Abstract
Accurate CT synthesis, sometimes called electron density estimation, from MRI is crucial for successful MRI-based radiotherapy planning and dose computation. Existing CT synthesis methods are able to synthesize normal tissues but are unable to accurately synthesize abnormal tissues (i.e., tumor), thus providing a suboptimal solution. We propose a multi-atlas-based hybrid synthesis approach that combines multi-atlas registration and patch-based synthesis to accurately synthesize both normal and abnormal tissues. Multi-parametric atlas MR images are registered to the target MR images by multi-channel deformable registration, from which the atlas CT images are deformed and fused by locally-weighted averaging using a structural similarity measure (SSIM). Synthetic MR images are also computed from the registered atlas MRIs by using the same weights used for the CT synthesis; these are compared to the target patient MRIs allowing for the assessment of the CT synthesis fidelity. Poor synthesis regions are automatically detected based on the fidelity measure and refined by a patch-based synthesis. The proposed approach was tested on brain cancer patient data, and showed a noticeable improvement for the tumor region.
Keywords: CT synthesis, multi-atlas registration, patch-based synthesis, MRI-based radiotherapy planning, multi-channel registration
1. INTRODUCTION
Magnetic resonance imaging (MRI) has been actively used in radiotherapy planning. Its superior soft tissue contrast offers improved visualization of tumor and organs at risk (OAR), and multi-parametric MRI facilitates the differentiation between normal and pathological tissues [1]. MRI can significantly improve the segmentation accuracy compared to CT-based approaches that suffer from large intra- and inter-observer variability due to the poor soft tissue contrast and image artifacts [2]. MRI is therefore considered the imaging modality of choice for detecting and staging tumors as well as for delineating structures in various sites such as the brain, head and neck (HN), and prostate [3]. These advantages have motivated efforts to develop MRI-based radiotherapy planning. However, MRI does not contain electron density information, and is therefore not intrinsically useable for radiation dose computation [4]. As a consequence, many clinics use MRI primarily for target detection and delineation in support of CT for radiotherapy treatment planning.
In recent years, there has been increasing efforts to carry out radiotherapy treatment planning and image guidance using MRI alone. By eliminating the CT scan, unnecessary x-ray dose to the patient is avoided and the MR-CT image fusion process is no longer needed. Consequently, the overall workflow is simplified and more cost-effective with improved target localization leading to potentially improved tumor control rates with reduced normal tissue toxicity. A major obstacle to achieving this goal is the lack of robust methods to accurately map MR image intensity to electron density which is necessary for dose computation. Currently, there are only a handful of MRI-based CT image synthesis methods, including tissue classification-based electron density assignment [5], single or multi-atlas registration approaches [6], and patch-based matching and machine learning approaches [7, 8]. These approaches have been applied mostly in the brain showing potential to synthesize normal tissue regions, but none of them are able to synthesize abnormal tissue regions such as tumors that are always present in radiotherapy cases.
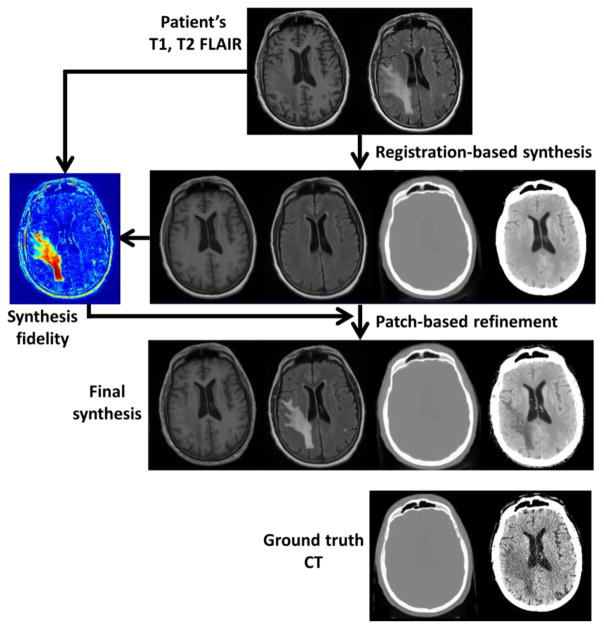
In this research, we propose to use a multi-atlas and multi-channel registration approach for CT synthesis to improve the synthesis accuracy while reducing the effect of registration errors. We also combine a patch-based image synthesis algorithm [9] to refine the synthetic CT image quality on regions where the atlas-based approach does not perform well due to significant geometrical differences, e.g., tumor regions. Figure 1 shows the overall workflow of our combined multi-atlas registration and patch-based refinement (i.e., hybrid) CT synthesis algorithm. The CT synthesis process consists of four key steps; (1) Multi-atlas registration, (2) initial CT synthesis by atlas intensity fusion, (3) iterative CT synthesis refinement, and (4) patch-based CT synthesis refinement. We now describe the details of each step.
Figure 1.
MRI-based CT synthesis workflow.
(Row 1) T1 and T2 FLAIR images of the patient.
(Row 2) Registration-based synthesis and synthesis fidelity map.
(Row 3) Final synthesis after patch-based refinement.
(Row 4) Ground truth CT of the patient for comparison.
CT images are displayed using two window ranges of [−1000, 1000] and [0, 50] HU. T1 and T2 FLAIR images are displayed using window ranges of [0, 1400]. The synthesis fidelity map uses cold (blue) to hot (red) color map with a window range of [0, 500].
2.1. Atlas database
The atlas consists of co-registered MR and CT images. Our method is not limited to a specific MR pulse sequence, but uses multi-parametric MR images such as standard T1 and T2-weighted MRIs as long as they are consistent with the target image data. In this study, T1 and T2 FLAIR (Fluid Attenuated Inversion Recovery) MR images were obtained using Siemens Magnetom Espree 1.5T scanner (Siemens Medical Solutions, Malvern, PA) and CT images were obtained using Philips Brilliance Big Bore scanner (Philips Medical Systems, Netherlands) under the routine clinical protocol from brain cancer patients treated by stereotactic-body radiation therapy (SBRT) or radiosurgery (SRS). The MR images were corrected for geometric distortions using a 3D correction algorithm available in Siemens console workstation (Syngo). The obtained MR images were registered to CT images (reference image for radiotherapy planning) in a semi-automatic way. For the patch-based refinement step, we separately created a tumor and soft tissue atlases that consist of 3×3×3 patch triplets extracted from T1, T2 FLAIR, and CT images. For consistent patch matching, atlas images were first resampled on a 0.7×0.7×1 mm3 grid before the patch extraction.
2.2. Multi-atlas registration
When MR images of a subject are obtained, the first step is to register the atlas and the subject MR images. It is then straightforward to map the atlas CT images to the subject image space by the computed deformations. We use a multi-channel registration to maximally utilize multi-parametric MR image information, thus improving the registration accuracy. Since our data set uses T1 and T2 FLAIR MR images (Figure 1, Row 1), we performed two-channel b-spline deformable registration with mutual information as the similarity metric [10]. In our data set, T1-weighted MRI has higher resolution and better image quality than the T2 FLAIR, and we empirically determined the relative weight for each channel as 0.7 (T1) and 0.3 (T2 FLAIR). Once the atlas T1 and T2 FLAIR images are registered to the target T1 and T2 FLAIR images, the atlas CT images are deformed to the subject image space using the computed deformation vector fields.
2.3. Atlas intensity fusion
To synthesize CT images from multiple registered atlas CT images, unlike Burgos et al. [6] who rank atlas patches based on local normalized cross-correlation (LNCC) and apply pre-determined weights to the selected number of top candidate intensities, we use a structural similarity measure (SSIM) [11] to measure the local similarity of two images. SSIM consists of three components, luminance l(x,y), contrast c(x,y), and structural s(x,y) similarity terms, and provides the following overall structural similarity score between two images x and y
| (1) |
where α>0, β>0, γ>0, and . In these equations, μx, μy, σx, σy are the mean and standard deviation of x and y, respectively, and we set α, β, γ, C1, C2, C3 values as recommended in [11]. SSIM is less sensitive to mean intensity shift and contrast stretching while being sensitive to structural differences, thus more robust to intensity inhomogeneities and inconsistencies which are commonly present in MR images. Since we have both T1 and T2 FLAIR images, the overall local similarity is computed by SSIM(T1atlas,T1subject)+SSIM(T2atlas,T2subject) based on patches centered on each voxel location. The measured local similarity is used to determine the relative weights, and the target CT image is synthesized by locally-weighted averaging. In this way, we do not select an arbitrary number of top candidate atlases (among all registered atlases) nor apply pre-determined weights for fusion. Instead, the weights are determined based on the actual similarity between the atlas and the target. Note that this weighted average is performed voxel-by-voxel with different relative weights (i.e., spatially-varying). This approach is superior to a single-atlas-based approach as each voxel is synthesized using the atlases that show the best local agreement to the subject, thus reducing the effect of registration errors.
2.4. Iterative CT synthesis refinement
Although the initial synthesis step synthesizes missing CT images with reasonable quality, bone and soft tissue boundaries may not be accurate as T1 and T2 FLAIR images do not carry sufficient information about bone. To further refine the bone–soft tissue boundary synthesis, we iteratively refine the CT synthesis by another multi-channel deformable registration in the same way as Burgos et al. [12]. The initial synthetic CT is included, and three-channel deformable registration with T1, T2 FLAIR, and CT is performed between the initially registered atlas and the subject. At this step, no affine registration is necessary, and the registration can be done faster than the initial atlas registration step. Once atlas registration is refined, the target images are synthesized in the same way as the initial synthesis (Figure 1, Row 2). Although multiple iterations might continue to improve the synthesis quality, a single iteration produced reasonable refinement for the bone–soft tissue boundaries.
2.5. CT synthesis fidelity
Previous studies have shown that multi-atlas registration-based approaches can produce good synthetic images [6]. However, these approaches cannot accurately synthesize regions where there are significant anatomical differences between the atlas and the subject, e.g., tumor regions. Even though our goal is to synthesize missing CT image intensities, we can synthesize T1 and T2 FLAIR images of the target as a by-product using the same weights used in CT synthesis. These syntheses are beneficial as they can be directly compared to the given T1 and T2 FLAIR images of the target subject to assess the registration-based synthesis fidelity (Figure 1, Row 2). The underlying idea is that the CT synthesis on a region where the similarity between the target T1/T2 FLAIR and the synthetic T1/T2 FLAIR images is low is likely to be inaccurate. Therefore, erroneously synthesized regions can be detected. We use sum of squared difference between the target T1/T2 FLAIR pair and the synthetic T1/T2 FLAIR pair to detect the erroneously synthesized regions.
2.6. Patch-based synthesis refinement
For the detected low fidelity synthesis regions, we refine the synthesis by using a patch-based synthesis approach (Figure 1, Row 3) [9]. We extract 3×3×3 cubic patches p1(x) and p2(x) centered at location x from T1 and T2 FLAIR images, respectively, and search K nearest neighbor patches of the stacked patch P(x) = [p1(x)T p2(x)T]T among the atlas patch dictionary using the squared L2-norm distance defined as . Then, the target synthetic CT value at location x will be computed by a similarity-weighted average as
| (3) |
and dk is the squared L2-norm distance of the kth patch. In our study, we use K = 5.
Patch-based synthesis is a time-consuming process as a large number of patches in the atlas dictionary have to be compared unless a patch-based intensity mapping is learned. Unlike other approaches that synthesize entire images by patch matching, our method synthesizes only small regions where registration-based synthesis fails. Therefore, our refinement process does not require significant computation time.
3. RESULTS
We applied our hybrid CT synthesis method on brain cancer patient data. Each patient data has a radiotherapy planning CT and T1-weighted and T2 FLAIR MRIs; these data are consistent with our atlas data. T1 and T2 FLAIR scans were taken at the same patient position as the CT scan, and rigidly registered to the CT. Finally, images were resampled to have a 0.7×0.7×1 mm3 resolution to be consistent with our atlas patch dictionary. A total of 20 atlas data sets were used, and a combined tumor (10,000 patch triplets) and soft tissue (100,000 patch triplets) patch dictionary was created from two atlas data sets. Atlas T1 and T2 FLAIR images were registered to the target patient’s T1 and T2 FLAIR images (Figure 1, Row 1) by two-channel b-spline deformable registration with mutual information as the similarity metric, and the atlas CT images were deformed and fused to reconstruct the synthetic CT images of the patient (Figure 1, Row 2). To assess the CT synthesis fidelity, we also synthesized the patient’s T1 and T2 FLAIR images (Figure 1, Row 2) using the same local weights used in CT synthesis. As shown in the example case in Figure 1, the initial multi-atlas registration-based synthesis produces synthetic images with reasonable quality for normal tissues. However, the tumor region cannot be accurately synthesized as the atlases do not carry sufficient information about the tumor, thus producing normal-looking tissue. This inaccuracy was automatically detected by the synthesis fidelity assessment step (Figure 1, Row 2) in which synthetic T1 and T2 FLAIR images are directly compared to the given T1 and T2 FLAIR images of the patient. We then refined the CT synthesis for the soft tissue regions with low synthesis fidelity (extracted regions with synthetic CT intensity between −100 and 300 HU, and T1/T2 FLAIR difference larger than 200) using the patch-based synthesis. In this example case, only 5% of the volume was refined by the patch-based synthesis. As shown in Figure 1, Row 3, the patch-based refinement significantly improved the synthesis in the tumor region where registration-based synthesis failed. In addition, refined synthetic T1 and T2 FLAIR images are given as a by-product of CT synthesis refinement, and can be used to assess the quality of the patch-based refinement process. As shown in Figure 1, Row 3, the patch-based refinement improved the synthesis of the T1 and T2 FLAIR, implying that the refinement was correct.
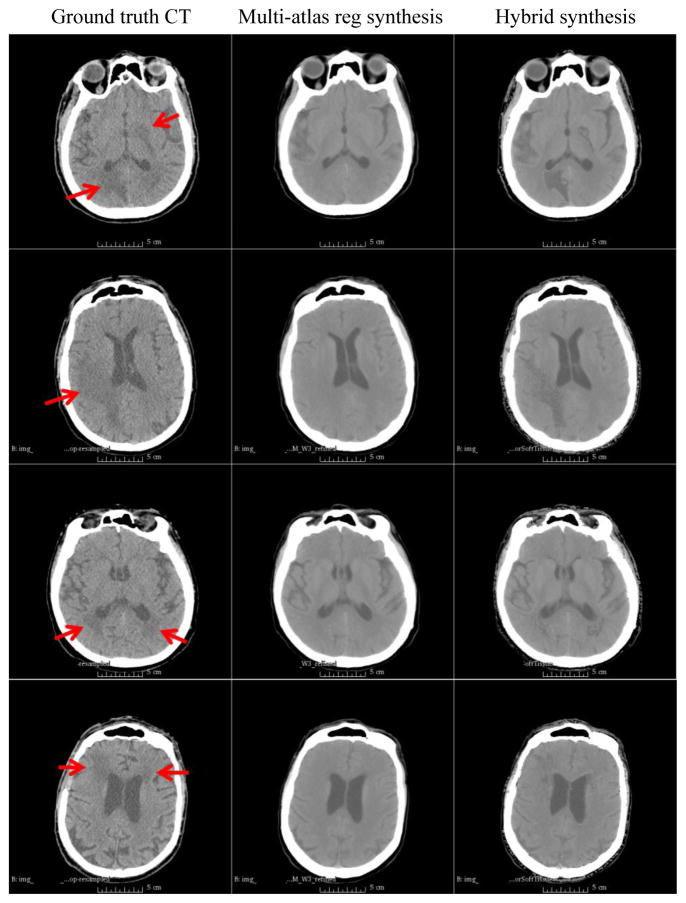
We processed a total of 4 cases, which includes the above example case. As shown in Figure 2, the proposed hybrid synthesis method was able to reliably synthesize both normal tissues and tumor regions. Table 1 summarizes absolute CT number (HU) differences between multi-atlas registration synthesis and hybrid synthesis compared to the ground truth CT. We compared the entire CT field of view (Overall) as well as tumor regions only (Tumor). The HU difference in the entire field of view was larger than the tumor regions because there were still mismatches on the boundaries of the bone and soft tissue or air. Due to the large CT number of the bone, slight boundary or intensity (within the bone) mismatches caused large HU differences. For tumor regions, there was noticeable synthesis improvement when there was relatively larger HU difference on tumor regions, i.e., cases 1 and 2. However, when the multi-atlas registration synthesis intensity was already close to that of the true CT, i.e., cases 3 and 4, the synthesis quality improvement was relatively minor.
Figure 2.
Examples of MRI-based CT synthesis. All images are displayed with a window range of [−50, 100] HU
(From top to bottom) Cases 1–4.
(Left) Patient’s ground truth CT.
(Middle) Multi-atlas registration synthesis.
(Right) Hybrid synthesis. Red arrows indicate tumor regions.
Table 1.
Mean±standard deviation absolute CT number differences between multi-atlas registration synthesis and hybrid synthesis compared to ground truth CT.
| Case | Overall | Tumor | ||
|---|---|---|---|---|
| Registration | Hybrid | Registration | Hybrid | |
| 1 | 86.02 ± 193.38 | 86.37 ± 193.60 | 11.21 ± 16.21 | 7.47 ± 14.40 |
| 2 | 75.29 ± 163.69 | 76.29 ± 164.01 | 11.06 ± 43.73 | 8.49 ± 43.27 |
| 3 | 91.27 ± 213.26 | 92.73 ± 213.82 | 5.98 ± 4.36 | 5.84 ± 4.45 |
| 4 | 78.80 ± 173.47 | 79.62 ± 173.86 | 8.76 ± 6.26 | 6.73 ± 5.30 |
4. CONCLUSIONS
We have proposed a hybrid MR to CT synthesis method that combines multi-atlas registration-based synthesis and a patch-based refinement. The key benefit of the proposed method is its capability of accurately synthesizing both normal tissues as well as abnormal regions. Especially, the initial multi-atlas registration-based CT synthesis can be naturally extended to multi-atlas autosegmentation of normal tissues with minimal additional computation, i.e., label fusion, which will significantly reduce the physician’s contouring burden in radiotherapy planning. The synthesis fidelity assessment step based on the synthesis of the given modalities (T1 and T2 FLAIR in this study) allows us to automatically detect erroneous synthesis regions for which the patch-based refinement can further improve the synthesis efficiently without involving a large number of regions. We showed promising synthesis results on brain cases here, and additional tests on larger data sets are under way. Although we focused on CT synthesis in this paper, our method can be applied to any modality synthesis with improved synthesis quality for which conventional registration-based and patch-based methods may fail.
Acknowledgments
This work was supported by the NIH/NIBIB under the grant R01EB017743.
References
- 1.Ng SH, Chang TC, Ko SF, Yen PS, Wan YL, Tang LM. Nasopharyngeal carcinoma: MRI and CT assessment. Neuroradiology. 1997;39(10):741–746. doi: 10.1007/s002340050499. [DOI] [PubMed] [Google Scholar]
- 2.Brouwer CL, Steenbakkers RJHM, van den Heuvel E, Duppen JC, Navran A, Bijl HP, Chouvalova O, Burlage FR, Meertens H, Langendijk JA, van ‘t Veld AA. 3D variation in delineation of head and neck organs at risk. Radiat Oncol. 2012;7(32):1–9. doi: 10.1186/1748-717X-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chong V. Oral cavity cancer. Cancer Imaging. 2005;5(A):S49–52. doi: 10.1102/1470-7330.2005.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jonsson JH, Karlsson MG, Karlsson M, Nyholm T. Treatment planning using MRI data: an analysis of the dose calculation accuracy for different treatment regions. Radiat Oncol. 2010;6:62. doi: 10.1186/1748-717X-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berker Y, Franke J, Salomon A, Palmowski M, Donker HCW, Temur Y, Mottaghy FM, Kuhl C, Izquierdo-Garcia D, Fayad ZA, Kiessling F, Schulz V. MRI-based attenuation correction for hybrid PET/MRI systems: a 4-class tissue segmentation technique using a combined ultrashort-echo-time/Dixon MRI sequence. J Nucl Med. 2012;53:796–804. doi: 10.2967/jnumed.111.092577. [DOI] [PubMed] [Google Scholar]
- 6.Burgos N, Cardoso MJ, Thielemans K, Modat M, Pedemonte S, Dickson J, Barnes A, Ahmed R, Mahoney CJ, Schott JM, Duncan JS, Atkinson D, Arridge SR, Hutton BF, Ourselin S. Attenuation correction synthesis for hybrid PET-MR scanners: application to brain studies. IEEE Trans Med Imag. 2014;33(12):2332–2341. doi: 10.1109/TMI.2014.2340135. [DOI] [PubMed] [Google Scholar]
- 7.Roy S, Carass A, Jog A, Prince JL, Lee J. MR to CT registration of brains using image synthesis. Proc of SPIE Medical Imaging. 2014;9034:903419. doi: 10.1117/12.2043954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roy S, Wang W-T, Carass A, Prince JL, Butman JA, Pham DL. PET attenuation correction using synthetic CT from ultrashort echo-time MR imaging. J Nucl Med. 2015;55:2071–2077. doi: 10.2967/jnumed.114.143958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andreasen D, Van Leemput K, Hansen RH, Andersen JAL, Edmund JM. Patch-based generation of a pseudo CT from conventional MRI sequences for MRI-only radiotherapy of the brain. Med Phys. 2015;42(4):1596–1605. doi: 10.1118/1.4914158. [DOI] [PubMed] [Google Scholar]
- 10.Klein S, Staring M, Murphy K, Viergever MA, Pluim JP. elastix: A toolbox for intensity-based medical image registration. IEEE Trans Med Imag. 2010;29(1):196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: from error visibility to structural similarity. IEEE Trans Imag Proc. 2004;13(4):600–612. doi: 10.1109/tip.2003.819861. [DOI] [PubMed] [Google Scholar]
- 12.Burgos N, Cardoso MJ, Guerreiro F, Veiga C, Modat M, McClelland J, Knopf A-C, Punwani S, Atkinson D, Arridge SR, Hutton BF, Ourselin S. Robust CT synthesis for radiotherapy planning: application to the head and neck region. MICCAI 2015, Part II, LNCS 9350. 2015:476–484. [Google Scholar]