Abstract
Introduction
It has been proposed that individual genetic predisposition may contribute to persistent apical periodontitis. Cytokines are associated with levels of inflammation and are involved in caries, pulpal, and periapical tissue destruction. We hypothesized that polymorphisms in cytokine genes may contribute to an individual’s increased susceptibility to apical tissue destruction in response to deep carious lesions.
Methods
Subjects with deep carious lesions, with or without periapical lesions (≥ 3 mm) were recruited at the University of Pittsburgh and the University of Texas at Houston. Genomic DNA samples of 316 patients were sorted into 2 groups: 136 cases with deep carious lesions and periapical lesions (cases), and 180 cases with deep carious lesions but no periapical lesions (controls). Nine single nucleotide polymorphisms in IL1B, IL6, TNF, RANK, RANKL and OPG genes were selected for genotyping. Genotypes were generated by endpoint analysis using Taqman chemistry in a real-time polymerase chain reaction instrument. Allele and genotype frequencies were compared among cases and controls using PLINK program. Ninety-three human periapical granulomas and 24 healthy periodontal ligament tissues collected post-operatively were used for mRNA expression analyses of IL1B.
Results
A SNP in IL1B (rs1143643) showed allelic (P=0.02) and genotypic (P=0.004) association with cases of deep caries and periapical lesions. We also observed altered transmission of IL1B marker haplotypes (P = 0.02) in these individuals. IL1B was highly expressed in granulomas (P<0.001).
Conclusions
Variations in IL1B may be associated with periapical lesion formation in individuals with untreated deep carious lesions. Future studies could help predict host susceptibility to developing periapical lesions.
Keywords: Apical periodontitis, cytokines, genetic polymorphisms
INTRODUCTION
Periapical inflammation occurs as a consequence of various insults to the dental pulp, including infection, physical and iatrogenic trauma (1). It is viewed as a dynamic encounter between microbial factors and host defenses at the interface between infected radicular pulp and periodontal ligament that results in local inflammation, resorption of hard tissues, destruction of other periapical tissues, and eventual formation of various histopathological categories of apical periodontitis, commonly referred to as periapical lesions (2). A network of stimulatory and inhibitory factors may influence the intensity of the defense and inflammatory responses and the balance between bone resorption and regeneration, resulting in lesion expansion or healing of apical periodontitis (3, 4).
In addition to local factors, genetic predisposition has been suggested as a differential etiologic factor for apical periodontitis development (5, 6, 7, 8, 9). Moreover, genetic susceptibility may influence host response to endodontic infection (9)
Cytokines play a major role in inflammatory and immune responses within the bone microenvironment. The balance between pro- and anti-inflammatory mediators determines the outcome of resorption in bone destructive diseases, including periapical granulomas (4, 10). The recognition of microbial structures such as lipopolysaccharides from Gram-negative bacteria through Toll-like receptors (TLRs) triggers intracellular signaling pathways that culminate in an inflammatory cytokine response and bone lesions (11). Previous studies have demonstrated that cytokines such as members of the TNF super family and interleukins are established agents in the pathogenesis of chronic inflammatory diseases including pulpitis and apical periodontitis (12, 13, 14, 15).
Here, we hypothesized that polymorphisms in cytokine genes may contribute to an individual’s increased susceptibility to apical tissue destruction in response to deep carious lesions. Hence, combined bacterial/host genotyping may provide an important tool in defining disease risk and targeting bacteria eradication to high-risk individuals
MATERIAL AND METHODS
Sample Population
Two different patient sources were used in this study. The first source was obtained from the University of Pittsburgh Dental Registry and DNA Repository (DRDR), which gathers clinical information and DNA samples from saliva from patients seeking treatment at the School of Dental Medicine and that agree to participate in the registry. Data is extracted from electronic patient records and linked to the DNA samples for genetic/clinical research and provided to investiagtors in deidenitifed format upon approved protocols.
The second source of patient samples was the Endodontic clinic at the University of Texas Health Science Center School of Dentistry at Houston. Patients were invited to participate if they met the selection criteria, and signed an informed consent document prior to data and sample collection. Both sources of patients derive from ongoing projects and new samples are collected routinely. For this study, samples were collected over a period of 5 years.
This study was approved by the University of Pittsburgh and the University of Texas Institutional Review Boards. All participants signed an informed consent and provided a saliva sample as source of genomic DNA.
For both datasets, individuals were selected based on their radiographic records showing deep carious lesions, involving at least 2/3 of the dentin depth, and periapical lesions ≥3 mm in diameter (cases), and individuals showing deep carious lesions but no periapical lesions (controls). All individuals selected for this study had thermal and electric pulpal vitality tests performed. Individuals presenting pulp necrosis and apical periodontitis were included In the ‘case’ group; individuals presenting vital pulps and no apical periodontitis were included as controls. Patients with any systemic conditions such as diabetes or other hormonal alterations that are related to exacerbated or uncontrolled inflammatory responses, patients with medical conditions requiring the use of systemic modifiers of bone metabolism or other assited drug therapy (i.e., systemic antibiotics, anti-inflammatory, hormonal therapy) during the lat 6 months before the study were excluded. At last, our sample population consisted of 316 white individuals with deep carious lesions: 136 case individuals (61 males, 75 females, average age 55 ± 3 SD) with deep carious lesions but no periapical lesions, and 180 control individuals (83 males, 97 females, average age 58 ± 8 SD) with deep carious lesions and periapical lesions.
Selection of Candidate Genes and Single Nucleotide Polymorphisms
We selected 9 single nucleotide polymorphisms spanning the IL1B, IL6, TNF, RANK, RANKL and OPG genes. Some of the SNPs were selected based on published reports [refs] and/or their locations within the genes. Additional SNPs were selected based on their likelihood to have functional consequences (i.e., located in the promoters, exons, or near exon/intron boundaries), or considered tag-SNPs as surrogates for the linkage disequilibrium blocks surrounding the candidate gene. We used information available at the NCBI dbSNP (http://www.ncbi.nlm.gov/SNP/) and HapMap Project (http://www.hapmap.org) databases to select polymorphisms. Details of studied genes and polymorphisms are presented in Table 1.
Table 1.
Association results for the SNPs investigated.
| Gene | Chr. | SNP Id. | Base Pair Position* | Alleles# | SNP function | MAF (cases) | MAF (controls) | P-value§ |
|---|---|---|---|---|---|---|---|---|
| IL10 | 1 | rs5743626 | 206944285 | C/T | Silent Mutation | 0.27 | 0.29 | 0.87 |
| IL1B | 2 | rs1143643 | 113588302 | C/T | Intron | 0.25 | 0.20 | 0.02 |
| IL1B | 2 | rs1143634 | 113590390 | C/T | Silent Mutation | 0.27 | 0.30 | 0.33 |
| IL1B | 2 | rs16062 | 113591081 | C/T | Silent Mutation | 0.43 | 0.44 | 0.51 |
| TNF | 6 | rs1800629 | 31543031 | A/G | Intron | 0.14 | 0.14 | 0.92 |
| IL6 | 7 | rs2069830 | 22767137 | C/T | Missense Mutation | 0.48 | 0.48 | 0.99 |
| OPG | 8 | rs1131380 | 119938762 | A/C | Missense Mutation | 0.25 | 0.27 | 0.18 |
| RANKL | 13 | rs12721445 | 43180881 | C/T | Missense Mutation | 0.46 | 0.45 | 0.93 |
| RANK | 18 | rs35589394 | 59992591 | C/G | Silent Mutation | 0.49 | 0.50 | 0.76 |
Chr. = Chromosome
according to NCBI GRCh37.p10 assembly
ancestral allele in bold
MAF = minor allele frequency
Chi-square test, significant if P≤0.05
Genotyping
Genomic DNA was extracted from saliva using established protocols. Genotypes were generated using Taqman chemistry (16). Reactions were carried out in 5-μL volumes in a ViiA7 Sequence Detection System (Applied Biosystems, Foster City, CA). Assays and reagents were supplied by Applied Biosystems. The results were analyzed using EDS software (Applied Biosystems, Foster City, CA). In order to ensure quality control of genotyping reactions, we used a non-template control (water instead of DNA) as negative control and a DNA sample of known genotype as positive control.
Association analyses
Allele and genotype frequencies for each polymorphism were compared among cases and controls using PLINK software version 1.06 (17). We used Bonferroni correction as implemented in PLINK to adjust for multiple testing. Haplotype analyses were performed for IL1B polymorphisms as implemented in PLINK.
Gene expression analyses
Ninety-three human periapical granulomas and 24 healthy periodontal ligament tissues collected post-operatively were used for mRNA expression analyses of IL1B in a quantitative real-time reverse transcription polymerase chain reaction. Periapical granulomas were collected from teeth referred to periapical surgery due to a failed endodontic treatment. Healthy periodontal ligament tissues were collected post-operatively from extracted teeth due to orthodontic reasons and used as controls in this study. All procedures were performed under IRB-approved protocols.
Total RNA was extracted from the tissue samples using TRIZOL (Life Technologies, Grand Island, NY), following manufacturer’s instructions. Next, the RNA pellet was dried under a vacuum and re-suspended in 50 mL of diethyl pyrocarbonate (DEPC)-treated water. The integrity of RNA samples was checked by analyzing 1μg of total RNA on a 1.2% (w/v) denaturing formaldehyde-agarose gel. After RNA extraction, complementary DNA was synthesized using 3 ug of RNA through a reverse transcription reaction using SuperScript III Reverse Trancriptase (Invitrogen, Carlsbad, CA). mRNA levels were measured using TaqMan gene expression assays (Invitrogen, Carlsbad, CA in a real time polymerase chain reaction in a Viia7 instrument (LifeTechnologies, Carlsbad, CA). 18S and GAPDH were used as endogenous reference genes for normalization
We also investigated the mRNA expression of RANKL and OPG in the periapical granulomas, in order to classify them as active/progressive or inactive/stable. The definition of active and passive was based on the expression of RANKL and OPG in each lesion (for details see Menezes et al., 2008). In brief, lesions showing higher RANKL:OPG ratio were categorized as active whereas lesions showing higher OPG:RANKL ratio were categorized as inactive (18). Experiments were performed in triplicates and repeated once.
Gene Expression Data Analysis
The mean cycle threshold (Ct) values from triplicate measurements of the target gene with normalization to the endogenous reference genes were calculated using the delta delta Ct method (19). Statistical analyses included analysis of variance (ANOVA), followed by Bonferroni correction in GraphPad Prism 6.0 (GraphPad Inc., San Diego, CA). A P-value ≤ 0.05 was considered statistically significant.
RESULTS
There was no evidence of deviation from Hardy-Weinberg equilibrium for any of the investigated polymorphisms between the groups (data not shown). Hardy-Weinberg Equilibrium (HWE) is used to describe the genotype distribution of a population when it is large, self-contained, and randomly mating. Testing for HWE in the control group is commonly used to detect genotyping errors in genetic association studies (20).
The results of the association analysis in the study groups are presented in Table 1. We assessed genotype and allele associations between the investigated genes and individuals affected with deep carious lesions and periapical disease. A SNP in IL1B (rs1143643) showed allelic [P=0.02; OR: 1.56 (1.07 – 2.28)] and genotypic (TT, P=0.004) association in the individuals with deep caries and periapical lesions. Haplotype analyses showed altered transmission of IL1B marker alleles in cases with deep carious lesions with periapical disease; the combination of T-A-G alleles for SNPs rs1143643-rs1143634-rs16062 were overrepresented in cases with deep carious lesions with periapical disease (P=0.02) (Table 2). No association was found with the remaining genes.
Table 2.
Results of haplotype analysis for IL1B SNPs in the studied groups.
| IL1 SNPs | HAPLOTYPE | Frequency cases | Frequency controls | P-value§ |
|---|---|---|---|---|
| rs1143643|rs1143634|rs16062 | TAA | 0.01 | 0.02 | 0.49 |
| rs1143643|rs1143634|rs16062 | TGG | 0.26 | 0.30 | 0.32 |
| rs1143643|rs1143634|rs16062 | TAG | 0.27 | 0.19 | 0.02 |
| rs1143643|rs1143634|rs16062 | CAG | 0.45 | 0.48 | 0.41 |
P≤0.05 indicates statistical difference (in bold).
IL1B mRNA expression was significantly higher in the periapical granulomas when compared to healthy periodontal ligaments samples (P<0.001). A markedly increased IL1B expression was also observed in the active lesions (RANKL>OPG) (P=0.01) (Figure 1).
Figure 1.
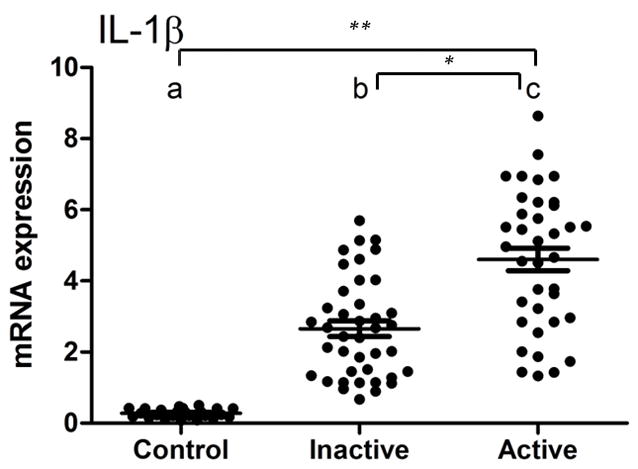
IL1B mRNA expression in active and inactive periapical granulomas in comparison to healthy periodontal ligament. IL1B expression was significantly higher in periapical granulomas when compared to control tissues (**P<0.001). A markedly increased IL1B expression was also observed in the active lesions (RANKL>OPG)(*P=0.01).
DISCUSSION
The present study examined the role of cytokine gene polymorphisms in individuals presenting deep caries with regards to an increased susceptibility to the development of apical periodontitis. Although it is widely accepted that microorganisms residing in the root canal system are the main cause of pulp necrosis and subsequent apical periodontitis (21), the observation that some individuals presenting with deep caries do not develop apical periodontitis is intriguing and warrants further investigation.
Recent evidence has suggested that individual genetic variance due to the presence of single nucleotide polymorphisms may play a role in apical periodontitis development (5, 6, 7, 8, 9). Cytokines play an important role in both physiological and pathological bone remodeling, as seen in common diseases such as osteoporosis, osteopetrosis, paget’s disease, rheumatoid arthritis and periodontal disease (22). Therefore cytokines are biologically relevant candidate genes for conditions characterized by defective bone remodeling. Indeed, single nucleotide polymorphisms in a variety of cytokines, including IL1B, IL6, IL10, TNFA, and in TLR4, an important component in the recognition and host response to lipopolysaccharide (LPS) from gram-negative bacteria, were associated with periodontal disease (23).
Primary endodontic contents can potentially stimulate macrophages to produce a wide variety of cytokines, which in turn establish different network interrelationships that are implicated in the development of endodontic diseases (24). IL-1B in particular, participates in the destruction of local tissues by stimulating bone resorption and collagenase production by fibroblasts (22). IL1B was previously associated with the pathogenesis of human periradicular lesions (14). In combination with TNF-α, IL-1B plays a role in the initiation and upregulation of the inflammatory response cascade in apical periodontitis (24). Polymorphisms in IL8 have been associated with higher risk of developing apical periodontitis although no association was found with polymorphisms in IL1B, IL12B and TNFA (5).
In the present study, we found significant association of IL1B in cases with deep caries and periapical lesions. Both allelic (allele T) and genotypic TT association was found between an intronic SNP in IL1B (rs1143643) in cases with deep caries and periapical lesions. This suggests that these individuals presenting one or two copies of the T allele in this IL1B SNP are at increased risk for developing periapical lesions. On the other hand, individuals that do not have the T allele might have a protective effect against periapical lesion development. Despite the fact that intronic SNPs are regarded as nonfunctional with unknown clinical significance, recent evidence has shown that mutations located in the introns of genes can interfere with splicing and cause aberrant spliced mRNA transcripts leading to non-functional proteins (25, 26).
The observation that specific IL1B allele haplotypes were overrepresented in individuals with deep caries and periapical lesions provides further support a likely role for IL1B in the pathogenesis of apical periodontitis, and may provide insights into future screening strategies and treatment planning options for endodontic patients. Our sample size of 136 cases and 180 controls was sufficient to overcome potential bias of associations occurring by chance, although may not directly extrapolate to the results in the general population, and additional studies are needed to confirm the association between IL1B and susceptibility to apical periodontitis development.
Our findings for the gene expression analyses provide additional evidence for the role of IL1B in the development of apical periodontitis. IL1B mRNA expression was significantly higher in human periapical granulomas compared to healthy periodontal ligament controls, suggesting an active role for IL1B in periapical disease tissues. This is in accordance with previous reports (14, 24). Furthermore, the increased expression of IL1B in lesions of active/progressive nature also highlights the importance of this particular cytokine in the development and/or progression of these human periapical lesions.
Overall, our results provide novel aspects for the involvement of IL1B in the development and progression of apical periodontitis in individuals with untreated deep carious lesions.
Acknowledgments
We are grateful for the participation of the many individuals in this study. This work was supported by NIH R01-DE18914 to ARV, FAPESP to GPG, and the American Association of Endodontists Foundation Research Grant to AL and RMS. AD is a second-year dental student and was supported by the UTSD Summer Research Program.
References
- 1.Stashenko P, Telles R, D’Souza R. Periapical inflammatory responses and their modulation. Crit Rev Oral Biol Med. 1998;9:498–521. doi: 10.1177/10454411980090040701. [DOI] [PubMed] [Google Scholar]
- 2.Nair PN. Pathogenesis of apical periodontitis and the causes of endodontic failures. Crit Rev Oral Biol Med. 2004;15:348–381. doi: 10.1177/154411130401500604. [DOI] [PubMed] [Google Scholar]
- 3.Márton IJ, Kiss C. Overlapping protective and destructive regulatory pathways in apical periodontitis. J Endod. 2014;40:155–163. doi: 10.1016/j.joen.2013.10.036. [DOI] [PubMed] [Google Scholar]
- 4.Graves DT, Oates T, Garlet GP. Review of osteoimmunology and the host response in endodontic and periodontal lesions. J Oral Microbiol. 2011;17:1–15. doi: 10.3402/jom.v3i0.5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amaya MP, Criado L, Blanco B, et al. Polymorphisms of pro-inflammatory cytokine genes and the risk for acute suppurative or chronic nonsuppurative apical periodontitis in a Colombian population. Int Endod J. 2013;46:71–78. doi: 10.1111/j.1365-2591.2012.02097.x. [DOI] [PubMed] [Google Scholar]
- 6.Rôças IN, Siqueira JF, Jr, Del Aguila CA, et al. Polymorphism of the CD14 and TLR4 genes and post-treatment apical periodontitis. J Endod. 2014;40:168–172. doi: 10.1016/j.joen.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Siqueira JF, Jr, Rocas IN, Provenzano JC, et al. Relationship between FCgamma receptor and interleukin-1 gene polymorphisms and post-treatment apical periodontitis. J Endod. 2009;35:1186–1192. doi: 10.1016/j.joen.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 8.Morsani JM, Aminoshariae A, Han YW, et al. Genetic predisposition to persistent apical periodontitis. J Endod. 2011;37:455–459. doi: 10.1016/j.joen.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menezes-Silva R, Khaliq S, Deeley K, et al. Genetic susceptibility to periapical disease: conditional contribution of MMP2 and MMP3 genes to the development of periapical lesions and healing response. J Endod. 2012;38:604–607. doi: 10.1016/j.joen.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva TA, Garlet GP, Fukada SY, et al. Chemokines in oral inflammatory diseases: apical periodontitis and periodontal disease. J Dent Res. 2007;86:309–319. doi: 10.1177/154405910708600403. [DOI] [PubMed] [Google Scholar]
- 11.Madianos PN, Bobetsis YA, Kinane DF. Generation of inflammatory stimuli: how bacteria set up inflammatory responses in the gingiva. J Clin Periodontol. 2005;32(suppl):57–71. doi: 10.1111/j.1600-051X.2005.00821.x. [DOI] [PubMed] [Google Scholar]
- 12.Safavi KE, Rossomando ER. Tumor necrosis factor identified in periapical tissue exudates of teeth with apical periodontitis. J Endod. 1991;17:12–14. doi: 10.1016/S0099-2399(07)80154-3. [DOI] [PubMed] [Google Scholar]
- 13.Menezes R, Bramante CM, da Silva Paiva KBS, et al. Receptor activator NFkB-ligand and osteoprotegerin protein expression in human periapical cysts and granulomas. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006;102:404–409. doi: 10.1016/j.tripleo.2005.10.054. [DOI] [PubMed] [Google Scholar]
- 14.Lim GC, Torabinejad M, Kettering J, Linkhardt TA, Finkelman RD. Interleukin 1-beta in symptomatic and asymptomatic human periradicular lesions. J Endod. 1994;20:225–227. doi: 10.1016/S0099-2399(06)80282-7. [DOI] [PubMed] [Google Scholar]
- 15.De Rossi A, Rocha LB, Rossi MA. Interferon-gamma, interleukin-10, Intercellular adhesion molecule-1, and chemokine receptor 5, but not interleukin-4, attenuate the development of periapical lesions. J Endod. 2008;34:31–38. doi: 10.1016/j.joen.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 16.Ranade K, Chang MS, Ting CT, et al. High-throughput genotyping with single nucleotide polymorphisms. Genome Res. 2001;11:1262–1268. doi: 10.1101/gr.157801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menezes R, Garlet TP, Letra A, et al. Differential patterns of receptor activator of nuclear factor kappa B ligand/osteoprotegerin expression in human periapical granulomas: possible association with progressive or stable nature of the lesions. J Endod. 2008;34:932–938. doi: 10.1016/j.joen.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Yu C, Zhang S, Zhou C, et al. A likelihood ratio test of population Hardy-Weinberg equilibrium for case-control studies. Genet Epidemiol. 2009;33:275–280. doi: 10.1002/gepi.20381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kakehashi S, Stanley HR, Fitzgerald RJ. The effects of surgical exposures of dental pulps in germ-free and conventional laboratory rats. Oral Surgery, Oral Medicine and Oral Pathology. 1965;20:340–349. doi: 10.1016/0030-4220(65)90166-0. [DOI] [PubMed] [Google Scholar]
- 22.Mundy GR. Cytokines and local factors which affect osteoclast function. Int J Cell Cloning. 1992;10:215–222. doi: 10.1002/stem.5530100404. [DOI] [PubMed] [Google Scholar]
- 23.Garlet GP, Trombone AP, Menezes R, et al. The use of chronic gingivitis as reference status increases the power and odds of periodontitis genetic studies: a proposal based in the exposure concept and clearer resistance and susceptibility phenotypes definition. J Clin Periodontol. 2012;39:323–332. doi: 10.1111/j.1600-051X.2012.01859.x. [DOI] [PubMed] [Google Scholar]
- 24.Martinho FC, Chiesa WM, Leite FR, et al. Correlation between clinical/radiographic features and inflammatory cytokine networks produced by macrophages stimulated with endodontic content. J Endod. 2012;38:740–745. doi: 10.1016/j.joen.2012.02.021. [DOI] [PubMed] [Google Scholar]
- 25.Petersen SM, Dandanell M, Rasmussen LJ, et al. Functional examination of MLH1, MSH2, and MSH6 intronic mutations identified in Danish colorectal cancer patients. BMC Med Genet. 2013;14:103. doi: 10.1186/1471-2350-14-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandell S, Schuit RJ, Bunyan DJ. An intronic polymorphic deletion in the PTEN gene: implications for molecular diagnostic testing. Br J Cancer. 2013;108:438–441. doi: 10.1038/bjc.2012.562. [DOI] [PMC free article] [PubMed] [Google Scholar]


