Abstract
Pharmacogenomics is the use of genomic and other “omic” information to individualize drug selection and drug use in order to avoid adverse drug reactions and to maximize drug efficacy. The science underlying pharmacogenomics has evolved rapidly over the 50 years since it was first suggested that genetics might influence drug response phenotypes. That process has occurred in parallel with advances in DNA sequencing and other molecular technologies, with striking increases in our understanding of the human genome. There are now many validated examples of the clinical utility of pharmacogenomics, and this type of clinical genomic information is increasingly being generated in clinical laboratories, incorporated into electronic health records (EHRs) and used to “tailor” or individualize drug therapy. This review will survey the origins and development of pharmacogenomics; it will address some of the challenges associated with the clinical implementation of pharmacogenomics; and it will attempt to foresee future advances in this important genomic discipline, one that almost certainly will be among the earliest and most widely adopted aspects of clinical genomics.
Keywords: Pharmacogenetics, Pharmacogenomics, Pharmaco-omics, Genomics, Adverse drug reactions, Drug efficacy, Precision medicine
INTRODUCTION
Pharmacogenomics is the study of the contribution of genomics and of other “omics” to individual variation in drug response phenotypes.1–3 That variation can range from inadequate therapeutic efficacy to serious, potentially life threatening adverse drug reactions. Pharmacogenomic information is increasingly being integrated into electronic health records (EHRs) and that information is rapidly becoming an important component of the “therapeutic encounter”. As a result, pharmacogenomics is the aspect of clinical genomics that will almost certainly see the earliest and broadest clinical implementation—with the potential to eventually touch the care of every patient everywhere. In subsequent paragraphs, we will briefly review the origins and development of this important aspect of “rational therapeutics”, touch briefly on the science underlying pharmacogenomics, address challenges associated with the clinical implementation of this aspect of genomic science and, finally, describe a vision for a future in which “pharmacogenomics” will have evolved into “pharmaco-omics” and will be an integral component of every medical drug-related therapeutic decision.
PHARMACOGENOMICS: ORIGINS AND DEVELOPMENT
The concept of “pharmacogenetics” was first put forward by the famed American geneticist Arno Moltulsky over a half century ago4 at a time when it was already becoming clear that variation in drug response could be due, at least in part, to the effects of genetic inheritance. Subsequently, a steady stream of reports appeared that described the contribution of genetics to variation in drug effect. Those examples most often involved pharmacokinetics (PK), i.e., factors that influence the concentration of drug that will ultimately reach the therapeutic target. Of those PK factors, most important was genetic variation in the expression and function of drug metabolizing enzymes, enzymes that can influence plasma drug concentrations. Included among early pioneers in pharmacogenomic research were Werner Kalow at the University of Toronto with his studies of butrylcholinesterase genetic variation and prolonged apnea after treatment with the muscle relaxant succinylcholine,5, 6 and David Price Evans at John’s Hopkins who pursued earlier reports7 of genetic variation in the N-acetylyation of the anti-tuberculosis drug isoniazid.8, 9 Subsequently, genetic variation in the thiopurine S-methyltransferase (TPMT) enzyme that was associated with potentially life-threatening myelosuppression after treatment with the antineoplastic and immunosuppressant agents mercaptopurine and azathioprine, substrates for metabolism by TPMT,10, 11 was reported, as was genetic variation in another drug metabolizing enzyme, cytochrome P-450 (CYP) 2D612, 13, that was associated with variation in plasma concentrations and the therapeutic effects of a variety of drugs.14, 15 These early examples were discovered at a time before genes were being cloned and sequenced, and they were most often shown to be genetic on the basis of family studies--using techniques similar in principal to those applied by Mendel in the preceding century when he performed his famous breeding studies with peas. However, primitive though they may appear when viewed from the perspective of the present “Post-Human Genome Project” world, these early examples have stood the test of time and are now well understood mechanistically. They also served as a powerful stimulus that set the stage for a series of systematic attempts to identify, study, and determine the clinical implications for drug response of genetic variation within whole families of genes encoding drug metabolizing enzymes, drug transporters and drug targets. Initially, the majority of those studies continued to focus on PK. One reason for that focus was the fact that variation in PK genes was often associated with relatively easily measured phenotypes, blood drug and drug metabolite concentrations, phenotypes that were often associated with variation in clinical drug response. It should be emphasized that even these early examples served to illustrate that pharmacogenetic variation could result in both serious adverse drug reactions and/or striking variation in drug efficacy. It should also be pointed out that drug metabolism may be required to “metabolically activate” a “prodrug”, a process that can also be significantly influenced by genetic variation in the biotransformation of the prodrug. We should also make it clear that metabolism is only one of many processes that can result in individual differences in drug response. For example, several severe adverse drug reactions can be predicted and prevented by knowledge of sequence variation involving HLA genes (http://www.allelefrequencies.net/hla-adr/default.asp).
During the late 1980s and throughout the 1990s—prior to completion of the Human Genome Project—pharmacogenomic research often involved cloning and sequencing genes encoding proteins that might contribute to variation in drug response phenotypes. Those studies demonstrated that much of the genetic variation in drug response that had been reported previously resulted from common sequence variation within or near genes that encoded the enzymes that had originally been studied as phenotypes, i.e., “high” or “low” levels of enzyme activity. The fact that pharmacogenomic variation was often due to common genetic polymorphisms will come up again subsequently when we cite evidence that every member of our species carries many potentially clinically actionable pharmacogenomic variants. The studies performed during the decade of the 1990s often focused on isoforms of the cytochrome P-450s (CYPs) as an extremely important family of drug metabolizing enzymes14, 15, but they also included other, so-called Phase II or conjugating drug metabolizing enzymes such as the sulfotransferases (SULTs),16, 17 methyltransferases (MTs) like TPMT18, 19 and UDP-glucuronosyltransferases (UGTs).20–22 Although research of that era predominately focused on gene cloning, gene sequencing and functional characterization of allozymes encoded by variant gene sequences, it was also during that time that we began to understand molecular mechanisms responsible for the functional effects of common genetic variation in those genes. For example, TPMT, as mentioned previously, catalyzes S-methylation, a reaction that inactivates thiopurine drugs such as azathioprine. The most common TPMT variant allele in European populations, TPMT*3A, with a minor allele frequency (MAF) of approximately 5%, resulting in one of every 300 European subjects being homozygous for this variant—is due to two nonsynonymous (ns) single nucleotide polymorphisms (SNPs) in the TPMT gene.23, 24 Those two changes in nucleotide sequence result in the alteration of two amino acids in the TPMT protein. The protein encoded by TPMT*3A is translated, but it misfolds and is rapidly degraded,25–27 with resultant lack of the enzyme and inability to metabolize thiopurines, leading to a ten-fold “overdose” of these cytotoxic agents when “standard” doses of thiopurines are administered to patients who are homozygous for TPMT*3A. Another example illustrating a different type of mechanism involved genetic variation in the TATA box in the promoter for the UGT1A1 gene that encodes a protein that metabolizes bilirubin as well as drugs such as the antineoplastic agent irinotecan. Most people have six “TA” elements in this TATA box, but some subjects have seven rather than six, and they display decreased expression of both UGT1A1 messenger RNA and protein—resulting in Gilbert’s Syndrome because of reduced bilirubin metabolism and, when they are exposed to irinotecan, an increased incidence of adverse drug reactions such as diarrhea and myelosuppression.21, 22 These examples and many others illustrate why, at the beginning of the 21st century, with completion of the Human Genome Project, the stage had been set to move forward quickly with both pharmacogenomic discovery and pharmacogenomic clinical implementation. In a display of the potential power of public-private collaboration, those efforts were enabled by and greatly stimulated in the United States by National Institutes of Health (NIH) funded Pharmacogenomics Research Network (PGRN) grants from by the National Institute of General Medical Sciences (NIGMS) and eMERGE grants from the National Human Genome Research Institute (NHGRI)—public funding that not only served to drive discovery by providing access to new and very expensive techniques such as genome-wide association studies (GWAS) and Next Generation Sequencing (NGS) but which also acted as a strong stimulus for the clinical implementation of pharmacogenomics. This brief overview should make it clear that pharmacogenomics as a discipline progressed in parallel with the maturation of genomic science during the latter decades of the twentieth century and the opening decades of the twenty first century.
The progress that has been made in identifying, characterizing and determining the clinical utility of pharmacogenomic variants since Motulsky first suggested that genetics might influence drug response is illustrated in striking fashion by the information listed in Table 1 which represents a modified version of the US FDA Pharmacogenomics website (https://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm) that lists drugs for which there are pharmacogenomic biomarkers with demonstrated clinical utility. It should be noted that Table 1 does not include drugs that are used primarily in oncology, but that information can also be found on the FDA website. The information listed in Table 1 also serves to emphasize that virtually every medical discipline is touched by pharmacogenomics, which immediately raises the question of how we can make this information available to practitioners in an easily accessible and usable fashion, topics addressed in subsequent paragraphs.
Table 1.
FDA Pharmacogenomic Biomarkers for Non-Cancer Therapeutics. (Modified from the FDA Website to remove antineoplastic agents).
| Drug | Therapeutic Area | Biomarker | |
|---|---|---|---|
| 1 | Abacavir | Infectious Diseases | HLA-B*57:01 |
| 2 | Amitriptyline | Psychiatry | CYP2D6 |
| 3 | Arformoterol | Pulmonary | UGT1A1, CYP2D6 |
| 4 | Aripiprazole | Psychiatry | CYP2D6 |
| 5 | Aripiprazole Lauroxil | Psychiatry | CYP2D6 |
| 6 | Atomoxetine | Psychiatry | CYP2D6 |
| 7 | Azathioprine | Rheumatology | TPMT |
| 8 | Boceprevir | Infectious Diseases | IFNL3 (IL28B) |
| 9 | Brexpiprazole | Psychiatry | CYP2D6 |
| 10 | Brivaracetam | Neurology | CYP2C19 |
| 11 | Carbamazepine | Neurology | HLA-B*15:02, HLA-A*31:01 |
| 12 | Carglumic Acid | Inborn Errors of Metabolism | NAGS |
| 13 | Carisoprodol | Rheumatology | CYP2C19 |
| 14 | Carvedilol | Cardiology | CYP2D6 |
| 15 | Celecoxib | Rheumatology | CYP2C9 |
| 16 | Cevimeline | Dental | CYP2D6 |
| 17 | Chloroquine | Infectious Diseases | G6PD |
| 18 | Chlorpropamide | Endocrinology | G6PD |
| 19 | Citalopram | Psychiatry | CYP2C19, CYP2D6 |
| 20 | Clobazam | Neurology | CYP2C19 |
| 21 | Clomipramine | Psychiatry | CYP2D6 |
| 22 | Clopidogrel | Cardiology | CYP2C19 |
| 23 | Clozapine | Psychiatry | CYP2D6 |
| 24 | Codeine | Anesthesiology | CYP2D6 |
| 25 | Daclatasvir | Infectious Diseases | IFNL3 (IL28B) |
| 26 | Dapsone | Dermatology, Infectious Diseases | G6PD |
| 27 | Darifenacin | Urology | CYP2D6 |
| 28 | Dasabuvir, Ombitasvir, Paritaprevir, and Ritonavir | Infectious Diseases | IFNL3 (IL28B) |
| 29 | Desipramine | Psychiatry | CYP2D6 |
| 30 | Dexlansoprazole | Gastroenterology | CYP2C19 |
| 31 | Dextromethorphan and Quinidine | Neurology | CYP2D6 |
| 32 | Diazepam | Neurology | CYP2C19 |
| 33 | Dolutegravir | Infectious Diseases | UGT1A1 |
| 34 | Doxepin | Psychiatry | CYP2D6, CYP2C19 |
| 35 | Dronabinol | Gastroenterology | CYP2C9 |
| 36 | Drospirenone and Ethinyl Estradiol | Gynecology | CYP2C19 |
| 37 | Duloxetine | Psychiatry | CYP2D6 |
| 38 | Efavirenz | Infectious Diseases | CYP2B6 |
| 39 | Elbasvir and Grazoprevir | Infectious Diseases | IFNL3 (IL28B) |
| 40 | Eliglustat | Inborn Errors of Metabolism | CYP2D6 |
| 41 | Elosulfase | Inborn Errors of Metabolism | GALNS |
| 42 | Eltrombopag | Hematology | F5 (Factor V Leiden), SERPINC1 (Antithrombin III) |
| 43 | Erythromycin and Sulfisoxazole | Infectious Diseases | G6PD |
| 44 | Escitalopram | Psychiatry | CYP2D6, CYP2C19 |
| 45 | Esomeprazole | Gastroenterology | CYP2C19 |
| 46 | Eteplirsen | Neurology | DMD |
| 47 | Fesoterodine | Urology | CYP2D6 |
| 48 | Flibanserin | Gynecology | CYP2C9, CYP2C19, CYP2D6 |
| 49 | Fluorouracil | Dermatology | DPYD |
| 50 | Fluoxetine | Psychiatry | CYP2D6 |
| 51 | Flurbiprofen | Rheumatology | CYP2C9 |
| 52 | Fluvoxamine | Psychiatry | CYP2D6 |
| 53 | Galantamine | Neurology | CYP2D6 |
| 54 | Glimepiride | Endocrinology | G6PD |
| 55 | Glipizide | Endocrinology | G6PD |
| 56 | Glyburide | Endocrinology | G6PD |
| 57 | Hydralazine | Cardiology | NAT1, NAT2 |
| 58 | Iloperidone | Psychiatry | CYP2D6 |
| 59 | Imipramine | Psychiatry | CYP2D6 |
| 60 | Indacaterol | Pulmonary | UGT1A1 |
| 61 | Isoniazid, Pyrazinamide, and Rifampin | Infectious Diseases | NAT1, NAT2 |
| 62 | Isosorbide Dinitrate | Cardiology | CYB5R1, CYB5R2, CYB5R3, CYB5R4 |
| 63 | Isosorbide Mononitrate | Cardiology | CYB5R1, CYB5R2, CYB5R3, CYB5R4 |
| 64 | Ivacaftor | Pulmonary | CFTR |
| 65 | Ivacaftor and Lumacaftor | Pulmonary | CFTR |
| 66 | Lacosamide | Neurology | CYP2C19 |
| 67 | Lansoprazole | Gastroenterology | CYP2C19 |
| 68 | Ledipasvir and Sofosbuvir | Infectious Diseases | IFNL3 (IL28B) |
| 69 | Lenalidomide | Hematology | del (5q) |
| 70 | Lesinurad | Rheumatology | CYP2C9 |
| 71 | Lidocaine and Prilocaine | Anesthesiology | G6PD |
| 72 | Mafenide | Infectious Diseases | G6PD |
| 73 | Methylene Blue | Hematology | G6PD |
| 74 | Metoclopramide | Gastroenterology | CYB5R1, CYB5R2, CYB5R3, CYB5R4, G6PD |
| 75 | Metoprolol | Cardiology | CYP2D6 |
| 76 | Modafinil | Psychiatry | CYP2D6 |
| 77 | Mycophenolic Acid | Transplantation | HPRT1 |
| 78 | Nalidixic Acid | Infectious Diseases | G6PD |
| 79 | Nebivolol | Cardiology | CYP2D6 |
| 80 | Nefazodone | Psychiatry | CYP2D6 |
| 81 | Nitrofurantoin | Infectious Diseases | G6PD |
| 82 | Nortriptyline | Psychiatry | CYP2D6 |
| 83 | Ombitasvir, Paritaprevir, and Ritonavir | Infectious Diseases | IFNL3 (IL28B) |
| 84 | Omeprazole | Gastroenterology | CYP2C19 |
| 85 | Ondansetron | Gastroenterology | CYP2D6 |
| 86 | Oxcarbazepine | Neurology | HLA-B*15:02 |
| 87 | Palonosetron | Gastroenterology | CYP2D6 |
| 88 | Pantoprazole | Gastroenterology | CYP2C19 |
| 89 | Parathyroid Hormone | Inborn Errors of Metabolism | CASR |
| 90 | Paroxetine | Psychiatry | CYP2D6 |
| 91 | Peginterferon Alfa-2b | Infectious Diseases | IFNL3 (IL28B) |
| 92 | Pegloticase | Rheumatology | G6PD |
| 93 | Perphenazine | Psychiatry | CYP2D6 |
| 94 | Phenytoin | Neurology | CYP2C9, CYP2C19, HLA-B*15:02 |
| 95 | Pimozide | Psychiatry | CYP2D6 |
| 96 | Piroxicam | Rheumatology | CYP2C9 |
| 97 | Prasugrel | Cardiology | CYP2C19, CYP2C9, CYP3A4, CYP2B6 |
| 98 | Primaquine | Infectious Diseases | G6PD, CYB5R1, CYB5R2, CYB5R3, CYB5R4 |
| 99 | Propafenone | Cardiology | CYP2D6 |
| 100 | Propranolol | Cardiology | CYP2D6 |
| 101 | Protriptyline | Psychiatry | CYP2D6 |
| 102 | Quinidine | Cardiology | CYP2D6 |
| 103 | Quinine Sulfate | Infectious Diseases | G6PD, CYP2D6 |
| 104 | Rabeprazole | Gastroenterology | CYP2C19 |
| 105 | Risperidone | Psychiatry | CYP2D6 |
| 106 | Rosuvastatin | Endocrinology | SLCO1B1 |
| 107 | Sevoflurane | Anesthesiology | RYR1 |
| 108 | Simeprevir | Infectious Diseases | IFNL3 (IL28B) |
| 109 | Sodium Nitrite | Toxicology | G6PD |
| 110 | Sofosbuvir | Infectious Diseases | IFNL3 (IL28B) |
| 111 | Sofosbuvir and Velpatasvir | Infectious Diseases | IFNL3 (IL28B) |
| 112 | Succimer | Hematology | G6PD |
| 113 | Sulfamethoxazole and Trimethoprim | Infectious Diseases | G6PD |
| 114 | Sulfasalazine | Gastroenterology | G6PD |
| 115 | Telaprevir | Infectious Diseases | IFNL3 (IL28B) |
| 116 | Tetrabenazine | Neurology | CYP2D6 |
| 117 | Thioridazine | Psychiatry | CYP2D6 |
| 118 | Ticagrelor | Cardiology | CYP2C19 |
| 119 | Tolterodine | Urology | CYP2D6 |
| 120 | Tramadol | Anesthesiology | CYP2D6 |
| 121 | Trimipramine | Psychiatry | CYP2D6 |
| 122 | Ustekinumab | Dermatology and Gastroenterology | IL12A, IL12B, IL23A |
| 123 | Valproic Acid | Neurology | POLG, ABL2, ASL, ASS1, CPS1, NAGS, OTC |
| 124 | Venlafaxine | Psychiatry | CYP2D6 |
| 125 | Voriconazole | Infectious Diseases | CYP2C19 |
| 126 | Vortioxetine | Psychiatry | CYP2D6 |
| 127 | Warfarin | Hematology | CYP2C9, VKORC1, PROS1, PROC |
PHARMACOGENOMICS: CLINICAL IMPLEMENTATION
The rapid growth of clinically relevant pharmacogenomic knowledge, as illustrated by the 127 drugs listed in Table 1, drugs used to treat patients in virtually every medical specialty, serves to highlight the challenges associated with pharmacogenomic implementation, one of which is that of making this information available to practitioners in a practical and easily understood fashion. To do that requires—among other important steps--objective, evidence-based guidelines and investment in the infrastructure required to make pharmacogenomic information accessible to physicians in a timely fashion. Physicians and other care-givers write prescriptions for drugs, not genes, so the approach that most institutions have taken is to focus on drug-gene pairs. Fortunately, the development of pharmacogenomics has occurred in parallel with the adoption of electronic health records (EHRs)—a development required for the storage of ever expanding genomic data as well as the tools required to instantly deliver that information to prescribers, preferably at the point of care, often while the prescription is being written. Of course, it always remains possible for a physician to order a specific pharmacogenomic test, either genotype-based—i.e., a test the queries only specific nucleotides that we currently know are of functional significance--or sequence-based, for a gene or genes known to be associated with variation in response to a specific drug or drugs. However, pharmacogenomic testing is moving increasingly to the use of panels of “pharmacogenes” that simultaneously test most of the genes that contribute to variation in response to commonly prescribed drugs for which there is evidence of pharmacogenomic clinical utility. To assist care-givers, many institutions have created automatic computer-based alerts that “fire” whenever a drug is prescribed for which a pharmacogenomic test might provide helpful information. For example, at the Mayo Clinic 17 drug-gene pair alerts currently fire when a prescription is first written for a drug that is included among those 17 drug-gene pairs (see Table 2). As at many medical centers, review and approval of the implementation of these alerts is the responsibility of a subcommittee of the Formulary Committee. Decision making with regard to the implementation of an alert depends on evidence-based guidelines that come from sources such as CPIC, the Clinical Pharmacogenetics Implementation Consortium (https://cpicpgx.org/guidelines/), a collaboration between the PGRN and the PharmGKB database or a similar European consortium, the Dutch Pharmacogenetics Working Group. It should be emphasized that, even though this review has generally focused on North American pharmacogenomic discovery and implementation efforts, these efforts are truly international in scope—as illustrated, for example, by the European “Ubiquitous Pharmacogenomics Consortium.”28 Current drug-gene pair alerts are primarily “reactive”, ie, they require that the physician—on the basis of his or her goals for the patient—order the genetic test in response to the alert. Although an important first step, reactive alerts represent only one step toward the eventual goal, which would involve having pharmacogenomic data for a specific patient “preemptively” available in the EHR so there will be no delay associated with waiting for a test result so the pharmacogenomic information can be incorporated into the clinical workflow seamlessly.
Table 2.
“Drug-Gene Pairs” included in Mayo Clinic Pharmacogenomic Electronic Alerts
| Drug | Gene(s) | |
|---|---|---|
| 1. | Abacavir | HLA-B*57:01 |
| 2. | Allopurinol | HLA-B*58:01 |
| 3. | Carbamazepine | HLA-B*15:02 and HLA-A*31:01 |
| 4. | Citalopram | CYP2C19 |
| 5. | Clopidogrel | CYP2C19 |
| 6. | Codeine | CYP2D6 |
| 7. | Escitalopram | CYP2C19 |
| 8. | Fluoxetine | CYP2D6 |
| 9. | Fluvoxamine | CYP2D6 |
| 10. | Paroxetine | CYP2D6 |
| 11. | Simvastatin | SLCO1B1 |
| 12. | Tacrolimus | CYP3A5 |
| 13. | Tamoxifen | CYP2D6 |
| 14. | Thiopurines | TPMT |
| 15. | Tramadol | CYP2D6 |
| 16. | Venlafaxine | CYP2D6 |
| 17. | Warfarin | CYP2C9 and VKORC1 |
Some academic medical centers are already experimenting with “preemptive” pharmacogenomic alerts. For example, the PGRN Centers, as a group, supported a “Deep Sequencing” facility that developed an NGS reagent that initially included 84 pharmacogenes that were “captured” and sequenced across exons and splice junctions—thus making it possible to obtain information with regard to both known, functionally significant variants as well as many “variants of unknown significance” (VUS). One of the challenges facing pharmacogenomics will be the development of high throughput methods that will make it possible to functionally characterize the very large number of VUS identified when we apply NGS to DNA samples from a large number of patients. For example, if we just select two common and intensively studied cytochrome P-450 (CYP) genes, CYP2C9 and CYP2C19, genes that encode enzymes that metabolize the anticoagulant agents warfarin and clopidogrel, respectively, we can compare what we currently know about these two highly studied genes with what we need to know. To determine what “we know”, we went to “The Human Cytochrome P450 Allele Nomenclature Database” at the Karolinska in Stockholm (http://www.cypalleles.ki.se). That canonical database for CYP genes and enzymes listed 165 missense variants for CYP2C9 and 189 for CYP2C19. Of those variants, only 29 CYP2C9 variants had been functionally validated—20 in a laboratory setting, and only 26 for CYP2C19—7 in a laboratory setting. Meanwhile in 60,000 DNA samples for which exome sequence data had been reported by the Broad Institute,33 there were 235 missense variants in CYP2C9 and 258 in CYP2C19. These numbers indicate that we need to develop methods to rapidly screen the function of these variant allozymes to determine what they might mean clinically. To place this task in context, a decade ago it would have been possible to justify a multi-year PhD thesis project for only a dozen variants in a single gene. This example represents a microcosm of the task that we face as more and more DNA sequence information becomes available. This coming tidal wave of DNA sequence information also raises the issue of the most efficient and cost-effective way to establish the clinical utility of novel pharmacogenomic information. It is clear that it will not be practical to perform a randomized clinical trial for all of these genomic variants, but the most practical way in which to do that remains a subject of continuing debate.
The NIH PGRN and eMERGE Networks have already begun to perform “pilot” studies designed to set the stage for more widespread clinical use of preemptive pharmacogenomic information. For example, each of the NIH eMERGE grant sites used the original “PGRN-seq” NGS reagent to sequence DNA samples from their biobanks. The Mayo Clinic, as one example, used this reagent to sequence 1013 DNA samples from the Mayo Biobank for local patients who had consented for their DNA to be used for research purposes. Patients participating in that “RIGHT” study—RIGHT drug at the RIGHT dose at the RIGHT time—now have DNA sequence information in the EHR for the 17 drug-gene pairs for which “reactive alerts” are currently firing at Mayo, but, in their case, if an alert fires, it will not inform the prescriber that a genetic test is available.29, 30 For these patients, the alert will instantly tell the care-giver the patients gene sequence with a clinical interpretation. For example, among those original 1013 subjects, if only five “common” pharmacogenes out of the 84 sequenced were included, 99.1% of the subjects had at least one actionable variant in at least one of those five genes—with many subjects having clinically actionable variant sequences in several of the five genes.31 This observation explains why, at the beginning of this overview, we made the statement that “pharmacogenomics is the aspect of clinical genomics that will almost certainly see the earliest and broadest clinical implementation—with the potential to eventually touch the care of every patient everywhere”. To follow up on those initial RIGHT study results, the Mayo Clinic Center for Individualized Medicine, in collaboration with the Baylor Human Genome Sequencing Center, is currently moving beyond the original 1013 biobank samples to consent and sequence 10,000 additional Mayo Biobank participants for a “RIGHT 10K” study designed to test the hypothesis that having preemptive pharmacogenomic information in the EHR might result in cost-effective health benefits for the patients involved—helping them to avoid adverse drug reactions and obtain maximum efficacy from drug therapy.
Finally, it goes without saying that the implementation of pharmacogenomics across a large academic medical center requires significant effort and significant resources. It is for that reason that we emphasized that NIH support provided by the PGRN and eMERGE grants acted as a valuable “catalyst” to move this aspect of genomic science to the bedside, but that this process must be a partnership with hospitals and medical centers, beginning with committed institutional leadership, engagement across multiple medical staffs that include physicians, nurses, allied health personnel and pharmacists, with significant investments required for the education for all of these groups, including patients, as well as infrastructural investment in information technology and the EHR. This may seem ambitious but—as outlined subsequently—we are only at the beginning of this process of discovery and implementation of genomics and other “omics” techniques to enhance drug therapy.
PHARMACOGENOMICS: FUTURE PROSPECTS
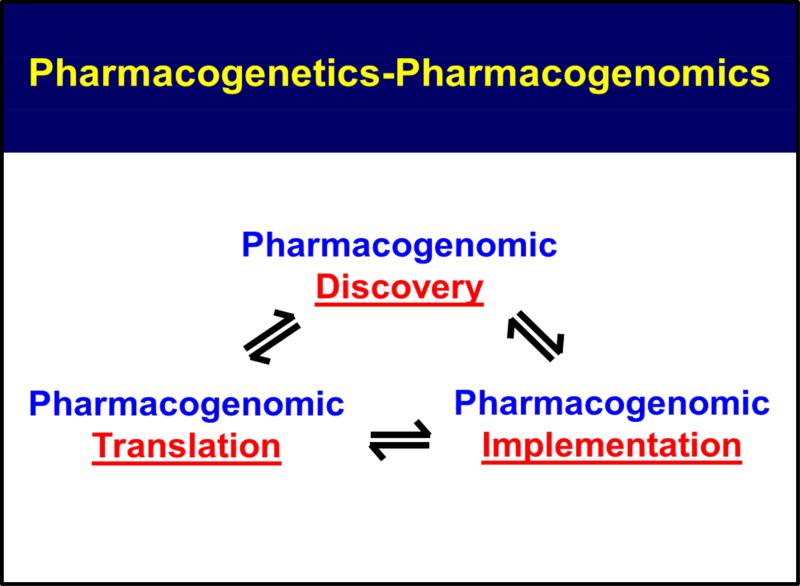
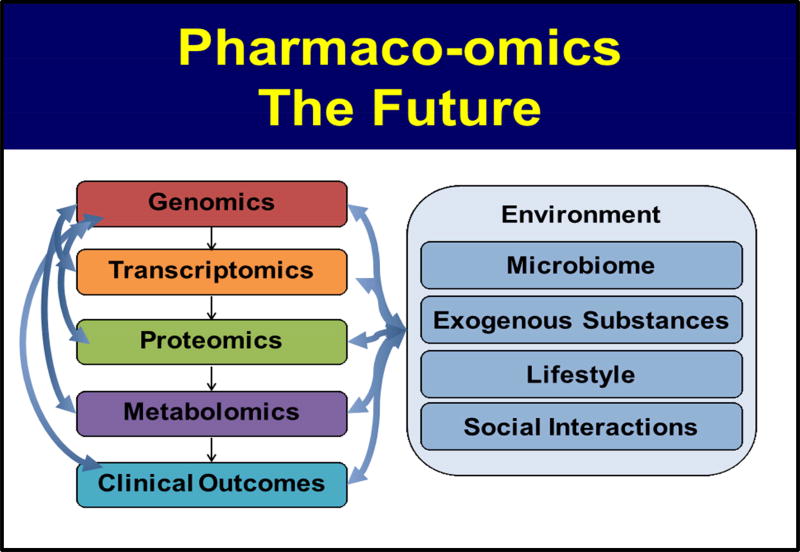
In this overview, we have described, briefly, the origins and development of pharmacogenomics as well as ongoing efforts to bring pharmacogenomics into the clinic and to make it a standard component of routine patient care. It might be useful to point out that the TPMT and CYP2D6 genetic polymorphisms that we are currently “implementing” clinically were discovered over 35 years ago10–13 and that both have been known to have clinical utility for over a quarter of a century. The association of statin-induced myopathy with a SNP in the SLCO1B1 gene was one of the early success stories for pharmacogenomic GWAS—but that occurred a decade ago.32 Therefore, the information that we are currently implementing in the clinic has been known for decades—or at least for a decade. Included among the questions that we should be asking ourselves now are what are the scientific challenges that face pharmacogenomics as we increasingly apply NGS and increasingly move genome-wide in our perspective as we attempt to apply genomics to “inform” drug selection and use; what other types of “omics” information might help to better inform the “therapeutic encounter”; how we can merge other “omic” information with genomic data; and what other types of information might help make drug selection and use as “rational” and as highly “individualized” as possible. Finally, we should point out that the processes of biomedical “discovery”, “translation” and “implementation” are not separate and distinct activities, but rather they are tightly intertwined and they inform each other, as illustrated graphically in Figure 1. In subsequent paragraphs, we will briefly outline challenges and possible future directions as genomic science is applied to drug response. That discussion will be followed by a description of recent efforts to move beyond genomics alone to include data from other “omics” disciplines, ie transcriptomics, epigenomics, proteomics and metabolomics, in combination with genomics (see Figure 2), to determine whether that might better inform our attempts to optimize and individualize drug therapy by moving beyond pharmacogenomics to what might be called “Pharmaco-omics”.
Figure 1.
Pharmacogenomic Discovery, Translation, Implementation
Figure 2.
Pharmacogenomics to Pharmaco-Omics
As pointed out previously, many of the earliest clinically relevant examples of pharmacogenomics involved genes that encoded drug metabolizing enzymes, drug transporters and drug targets, proteins that might logically be expected to influence drug response. Furthermore, there was also an emphasis on ns SNPs, nucleotide sequence changes in the portion of the gene that encodes protein—resulting in altered protein amino acid sequence. However, it is becoming increasingly clear now that a large number of GWAS for drug response have been performed, with both drug efficacy and adverse drug reactions as phenotypes, that—even though SNPs within the open reading frame of a gene can have a striking effect on function—many of the “top hit” SNPs identified during GWA studies are outside of the coding portion of the gene. Those SNPs often alter transcription and, as a result, they alter gene expression. That can occur, for example, if the SNP either creates or disrupts a transcription factor binding site, a DNA sequence that the transcription factor “recognizes” and binds to in the promoter of the gene or in so-called “enhancer” elements, DNA sequence motifs that can be located hundreds of thousands of base pairs away from the gene. There are now databases that can be used predict whether a SNP might create or disrupt a transcription factor binding site and there is also the GTEx database (https://www.gtexportal.org/home/), which, in a tissue-specific fashion, provides information with regard to whether a specific SNP might be an “expression quantitative trait locus” (eQTL), ie whether that SNP is associated with variation in the level of mRNA expression. To complicate the situation even further, there is recent evidence that SNPs located hundreds of base pairs away from sites of transcription factor binding can have a profound effect on the binding of the transcription factor and on subsequent gene expression.33, 34 Since exome sequencing only covers about 1.5% of the genome, in the future we will need to move beyond the portion of the gene that encodes protein if we want to understand the functional implications of genetic polymorphisms. There can be no doubt that we are currently missing a great deal of information that might be important for our understanding of variation in drug response. Therefore, new, high throughput approaches will be needed to help us understand and use that information because we are being inundated by a tidal wave of data with regard to genomic sequence variation.35 The issue of the extremely large number of ns SNP variants within the coding regions of genes and how we can quickly and accurately predict their functional implications that was mentioned previously is just one example of the fact that we are still only at the beginning of our understanding of the relationship of genomic sequence variation with clinically relevant variation in drug response phenotypes. There is no doubt that we will soon be dealing not with hundreds of potentially functionally significant variants for each gene, but rather with thousands. Even if we develop high throughput methods to determine which gene sequence or structural variants have functional implications, there is no way that a human being could possibly remember all of them, so sophisticated computerized systems will be a necessity—as will systems to convey this information to the health care team caring for the patient in a form that can be easily and quickly understood and used. If that is true of only genomics alone, how will we approach the integration of other “omics” information with genomics, information with regard to the transcriptome, the epigenome, the proteome, the metabolome and the microbiome?
The process of joining multiple “omics” datasets to us help make better informed therapeutic decisions represents a significant challenge. However, it is now commonplace to use both exome and RNA-seq data, ie to join genomics and transcriptomics, for a tumor to gain greater insight into underlying—potentially “druggable”--therapeutic targets. That process is also “pharmacogenomics”. Psychiatry represents another clinical discipline in which multiple “omics” have been joined. It is probably fair to say that the application of genomics alone has been somewhat disappointing in psychiatry—perhaps, at least in part, because the phenotypes in psychiatry have not yet been closely related to the underlying biology in same way that they have been in many other medical specialities. In an attempt to overcome that limitation, there have been recent attempts to “inform” genomic analyses by beginning with metabolomics data, determine which metabolite is most highly associated with the psychiatric phenotype (eg., Hamilton D Scores for patients with Major Depressive Disorder), perform a GWAS to identify genes associated with the concentration of the metabolite and then functionally validate the genes/SNPs that were identified during the GWAS.36 These examples have begun to show us that, in the future, biomarkers for drug response might well be composed jointly of genomic data, transcriptomic data and metabolomic data, ie that we will be using “pharmaco-omics” to help us individualize and optimize drug therapy.
CONCLUSIONS
Pharmacogenomics is the application of genomic—and other “omic” information--to help guide, inform and individualize drug therapy. In the decades since Arno Motulsky first put forward the concept of pharmacogenetics, striking progress has been made and there can no longer be any doubt that drug efficacy and the occurrence of adverse drug reactions can both be influenced by genomics or that genomic information can be used to help maximize efficacy and minimize the occurrence of adverse drug reactions. That progress has occurred in parallel with the striking advances that have occurred in human genomics during the past half century. As outlined in preceding paragraphs, pharmacogenomics and pharmaco-omics are still young disciplines, but they are already moving into the clinic and are already being used to help physicians and other healthcare team members to make better and more highly individualized therapeutic decisions. A great remains to be done, and we are only at the beginning of the process of bringing this aspect of genomics to the bedside, but we will end this overview as we began. Pharmacogenomics is the aspect of clinical genomics that will almost certainly see the earliest and broadest clinical implementation—with the potential to eventually touch the care of every patient everywhere.
Figure 3.
Pharmaco-Omics, The Future
Acknowledgments
Supported, in part, by NIH grants U19 GM61388 (LW and RMW), RO1 CA196648 (LW), U54 GM114838 (RMW), RO1 GM28157 (RMW), NSF1624615 (LW), P50 CA116201 (Breast Cancer SPORE) (LW), the Breast Cancer Research Foundation and the Mayo Center for Individualized Medicine.
ABBREVIATIONS
- PK
Pharmacokinetics
- NAT
N-Acetyltransferase
- TPMT
Thiopurine S-methyltransferase
- CYP
Cytochrome P-450
- SULT
Sulfotransferase
- MT
Methyltransferase
- UGT
UDP-Glucuronosyltransferase
- MAF
Minor allele frequency
- ns
Nonsynonymous
- NIH
National Institutes of Health
- FDA
Food and Drug Administration
- EHR
Electronic Health Record
- VUS
Variants of Unknown Significance
- NGS
Next Generation Sequencing
- SNP
Single Nucleotide Polymorphism
- GWAS
Genome-wide Association Study
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES
Drs. Wang and Weinshilboum are co-founders and stockholders in One
References
- 1.Weinshilboum RM, Wang L. Pharmacogenetics and Pharmacogenomics: Development, Science, and Translation. Annual Rev Genomics Hum Genet. 2006;7:223–245. doi: 10.1146/annurev.genom.6.080604.162315. [DOI] [PubMed] [Google Scholar]
- 2.Wang L, McLeod HL, Weinshilboum RM. Genomics and Drug Response. N Engl J Med. 2011;364:1144–1153. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giacomini KM, Yee SW, Ratain MG, Weinshilboum RM, Kamatani N, Nakamura Y. Pharmacogenomics and patient care: one size does not fit all. Sci Transl Med. 2012 Sep 26;4(153) doi: 10.1126/scitranslmed.3003471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motulsky AG. Drug reactions enzymes and biochemical genetics. JAMA. 1957;165:835–837. doi: 10.1001/jama.1957.72980250010016. [DOI] [PubMed] [Google Scholar]
- 5.Kalow W. Familial incidence of low pseudocholinesterase level. The Lancet. 1956;2:576–577. doi: 10.1016/s0140-6736(56)90869-8. [DOI] [PubMed] [Google Scholar]
- 6.Kalow W, Staron N. On distribution and inheritance of atypical forms of human serum cholinesterase, as indicated by dibucaine numbers. Canadian J Biochem Physiol. 1957;35:1305–1320. [PubMed] [Google Scholar]
- 7.Bonicke R, Lisboa BP. Uber die Erbbedingtheit der intraindividuellen Konstanz der Isoniazidaus-scheidung beim Menschen. Naturwissenshaften. 1957;44:314. [Google Scholar]
- 8.Evans DAP, Manley KA, McKusick VA. Genetic Control of Isoniazid Metabolism in Man. Br Med J. 1960;2:461–485. doi: 10.1136/bmj.2.5197.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans DAP, Storey PB, McKusick VA. Further observations on the determination of isoniazid inactivator phenotype. Bulletin Johns Hopkins Hospital. 1961;108:60–66. [PubMed] [Google Scholar]
- 10.Weinshilboum RM, Sladek SL. Mercaptopurine pharmacogenetics: monogenic inheritance of erythrocyte thiopurine methyltransferase activity. Am J Hum Genet. 1980 Sep;32(5):651–62. [PMC free article] [PubMed] [Google Scholar]
- 11.Lennard L, Van Loon JA, Weinshilboum RM. Pharmacogenetics of acute azathioprine toxicity: relationship to thiopurine methyltransferase genetic polymorphism. Clin Pharmacol Ther. 1989 Aug;46(2):149–54. doi: 10.1038/clpt.1989.119. [DOI] [PubMed] [Google Scholar]
- 12.Mahgoub A, Dring LG, Idle JR, Lancaster R, Smith RL. Polymorphic hydroxylation of debrisoquine in man. Lancet. 1977;2:584–586. doi: 10.1016/s0140-6736(77)91430-1. [DOI] [PubMed] [Google Scholar]
- 13.Eichelbaum M, Spannbrucker N, Dengler HJ. A probable genetic defect of the metabolism of sparteine. Biological oxidation of Nitrogen. 1978:113–118. [PubMed] [Google Scholar]
- 14.Gonzalez FJ, Skoda RC, Kimura S, Umeno M, Zanger UM, Nebert DW, Gelboin HV, Hardwick JP, Meyer UA. Characterization of the common genetic defect in humans deficient in debrisoquine metabolism. Nature. 1988 Feb 4;331(6155):442–446. doi: 10.1038/331442a0. [DOI] [PubMed] [Google Scholar]
- 15.Broly F, Gaedigk A, Heim M, Eichelbaum M, Morike K, Meyer UA. Debrisoquine/sparteine hydroxylation genotype and phenotype: analysis of common mutations and alleles of CYP2D6 in a European population. DNA Cell Biol. 1991 Oct;10(8):545–558. doi: 10.1089/dna.1991.10.545. [DOI] [PubMed] [Google Scholar]
- 16.Weinshilboum R, Aksoy I. Sulfation pharmacogenetics in humans. Chem Biol Interact. 1994 Jun;92(1–3):233–246. doi: 10.1016/0009-2797(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 17.Weinshilboum RM, Otterness DM, Aksoy IA, Wood TC, Her C, Raftogianis RB. Sulfation and sulfotransferases 1: Sulfotransferase molecular biology: cDNAs and genes. FASEB J. 1997 Jan;11(1):3–14. [PubMed] [Google Scholar]
- 18.Weinshilboum RM. Methylation pharmacogenetics: thiopurine methyltransferase as a model system. Xenobiotica. 1992 Sep-Oct;22(9–10):1055–1071. doi: 10.3109/00498259209051860. [DOI] [PubMed] [Google Scholar]
- 19.Weinshilboum RM, Otterness DM, Szumlanski CL. Methylation pharmacogenetics: catechol O-methyltransferase, thiopurine methyltransferase, and histamine N-methyltransferase. Annu Rev Pharmacol Toxicol. 1999;39:19–52. doi: 10.1146/annurev.pharmtox.39.1.19. [DOI] [PubMed] [Google Scholar]
- 20.Owens IS, Ritter JK. Gene structure at the human UGT1 locus creates diversity in isozyme structure, substrate specificity, and regulation. Prog Nucleic Acid Res Mol Biol. 1995;51:305–338. doi: 10.1016/s0079-6603(08)60882-x. [DOI] [PubMed] [Google Scholar]
- 21.Bosma PJ, Chowdhury JR, Bakker C, Gantla S, de Boer A, Oostra BA, Lindhout D, Tytgat GN, Jansen PL, Oude Elferink RP, et al. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome. N Engl J Med. 1995 Nov 2;333(18):1171–1175. doi: 10.1056/NEJM199511023331802. [DOI] [PubMed] [Google Scholar]
- 22.Iyer L, King CD, Whitington PF, Green MD, Roy SK, Tephly TR, Coffman BL, Ratain MJ. Genetic predisposition to the metabolism of irinotecan (CPT-11). Role of uridine diphosphate glucuronosyltransferase isoform 1A1 in the glucuronidation of its active metabolite (SN-38) in human liver microsomes. J Clin Invest. 1998 Feb 15;101(4):847–854. doi: 10.1172/JCI915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szumlanski C, Otterness D, Her C, Lee D, Brandriff B, Kelsell D, Spurr N, Wieben E, Weinshilboum R. Thiopurine Methyltransferase Pharmacogenetics: Human Gene Cloning and Characterization of a Common Polymorphism. DNA Cell Biol. 1996 Jan;15(1):17–30. doi: 10.1089/dna.1996.15.17. [DOI] [PubMed] [Google Scholar]
- 24.Tai HL, Krynetski EY, Yates CR, Loennechen T, Fessing MY, Krynetskaia NF, Evans WE. Thiopurine S-methyltransferase deficiency: two nucleotide transitions define the most prevalent mutant allele associated with loss of catalytic activity in Caucasians. Am J Hum Genet. 1996 Apr;58(4):694–702. [PMC free article] [PubMed] [Google Scholar]
- 25.Tai HL, Fessing MY, Bonten EJ, Yanishevsky Y, d’Azzo A, Krynetski EY, Evans WE. Enhanced proteasomal degradation of mutant human thiopurine S-methyltransferase (TPMT) in mammalian cells: mechanism for TPMT protein deficiency inherited by TPMT*2, TPMT*3A, TPMT*3B or TPMT*3C. Pharmacogenetics. 1999 Oct;9(5):641–650. doi: 10.1097/01213011-199910000-00011. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Sullivan W, Toft D, Weinshilboum R. Thiopurine S-methyltransferase pharmacogenetics: chaperone protein association and allozyme degradation. Pharmacogenetics. 2003 Sep;13(9):555–564. doi: 10.1097/01.fpc.0000054124.14659.99. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Nguyen TV, McLaughlin RW, Sikkink LA, Ramirez-Alvarado M, Weinshilboum RM. Human thiopurine S-methyltransferase pharmacogenetics: Variant allozyme misfolding and aggresome formation. Proc Natl Acad of Sci USA. 2005 Jun 28;102(26):9394–99. doi: 10.1073/pnas.0502352102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van der Wouden CH, Cambon-Thomsen A, Cecchin E, Cheung KC, Dávila-Fajardo CL, Deneer VH, Dolžan V, Ingelman-Sundberg M, Jönsson S, Karlsson MO, Kriek M, Mitropoulou C, Patrinos GP, Pirmohamed M, Samwald M, Schaeffeler E, Schwab M, Steinberger D, Stingl J, Sunder-Plassmann G, Toffoli G, Turner RM, van Rhenen MH, Swen JJ, Guchelaar HJ. Implementing Pharmacogenomics in Europe: Design and Implementation Strategy of the Ubiquitous Pharmacogenomics Consortium. Clin Pharmacolo Ther. 2017 Mar;101(3):341–358. doi: 10.1002/cpt.602. [DOI] [PubMed] [Google Scholar]
- 29.Olson JE, Rohrer Vitek CR, Bell EJ, McGree ME, Jacobson DJ, St Sauver JL, Caraballo PJ, Griffin JM, Roger VL, Bielinski SJ. Participant-perceived understanding and perspectives on pharmacogenomics: the Mayo Clinic RIGHT protocol (Right Drug, Right Dose, Right Time) Genet Med. 2017 Jan 5; doi: 10.1038/gim.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caraballo PJ, Bielinski SJ, St Sauver JL, Weinshilboum RM. Electronic Medical Record-Integrated Pharmacogenomics and Related Clinical Decision Support Concepts. Clin Pharmacol Ther. 2017 Apr 8; doi: 10.1002/cpt.707. [DOI] [PubMed] [Google Scholar]
- 31.Ji Y, Skierka JM, Blommel JH, Moore BE, VanCuyk DL, Bruflat JK, Peterson LM, Veldhuizen TL, Fadra N, Peterson SE, Lagerstedt SA, Train LJ, Baudhuin LM, Klee EW, Ferber MJ, Bielinski SJ, Caraballo PJ, Weinshilboum RM, Black JL., 3rd J Mol Diagn. 2016 May;18(3):438–445. doi: 10.1016/j.jmoldx.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.SEARCH Collaborative Group. Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, Gut I, Lathrop M, Collins R. SLCO1B1 variants and statin-induced myopathy--a genomewide study. N Engl J Med. 2008 Aug 21;359(8):789–99. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 33.Ingle JN, Liu M, Wickerham DL, Schaid DJ, Wang L, Mushiroda T, Kubo M, Costantino JP, Vogel VG, Paik S, Goetz MP, Ames MM, Jenkins GD, Batzler A, Carlson EE, Flockhart DA, Wolmark N, Nakamura Y, Weinshilboum RM. Selective estrogen receptor modulators and pharmacogenomic variation in ZNF423 regulation of BRCA1 expression: individualized breast cancer prevention. Cancer Discov. 2013 Jul;3(7):812–825. doi: 10.1158/2159-8290.CD-13-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho MF, Bongartz T, Liu M, Kalari KR, Goss PE, Shepherd LE, Goetz MP, Kubo M, Ingle JN, Wang L, Weinshilboum RM. Estrogen, SNP-Dependent Chemokine Expression and Selective Estrogen Receptor Modulator Regulation. Mol Endocrinol. 2016 Mar;30(3):382–398. doi: 10.1210/me.2015-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lek Monkol, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016 Aug;536.7616(2016):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta M, Neavin D, Liu D, Biernacka J, Hall-Flavin D, Bobo WV, Frye MA, Skime M, Jenkins GD, Batzler A, Kalari K, Matson W, Bhasin SS, Zhu H, Mushiroda T, Nakamura Y, Kubo M, Wang L, Kaddurah-Daouk R, Weinshilboum RM. TSPAN5, ERICH3 and selective serotonin reuptake inhibitors in major depressive disorder: pharmacometabolomics-informed pharmacogenomics. Mol Psychiatry. 2016 Dec;21(12):1717–1725. doi: 10.1038/mp.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]