Supplemental Digital Content is available in the text
Keywords: fragile histidine triad, genome-wide association study, maximal voluntary ventilation, single nucleotide variant, spirometry parameter
Abstract
Genome-wide association studies (GWAS) for spirometry parameters have been limited to forced vital capacity (FVC), forced expiratory volume in 1 second (FEV1), and their ratio. This study examined to identify genetic variants associated with maximal voluntary ventilation (MVV), an important spirometry parameter presenting inspiratory muscle strength.
A total of 8842 Korean subjects participated in the Korean Association REsource Consortium were used to identify nucleotide variants associated with MVV and other spirometry parameters through a GWAS. Genetic associations were determined by employing a mixed model that can control background polygenic effects.
The analysis revealed 3 nucleotide variants associated with MVV (P < 5 × 10−8). One (rs1496255) was also associated with FVC and FEV1. The other 2 variants were identified only for MVV and located in the genes of LOC102724340 (rs41434646) and FHIT (rs9833533). In particular, FHIT represses transcriptional activity of β-catenin, a critical protein for growth of skeletal muscle, and thus might have influenced the level of MVV.
The current study revealed 2 novel nucleotide variants as genetic association signals for MVV. The association signals were suggested specific for neuromuscular diseases with a restrictive ventilatory impairment. Further studies are required to understand underlying mechanisms for their influence to restrictive lung diseases.
1. Introduction
Parameters in spirometry reflect physiological state of lung functions and predict many lung diseases that greatly contribute to morbidity and mortality.[1] There have been great efforts to understand genetics of the spirometry parameters since spirometry parameters have been shown large heritability; 0.85 for forced expiratory volume at timed intervals of 1 second (FEV1), 0.91 for forced vital capacity (FVC), and 0.45 for the ratio of FEV1 to FVC (FEV1/FVC).[2–4] Recently, genetic factors were identified by genome-wide association studies (GWAS). For example, meta-analyses with Europeans participated in the SpiroMeta and CHARGE consortia showed genetic associations of 27 nucleotide variants with FEV1, FVC, or FEV1/FVC.[5,6] These genetic studies limited to the parameters of FVC, FEV1, and FEV1/FVC, which are used to distinguish restrictive and obstructive lung diseases. Additional parameters are required to determine more specific diseases. The FEF25–75, mean forced expiratory flow of FVC ranged from 25% to 75%, can help differentiate small airway diseases from obstructive lung diseases.[7,8] Maximal voluntary ventilation (MVV) shows inspiratory muscle strength as the maximum volume of air inspired and expired over a specified period of time.[8–11] Thus, this parameter is sensitive to ventilatory muscle strength, reflecting volume change and airway resistance. In particular, large values of MVV have been associated with increased risk of preoperative complications and postoperative mortality for a variety of surgeries (e.g., surgery for cervical spondylotic myelopathy, abdominal surgery, and thoracic surgery).[12–14]
Genetic studies on these additional parameters have been hardly found. However, their genetic factors should be independently investigated because of genetic heterogeneity among spirometry parameters, for example, genetic correlation between basic parameters and supportive parameters ranged from 0.42 to 0.70 in a previous twin study.[15] Identifying genetic factors for MVV may be helpful to understand pathological mechanisms different from those of FEV1 and FVC. The objective of this study was to identify genetic variants associated with MVV and FEF25–75 in a Korean population.
2. Materials and methods
2.1. Subjects and genotypes
This GWAS used subjects recruited by the Korean Association REsource (KARE) Consortium. They were collected on the basis of cohorts in Ansan and Ansung, Gyeonggi-do, Korea.[16] Ansan is an urban area, and Ansung is a rural area. Ethical approval was obtained from the institutional review board of the Korea National Institute of Health, and all participants provided written informed consent. Their genotypes were obtained using the Affymetrix Genome-Wide Human SNP Array 5.0 (Affymetrix, Inc., Santa Clara, CA) and the algorithm of Bayesian robust linear modeling using Mahalanobis distance (BRLMM).[16,17] Quality assurance filtering was conducted to exclude subjects with genotype call rate <95%, sex inconsistency, or cryptic relatedness (identical by state value >0.80), and 8842 subjects were available for association analysis. We filtered out nucleotide variants with genotype call rate <0.95, Hardy-Weinberg disequilibrium (HWE; P < 1 × 10–6), or minor allele frequency (MAF) <0.01, and as a result, 352,228 nucleotide variants were remained.
Genotypes were also imputed with the Japanese and Chinese HapMap phase 2 haplotype panel (release 23) using IMPUTE software program (version 2, http://mathgen.stats.ox.ac.uk/impute). After removing nucleotide variants with MAF < 0.01 or genotype call rate <0.95, there were 2,124,148 imputed variants (r2 ≥ 0.3). A total of 2,476,376 nucleotide variants were analyzed in this study.
2.2. Spirometry
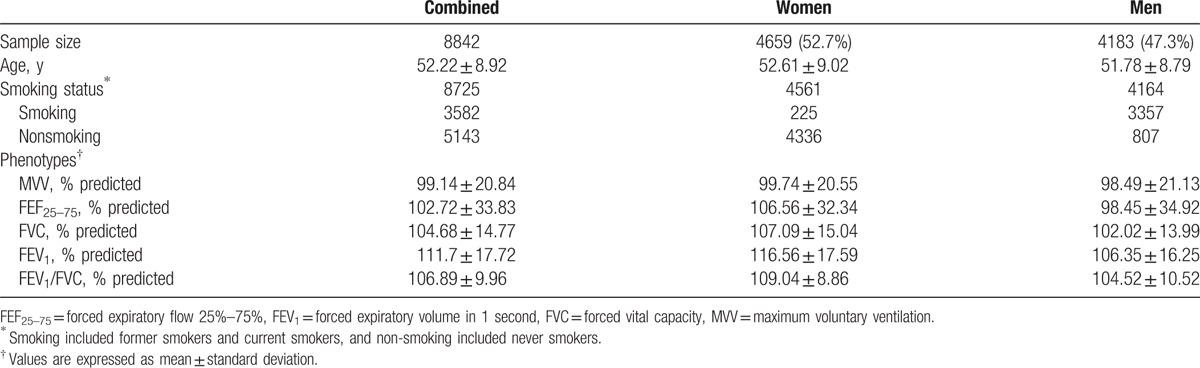
Five spirometry indices for lung function were used as phenotypes in this study. FVC was measured in liters for the maximum amount of air forcibly exhaled from a maximal inspiration. FEV1 was measured also in liters as the maximum amount of air exhaled for one second of a forced expiration from a full inspiration. FEV1/FVC was also used as an index variable. FEF 25–75 was also measured as the average forced expiratory flow during the mid-portion (25 ∼ 75%) of the FVC. MVV was measured in liters per minute as the maximum volume of air exhaled for 12 seconds.[11] Entire procedures for measuring parameters were explained to subjects, and the parameters were carefully measured under the guidance of technicians. Subjects breathed as fast and as deeply as possible. All the measurements were repeated at least 3 times to obtain more reliable values. The variables were all transferred into the ratio of actual values divided by the corresponding normal values predicted by age, sex, and height, and the standardized values were analyzed as phenotypes in the present study. Means of the 5 indices for lung function are presented in Table 1.
Table 1.
Baseline characteristics of the study subjects.
2.3. Genetic association analysis
Genetic association analysis was performed employing a mixed model with random polygenic effects to avoid population stratification.[18] Leaving-one-chromosome-out approach was used to avoid underestimating genetic association.[19]
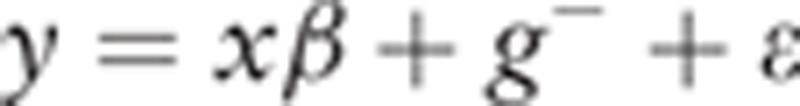 |
where y is the vector of spirometry parameters; β is the vector of fixed effects for region, sex, smoking status, and the candidate nucleotide variant; and x is the design matrix for β. For the candidate nucleotide variant effect, elements of x are 0, 1, and 2 for the homozygote of the minor allele, heterozygote, and homozygote of the major allele, respectively. g− is the vector of random polygenic effects explained by the genome except for the chromosome housing the candidate nucleotide variant 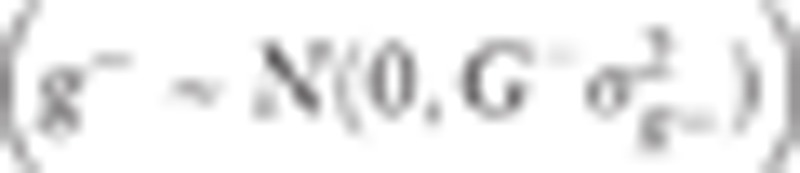 , where G− is the genomic relationship matrix (GRM), and
, where G− is the genomic relationship matrix (GRM), and 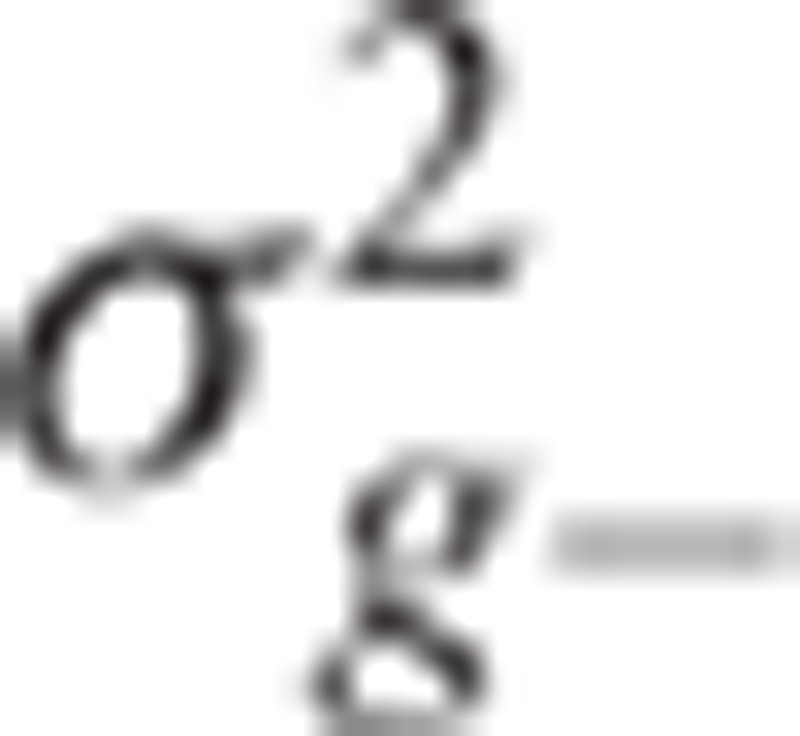 is the polygenic variance component. This polygenic variance component should be re-estimated whenever the specific chromosome excluded from the calculation of GRM is changed. Elements of the GRM are coefficients of pairwise genetic relationship coefficients. The genetic relationship coefficient between 2 individuals was calculated using genotypes of variants in linkage equilibrium (r2 < 0.8) as following:
is the polygenic variance component. This polygenic variance component should be re-estimated whenever the specific chromosome excluded from the calculation of GRM is changed. Elements of the GRM are coefficients of pairwise genetic relationship coefficients. The genetic relationship coefficient between 2 individuals was calculated using genotypes of variants in linkage equilibrium (r2 < 0.8) as following:
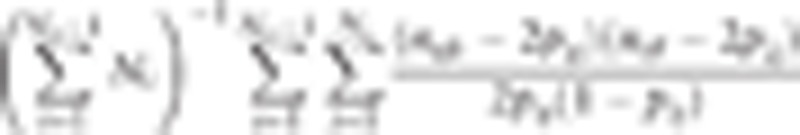 |
where Nc is the number of chromosomes, Ni is the number of variants in the ith chromosome, pij is the frequency of the minor allele at the jth variant in the ith chromosome, and nijk (nijl) is the number (0, 1, or 2) of the minor allele at the jth variant in the ith chromosome for the kth (lth) individual. ε is the vector of random residuals 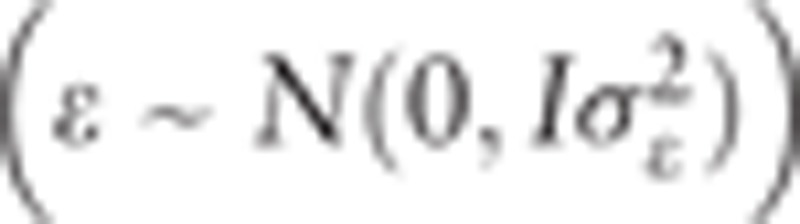 , where
, where 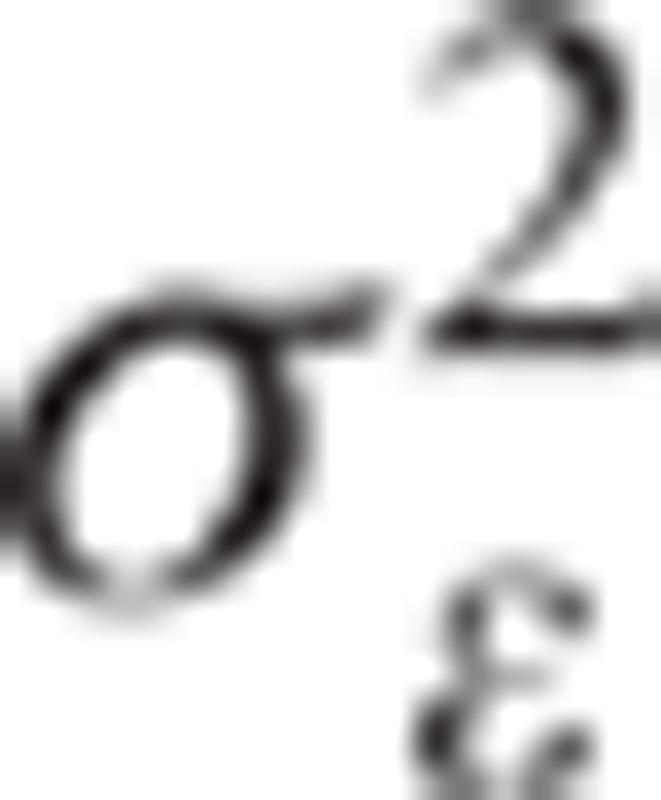 is the residual variance component, and I is the identity matrix. To solve candidate nucleotide variant effect, the polygenic and residual variance components were estimated using restricted maximum likelihood (REML). The variance components were first estimated by EM-REML, and then the EM-REML estimates were used as initial values to obtain their AI-REML estimates. The fixed variant effect was then estimated with the variance component estimates under the mixed model equations. The statistical analysis was conducted using the Genome-wide Complex Trait Analysis (GCTA) freeware.[20] Multiple testing was applied to the genetic association analyses using the significance threshold of P = 5 × 10−8.
is the residual variance component, and I is the identity matrix. To solve candidate nucleotide variant effect, the polygenic and residual variance components were estimated using restricted maximum likelihood (REML). The variance components were first estimated by EM-REML, and then the EM-REML estimates were used as initial values to obtain their AI-REML estimates. The fixed variant effect was then estimated with the variance component estimates under the mixed model equations. The statistical analysis was conducted using the Genome-wide Complex Trait Analysis (GCTA) freeware.[20] Multiple testing was applied to the genetic association analyses using the significance threshold of P = 5 × 10−8.
3. Results
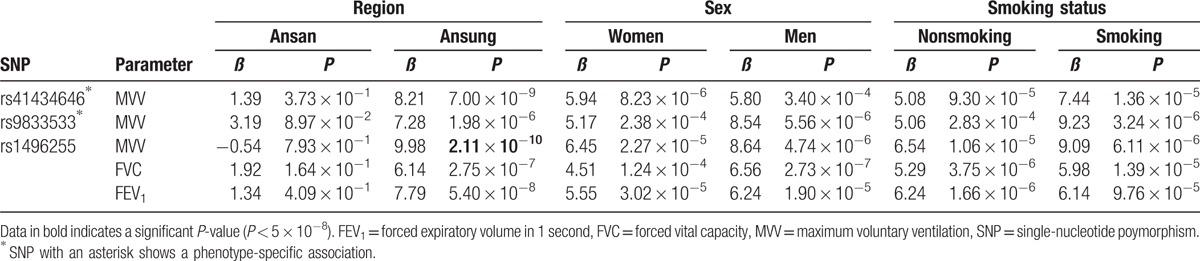
The genome-wide association analysis for spirometry parameters revealed 3 association signals for MVV (P < 5 × 10−8), but none for FEF25–75 (P > 5 × 10−8; Table 2, Figs. 1 and 2). In particular, 2 nucleotide variants (rs41434646 and rs9833533) were significantly associated only with MVV, whereas rs1496255 was also associated with FVC and FEV1.
Table 2.
Associations of SNPs with spirometry parameters∗.
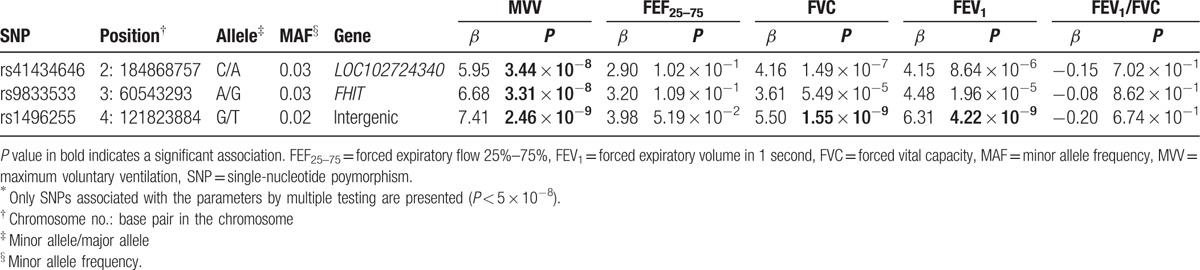
Figure 1.
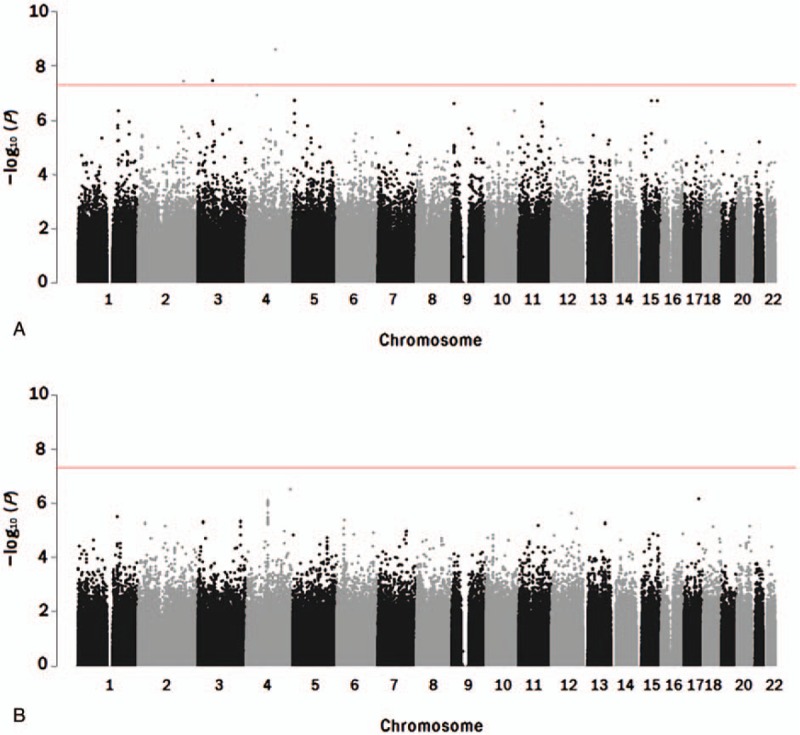
Manhattan plots of genome-wide association analysis for spirometry parameters (A: MVV; B: FEF25–75). The horizontal bar indicates the significance threshold (P = 5 × 10−8) for multiple testing.
Figure 2.
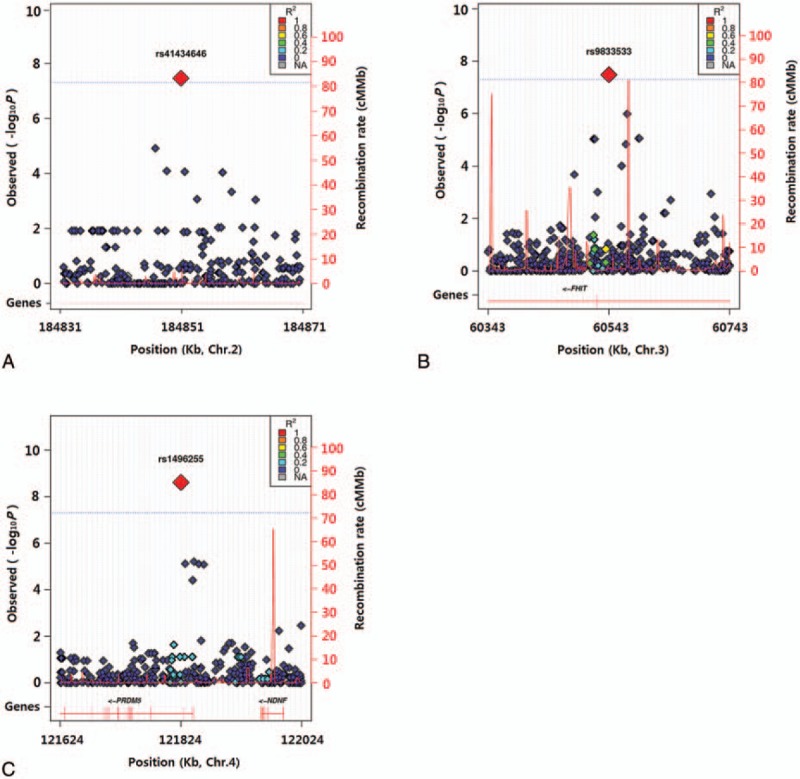
Regional plots for genetic associations of maximal voluntary ventilation with rs41434646 at 2q32.1 (A), rs9833533 at 3p14.2 (B), and rs1496255 at 4q27 (C). Linkage disequilibrium with rs41434646, rs9833533, or rs1496255 is presented in color based on r2.
Further association analysis using data partitioned by region, sex, or smoking status showed heterogeneity of the identified associations (Table 3). Although 2 associations were identified in Ansung (P < 5 × 10−8), none were observed in Ansan (P > 5 × 10−8). Any associations were not observed using data partitioned by sex or smoking status (P > 5 × 10−8).
Table 3.
Associations of single-nucleotide polymorphisms with spirometry parameters using data partitioned by region, sex, or smoking status.
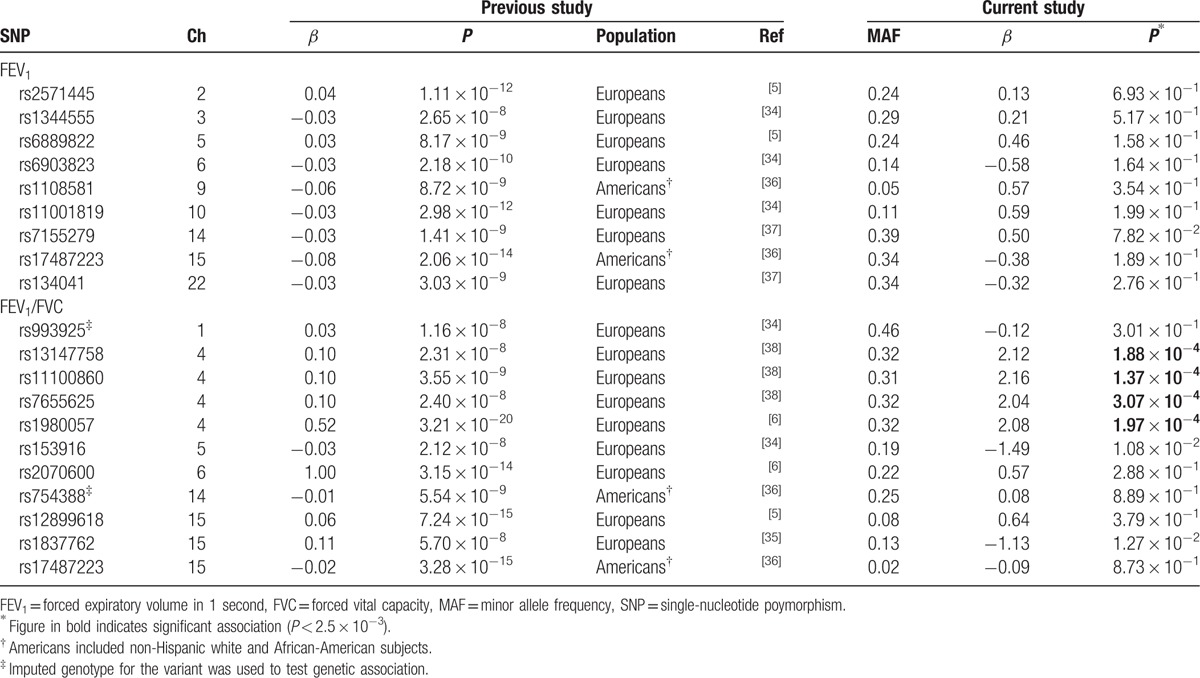
A replication analysis using nucleotide variants previously identified for spirometry parameters showed that associations of 4 nucleotide variants with FEV1/FVC were replicated (P < 2.5 × 10−3, Table 4). They were all intergenic nucleotide variants 81 ∼ 107Kb upstream of the gene encoding hedgehog interacting protein (HHIP) on chromosome 4, showing strong linkage disequilibrium (r2 > 0.95).
Table 4.
Association of nucleotide variants identified in previous studies with FEV1 and FEV1/FVC.
4. Discussion
The MVV had been utilized as the major spirometry parameter before FEV1 was demonstrated as a powerful prognostic indicator.[21] The measurement of MVV has been greatly decreased, and this made it hard to examine its genetic factors in association studies in which a large number of samples are required. Nevertheless, MVV can be an essential indicator of inspiratory airway obstruction and impaired neuromuscular function.[22] Lung volumes are reduced by the degree of respiratory muscle weakness, often yielding neuromuscular diseases with a restrictive ventilatory impairment.[23] Thus, a major airway lesion or a neuromuscular disorder can be suspected with a low MVV (<80%), although FEV1 is observed within the normal range.[8] Biological mechanisms and corresponding genetic factors affecting ventilator capacity might be different from those affecting FEV1.[9–11] The current study found 3 genetic variants associated with MVV, and 2 of them were identified only for MVV. One variant (rs41434646) is located in the uncharacterized gene of LOC102724340. The other (rs9833533) was an intronic nucleotide variant in the gene encoding fragile histidine triad (FHIT) at 3p14.2. We suspect that FHIT can influence the level of MVV. This might be supported by previous studies in which FHIT represses transcriptional activity of β-catenin that is essential for physiological growth of skeletal muscle.[24–26] Functional investigation of the association signal using the RegulomeDB (http://www.regulomedb.org) revealed that the rs9833533 was identified as a DNase peak and was bound by transcription factors such as CCCTC-binding factor (CTCF), SAM-pointed domain containing ETS transcription factor (SPDEF), and RAD21 cohesin complex component (RAD21). Previous studies showed that the transcription factors might be critical in muscle cells. CTCF may modulate myogenesis through regulating muscle-specific gene expression.[27] SPDEF represses β-catenin transcriptional activity.[28] RAD21 is important to CTCF-mediated chromatin interactions, and its displacement was observed with MyoD binding by disrupting chromatin loop.[29,30]
The present study also revealed the heterogeneity of some genetic associations by region, showing association signals in a rural area (Ansung), but not in an urban area (Ansan). This implied that the spirometry parameters were influenced by interaction effects between the genetic variants and the region-associated environments. Living in the urban area with a high population density may decrease lung functions and deteriorate respiratory system.[31,32] Nevertheless, the present study found gene-by-region interaction for the first time.
All the association signals identified using combined data were not significant (P > 5 × 10−8) using data partitioned by sex or smoking status. The identified signals might be contributed by both males and females and also by smoking and non-smoking. Statistical power decreased by partitioning data. Further studies with a larger sample size would help understand genetic effects by sex or smoking status.
The present study confirmed a spirometry parameter (FEV1/FVC) GWAS signal upstream of HHIP that is critical to airway epithelial repair as a regulator of the hedgehog signaling pathway.[33] Further association studies with a larger sample size of Koreans should be conducted to determine whether nonreplicated associations are caused by ethnic heterogeneity or by false-negative associations. Replicating the identified genetic associations with MVV in such independent studies is an essential step to overcome another limitation of the present study.
In this GWAS, efforts were made to avoid spurious genetic associations. Results of all the associations in this study were obtained after a series of quality controls as explained above. In particular, we could not find any outliers from a principal component analysis among the subjects included in the association analysis (Supplementary Figure S1). Furthermore, the mixed model employed in the present study further explained polygenic effects that were treated as errors in fixed model analysis.
The current GWAS identified 2 novel nucleotide variants associated with MVV. They were genetic associations with lung disease, which could not be identified from GWAS for other spirometry parameters. And they were specific for neuromuscular diseases with a restrictive ventilatory impairment. Further studies are in need to understand their underlying mechanism to affect susceptibility to restrictive lung diseases.
Supplementary Material
Footnotes
Abbreviations: FEF25–75 = forced expiratory flow 25–75%, FEV1 = forced expiratory volume in 1 second, FEV1/FVC ratio = ratio of FEV1 to FVC, FHIT = fragile histidine triad, FVC = forced vital capacity, GCTA = Genome-wide Complex Trait Analysis, GWAS = genome-wide association study, LD = linkage disequilibrium, MVV = maximal voluntary ventilation.
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
This study was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) founded by the Ministry of Education, Science, and Technology (Grant No. 2012002096). We thank the National Institute of Health in Korea for providing the genotypic and epidemiological data to the KARE Analysis Consortium.
The authors report no conflicts of interest.
Supplemental Digital Content is available for this article.
References
- [1].Ferkol T, Schraufnagel D. The global burden of respiratory disease. Ann Am Thorac Soc 2014;11:404–6. [DOI] [PubMed] [Google Scholar]
- [2].Gibson JB, Martin NG, Oakeshott JG, et al. Lung function in an Australian population: contributions of polygenic factors and the Pi locus to individual differences in lung function in a sample of twins. Ann Hum Biol 1983;10:547–56. [DOI] [PubMed] [Google Scholar]
- [3].Hubert HB, Fabsitz RR, Feinleib M, et al. Genetic and environmental influences on pulmonary function in adult twins. Am Rev Respir Dis 1982;125:409–15. [DOI] [PubMed] [Google Scholar]
- [4].Wilk JB, Djousse L, Arnett DK, et al. Evidence for major genes influencing pulmonary function in the NHLBI family heart study. Genet Epidemiol 2000;19:81–94. [DOI] [PubMed] [Google Scholar]
- [5].Repapi E, Sayers I, Wain LV, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet 2010;42:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hancock DB, Eijgelsheim M, Wilk JB, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet 2010;42:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sherter CB, Connolly JJ, Schilder DP. The significance of volume-adjusting the maximal midexpiratory flow in assessing the response to a bronchodilator drug. Chest 1978;73:568–71. [DOI] [PubMed] [Google Scholar]
- [8].Barreiro TJ, Perillo I. An approach to interpreting spirometry. Am Fam Physician 2004;69:1107–14. [PubMed] [Google Scholar]
- [9].Aldrich TK, Arora NS, Rochester DF. The influence of airway obstruction and respiratory muscle strength on maximal voluntary ventilation in lung disease. Am Rev Respir Dis 1982;126:195–9. [DOI] [PubMed] [Google Scholar]
- [10].Pellegrino R, Viegi G, Brusasco V, et al. Interpretative strategies for lung function tests. Eur Respir J 2005;26:948–68. [DOI] [PubMed] [Google Scholar]
- [11].Miller MR, Hankinson J, Brusasco V, et al. Standardisation of spirometry. Eur Respir J 2005;26:319–38. [DOI] [PubMed] [Google Scholar]
- [12].Datta D, Lahiri B. Preoperative evaluation of patients undergoing lung resection surgery. Chest 2003;123:2096–103. [DOI] [PubMed] [Google Scholar]
- [13].Nomura T, Tani T, Ikeuchi M, et al. Maximum voluntary ventilation as a sensitive measure to monitor the ventilatory function in cervical spondylotic myelopathy. Spinal Cord 2012;50:328–32. [DOI] [PubMed] [Google Scholar]
- [14].Mottram CD. Ruppels's Manual of pulmonary function testing. Elsevier 2013;65–7. [Google Scholar]
- [15].Vasilopoulos T, Grant MD, Franz CE, et al. Shared and distinct genetic influences among different measures of pulmonary function. Behav Genet 2013;43:141–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cho YS, Go MJ, Kim YJ, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet 2009;41:527–34. [DOI] [PubMed] [Google Scholar]
- [17].Rabbee N, Speed TP. A genotype calling algorithm for affymetrix SNP arrays. Bioinformatics 2006;22:7–12. [DOI] [PubMed] [Google Scholar]
- [18].Shin J, Lee C. A mixed model reduces spurious genetic associations produced by population stratification in genome-wide association studies. Genomics 2015;105:191–6. [DOI] [PubMed] [Google Scholar]
- [19].Lippert C, Listgarten J, Liu Y, et al. FaST linear mixed models for genome-wide association studies. Nat Methods 2011;8:833–5. [DOI] [PubMed] [Google Scholar]
- [20].Yang J, Lee SH, Goddard ME, et al. GCTA: a tool for genome- wide complex trait analysis. Am J Hum Genet 2011;88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kannel WB, Lew EA, Hubert HB, et al. The value of measuring vital capacity for prognostic purposes. Trans Assoc Life Insure Med Dir Am 1980;64:66–83. [PubMed] [Google Scholar]
- [22].García-Pachón E, Casan P, Sanchis J. Indices of upper airway obstruction in patients with simultaneous chronic airflow limitation. Respiration 1994;61:121–5. [DOI] [PubMed] [Google Scholar]
- [23].De Troyer A, Borenstein S, Cordier R. Analysis of lung volume restriction in patients with respiratory muscle weakness. Thorax 1980;35:603–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Weiske J, Albring KF, Huber O. The tumor suppressor Fhit acts as a repressor of beta-catenin transcriptional activity. Proc Natl Acad Sci USA 2007;104:20344–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Huber O, Weiske J. Beta-catenin takes a HIT. Cell Cycle 2008;7:1326–31. [DOI] [PubMed] [Google Scholar]
- [26].Armstrong DD, Wong VL, Esser KA. Expression of beta-catenin is necessary for physiological growth of adult skeletal muscle. Am J Physiol Cell Physiol 2006;291:C185–188. [DOI] [PubMed] [Google Scholar]
- [27].Delgado-Olguín P, Brand-Arzamendi K, Scott IC, et al. CTCF promotes muscle differentiation by modulating the activity of myogenic regulatory factors. J Biol Chem 2011;286:12483–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Noah TK, Lo YH, Price A, et al. SPDEF functions as a colorectal tumor suppressor by inhibiting (-catenin activity. Gastroenterology 2013;144:1012–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dorsett D. Cohesin: genomic insights into controlling gene transcription and development. Curr Opin Genet Dev 2011;21:199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Battistelli C, Busanello A, Maione R. Functional interplay between MyoD and CTCF in regulating long-range chromatin interactions during differentiation. J Cell Sci 2014;127:3757–67. [DOI] [PubMed] [Google Scholar]
- [31].Zijlema WL, Klijs B, Stolk RP, et al. (Un)Healthy in the City: Respiratory, Cardiometabolic and Mental Health Associated with Urbanity. PLoS One 2015;10:e0143910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bröms K, Norbäck D, Eriksson M, et al. Effect of degree of urbanisation on age and sex-specific asthma prevalence in Swedish preschool children. BMC Public Health 2009;9:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Young RP, Whittington CF, Hopkins RJ, et al. Chromosome 4q31 locus in COPD is also associated with lung cancer. Eur Respir J 2010;36:1375–82. [DOI] [PubMed] [Google Scholar]
- [34].Soler Artigas M, Loth DW, Wain LV, et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet 2011;43:1082–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yao TC, Du G, Han L, et al. Genome-wide association study of lung function phenotypes in a founder population. J Allergy Clin Immunol 2014;133:248–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lutz SM, Cho MH, Young K, et al. A genome-wide association study identifies risk loci for spirometric measures among smokers of European and African ancestry. BMC Genet 2015;16:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Soler Artigas M, Wain LV, Miller S, et al. Sixteen new lung function signals identified through 1000 Genomes Project reference panel imputation. Nat Commun 2015;6:8658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wilk JB, Chen TH, Gottlieb DJ, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet 2009;5:e1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


