosphate is an example of a membrane-bound type I TC.22 Pyr4 from the pyripyropene biosynthetic pathway16 was the first membrane-bound type II TC characterized from fungi. All subsequently characterized Pyr4 homologs from indole-diterpene and meroterpenoid pathways work in tandem with a flavin-dependent monooxygenase to cyclize the linear terpene unit in a two-step reaction, during which the “disappearing” epoxide serves as the site of proton-initiated cyclization steps.12 In contrast, MacJ identified here represents the first example of a membrane-bound type II cyclase that can cyclize a terpene through direct protonation of the terminal olefinic bond without requiring the epoxidation step. Our phylogenetic analysis showed that many homologs of MacJ of unknown function are found in the same subclade as MacJ19, suggesting new cyclized meroterpenoid natural products may be mined from these pathways.
In conclusion, we identified the biosynthetic gene cluster of the epoxycyclohexenone macrophorins using genome mining approaches. The mac cluster contains a single gene insertion compared to the yan cluster that produces the linear compound yanuthone. MacJ was confirmed to be the first example of an integral membrane terpene cyclase that can cyclize terpenes through direct olefinic bond protonation. MacJ was shown to catalyze a late-stage cyclization reaction and could potentially be used as a biocatalyst for synthesizing diverse macrophorin analogs.
Supplementary Material
Acknowledgments
This work was supported by the NIH 1R35GM118056 to Y.T.; and National Natural Science Foundation of China (21542001), the Shandong Provincial Natural Science Fund for Distinguished Young Scholars (JQ201422), AoShan Talents Program Supported by Qingdao National Laboratory for Marine Science and Technology (2015ASTP-ES09), NSFC-Shandong Joint Fund for Marine Science Research Centers (U1606403), Scientific and Technological Innovation Project Financially Supported by Qingdao National Laboratory for Marine Science and Technology (2015ASKJ02) to D.L..
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website.
Experimental details and spectroscopic data (PDF)
References
- 1.(a) Marco-Contelles J, Molina MT, Anjum S. Chem Rev. 2004;104:2857. doi: 10.1021/cr980013j. [DOI] [PubMed] [Google Scholar]; (b) Marcos IS, Conde A, Moro RF, Basabe P, Diez D, Urones JG. Mini-Rev Org Chem. 2010;7:230. [Google Scholar]; (c) Sunassee SN, Davies-Coleman MT. Nat Prod Rep. 2012;29:513. doi: 10.1039/c2np00086e. [DOI] [PubMed] [Google Scholar]
- 2.Bugni TS, Abbanat D, Bernan VS, Maiese WM, Greenstein M, Van Wagoner RM, Ireland CM. J Org Chem. 2000;65:7195. doi: 10.1021/jo0006831. [DOI] [PubMed] [Google Scholar]
- 3.Anderson MG, Jarman TB, Rickards RW. J Antibiot (Tokyo) 1995;48:391. doi: 10.7164/antibiotics.48.391. [DOI] [PubMed] [Google Scholar]
- 4.Garlaschelli L, Magistrali E, Vidari G, Zuffardi O. Tetrahedron Lett. 1995;36:5633. [Google Scholar]
- 5.(a) Sassa T, Yoshikoshi H. Agr Biol Chem (Tokyo) 1983;47:187. [Google Scholar]; (b) Sassa T, Ishizaki A, Nukina M, Ikeda M, Sugiyama T. Biosci Biotech Bioch. 1998;62:2260. doi: 10.1271/bbb.62.2260. [DOI] [PubMed] [Google Scholar]; (c) Fujimoto H, Nakamura E, Kim YP, Okuyama E, Ishibashi M, Sassa T. J Nat Prod. 2001;64:1234. doi: 10.1021/np010152n. [DOI] [PubMed] [Google Scholar]; (d) Zhang L, Shen Y, Wang F, Leng Y, Liu JK. Phytochemistry. 2010;71:100. doi: 10.1016/j.phytochem.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 6.Mohamed IE, Gross H, Pontius A, Kehraus S, Krick A, Kelter G, Maier A, Fiebig HH, Konig GM. Org Lett. 2009;11:5014. doi: 10.1021/ol901996g. [DOI] [PubMed] [Google Scholar]
- 7.(a) Stahl P, Kissau L, Mazitschek R, Huwe A, Furet P, Giannis A, Waldmann H. J Am Chem Soc. 2001;123:11586. doi: 10.1021/ja011413i. [DOI] [PubMed] [Google Scholar]; (b) Hua DH, Huang X, Chen Y, Battina SK, Tamura M, Noh SK, Koo SI, Namatame I, Tomoda H, Perchellet EM, Perchellet JP. J Org Chem. 2004;69:6065. doi: 10.1021/jo0491399. [DOI] [PubMed] [Google Scholar]; (c) Sakurai J, Oguchi T, Watanabe K, Abe H, Kanno S, Ishikawa M, Katoh T. Chemistry. 2008;14:829. doi: 10.1002/chem.200701386. [DOI] [PubMed] [Google Scholar]; (d) Gordaliza M. Marine drugs. 2012;10:358. doi: 10.3390/md10020358. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Fukui Y, Narita K, Katoh T. Chemistry. 2014;20:2436. doi: 10.1002/chem.201304809. [DOI] [PubMed] [Google Scholar]; (f) Markwell-Heys AW, Kuan KK, George JH. Org Lett. 2015;17:4228. doi: 10.1021/acs.orglett.5b01973. [DOI] [PubMed] [Google Scholar]
- 8.Holm DK, Petersen LM, Klitgaard A, Knudsen PB, Jarczynska ZD, Nielsen KF, Gotfredsen CH, Larsen TO, Mortensen UH. Cell Chem Biol. 2014;21:519. doi: 10.1016/j.chembiol.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 9.Gao Y, Honzatko RB, Peters RJ. Nat Prod Rep. 2012;29:1153. doi: 10.1039/c2np20059g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Hammer SC, Marjanovic A, Dominicus JM, Nestl BM, Hauer B. Nat Chem Biol. 2015;11:121. doi: 10.1038/nchembio.1719. [DOI] [PubMed] [Google Scholar]; (b) Wendt KU, Poralla K, Schulz GE. Science. 1997;277:1811. doi: 10.1126/science.277.5333.1811. [DOI] [PubMed] [Google Scholar]
- 11.Thoma R, Schulz-Gasch T, D’Arcy B, Benz J, Aebi J, Dehmlow H, Hennig M, Stihle M, Ruf A. Nature. 2004;432:118. doi: 10.1038/nature02993. [DOI] [PubMed] [Google Scholar]
- 12.(a) Baunach M, Franke J, Hertweck C. Angew Chem Int Ed Engl. 2015;54:2604. doi: 10.1002/anie.201407883. [DOI] [PubMed] [Google Scholar]; (b) Matsuda Y, Awakawa T, Mori T, Abe I. Curr Opin Chem Biol. 2016;31:1. doi: 10.1016/j.cbpa.2015.11.001. [DOI] [PubMed] [Google Scholar]; (c) Matsuda Y, Abe I. Nat Prod Rep. 2016;33:26. doi: 10.1039/c5np00090d. [DOI] [PubMed] [Google Scholar]; (d) Tang MC, Zou Y, Watanabe K, Walsh CT, Tang Y. Chem Rev. 2017;117:5226. doi: 10.1021/acs.chemrev.6b00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Kawasaki T, Kuzuyama T, Kuwamori Y, Matsuura N, Itoh N, Furihata K, Seto H, Dairi T. J Antibiot (Tokyo) 2004;57:739. doi: 10.7164/antibiotics.57.739. [DOI] [PubMed] [Google Scholar]; (b) Koksal M, Hu H, Coates RM, Peters RJ, Christianson DW. Nat Chem Biol. 2011;7:431. doi: 10.1038/nchembio.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moosmann P, Ueoka R, Grauso L, Mangoni A, Morinaka BI, Gugger M, Piel J. Angew Chem Int Ed Engl. 2017;56:4987. doi: 10.1002/anie.201611617. [DOI] [PubMed] [Google Scholar]
- 15.Matsuda Y, Wakimoto T, Mori T, Awakawa T, Abe I. J Am Chem Soc. 2014;136:15326. doi: 10.1021/ja508127q. [DOI] [PubMed] [Google Scholar]
- 16.Itoh T, Tokunaga K, Matsuda Y, Fujii I, Abe I, Ebizuka Y, Kushiro T. Nat Chem. 2010;2:858. doi: 10.1038/nchem.764. [DOI] [PubMed] [Google Scholar]
- 17.Matsuda Y, Awakawa T, Itoh T, Wakimoto T, Fujii I, Ebizuka Y, Abe I. Chembiochem. 2012;13:1738. doi: 10.1002/cbic.201200369. [DOI] [PubMed] [Google Scholar]
- 18.(a) Tagami K, Minami A, Fujii R, Liu C, Tanaka M, Gomi K, Dairi T, Oikawa H. Chembiochem. 2014;15:2076. doi: 10.1002/cbic.201402195. [DOI] [PubMed] [Google Scholar]; (b) Zhang S, Monahan BJ, Tkacz JS, Scott B. Appl Environ Microbiol. 2004;70:6875. doi: 10.1128/AEM.70.11.6875-6883.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tagami K, Liu C, Minami A, Noike M, Isaka T, Fueki S, Shichijo Y, Toshima H, Gomi K, Dairi T, Oikawa H. J Am Chem Soc. 2013;135:1260. doi: 10.1021/ja3116636. [DOI] [PubMed] [Google Scholar]
- 20.Tang MC, Lin HC, Li DH, Zou Y, Li J, Xu W, Cacho RA, Hillenmeyer ME, Garg NK, Tang Y. J Am Chem Soc. 2015;137:13724. doi: 10.1021/jacs.5b06108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.(a) Tsunematsu Y, Ishikawa N, Wakana D, Goda Y, Noguchi H, Moriya H, Hotta K, Watanabe K. Nat Chem Biol. 2013;9:818. doi: 10.1038/nchembio.1366. [DOI] [PubMed] [Google Scholar]; (b) Sato M, Dander JE, Sato C, Hung YS, Gao SS, Tang MC, Hang L, Winter JM, Garg NK, Watanabe K, Tang Y. J Am Chem Soc. 2017;139:5317. doi: 10.1021/jacs.7b02432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin HC, Chooi YH, Dhingra S, Xu W, Calvo AM, Tang Y. J Am Chem Soc. 2013;135:4616. doi: 10.1021/ja312503y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.