Abstract
Intestinal colonization by bacteria of oral origin has been correlated with several negative health outcomes, including inflammatory bowel disease. However, a causal role of oral bacteria ectopically colonizing the intestine remains unclear. Using gnotobiotic techniques, we show that strains of Klebsiella spp. isolated from the salivary microbiota are strong inducers of T helper 1 (TH1) cells when they colonize in the gut. These Klebsiella strains are resistant to multiple antibiotics, tend to colonize when the intestinal microbiota is dysbiotic, and elicit a severe gut inflammation in the context of a genetically susceptible host. Our findings suggest that the oral cavity may serve as a reservoir for potential intestinal pathobionts that can exacerbate intestinal disease.
The average person generates and ingests ∼1.5 liters of saliva per day, containing an enormous number of oral resident bacteria (1, 2). Ingested oral bacteria poorly colonize the healthy intestine (3); however, increased levels of microbes of oral origin have been reported in the gut microbiota of patients with several diseases, including inflammatory bowel disease (IBD) (4), HIV infection (5, 6), liver cirrhosis (7, 8), and colon cancer (9). For instance, oral bacteria, such as Veillonellaceae and Fusobacteriaceae, in the intestinal mucosal microbiota strongly correlates with disease status in Crohn's disease (CD) (4). Mining of our in-house datasets of 16S ribosomal RNA (rRNA) gene sequences revealed that several bacterial taxa—including species belonging to Rothia, Streptococcus, Neisseria, Prevotella, and Gemella (Table S1A), all of which are aerotolerant and typically members of the oral microbiota—were significantly more abundant in the fecal microbiota of patients with ulcerative colitis (UC), primary sclerosing cholangitis (PSC), gastroesophageal reflux disease (GERD) being treated by long-term proton pump inhibitor therapy, and alcoholism, compared with that of healthy controls (Fig. 1A and table S1B). Thus, we hypothesized that a subset of oral microbiota may ectopically colonize and persist in the intestine under certain circumstances to aberrantly activate the intestinal immune system resulting in chronic inflammatory diseases.
Fig. 1. Isolation of a TH1-cell–inducing, multiple-antibiotic–resistant, and proinflammatory Klebsiella pneumoniae strain from the microbiota of human saliva.
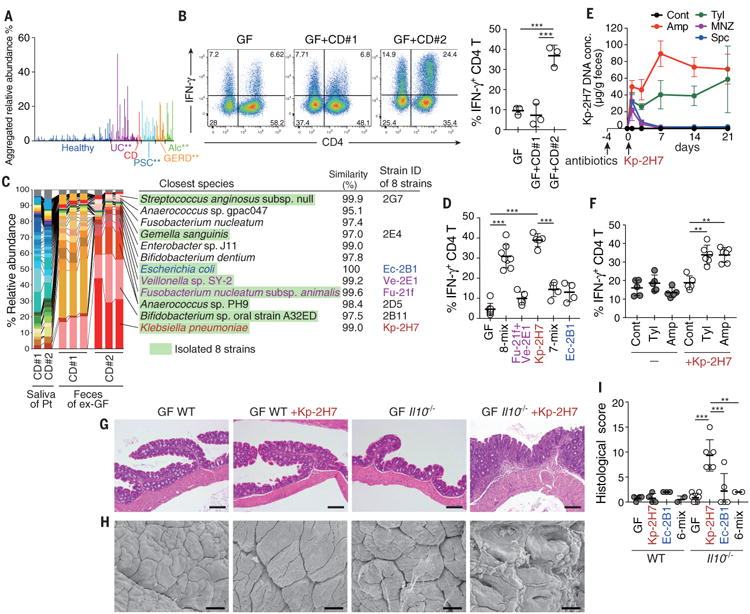
(A) Aggregated relative abundance of operational taxonomic units (OTUs) typical of human oral microbiota in the fecal microbiota of healthy individuals (n=150), patients with ulcerative colitis (UC; n=51), Crohn's disease (CD; n=7), primary sclerosing cholangitis (PSC; n=27), gastroesophageal reflux disease (GERD; n=18), and alcoholism (Alc; n=16). **P < 0.001; Wilcoxon rank-sum test (fig. S1B). (B) Representative FACS plots (left) and frequencies of IFN-γ+ cells (right) among colonic LP CD4+TCRβ+ T cells from ex-germ free (exGF) B6 mice inoculated with saliva samples from patients with CD. Each point (right) represents an individual mouse. (C) Pyrosequencing of 16S rRNA genes from the saliva microbiota of patients (Pt) and from the resulting fecal microbiota of exGF mice (n = 3 per group). Quality filter–passed sequences were classified into OTUs on the basis of sequence similarity (96% identity), and the relative abundance of OTUs and closest known species for each OTU are shown. OTUs corresponding to the eight isolated strains are marked in green. (D) The percentage of TH1 cells in the colonic LP of exGF B6 mice colonized with 8-mix, Fu-21f+Ve-2E1, Kp-2H7, 7-mix, or Ec-2B1. (E and F) SPF B6 mice were untreated (Cont) or continuously treated with antibiotics in drinking water, starting 4 days before oral administration of 2 × 108 colony-forming units (CFU) of Kp-2H7. The relative abundance of Kp-2H7 DNA over time in fecal samples was determined by qPCR (E). The percentage of TH1 cells among colonic LP CD4+ cells was analyzed by flow cytometry on day 21 after Kp-2H7 administration (F). Amp, ampicillin; Tyl, tylosin; Spc, spectinomycin; MNZ, metronidazole. (G to I) Representative hematoxylin and eosin staining (G), representative scanning electron micrograph (SEM) (H), and histological colitis scores (I) of the proximal colon of Kp-2H7-, Ec-2B1-, or 6-mix-colonized WT or Il10-/- mice. Scale bars, 200 μm (G) and 30 μm (H). Symbols represent individual mice. Error bars indicate means ± SD. **P < 0.01; ***P < 0.001; one-way analysis of variance (ANOVA) with post hoc Turkey's test. Data represent at least two independent experiments with similar results.
To search the human oral microbiota for bacterial strains showing strong immune-stimulatory activities upon intestinal colonization, we transplanted saliva samples from two patients with CD into C57BL/6 (B6) germ-free (GF) mice by gavage. Each group of mice was housed in separate gnotobiotic isolators for 6 weeks, at which time small intestinal and colonic lamina propria (LP) immune cells were examined. In mice receiving a saliva sample from CD patient #1 (GF+CD#1 mice), there were no significant changes in the intestinal T cells (Fig. 1B). In contrast, in the group that received a saliva sample from CD patient #2 (GF+CD#2 mice), we noticed a marked accumulation of interferon-γ+ (IFN-γ+) CD4+ T cells [T helper 1 (TH1) cells] in the intestinal LP (Fig. 1B). Using 16S rRNA gene sequencing, we compared the community composition of the saliva microbiota before administration into GF mice and the fecal microbiota of the colonized animals (Fig. 1C). Although the saliva samples of both patients contained similar microbial communities, the fecal microbiota compositions differed markedly between GF+CD#2 mice and GF+CD#1 mice (Fig. 1C). Importantly, most of the bacterial species observed in the fecal microbiota of the mice had been minor components of the salivary microbiota (Fig. 1C). These results indicate that bacterial species that constitute a small fraction of the oral microbiota can expand and colonize the gut, and a subset of these oral species can induce the accumulation of intestinal TH1 cells.
To isolate TH1-cell-inducing bacteria, we anaerobically cultured cecal contents from GF+CD#2 mice using several culture media and picked 224 colonies with different colony appearances. Sequencing of the 16S rRNA genes revealed that these colonies contained eight strains from diverse genera, including Gemella, Bifidobacterium, Streptococcus, Escherichia, Fusobacterium, Veillonella, Anaerococcus, and Klebsiella, and broadly represented the major members of the gut microbiota colonizing GF+CD#2 mice (Fig. 1C). To examine whether these isolated strains had TH1 cell-inducing capability, we cultured all eight of them and introduced them as a mixture (8-mix) into GF mice. We observed efficient induction of TH1 cells in the colonic LP of these mice, with a magnitude comparable to that observed in GF+CD#2 mice (compare Fig. 1B and D). Because Fusobacterium and Veillonella have been implicated in IBD pathogenesis (4), we colonized mice with these two strains (strain IDs Fu-21f and Ve-2E1, respectively); however, this resulted in only marginal elevation of TH1 cell frequency (Fig. 1D). We tested Klebsiella pneumoniae 2H7 (Kp-2H7) because it was the most prominent component of the GF+CD#2 microbiota (Fig. 1C). Oral administration of Kp-2H7 alone significantly induced TH1 cells, whereas a mixture of the remaining seven strains (7-mix) failed to do so (Fig. 1D), indicating that the Kp-2H7 strain was the major contributor to the accumulation of TH1 cells observed in GF+CD#2 mice. The effect of Kp-2H7 was relatively specific for TH1 cells (fig. S1A), which were negative for interleukin-17 (IL-17), RORγt, and Foxp3 but positive for T-bet and CD44 (fig. S1B). Kp-2H7 mainly colonized the colon and cecum (fig. S1C), reflecting greater TH1 cell induction in the colon than in the small intestine (fig. S1D). There was no increase in the percentage of TH1 cells in the oral tissues (palate and tongue) of B6 GF+Kp-2H7 mice (fig. S1E). The increase in TH1 cells was observed in IQI/Jic mice and B6 mice, but not in BALB/c mice (fig. S1F), implying interplay between host genotype and Kp-2H7 for colonic TH1 cell induction.
Klebsiella spp. often acquire resistance to multiple antibiotics and can be a cause of healthcare–associated infection (10-12). Our isolate Kp-2H7 was resistant to multiple antibiotics, including ampicillin (Amp), tylosin (Tyl), spectinomycin (Spc), and metronidazole (MNZ) (fig. S2). Specific-pathogen free (SPF) mice were untreated or continuously treated with Amp, Tyl, Spc, or MNZ in drinking water starting 4 days before oral gavage with Kp-2H7. Antibiotic-naïve mice were resistant to colonization by Kp-2H7, but Amp or Tyl treatment allowed Kp-2H7 to persist in the intestine (Fig. 1E). Persistent colonization with Kp-2H7 was accompanied by a significant increase in the frequency of colonic TH1 cells (Fig. 1F). In contrast to Amp and Tyl treatments, MNZ and Spc treatments did not allow Kp-2H7 colonization, although the Kp-2H7 strain is resistant to both of these antibiotics (Fig. 1E). These results suggest that antibiotic exposure potentiates orally derived Kp-2H7 colonization by disrupting the colonization resistance provided by specific members of the gut microbiota that are Amp- and Tyl-sensitive but MNZ- and Spc-resistant.
Despite induction of TH1 cells, Kp-2H7 colonization did not induce any inflammatory changes in the intestine of wild-type (WT) hosts—either GF mice or Amp-treated SPF mice (Fig. 1G to I, and fig. S3, A and B). Because microbial and host genetic factors both contribute to the pathogenesis of IBD (13), we tested the influence of Kp-2H7 colonization in colitis-prone Il10-/- mice. GF WT and GF Il10-/- mice were orally administered Kp-2H7, Escherichia coli 2B1 (Ec-2B1), or a mixture of the six other strains (6-mix). K. pneumoniae and E. coli are both in the family Enterobacteriaceae, which has been implicated in IBD pathogenesis (4, 14). One week after colonization, more potent induction of colonic TH1 cells was observed in Il10-/-+Kp-2H7 mice than in the other groups of mice (fig. S3C). In addition, there was a greater induction of colonic LP IL-17+IFN-γ+ CD4+ T cells (fig. S3C) and epithelial tumor necrosis factor–α mRNA expression (fig. S3D) in the Il10-/-+Kp-2H7 mice. Histological analysis revealed that Kp-2H7 induced more severe inflammation than Ec-2B1 or the 6-mix in the proximal colon of Il10-/- mice (Fig. 1, G to I, and fig. S3E). Colitis exacerbation was similarly observed in Amp-treated SPF Il10-/- mice colonized with Kp-2H7 (fig. S3, A and B). These results suggest that Kp-2H7 acts as a gut pathobiont in the context of a genetically susceptible host. When Kp-2H7 was intratracheally injected into the lung, a TH17 response predominated over a TH1 response in lung T cells, consistent with previous reports (15, 16), and this response was accompanied by severe lung pathology, even in genetically normal mice (fig. S4). Therfore, the effect of Kp-2H7 on TH1 induction thus appears to be a gut-specific response.
To investigate the mechanism of Kp-2H7–mediated TH1 cell induction, we orally administered heat-killed Kp-2H7 to GF WT B6 mice in drinking water for 3 weeks. Heat-killed bacteria had no effect on the frequency of TH1 cells (Fig. 2A). Fluorescence in situ hybridization (FISH) of the colon of mice colonized with Kp-2H7 revealed the presence of Kp-2H7 on the mucus layer, with no evidence of adhesion or invasion of bacteria on or into the epithelial layer (Fig. 2B). Phylogenetically distinct human, mouse, and environmental K. pneumoniae strains (fig. S5A and Table S2) showed considerable variability in the ability to elicit colonic TH1 cell induction when monocolonized in B6 WT mice (Fig. 2C). In particular, KCTC2242 and BAA2552 did not affect, or only weakly affected, TH1 cell frequency. The intensity of induction was independent of bacterial load and was not accompanied by inflammation (fig. S5B). There was no correlation between TH1 cell induction and multilocus sequence typing, K-typing, orphylogeny (fig. S5A and Table S2). Comparative analysis of the whole genomes revealed that 61 orthologous groups of genes were positively correlated with the TH1 induction ability, including genes predicted to encode hemolysin-coregulated protein (Hcp) and enzymes involved in fructose-, galactitol-, mannose-, and long-chain fatty acid–related uptake and metabolic pathways (Fig. 2D and Table S3). These genes have been reported to be enriched in the fecal microbiome of patients with inflammatory diseases and have been suggested to have immunomodulatory effects, and they thereby may contribute to the induction of TH1 cells (8, 14, 17-19).
Fig. 2. Strain-specific induction of TH1 cells by K. pneumoniae.
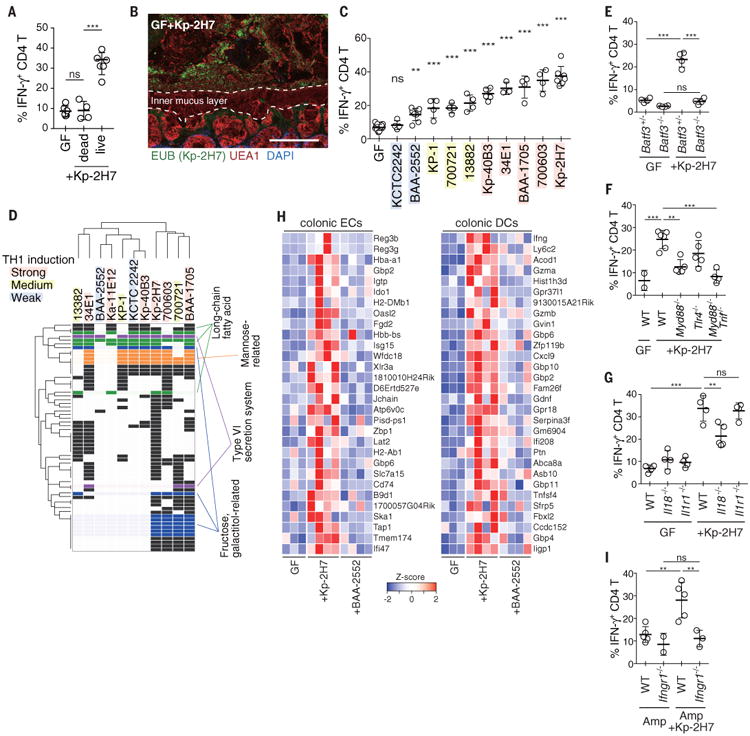
(A) Frequency of TH1 cells in the colon of GF mice orally administered heat-killed Kp-2H7 in drinking water for 3 weeks (Kp-2H7 dead). (B) Staining with DAPI (4′,6-diamidino-2-phenylindole; blue), EUB338 FISH probe (green), and Ulex europaeus agglutinin 1 (UEA1; red) of the proximal colon of mice monocolonized with Kp-2H7. Scale bar, 100 μm. (C) GF mice were monocolonized with the indicated K. pneumoniae strain, and colonic TH1 cells were analyzed after 3 weeks. Frequencies of IFN-γ+ colonic LP CD4+TCRβ+ T cells are depicted. Colored shading is as in (D). (D) Comparative analysis of whole genomes of the tested Klebsiella strains revealed 61 orthologous groups correlating with TH1-inducing activity. Detailed information about gene ID for each row, is given table S3 and fig. S13. (E to G) The percentage of TH1 cells in the colon of WT, Batf3-/-, Myd88-/-, Tlr4-/-, Myd88-/-Trif-/-, Il18-/-, and Il1r1-/- mice monocolonized with Kp-2H7. (H) Differential gene expression in the colonic ECs and DCs from WT mice monocolonized with Kp-2H7 or BAA-2552 for 1 week. Heatmap colors represent the z-score normalized fragments per kilobase of exon per million fragments mapped (FPKM) values for each gene. (I) SPF WT and Ifngr1-/- mice were treated with Amp and gavaged with Kp-2H7 as in Fig. 1E. The percentage of TH1 cells among colonic LP CD4+ cells is depicted. Symbols represent individual mice. Error bars indicate means ± SD. **P < 0.01; ***P < 0.001; ns, not significant (P > 0.05); one-way ANOVA with post hoc Turkey's test. Data represent at least two independent experiments with similar results.
Mice deficient in basic leucine zipper transcription factor ATF-like 3 (Batf3-/-), which lack the intestinal CD11b-CD103+ dendritic cell (DC) subset, failed to mount a colonic TH1 cell response (Fig. 2E). Assessment of the antigen specificity of colonic TH1 cells induced by Kp-2H7 revealed that a substantial fraction recognized Klebsiella antigens (fig. S6), including OmpX (outer membrane protein X) (20). Colonic TH1 induction by Kp-2H7 monocolonization was significantly suppressed in Myd88-/- and Myd88-/-Trif-/- mice and partially attenuated in Tlr4-/- mice and Il18-/- mice, but occurred normally in Il1r1-/- mice (Fig. 2, F and G). Increased production of IL-18 by colonic epithelial cells (ECs) was detected after monocolonization with Kp-2H7 (fig. S6B). Together these results suggest contributions from Toll-like receptors (TLRs) and IL-18 signaling to the DC-mediated Klebsiella antigen-specific TH1 cell induction.
RNA-sequencing (RNA-seq) data for colonic ECs and CD11c+ DCs isolated from WT mice monocolonized with Kp-2H7 or BAA2552 for 1 week, and from WT, Myd88-/-, Myd88-/-Trif-/-, and Tlr4-/- mice monocolonized with Kp-2H7 for 3 weeks, were compared with data for GF mice (Fig. 2H and fig. S7 and S8). IFN-inducible (IFI) genes—such as those encoding guanylate-binding proteins (GBPs), chemokine (C-X-C motif) ligand 9 (Cxcl9), major histocompatibility complex–related molecules (MHCs; e.g., H2-DMb1, H2-Ab1 and Tap1), and dual oxidase 2 (Duox2) were significantly upregulated in colonic ECs and DCs from GF WT+Kp-2H7 mice. Expression differences were confirmed by quantitative polymerase chain reaction (qPCR) analysis (fig. S9A). The upregulation of IFI genes in colonic ECs of GF+Kp-2H7 mice began within 3 days of colonization (fig. S9B), when TH1 induction was limited (fig. S9C). Bacteria-free cecal suspensions from GF+Kp-2H7 mice upregulated IFI genes in a colonic EC line in vitro (fig. S10), suggesting direct effects of bacterial products on the induction of IFI genes. In the later phase of Kp-2H7 colonization, IFI genes were further upregulated, coinciding with the increase of TH1 cells (fig. S9, A to C). GBPs are known to function as microbial receptors (21). MHCs and Cxcl9 mediate development and recruitment of TH1 cells. Duox2 mediates production of hydrogen peroxide, which may facilitate further colonization of reactive oxygen–tolerant Klebsiella spp (22, 23). IFN-γ receptor 1-deficient (IFNγR1-/-) mice mounted a defective TH1 cell response upon colonization with Kp-2H7 (Fig. 2I). These results suggest that a feed forward loop involving IFN-γ and IFIs was created among ECs, DCs and T cells for sustained accumulation of TH1 cells.
To confirm the link between oral-derived bacteria and TH1 cell induction, we obtained additional saliva samples from two healthy donors (He#1 and He#2) and two patients with active UC (UC#1 and UC#2) and orally administered these samples to GF WT B6 mice. TH1 cells accumulated in the colonic LP of mice inoculated with a saliva sample from UC patient #2 (GF+UC#2 mice), comparably to the accumulation observed in GF+CD#2 mice (Fig. 3A). We cultured cecal contents from GF+UC#2 mice and isolated 13 strains, which resembled the microbiota composition of GF+UC#2 mice (Fig. 3B). Oral administration of the 13 strains (13-mix) into GF mice fully replicated the phenotype observed in GF+UC#2 mice in terms of colonic TH1-cell induction (Fig. 3C). Among the 13 strains, Enterococcus faecium 11A1 (Ef-11A1) and Klebsiella aeromobilis 11E12 (Ka-11E12) drew our attention, because both species have been implicated in IBD pathogenesis and reported to be important multidrug-resistant pathobionts (24-27). GF mice were gavaged either with Ef-11A1, Ka-11E12, or a mixture of the 11 other strains (11-mix). Ka-11E12 induced TH1 cells in the colon comparably to the 13-mix, whereas Ef-11A1 and the 11-mix failed to do so (Fig. 3C). Therefore, Ka-11E12 was most likely the major driver for the induction of TH1 cells observed in GF+UC#2 mice, even as a minor member of the gut microbiota (Fig. 3B). Ka-11E12 was also resistant to multiple antibiotics (fig. S11, A and B) and persisted in the intestine of vancomycin- or Tyl-treated SPF mice (Fig. 3D). Ka-11E12 colonization resulted in severe inflammation in Il10-/- mice (fig. S11, C and D, and Fig. 3E), suggesting that the orally derived K. aeromobilis strain may act similarly to Kp-2H7. Likewise, Ka-11E12 did not attach to the EC surface as shown by FISH (Fig. 3F). However, genome sequence analysis and electron microscopy (EM) revealed that, in contrast to Kp-2H7, Ka-11E12 had a flagellar assembly system (Table S4 and fig. S12A) and was highly motile and stimulatory for TLR5 (fig. S12, B and C). Therefore, distinct mechanisms may drive TH1 cell induction by different Klebsiella strains.
Fig. 3.
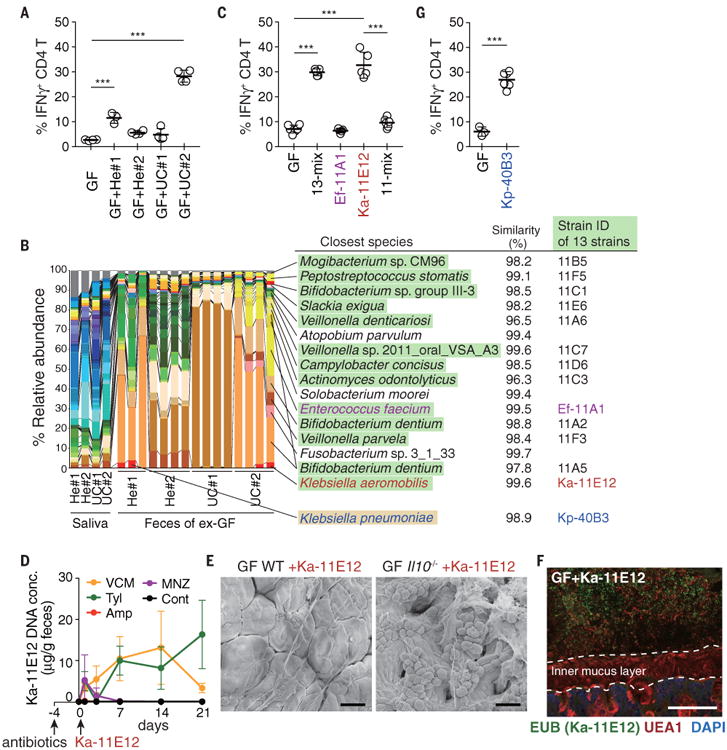
Klebsiellaaeromobilis is another TH1cell–inducing oral cavity-derived bacterial species. (A) Frequencies of IFNγ+ within colonic LP CD4+TCRβ+ T cells from exGF mice inoculated with saliva samples from healthy donors and UC patients. (B) Pyrosequencing of 16S rRNA genes of the saliva microbiota of healthy controls and patients with UC and of the resulting fecal microbiota of exGF mice (n = 3 to 4 mice per group). The relative abundance of OTUs and closest known species for each OTU are shown. OTUs corresponding to the isolated 13 strains and Kp-40B3 are marked in green and yellow, respectively. (C) The percentage of TH1 cells in the colonic LP of B6 mice colonized with 13-mix, Ef-11A1, Ka-11E12, or 11-mix. (D) SPF B6 mice were untreated or continuously treated with antibiotics in the drinking water starting 4 days before oral administration of 2 × 108 CFU of Ka-11E12. Ka-11E12 abundance in fecal samples was determined by qPCR. (E) Representative SEM images of the colon of Ka-11E12–monocolonized WT or Il10-/- mice. (F) Staining with DAPI (blue), EUB338 FISH probe (green), and UEA1 (red) of the colon of mice monocolonized with Ka-11E12. Scale bars, 30 μm (E) and 100 μm (F). (G) The percentage of TH1 cells in the colonic LP of B6 mice colonized with Kp-40B3. Each point in (A), (C), and (G) represents an individual mouse (thick bars, means); points in (D) represent means of a group of five or six mice. Error bars, SD. ***P < 0.001; one-way ANOVA with post hoc Tukey's test [(A) and (C)] and two-tailed unpaired Student's t test (G).
Somewhat unexpectedlly, the microbiota sample from one healthy donor (He#1) also induced a substantial increase in TH1 cells (Fig. 3A). In this case, the fecal microbiome contained a K. pneumoniae sequence. We isolated a K. pneumoniae strain, Kp-40B3 (Fig. 3B). This strain, Kp-40B3, induced marked TH1 cell accumulation in the colon of monocolonized mice (Fig. 3G). Whole genome sequence analysis revealed that Kp-40B3 possessed several genes that correlated with Kp-2H7-mediated TH1 cell induction (Fig. 2D). These results suggest that Klebsiella spp. with TH1 cell induction capability may exist in the oral cavity of not only IBD patients but also healthy humans.
We mined the 16S rRNA gene sequencing datasets used in the analysis in Fig. 1A and found that the relative abundance of members of Klebsiella was significantly higher in patients with CD (P=0.0157), PSC (P=0.0309), and alcoholism (P<0.0001) compared with that in healthy controls (Fig. 4A). We extended our analysis to the metagenome databases of intestinal microbiota of IBD patients and non-IBD controls from the Prospective Registry in IBD Study at Massachusetts General Hospital (PRISM) (14), and the CD cohort from the University of Pennsylvania (UPenn cohort) (28). The aggregated relative abundance of Klebsiella species was significantly higher in patients with IBD (P<0.01) (Fig. 4B). The genes that were correlated with Kp-2H7–mediated TH1 induction (Fig. 2D) were enriched in the fecal microbiome of IBD patients who carried Klebsiella species, but not in non-IBD individuals (Fig. 4, C and D; P=0.00988, Mann-Whitney U test).
Fig. 4. Association of Klebsiella species and TH1-related genes with IBD.
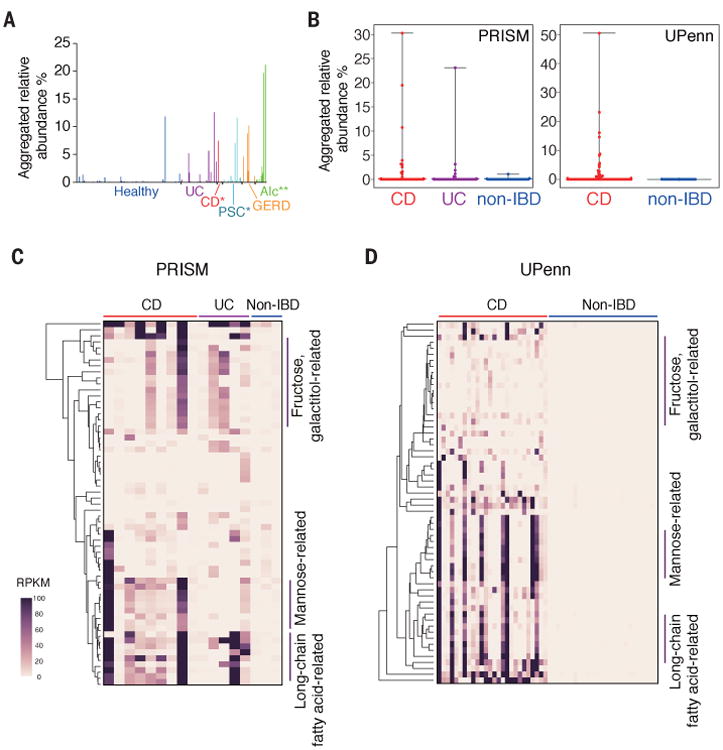
(A) Aggregated relative abundance of OTUs assigned to the genus Klebsiella in the samples from healthy donors and patients with the indicated diseases. *P < 0.05; **P < 0.01; Wilcoxon rank-sum test. (B to D) The reads of individual samples from PRISM and UPenn cohorts were mapped to Klebsiella species (B) and to the gene sequences correlated with TH1 induction [(C) and (D)]. Detailed information about gene ID for each row is given in table S3 and fig. S13. Bars in (B) represent mean values, with error bars indicating the range. Heatmaps in [(C) and (D)] show values of reads per kilobase per million reads (RPKM) for the TH1-related genes.
Ectopic colonization of the colon by orally derived Klebsiella spp. is associated with aberrant activation of the immune system. The lower gastrointestinal tract is the expected niche for Klebsiella spp., where they constitute a minor population that can proliferate after antibiotic treatment and other conditions (24). Our data suggest that the oral cavity may serve as another reservoir for Klebsiella pathobionts. Indeed, the oral microbiota contains the highest relative abundance of Enterobacteriaceae compared with other mucosal sites (29). The inflammatory state in IBD may render the intestine more permissive to aerotolerant oral-derived bacteria than the steady-state intestine, and ongoing colonization by oral bacteria may help perpetuate gut microbiota dysbiosis and chronic inflammation. Overrepresentation of Klebsiella spp. in the gut microbiota, coupled with elevated levels of serum antibodies against Klebsiella antigens, has been reported in patients with IBD (30, 31). In mouse models of IBD, such as T-bet-/-Rag2-/- mice, K. pneumoniae is known to proliferate and play an important role in triggering disease (32). Furthermore, persistence of Klebsiella spp. is also observed in several other diseases (Fig. 4A) (33). Thus, our findings indicate that targeting oral-derived bacteria, particularly Klebsiella, could provide a therapeutic strategy to correct IBD and many other disease conditions.
Our results highlight the disease potential of antibiotic resistant, pro-inflammatory Klebsiella in chronic human disease. K. pneumoniae and K. aeromobilis commonly cause hospital infections (10-12, 24, 25). Our isolates were resistant to several antibiotics, and treatment with antibiotics allowed their colonization in the intestine of SPF mice. The colonization efficiency of Kp-2H7 and Ka-11E12 varied, depending on the spectrum of antibiotics used as treatment. Therefore, identifying members of the normal gut microbiota that can provide colonization resistance against orally derived bacteria could foster future avenues for the development of effective treatments for multidrug-resistant bacteria and chronic inflammation.
Supplementary Material
Table S1A. OTUs typically present in the human salivary microbiota. OTUs that were present at >1% average relative abundance in the salivary microbiome of 89 healthy individuals were defined as typical members of the oral microbiota (marked in red).
Table S1B. Statistical significance of abundance difference of typical members of the oral microbiota in the fecal microbiota between healthy and diseased individuals. Aggregated relative abundance of OTUs typical of human oral microbiota was calculated for the fecal microbiota of healthy individuals, patients with UC, CD, PSC, GERD, and alcoholism. Sample variances and p-value for Wilcoxon Signed-Rank test are shown in upper table. Steel's and Dunn's multiple comparison test with healthy donors are shown in middle and lower tables, respectively. See also Fig. 1A.
Table S2. Phylogenetic relationship among K. pneumoniae strains used in this study. Origin, MLST profiles, sequence-based capsular (K) typing (genotyping of wzi and wzc genes) of K. pneumoniae strains used in this study.
Table S3. Genes positively correlated with the TH1 induction ability. Comparative analysis of the whole genomes of the tested Klebsiella strains revealed that 60 orthologous groups of genes were positively correlated with the TH1 induction ability. The presence (>80% coverage and >80% sequence identity) of 61 orthologs in Klebsiella strains is marked in red. The sequences were mapped to the KEGG Orthology (KO), and also Blastx searched against NCBI non-redundant (NR) protein database to retrieve putative functional classifications. Long-chain fatty acid-, type VI secretion system, galactitol-, fructose- and mannose-related genes are marked in green, purple, blue, yellow and orange, respectively.
Table S4. Putative genes in the genome of Ka-11E12 strain that are associated with flagellar assembly system. Flagellar assembly-associated genes in each genome of Kp-2H7, Kp-40B3 and Ka-11E12 were searched using BLASTP against Kyoto Encyclopedia or Genes and Genomes (KEGG) database. Filled box (red) indicates the presence of gene (the e-value cut off of 1.0e-10, the identity >30% and the length coverage >60%).
Acknowledgments
K.A. and K.H. acknowledges funding from the Uehara Memorial Foundation, the Takeda Science Foundation, the Mitsubishi Foundation, Core Research for Evolutionary Medical Science and Technology, and Leading Advanced Projects for Medical Innovation, a program of the Japan Agency for Medical Research and Development. C.L. and R.J.X. acknowledge funding from the U.S. NIH (grants DK043351 and DK92405), the Helmsley Charitable Trust, and the Crohn's & Colitis Foundation. We thank P. Wilmes, J. Baginska, M. Wakazaki, O. Ohara, and Y. Arakawa for their technical support, and P. Burrows for helpful comments. M.H., H.M., and Y.K. conceived the research and performed initial experiments; K.H. planned experiments, analyzed data, and wrote the manuscript together with K.A., C.L., and R.J.X.; K.A., T.K., S.N., Y.K., I.M., K.Y., E.W., T.T., and C.A.T performed gnotobiotic studies, immunological analyses, and bacterial cultures.; C.L, D.G., R.C. J., J.S., R.J.X, E.E., W.S., H.S.S., and M.H. performed bacterial sequence and microbiome analyses; M.S., K.T., and S.N. performed EM analyses; H.Y. provided clinical samples; and K.C., J.K.K., and S.A.R. provided essential materials and contributed to data discussions. J.K.K. is a recipient of Public Health Service grant R37 HL079142. K.H. is a scientific advisory board member of Vedanta Biosciences. All data and code to understand and assess the conclusions of this research are available in the main text, the supplementary materials, and the indicated repositories. Sequences of the genome of Klebsiella spp. and the 16S rRNA sequence data set are deposited in the DNA Data Bank of Japan under accession numbers PRJDB5883-5886 and PRJDB5967, respectively. The raw and processed RNA-seq data are deposited in the National Center for Biotechnology Information's Gene Expression Omnibus under accession number GSE23056. Klebsiella strains are available under a material transfer agreement with Keio University.
References
- 1.Nasidze I, Li J, Quinque D, Tang K, Stoneking M. Global diversity in the human salivary microbiome. Genome research. 2009;19:636–643. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Humphrey SP, Williamson RT. A review of saliva: normal composition, flow, and function. The Journal of prosthetic dentistry. 2001;85:162–169. doi: 10.1067/mpr.2001.113778. [DOI] [PubMed] [Google Scholar]
- 3.Seedorf H, et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell. 2014;159:253–266. doi: 10.1016/j.cell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell host & microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lozupone CA, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell host & microbe. 2013;14:329–339. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vujkovic-Cvijin I, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Science translational medicine. 2013;5:193ra191. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin N, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, et al. Dysbiosis of small intestinal microbiota in liver cirrhosis and its association with etiology. Scientific reports. 2016;6:34055. doi: 10.1038/srep34055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell host & microbe. 2014;15:317–328. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snitkin ES, et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Science translational medicine. 2012;4:148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holt KE, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E3574–3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pamer EG. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science. 2016;352:535–538. doi: 10.1126/science.aad9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu H, et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016;352:1116–1120. doi: 10.1126/science.aad9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morgan XC, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome biology. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen K, et al. Th17 cells mediate clade-specific, serotype-independent mucosal immunity. Immunity. 2011;35:997–1009. doi: 10.1016/j.immuni.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong H, et al. Innate Lymphocyte/Ly6C(hi) Monocyte Crosstalk Promotes Klebsiella Pneumoniae Clearance. Cell. 2016;165:679–689. doi: 10.1016/j.cell.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Records AR. The type VI secretion system: a multipurpose delivery system with a phage-like machinery. Molecular plant-microbe interactions. 2011;24:751–757. doi: 10.1094/MPMI-11-10-0262. [DOI] [PubMed] [Google Scholar]
- 18.Fukuda S, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 19.Thorburn AN, Macia L, Mackay CR. Diet, metabolites, and “western-lifestyle” inflammatory diseases. Immunity. 2014;40:833–842. doi: 10.1016/j.immuni.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 20.Chen K, et al. Recombinant outer membrane protein: a potential candidate for Th17 based vaccine against Klebsiella pneumoniae. (VAC7P.967) The Journal of Immunology. 2014;192:141.112. [Google Scholar]
- 21.Kim BH, et al. Interferon-induced guanylate-binding proteins in inflammasome activation and host defense. Nat Immunol. 2016;17:481–489. doi: 10.1038/ni.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winter SE, Lopez CA, Baumler AJ. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 2013;14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haberman Y, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014;124:3617–3633. doi: 10.1172/JCI75436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taur Y, Pamer EG. The intestinal microbiota and susceptibility to infection in immunocompromised patients. Current opinion in infectious diseases. 2013;26:332–337. doi: 10.1097/QCO.0b013e3283630dd3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davin-Regli A, Pages JM. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Frontiers in microbiology. 2015;6:392. doi: 10.3389/fmicb.2015.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mondot S, et al. Highlighting new phylogenetic specificities of Crohn's disease microbiota. Inflammatory bowel diseases. 2011;17:185–192. doi: 10.1002/ibd.21436. [DOI] [PubMed] [Google Scholar]
- 27.Diene SM, et al. The rhizome of the multidrug-resistant Enterobacter aerogenes genome reveals how new “killer bugs” are created because of a sympatric lifestyle. Molecular biology and evolution. 2013;30:369–383. doi: 10.1093/molbev/mss236. [DOI] [PubMed] [Google Scholar]
- 28.Lewis JD, et al. Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn's Disease. Cell host & microbe. 2015;18:489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends in biotechnology. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 30.Tiwana H, et al. Characterization of the humoral immune response to Klebsiella species in inflammatory bowel disease and ankylosing spondylitis. British journal of rheumatology. 1998;37:525–531. doi: 10.1093/rheumatology/37.5.525. [DOI] [PubMed] [Google Scholar]
- 31.Rashid T, Ebringer A, Wilson C. The role of Klebsiella in Crohn's disease with a potential for the use of antimicrobial measures. International journal of rheumatology. 2013;2013:610393. doi: 10.1155/2013/610393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell host & microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebringer A, Rashid T, Tiwana H, Wilson C. A possible link between Crohn's disease and ankylosing spondylitis via Klebsiella infections. Clinical rheumatology. 2007;26:289–297. doi: 10.1007/s10067-006-0391-2. [DOI] [PubMed] [Google Scholar]
- 34.Said HS, et al. Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA research. 2014;21:15–25. doi: 10.1093/dnares/dst037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee KW, et al. Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. The ISME journal. 2014;8:894–907. doi: 10.1038/ismej.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 37.Ondov BD, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome biology. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brisse S, et al. wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. Journal of clinical microbiology. 2013;51:4073–4078. doi: 10.1128/JCM.01924-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan YJ, et al. Capsular types of Klebsiella pneumoniae revisited by wzc sequencing. PloS one. 2013;8:e80670. doi: 10.1371/journal.pone.0080670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1A. OTUs typically present in the human salivary microbiota. OTUs that were present at >1% average relative abundance in the salivary microbiome of 89 healthy individuals were defined as typical members of the oral microbiota (marked in red).
Table S1B. Statistical significance of abundance difference of typical members of the oral microbiota in the fecal microbiota between healthy and diseased individuals. Aggregated relative abundance of OTUs typical of human oral microbiota was calculated for the fecal microbiota of healthy individuals, patients with UC, CD, PSC, GERD, and alcoholism. Sample variances and p-value for Wilcoxon Signed-Rank test are shown in upper table. Steel's and Dunn's multiple comparison test with healthy donors are shown in middle and lower tables, respectively. See also Fig. 1A.
Table S2. Phylogenetic relationship among K. pneumoniae strains used in this study. Origin, MLST profiles, sequence-based capsular (K) typing (genotyping of wzi and wzc genes) of K. pneumoniae strains used in this study.
Table S3. Genes positively correlated with the TH1 induction ability. Comparative analysis of the whole genomes of the tested Klebsiella strains revealed that 60 orthologous groups of genes were positively correlated with the TH1 induction ability. The presence (>80% coverage and >80% sequence identity) of 61 orthologs in Klebsiella strains is marked in red. The sequences were mapped to the KEGG Orthology (KO), and also Blastx searched against NCBI non-redundant (NR) protein database to retrieve putative functional classifications. Long-chain fatty acid-, type VI secretion system, galactitol-, fructose- and mannose-related genes are marked in green, purple, blue, yellow and orange, respectively.
Table S4. Putative genes in the genome of Ka-11E12 strain that are associated with flagellar assembly system. Flagellar assembly-associated genes in each genome of Kp-2H7, Kp-40B3 and Ka-11E12 were searched using BLASTP against Kyoto Encyclopedia or Genes and Genomes (KEGG) database. Filled box (red) indicates the presence of gene (the e-value cut off of 1.0e-10, the identity >30% and the length coverage >60%).


