Abstract
The purpose of this study was to investigate the association of tumor tissue and plasma miR-146a/b expressions with the clinicopathological properties and overall survival (OS) in surgical patients with intrahepatic cholangiocarcinomas (ICC).
Eighty-seven patients with ICC were enrolled. Tumor tissue and plasma sample were collected and miR-146a/b expressions were assessed by quantitative polymerase chain reaction (qPCR). The median follow-up duration was 31 months, and the last follow-up date was January 2017.
miR-146a (P < .001) and miR-146b (P = .006) expressions in tumor tissue were positively associated with that in plasma. Tissue miR-146a was negatively correlated with age (P = .036), poor differentiation (P = .020), N stage (P = .020), and TNM stage (P = .007), as well as ECOG performance (P = .008), whereas plasma miR-146a was inversely associated with N stage (P = .003), TNM stage (P = .003), and ECOG performance (P = .011). Moreover, tissue miR-146b was negatively correlated with gender (P = .043) and T stage (P = .047). Kaplan-Meier curves suggested that high expression of tissue miR-146a (P < .001) and plasma miR-146a (P = .029) were correlated with prolonged OS. Nevertheless, no association of miR-146b expression in tumor tissue (P = .187) and plasma (P = .336) with OS was discovered. Univariate analysis indicated that both tissue miR-146a (P < .001) and plasma miR-146a (P = .035) could predict better OS, whereas multivariate analysis revealed that only tissue miR-146a (P = .001) high expression was an independent factor for prolonged OS.
Both plasma and tissue miR-146a expression correlated with favorable OS, whereas only tissue miR-146a was an independent prognostic biomarker in surgical patients with ICC.
Keywords: intrahepatic cholangiocarcinoma, miR-146a/b, overall survival, prolonged, surgical patients
1. Introduction
Intrahepatic cholangiocarcinomas (ICC), with the second highest incidence of all liver cancer accounting for the proportion of 5% to 20%, is a critical threat imperiling people's health worldwide.[1,2] In the last 2 decades, accumulating evidence revealed that the morbidity and mortality caused by ICC are increasing gradually over the world.[3] Multiple elements are involved in the development and progression of ICC, among which the main risk factors consist of cholangiolithiasis, hepatitis B virus (HBV) infection, and Clonorchis sinensis infection, as well as primary sclerosing cholangitis (PSC).[3–5] Despite great improvements of early diagnosis, optimal treatments (eg, surgery, chemotherapy, and radiotherapy), as well as patients’ care, the prognosis of ICC is still far from satisfaction, the 5-year survival rate of ICC patients with complete resection is only 20% to 40%.[6,7]
microRNAs (miRNAs), belonging to a great family of small endogenous non-coding RNAs, regulate target genes and protein expression via binding to the 3′-untranslated regions (3′-UTRs) directly.[8] miRNAs contribute to multiple molecule activities, such as cell proliferation, differentiation, migration, and apoptosis.[9,10] Accumulating evidence has defined miRNAs as biomarkers in various carcinomas, including colorectal cancer, gastric cancer, breast cancer, and lung cancer.[11–14] microRNA-146 family (miR-146) consists of miR-146a and miR-146b, both of them have the similar sequences in the mature miRNAs excluding 2 bases toward the 3′-end.[15] Dysregulated miR-146 expression has been described in a variety of malignant tumors, such as gastric cancer, breast cancer, or even hepatocellular cancer.[16–18] However, few studies about the effects of miR-146 expression on the prognosis of ICC patients have been explored. Therefore, the purpose of this study was to investigate the association of tumor tissue and plasma miR-146 a/b expressions with the clinicopathological properties and overall survival (OS) in surgical patients with ICC.
2. Methods
2.1. Patients
Eighty-seven patients with ICC underwent surgery at Department of Hepatobiliary & Pancreatic Surgery in The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology, from May 2012 to April 2014 were recruited in this prospective cohort study. The diagnosis of ICC was confirmed by clinical features, pathological determination, and/or radiology (computerized tomography [CT], magnetic resonance imaging [MRI], or endoscopic retrograde cholangiopancreatography [ERCP]). Patients with other tumors (solid or blood), inflammatory diseases, severe infection history, cognitive impairment, life expectancy <6 months were excluded from this study.
No patient received neo-adjuvant systemic therapy before the operation, and after the surgery, all the patients received adjuvant therapy with catheter radiofrequency ablation, or chemotherapy by gemcitabine plus cis-platinum, and/or radiotherapy.
This study was approved by Ethics Committee of The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology; the written informed consents were obtained from all patients.
2.2. Clinicopathological features and follow-ups
Clinical and pathological properties of ICC patients at baseline were collected including age, gender, hepatitis B surface antigen (HBsAg) status, hepatic C virus (HCV) status, smoke, drink, differentiation, TNM stage, and ECOG performance status. The median follow-up duration was 31 months, and the last follow-up date was January 2017. OS was calculated from the date of surgery to patients’ death or last follow-up.
2.3. Sample collection and RNA extraction
Tumor tissue samples were obtained during the surgery. Blood samples were collected before the operation and plasma were separated. Total RNA was then extracted from the samples with TRIzol reagent (Invitrogen) according to the instructions of the manufacturer.
2.4. miR-146a/b determination by qPCR
Total RNA was extracted from tumor tissue and plasma samples by Trizol LS kit (TaKaRa, Japan), and RNA was subjected to reverse transcription by utilizing the PrimerScript Real-time reagent kit (TaKaRa, Japan). Subsequently, miR-146 a/b expressions were quantitated by SYBR Premix Ex TaqTM II (TaKaRa, Japan). The primer sequences (BGI, China) for the primer source were performed as follows: hsa-miR-146a-5p RT primer: 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAACCCATG-3′; hsa-miR-146a-5p Forward primer: 5′-ACACTCCAGCTGGGTGAGAACTGAATTCCATG-3′; hsa-miR-146a-5p Revert: 5′-TGTCGTGGAGTCGGCAATTC-3′; hsa-miR-146b-5p RT primer; 5′-ACACTCCAGCTGGGTGAGAACTGAATTCCATG-3′; hsa-miR-146b-5p Forward primer: 5′-ACACTCCAGCTGGGTGAGAACTGAATTCCATA-3′; hsa-miR-146b-5p Revert: 5′-TGTCGTGGAGTCGGCAATTC-3′. The expression levels of miR-146a/b were calculated by using the 2-△△t method and U6 served as the internal reference.
2.5. Statistics
Statistics was carried out using SPSS 22.0 (IBM) and 2010 office software (Microsoft). Correlation of miR-146a/b expression in tissue and plasma sample, as well as miR-146a/b expression with clinicopathological features were determined by Spearman test. Kaplan-Meier curves and log-rank test were used to compare OS in different groups. Univariate and multivariate Cox regression were used to evaluate the baseline predictive factors for OS in ICC surgical patients. P <.05 was considered significant.
3. Results
3.1. Patients characteristics
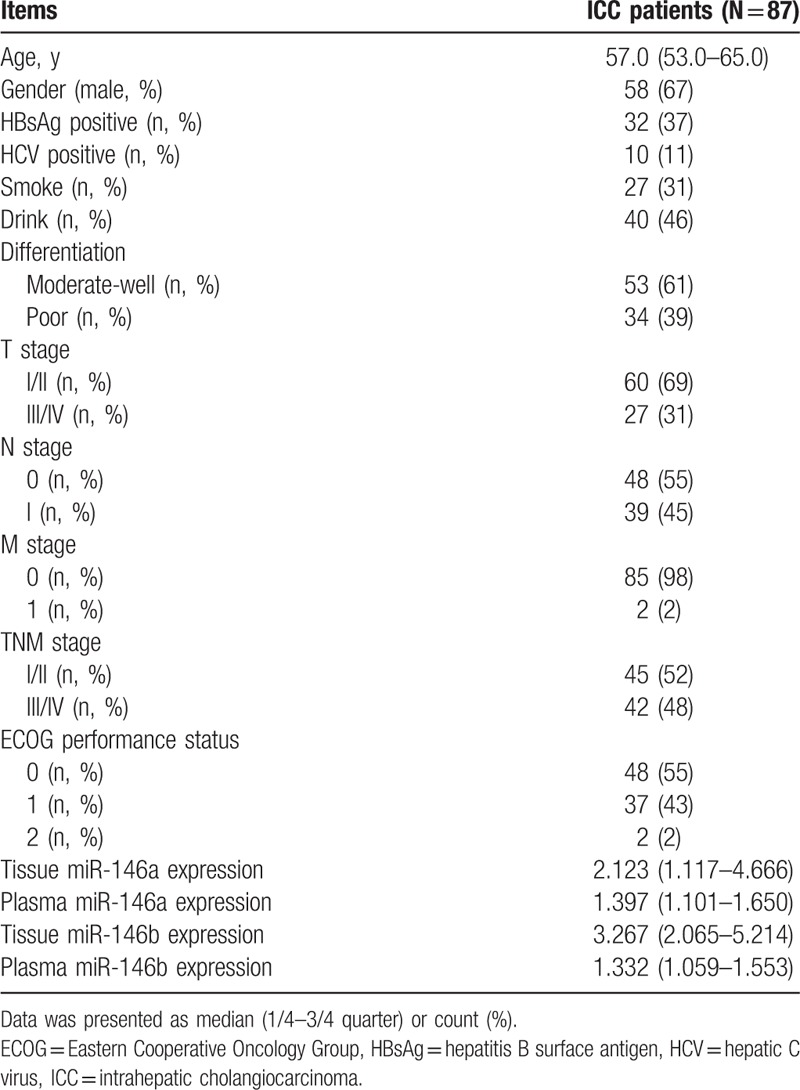
The median age was 57.0 (53.0–65.0) years, male and female were 58 (67%) and 29 (33%) (Table 1). Thirty-two (37%) patients were HBsAg positive and 10 (11%) patients were HCV positive, and 27 (31%) patients were with smoke history and 40 (46%) patients were with drink history. Meanwhile, there were 53 (61%) patients with moderate-well differentiation, but 34 (39%) patients with poor differentiation. The number of patients in I/II TNM stage was 45 (52%), and in III/IV TNM stage was 42 (48%). Other clinicopathological characteristics of ICC patients are shown in Table 1.
Table 1.
Baseline clinicopathological characteristics of ICC patients (N = 87).
3.2. Correlation between miR-146a/b in tumor tissue and plasma
Correlations of miR-146a/b in tumor tissue and plasma were evaluated in Figure 1. miR-146a (R = 0.376, P < .001, Fig. 1A) and miR-146b (R = 0.291, P = .006, Fig. 1B) expressions in tumor tissue were positively associated with that in plasma.
Figure 1.
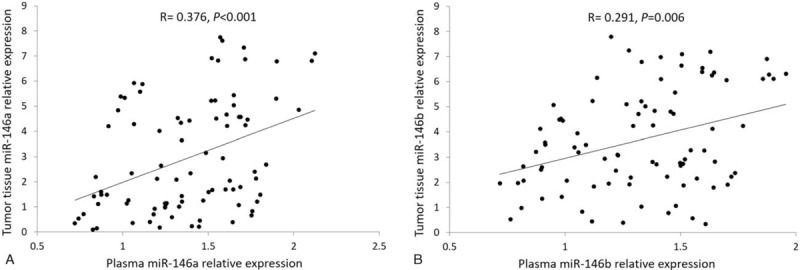
Correlation between miR-146a/b expressions in plasma and in tumor tissues. (A) Correlation between miR-146a expressions in plasma and in tumor tissues. (B) Correlation between miR-146b expressions in plasma and in tumor tissues. Spearman test was used to analyze the correlation of miR-146a/b expression in plasma with that in tumor tissue. P <.05 was considered significant.
3.3. Correlation of miR-146a/b with clinical and pathological features
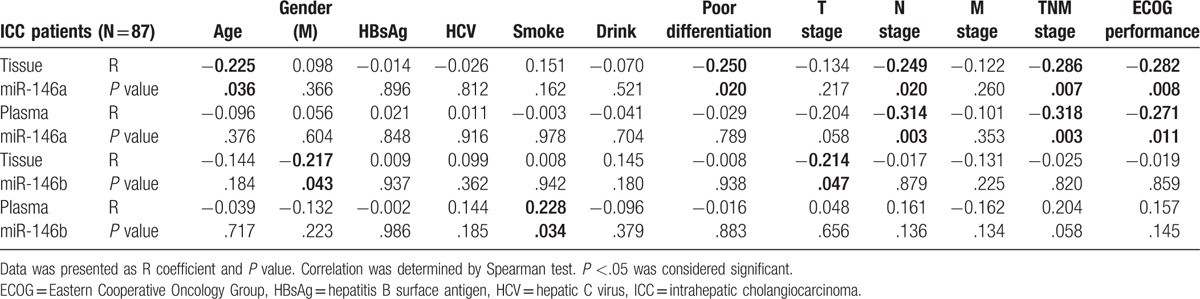
As listed in Table 1, miR-146a expressions in tissue and plasma samples were observed to be 2.123 (1.117–4.666) and 1.397 (1.101–1.650), respectively, whereas miR-146b expressions were 3.267 (2.065–5.214) in tissue samples, and 1.332 (1.059–1.553) in plasma samples. As presented in Table 2, tissue miR-146a was negatively correlated with age (R = −0.225, P = .036), poor differentiation (R = −0.250, P = .020), N stage (R = −0.249, P = .020), and TNM stage (R = −0.286, P = .007), as well as ECOG performance (R = −0.282, P = .008), whereas plasma miR-146a was inversely associated with N stage (R = −0.314, P = .003), TNM stage (R = −0.318, P = .003), and ECOG performance (R = −0.271, P = .011). Moreover, tissue miR-146b was negatively correlated with gender (R = −0.217, P = .043) and T stage (R = −0.214, P = .047), and plasma miR-146b was negatively associated with smoke (R = 0.228, P = .034).
Table 2.
Correlation of miR-146a/b expression with clinicopathological features.
3.4. Tissue/plasma miR-146a correlated with prolonged OS but not miR-146b
In terms of miR-146a, its high expression in both tumor tissue (P < .001, Fig. 2A) and plasma (P = .029, Fig. 2B) were correlated with prolonged OS compared with low expression. Nevertheless, no association of miR-146b expression in tumor tissue (P = .187, Fig. 2C) or plasma (P = .336, Fig. 2D) with OS was discovered in the present study.
Figure 2.
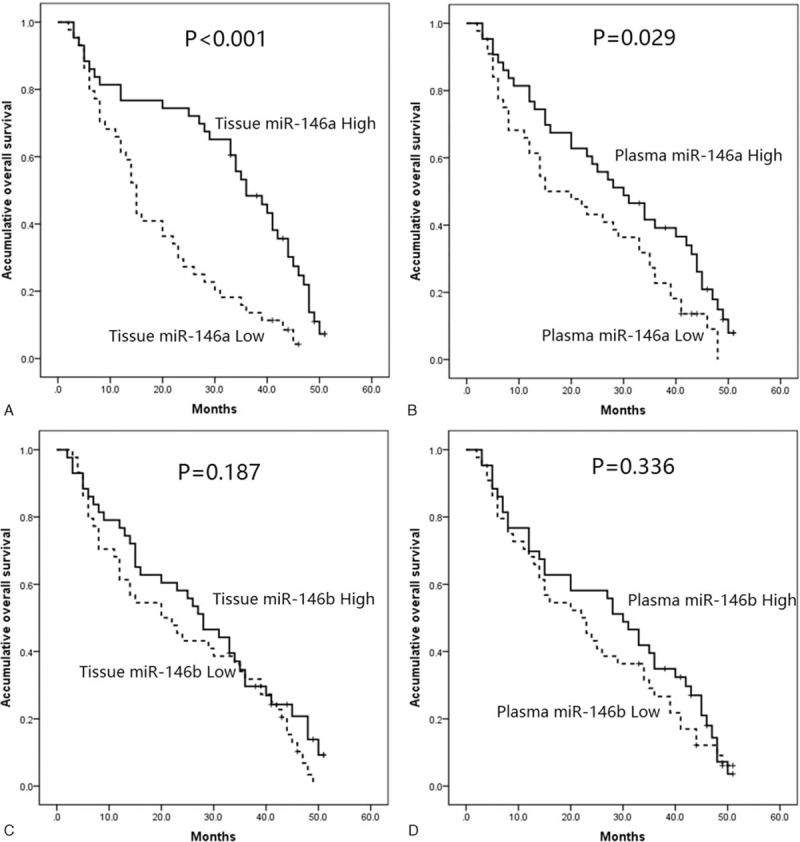
Association of plasma and tumor tissue miR-146a/b levels with OS. (A) Association of miR-146a with OS in tissue sample. (B) Association of miR-146a with OS in plasma sample. (C) Association of miR-146b with OS in tissue sample. (D) Association of miR-146b with OS in plasma sample. Kaplan-Meier curves and log-rank test were used to evaluate the correlation of plasma and tumor tissue miR-146a/b levels with OS. P <.05 was considered significant.
3.5. Tissue miR-146a independently predicted longer OS
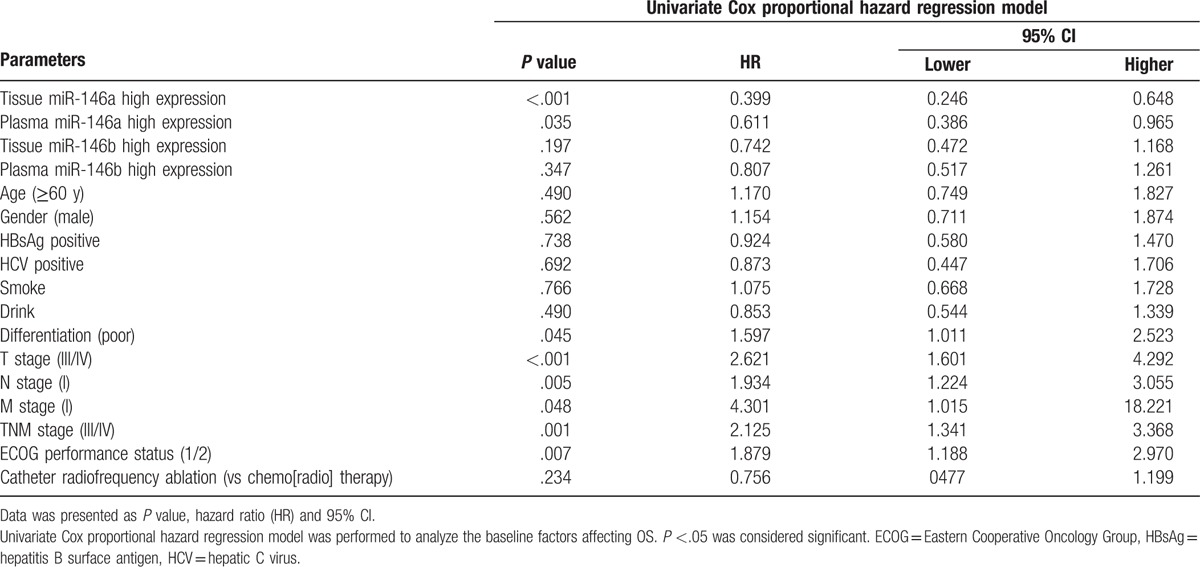
Univariate Cox proportional hazards regression was used to evaluate the baseline predictive factors for OS in ICC surgical patients, as presented in Table 3, which indicated that miR-146a high expression in tissue (P < .001) and in plasma (P = .035) were associated with better OS. Besides, poor differentiation (P = .045), higher T stage (III/IV) (P < .001), higher N stage (I) (P = .005), higher M stage (I) (P = .048), increased TNM stage (III/IV) (P = .001), and elevated ECOG performance status (1/2) (P = .007) were correlated with worse OS in surgical patients with ICC. Furthermore, ICC patients treated with catheter radiofrequency ablation compared with chemo(radio) therapy seemed to be associated with better OS, but without statistical significance (P = .234).
Table 3.
Factors at baseline predicting OS by univariate Cox analysis.
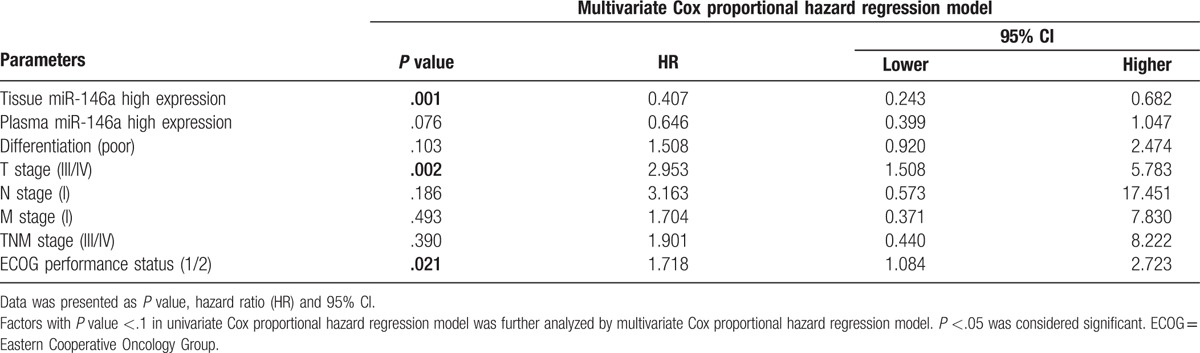
All factors with a P value <.1 were further analyzed by the multivariate Cox proportional hazards regression model. It revealed that the high expression of tissue miR-146a (P = .001) but not plasma miR-146a (P = .076) was an independent factor for prolonged OS, whereas higher T stage (III/IV) (P = .002) and evaluated ECOG performance status (1/2) (P = .021) could predict worse OS in ICC surgical patients independently (Table 4).
Table 4.
Independent factors at baseline predicting OS by multivariate Cox analysis.
4. Discussion
In the present study, we found that both miR-146a and miR-146b expressions in tumor tissue were positively associated with that in plasma; and tumor tissue miR-146a was negatively correlated with age, poor differentiation, N stage, and TNM stage, as well as ECOG performance, whereas plasma miR-146a was inversely associated with N stage, TNM stage, and ECOG performance. As to miR-146b, only tumor tissue was negatively associated with gender and T stage. Both tissue miR-146a and plasma miR-146a were associated with better OS, whereas only tissue miR-146a high expression was an independent factor for prolonged OS of ICC patients, and T stage (III/IV)) and ECOG performance status (1/2) predict worse OS independently.
ICC, one of the multifactorial malignancies, originates from the epithelial cells of the intrahepatic bile duct beyond second-order branches.[19] The most common pathological process of ICC is chronic inflammation and fibrosis of bile duct, which could induce epithelial cells apoptosis and proliferation. Subsequently, this repeated process lead to aberrant molecule activities, stimulating tumorigenesis.[1]
miR-146a, encoded on chromosome 5(5q33.3),[16] has been disclosed as a potential tumor repressor in several malignant tumors. Upregulation of miR-146a inhibits lung cancer cell proliferation and differentiation via mediating the macrophage migration inhibitory factor (MIF) gene and NF-κB signaling pathway.[20] Also, its overexpression represses gastric cancer cell migration by reducing the UHRF1 level via targeting its 3′-UTR, resulting in the inhibition of tumor invasion and metastasis subsequently.[21] Additionally, miR-146a expression has been observed to be positively associated with antitumor immune systems by targeting signal transducer and activator of transcription 3 (STAT3) in human hepatocellular carcinoma (HCC) cells.[22] Analysis of miR-146a in triple-negative breast cancer (TNBC) tumors has been observed to predict better OS by regulating breast cancer 1 (BRCA1) expression.[23] As to tissue miR-146, it is considered as a suppressor of HCC, negatively associated with the clinical TNM stage, metastasis, and portal vein tumor embolus, as well as a number of tumor nodes.[24] Furthermore, increased expression of tissue miR-146a has been confirmed to predict a higher survival rate of gastric cancer, esophageal squamous cell carcinoma and non-small cell lung cancer (NSCLC), as well as renal cell carcinoma (RCC).[25–28] These researches suggest that miR-146a acts as tumor suppressor on malignant tumors, and it is correlated with better prognosis. Partially in line with these results, our study found that tumor tissue miR-146a was negatively correlated with poor differentiation, N stage, and TNM stage, as well as ECOG performance, whereas plasma miR-146a was inversely associated with N stage, TNM stage, and ECOG performance. This might arise from the inhibition of cell proliferation, differentiation, and migration by miR-146a through regulating several pathways.[20,23] In addition, we also found that high expressions of tissue miR-146a and plasma miR-146a were associated with better OS, whereas only tissue miR-146a high expression was an independent factor for prolonged OS of ICC patients. There are 2 possible reasons. First, miR-146a inhibits tumor growth and metastasis by mediating various molecule activities[21,22]; second, miR-146a increases the sensitivity of chemotherapy, radiotherapy, and targeted therapy, decreasing recurrence of ICC patients.[29–32]
miR-146b, consisting of miR-146b-5p and miR-146b-3p, is located on human chromosome 10 at position q24.32.[33] Published studies have defined miR-146b as a tumor inhibitor in various carcinomas, including esophageal cancer, gallbladder cancer and thyroid cancer.[34–36] A larger number of studies have elucidated the role of miR-146b in carcinoma cell lines. In a study of esophageal cancer cell, miR-146b acted as a tumor suppressor by targeting CCAAT/enhancer-binding proteins β liver-enriched transcriptional activator protein 2 (C/EBPβ LAP2) to inducing cell proliferation and repressing cell apoptosis.[36] In addition, miR-146b expression was associated with the TNM stage, liver metastasis and differentiated degree, and its overexpression showed better OS in patients with gallbladder cancer (GBC).[35] These studies illustrates that miR-146b serves as tumor suppressor on the prognosis of carcinomas. On the other hand, miR-146b expression could also act as tumor promoter to inducing thyroid cancer cell migration and invasion by downregulating zinc and ring finger 3 (ZNRF3).[37] However, our study showed that no association of plasma and tissue miR-146b with OS in ICC surgical patients. The possible reasons are that the dual effects of miR-146b on both promotion and repression of malignant tumors. Meanwhile, we also observed that tumor tissue miR-146b was only negatively correlated with T stage, this might result from the repression of cell proliferation and apoptosis via targeting several genes.[36,38]
Several limitations still existed in this study. The sample size was relatively small. The plasma miR-146a/b expression of healthy people was not evaluated to compare with ICC patients. The follow-up duration was relatively short with median value 31 months. Further study with larger sample size and longer follow-up duration is necessary to be carried out in the future.
In conclusion, both plasma and tissue miR-146a expression correlated with favorable OS, whereas only tissue miR-146a was an independent prognostic biomarker in surgical patients with ICC.
Footnotes
Abbreviations: 3′-UTRs = 3′-untranslated regions, C/EBPβ LAP2 = CCAAT/enhancer-binding proteins β liver-enriched transcriptional activator protein 2, CT = computerized tomography, ERCP = endoscopic retrograde cholangiopancreatography, HBsAg = hepatitis B surface antigen, HBV = hepatitis B virus, HCC = hepatocellular carcinoma, HCV = hepatic C virus, ICC = intrahepatic cholangiocarcinoma, MIF = macrophage migration inhibitory factor, MRI = magnetic resonance imaging, NSCLC = non-small cell lung cancer, OS = overall survival, PSC = primary sclerosing cholangitis, qPCR = quantitative polymerase chain reaction, RCC = renal cell carcinoma, STAT3 = signal transducer and activator of transcription 3, TNBC = triple-negative breast cancer, ZNRF3 = zinc and ring finger 3.
RXZ and ZZ contributed equally to this work.
The authors have no conflicts of interest to disclose.
References
- [1].Vijgen S, Terris B, Rubbia-Brandt L. Pathology of intrahepatic cholangiocarcinoma. Hepatobiliary Surg Nutr 2017;6:22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Padia SA. Intrahepatic cholangiocarcinoma. Tech Vasc Interv Radiol 2015;18:227–35. [DOI] [PubMed] [Google Scholar]
- [3].Kirstein MM, Vogel A. Epidemiology and risk factors of cholangiocarcinoma. Visc Med 2016;32:395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Luk’yanchenko AV, Medvedeva BM, Shabanov MA, et al. Intrahepatic cholangiocarcinoma. Vestn Rentgenol Radiol 2015;52–63. [PubMed] [Google Scholar]
- [5].Ghouri YA, Mian I, Blechacz B. Cancer review: cholangiocarcinoma. J Carcinog 2015;14:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Oliveira IS, Kilcoyne A, Everett JM, et al. Cholangiocarcinoma: classification, diagnosis, staging, imaging features, and management. Abdom Radiol (NY) 2017;42:1637–49. [DOI] [PubMed] [Google Scholar]
- [7].Doussot A, Groot-Koerkamp B, Wiggers JK, et al. Outcomes after resection of intrahepatic cholangiocarcinoma: external validation and comparison of prognostic models. J Am Coll Surg 2015;221:452–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Li J, Tan S, Kooger R, et al. MicroRNAs as novel biological targets for detection and regulation. Chem Soc Rev 2014;43:506–17. [DOI] [PubMed] [Google Scholar]
- [9].Dong H, Lei J, Ding L, et al. MicroRNA: function, detection, and bioanalysis. Chem Rev 2013;113:6207–33. [DOI] [PubMed] [Google Scholar]
- [10].Rottiers V, Näär AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol 2012;13:239–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nagy ZB, Wichmann B, Kalmár A, et al. Colorectal adenoma and carcinoma specific miRNA profiles in biopsy and their expression in plasma specimens. Clin Epigenetics 2017;9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liu JN, Shangguan YM. Long non-coding RNA CARLo-5 upregulation associates with poor prognosis in patients suffering gastric cancer. Eur Rev Med Pharmacol Sci 2017;21:530–4. [PubMed] [Google Scholar]
- [13].Dvinge H, Git A, Gräf S, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature 2013;497:378–82. [DOI] [PubMed] [Google Scholar]
- [14].Florczuk M, Szpechcinski A, Chorostowska-Wynimko J. miRNAs as biomarkers and therapeutic targets in non-small cell lung cancer: current perspectives. Target Oncol 2017;12:179–200. [DOI] [PubMed] [Google Scholar]
- [15].Hao Y, Zhou Q, Ma J, et al. miR-146a is upregulated during retinal pigment epithelium (RPE)/choroid aging in mice and represses IL-6 and VEGF-A expression in RPE cells. J Clin Exp Ophthalmol 2016;7: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li Y, Zhao L, Shi B, et al. Functions of miR-146a and miR-222 in tumor-associated macrophages in breast cancer. Sci Rep 2015;5:18648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xia ZG, Yin HF, Long Y, et al. Genetic variant of miR-146a rs2910164 C>G and gastric cancer susceptibility. Oncotarget 2016;7:34316–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen J, Cao X, Zhang H. MiR-146a rs2910164 polymorphism is associated with hepatocellular carcinoma: a meta-analysis. Int J Clin Exp Med 2015;8:15852–6. [PMC free article] [PubMed] [Google Scholar]
- [19].Ronnekleiv-Kelly SM, Pawlik TM. Staging of intrahepatic cholangiocarcinoma. Hepatobiliary Surg Nutr 2017;6:35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang WM, Liu JC. Effect and molecular mechanism of mir-146a on proliferation of lung cancer cells by targeting and regulating MIF gene. Asian Pac J Trop Med 2016;9:806–11. [DOI] [PubMed] [Google Scholar]
- [21].Zhou L, Zhao X, Han Y, et al. Regulation of UHRF1 by miR-146a/b modulates gastric cancer invasion and metastasis. FASEB J 2013;27:4929–39. [DOI] [PubMed] [Google Scholar]
- [22].Sun X, Zhang J, Hou Z, et al. miR-146a is directly regulated by STAT3 in human hepatocellular carcinoma cells and involved in anti-tumor immune suppression. Cell Cycle 2015;14:243–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zavala V, Pérez-Moreno E, Tapia T, et al. miR-146a and miR-638 in BRCA1-deficient triple negative breast cancer tumors, as potential biomarkers for improved overall survival. Cancer Biomark 2016;16:99–107. [DOI] [PubMed] [Google Scholar]
- [24].Rong M, He R, Dang Y, et al. Expression and clinicopathological significance of miR-146a in hepatocellular carcinoma tissues. Ups J Med Sci 2014;119:19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ahn DH, Rah H, Choi YK, et al. Association of the miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G polymorphisms with gastric cancer risk and survival in the Korean population. Mol Carcinog 2013;52(suppl 1):E39–51. [DOI] [PubMed] [Google Scholar]
- [26].Wang C, Guan S, Liu F, et al. Prognostic and diagnostic potential of miR-146a in oesophageal squamous cell carcinoma. Br J Cancer 2016;114:290–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wu C, Cao Y, He Z, et al. Serum levels of miR-19b and miR-146a as prognostic biomarkers for non-small cell lung cancer. Tohoku J Exp Med 2014;232:85–95. [DOI] [PubMed] [Google Scholar]
- [28].Huang Z, Lu Z, Tian J, et al. Effect of a functional polymorphism in the pre-miR-146a gene on the risk and prognosis of renal cell carcinoma. Mol Med Rep 2015;12:6997–7004. [DOI] [PubMed] [Google Scholar]
- [29].Chen M, Zhou ZY, Chen JG, et al. Effect of miR-146a polymorphism on biochemical recurrence risk after radical prostatectomy in southern Chinese population. Genet Mol Res 2014;13:10615–21. [DOI] [PubMed] [Google Scholar]
- [30].Cui Y, She K, Tian D, et al. miR-146a inhibits proliferation and enhances chemosensitivity in epithelial ovarian cancer via reduction of SOD2. Oncol Res 2016;23:275–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Liao YQ, Liao YL, Li J, et al. Polymorphism in miR-146a associated with clinical characteristics and outcomes in gastric cancer patients treated with adjuvant oxaliplatin and fluoropyrimidines. Onco Targets Ther 2015;8:2627–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sun B, Xie C, Zheng T, et al. Selecting molecular therapeutic drug targets based on the expression profiles of intrahepatic cholangiocarcinomas and miRNA-mRNA regulatory networks. Oncol Rep 2016;35:382–90. [DOI] [PubMed] [Google Scholar]
- [33].Bhaumik D, Scott GK, Schokrpur S, et al. MicroRNAs miR-146a/b negatively modulate the senescence-associated inflammatory mediators IL-6 and IL-8. Aging (Albany NY) 2009;1:402–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Geraldo MV, Yamashita AS, Kimura ET. MicroRNA miR-146b-5p regulates signal transduction of TGF-beta by repressing SMAD4 in thyroid cancer. Oncogene 2012;31:1910–22. [DOI] [PubMed] [Google Scholar]
- [35].Lv YP, Shi W, Liu HX, et al. Identification of miR-146b-5p in tissues as a novel biomarker for prognosis of gallbladder carcinoma. Eur Rev Med Pharmacol Sci 2017;21:518–22. [PubMed] [Google Scholar]
- [36].Li J, Shan F, Xiong G, et al. Transcriptional regulation of miR-146b by C/EBPbeta LAP2 in esophageal cancer cells. Biochem Biophys Res Commun 2014;446:267–71. [DOI] [PubMed] [Google Scholar]
- [37].Qiu W, Yang Z, Fan Y, et al. ZNRF3 is downregulated in papillary thyroid carcinoma and suppresses the proliferation and invasion of papillary thyroid cancer cells. Tumour Biol 2016;37:12665–72. [DOI] [PubMed] [Google Scholar]
- [38].Lin F, Wang X, Jie Z, et al. Inhibitory effects of miR-146b-5p on cell migration and invasion of pancreatic cancer by targeting MMP16. J Huazhong Univ Sci Technolog Med Sci 2011;31:509–14. [DOI] [PubMed] [Google Scholar]


