Abstract
Nucleotide excision repair in Escherichia coli is stimulated by transcription, specifically in the transcribed strand. Previously, it was shown that this transcription-coupled repair (TCR) is mediated by the Mfd translocase. Recently, it was proposed that in fact the majority of TCR in E. coli is catalyzed by a second pathway (“backtracking-mediated TCR”) that is dependent on the UvrD helicase and the guanosine pentaphosphate (ppGpp) alarmone/stringent response regulator. Recently, we reported that as measured by the excision repair–sequencing (XR-seq), UvrD plays no role in TCR genome-wide. Here, we tested the role of ppGpp and UvrD in TCR genome-wide and in the lacZ operon using the XR-seq method, which directly measures repair. We found that the mfd mutation abolishes TCR genome-wide and in the lacZ operon. In contrast, the relA−spoT− mutant deficient in ppGpp synthesis carries out normal TCR. We conclude that UvrD and ppGpp play no role in TCR in E. coli.
Keywords: DNA repair, DNA sequencing, Escherichia coli (E. coli), genomics, nucleotide excision repair, Mfd, UvrD, lac operon, ppGpp, transcription-coupled repair
Introduction
In Escherichia coli and humans, DNA damage caused by UV light and certain chemicals such as cisplatin, psoralen, and benzo[a]pyrene is repaired by the nucleotide excision repair enzyme system (1, 2). The core repair system consists of three proteins (UvrA, UvrB, and UvrC) in E. coli, and six repair factors encompassing 16 polypeptides in humans. The damage is removed in the form of 12–13-nucleotide-long oligomers in E. coli and 26–27-nucleotide-long oligomers in humans by concerted dual incisions bracketing the lesion (2). This core repair is affected by multiple factors in vivo that impact the rate of repair and the catalytic turnover of the enzyme systems. Among those factors that impact the rate of repair, it was found that transcription plays a unique role (3–5). Both in humans and in E. coli, transcription stimulates repair in the transcribed strand (TS) but not in the nontranscribed strand (NTS),3 and this phenomenon has been named transcription-coupled repair (TCR).
Using in vitro assays with cell-free extracts and with purified UvrA, UvrB, and UvrC proteins, a protein essential for TCR in E. coli was purified and named transcription–repair coupling factor (TRCF) (6, 7). Genetic analysis revealed that TRCF is encoded by the mfd gene (8, 9), which had been previously identified as a gene responsible for strand-specific repair of certain tRNA genes (10). Following purification of the Mfd/TRCF protein, a defined in vitro system consisting of UvrA, UvrB, and UvrC, Mfd, RNA polymerase, UV-irradiated DNA, ATP, and four ribonucleoside tri-phosphates was developed (11, 12). The magnitude of TCR in this system was comparable with the TCR in vivo (3, 7, 11). Based on structural and functional characterizations of Mfd and TCR (13, 14), the following model was proposed for TCR in E. coli. Mfd has affinities for both UvrA and RNAP. When RNAP arrests at a damage site, the stalled RNAP is recognized by Mfd translocase, which displaces the stalled RNAP while recruiting the UvrA2–UvrB1 complex to the damage site in a concerted reaction. Because damage recognition is the rate-limiting step in excision repair, these activities of Mfd accelerate the rate of repair of the transcribed strand ∼5-fold. This model was subsequently confirmed and refined by additional biochemical studies (15, 16) as well as analyses by X-ray crystallography (17) and single-molecule experiments (18).
Recently, an alternative model called the “backtracking pathway” was proposed, and it was suggested that this is the predominant pathway of TCR (19). According to this model, the UvrD helicase dimer stabilized by the stress alarmone ppGpp (20) recognizes stalled RNAP, promotes backtracking of RNAP from the damage site, and at the same time recruits the UvrA2–UvrB1 complex by its interaction with UvrB (21) and thus accelerates the rate of damage removal without release of the RNA, which may then be elongated by RNAP following repair synthesis and ligation. Perhaps the strongest evidence for this model was the reported absence of TCR in a ppGpp-deficient strain using the inducible lacZ operon for TCR readout (19).
To investigate the relative contributions of the Mfd pathway and the so-called backtracking pathway, we wished to use the highly sensitive XR-seq method (22, 23) to analyze TCR in E. coli strains of various genetic background. Previously, using XR-seq, we found that genome-wide TCR was mediated by Mfd but found no evidence for a role of UvrD in TCR (23). Instead XR-seq analysis confirmed the role of UvrD in catalytic turnover of the Uvr(A)BC excision nuclease, which had been deduced from in vitro studies (24, 25). Here, we investigated the role of ppGpp in TCR both genome-wide and in the inducible lacZ operon. We found that both genome-wide TCR as well as inducible lacZ TCR are dependent on Mfd but are not affected by the absence or presence of ppGpp.
Results
Genome-wide effects of Mfd, UvrD, and ppGpp on transcription-coupled repair
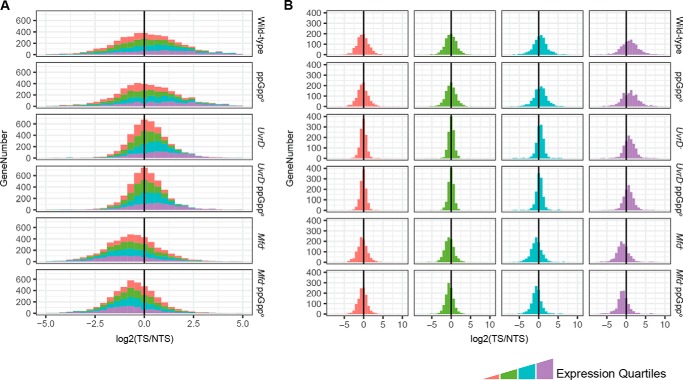
To determine the contributions of Mfd, UvrD, and ppGpp on TCR on the genomic scale, we conducted XR-seq on wild-type E. coli as well as E. coli strains with mfd−, uvrD−, and relA−spoT− mutations. The results in the forms of histograms are shown in Fig. 1. As noted previously, in the wild type, although in the majority of genes, the TS is repaired at a faster rate than the NTS, in many genes, the NTS is repaired more efficiently. Moreover, this trend is observed in all genetic backgrounds tested. This unexpected phenomenon has been ascribed to the complex transcriptional landscape of the E. coli genome, which encompasses numerous annotated and unannotated antisense transcripts and “transcriptional noise” due to spurious promoter-like sequences capable of initiating transcription (26–28). In addition, UV damage hotspots may contribute to this anomalous repair pattern (23).
Figure 1.
The effects of mutations on genome-wide TCR profiles. The TCR values, measured as the TS/NTS repair signal of all the E. coli genes, grouped and colored by expression quartiles, are plotted as stacked (A) and separated (B) histograms. The x axis shows log2-scaled TS/NTS values normalized by the TT content of each strand per gene. The y axis shows the total gene number. In the stacked histogram (A), the x axis scale was focused (−5 to 5) to show the majority of the genes and misses the outliers that can be viewed in panel B.
From the perspective of the molecular mechanism of TCR and the roles of various factors in this phenomenon, we note the following facts evident in Fig. 1. First, in the mfd− mutant, the TS/NTS repair ratio is drastically reduced and the NTS has become the preferentially repaired strand because of the inhibition of repair by stalled RNAP in the TS, especially in the highly transcribed genes (Fig. 1B). Second, in agreement with a previous finding (23), in the uvrD−strain, either TCR was unchanged or the TS/NTS ratio was amplified, although overall repair was drastically reduced in both strands. Finally, the genome-wide TS/NTS histogram of relA−spoT− (ppGppo) is essentially identical to that of the wild-type strain, indicating that the ppGpp alarmone has no role in TCR.
Analysis of the effect of ppGpp on TCR at gene resolution
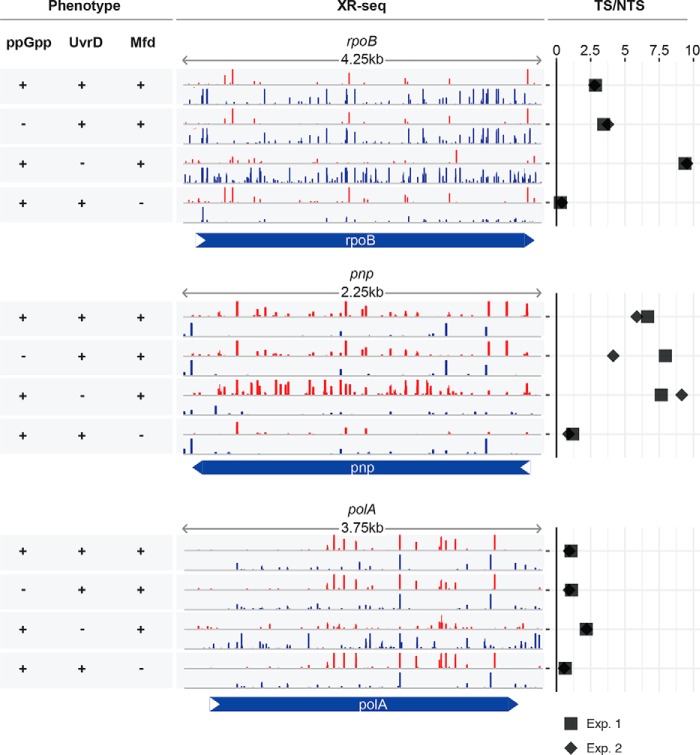
Although Fig. 1 shows no indication that ppGpp plays a role in TCR, the confounding factors of genome-wide analysis (antisense transcripts, spurious transcripts, overlapping genes) preclude a definitive conclusion vis à vis the role of ppGpp in TCR. Hence, we decided to analyze the repair pattern of a few sentinel genes with high levels of transcription and no known antisense transcription or other complicating factors to more precisely define the putative role of ppGpp in TCR. We chose RNA polymerase β subunit (rpoB), polyribonucleotide nucleotidyltransferase (pnp), and DNA polymerase I (polA) as representatives of genes expressed at high levels. Fig. 2 shows that there is TCR in wild-type cells in all three genes. In the mfd− mutant, the TCR is abolished in all three operons, as expected. Of significance, in the highly transcribed rpoB gene, the TS/NTS repair ratio is reversed by a factor of about 10 in favor of the NTS because in the absence of Mfd, the high-density RNAPs stalled at damage in the TS inhibit repair in this strand but do not affect repair in the NTS (6). This effect is less obvious or absent in genes with lower transcription rates. The uvrD− mutation, as reported previously (23), reduces the level of overall repair and either enhances TCR (rpoB) or does not significantly affect the TS/NTS repair ratio. Importantly, the TCR profiles of all three genes in the ppGppo strain are indistinguishable from the wild-type strain, consistent with the conclusion drawn from the genome-wide analyses of TCR.
Figure 2.
The contribution of ppGpp, UvrD, and Mfd to the XR-seq profiles in the highly expressed genes. The repair screenshots show normalized read numbers for both DNA strands of the RNA polymerase β subunit (rpoB), polyribonucleotide nucleotidyltransferase (pnp), and DNA polymerase I (polA) genes from top to bottom. The rows with red bars correspond to the TS of pnp and NTS of rpoB and polA. The rows with blue bars correspond to the TS of rpoB and polA and NTS of pnp. The y axis was scaled to show 50 RPM (reads per million mapped reads), and some hotspots with high repair values remain off-scale. The screen shots are derived from the data set of the first experiment. The TS/NTS ratios of the two experiments (Exp. 1 and Exp. 2) are horizontally plotted in the right-hand panes. +, protein or alarmone present; −, absent.
Effect of ppGpp on transcription-coupled repair on an inducible operon
Traditionally, in vivo TCR experiments in E. coli have been done using the lac operon for readout (4). E. coli cells grown in glucose-containing medium do not express lacZ, but the operon can be turned on by the addition of IPTG to induce transcription to varying degrees and thus study the effects of various levels of transcription in a relatively well-defined system. With these considerations, we then analyzed TCR in the lacZ operon in wild-type, mfd−, relA−spoT−, and uvrD− strains under conditions of no induction and full induction. Fig. 3 shows screen shots of XR-seq results for the relevant strains under conditions of various levels of transcriptional activity. The results with the wild-type strain are consistent with a previous report (6) and with the in vitro data (7, 8, 11) that under conditions of full induction, the TS is repaired at about a 4.6-fold faster rate than the nontranscribed strand. This preferential repair is abolished in the mfd− mutant, and the TS/NTS repair ratio is reversed in favor of the NTS, as has been observed in the reconstituted TCR repair system (8, 11). In the uvrD− mutant, overall repair is reduced because of the lack of catalytic turnover of Uvr(A)BC excision nuclease. However, the TS/NTS is either unchanged or slightly enhanced because of the retention of the UvrB–UvrC complex on the TS following the dual incision and in agreement with the model that UvrD plays a role in post-incision steps of excision repair but not in the pre-incision steps (24, 25), in the absence or presence of transcription. Finally, the ppGppo (relA−spoT−) is not distinguishable from the wild-type strain in lacZ: the TS is repaired at about 4.3-fold faster than the NTS. Quantitative data points are shown in Fig. 3. A minor difference between WT (4.6-fold) and ppGppo (4.3-fold) is likely because of the diminished transcription levels in the absence of ppGpp as indicated by the β-galactosidase activities (Fig. 3).
Figure 3.
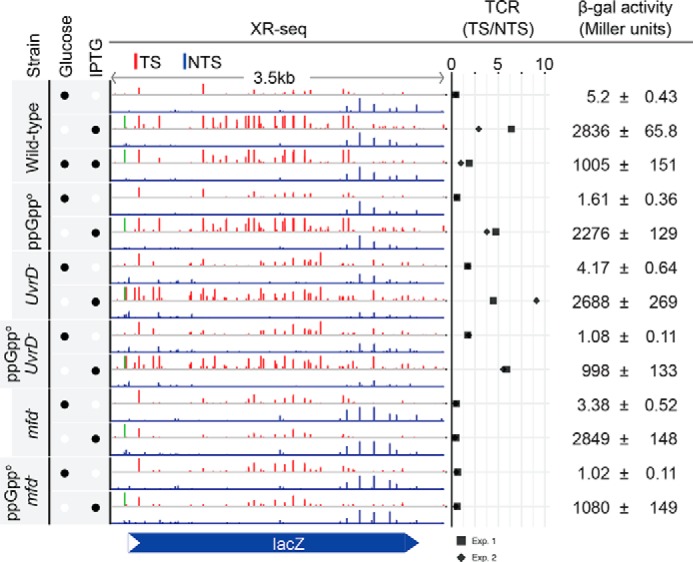
TCR profiles of the lacZ gene for six E. coli strains with and without IPTG induction. The presence of glucose and IPTG is indicated by the filled black circles in the left column. The XR-seq bars (red for TS and blue for NTS) exhibit the repair levels on the y axis, which is scaled to 50 RPM (reads per million mapped reads). The TS/NTS ratios, which quantitatively represent TCR, from two experiments (Exp. 1 and Exp. 2) are horizontally plotted. β-Galactosidase (β-gal) activities are shown in Miller units with the standard deviations. The XR-seq signal shown in green represents TCR independent of Mfd. It is within the lac operator O1 and indicates inhibition of repair by bound lac repressor, which cannot be overcome by Mfd.
We considered the possibility that TCR may have a biphasic character in which the early mode mediated by Mfd is followed by a second repair mode that becomes active upon induction of UvrD protein and the ppGpp alarmone. Hence we conducted a time-course XR-seq experiment in the UvrD− background with and without IPTG induction and analyzed repair in lacZ. The drawback of UvrD− strains is the fact that there is a slow catalytic turnover of the Uvr(A)BC proteins and slow release and degradation of the excised oligonucleotide. Therefore, an XR-seq profile at a certain time essentially represents cumulative rather than ongoing repair. To overcome this drawback, we performed subtractive analysis to determine the incremental change in repair from one time point to the next (see “Experimental procedures”). As seen in Fig. 4, the TS is preferentially repaired only when the operon is actively transcribed, even at later time points. This observation shows that TCR is UvrD-independent at all stages, consistent with the notion that Mfd/TRCF is solely responsible for coupling transcription to repair in E. coli.
Figure 4.
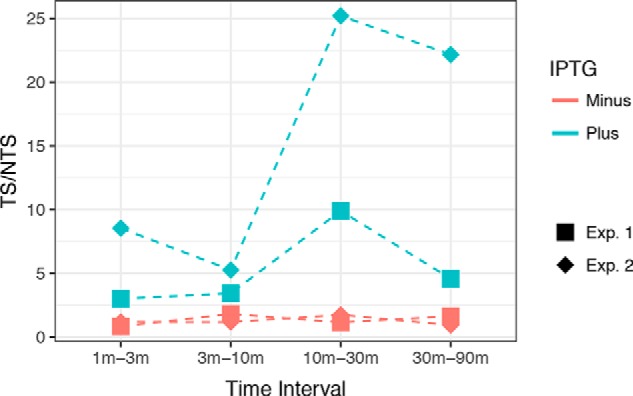
Time-course subtractive analysis of the TS/NTS in lacZ gene. Results were obtained with E. coli uvrD− phr− cells irradiated with 60 J/m2 UV and allowed to repair for 1, 3, 10, 30, and 90 min. On the y axis, each data point represents a TS/NTS value derived from the change in reads seen at successive time points. The x axis shows the time intervals; for example, the initial time-point (1 min) reads were subtracted from 3-min reads to obtain the reads plotted for the 1-min to 3-min time interval. Each data point from two experiments (Exp. 1 and Exp. 2) is plotted separately. Blue and red colors indicate plus and minus IPTG induction, respectively.
Discussion
Following the identification of Mfd as the protein responsible for preferential repair of UV-induced cyclobutane pyrimidine dimers in the anticodon of some tRNA genes and for being essential for transcription-coupled repair in several in vitro assays, it was concluded that Mfd was solely responsible for specifically stimulating excision repair in the transcribed strand (11, 12). Thus, it was concluded that Mfd and the biochemically identified transcription–repair coupling factor were identical and hence Mfd and TRCF were used interchangeably. In vivo mutagenesis experiments (29) and structural (16, 17) and single-molecule (18) studies confirmed and refined the original model.
However, several studies reported results suggesting either that factors in addition to Mfd and Uvr(A)BC were required for TCR or that there were multiple pathways for TCR. First, in a high-resolution study of the initially transcribed region of lacZ, it was reported that template strand repair stimulated by IPTG was Mfd-independent (30). Our results overall are not consistent with these findings; however, we did observe IPTG-dependent, Mfd-independent repair at one site, located in the operator-binding site (O1) in the initially transcribed region (shown in green in Fig. 3). Thus, binding of the lac repressor inhibits repair. Second, it was reported that mutL−and mutS− mutations abolished TCR of the lacZ operon in a manner identical to the mfd mutation (31). However, in vitro experiments with cell-free extracts revealed that although mfd− extract was deficient in TCR, both mutL− and mutS− extracts performed normal TCR, and thus the in vivo findings implicating these genes were ascribed to the limitations in the in vivo assay (13). Finally, a third series of studies using an indirect in vitro repair assay, and the lac-inducible system in uvrD−, ppGppo, or uvrD− ppGppo backgrounds, concluded that UvrD with the aid of damage-induced ppGpp led to backtracking of RNAP stalled at damage, allowing the Uvr(A)BC to repair damage and enabling RNAP to elongate the truncated transcript (19). We are unable to confirm these results. We have no explanation for the data in the report that links ppGpp to TCR. Importantly, using the canonical lacZ-inducible operon, we show that TCR in this system is absolutely dependent on Mfd and that its magnitude as defined by the TS/NTS repair ratio is unaffected by ppGpp and either unaffected or enhanced by the absence of UvrD because of the lack of catalytic turnover of Uvr(A)BC excision nuclease. We conclude that Mfd/TRCF is the sole protein that couples transcription to repair in E. coli.
Experimental procedures
Strains are described in supplemental Table S1. Double mutant ΔrelA25GΔspoT203 strains are unable to synthesize ppGpp and are designated ppGppo. Cultures were grown in LB (10 g of Tryptone, 5 g of yeast extract, 10 g of NaCl/liter). For experiments, overnight liquid cultures (MG1655-derived strains) or colonies grown overnight on LB-agar (CF10237-derived strains) were used to inoculate LB supplemented with glucose (0.4%) and/or IPTG (1 mm) where indicated. Experimental cultures were grown at 37 °C to an OD of ∼0.4, which, at the dilutions used, took over an hour.
For measuring β-galactosidase, 0.2- or 0.3-ml volumes of culture were put on ice and pelleted, and then pellets were frozen at −80 °C. β-Galactosidase activity was measured as described (32) using the procedure in which 20 μl of SDS (0.1%) and 20 μl of chloroform were mixed with resuspended cells.
For the XR-seq assay, cells grown to an OD of ∼0.36 were cooled to room temperature by swirling in a water bath at about 15 °C. 15-ml volumes were transferred to R-150 dishes and irradiated at room temperature with UV at 60 J/m2 with agitation. Cell survival under these conditions was 88% (40 J/m2) and 72% (120 J/m2) for MGP cells and 70% (40 J/m2) and 41% (120 J/m2) for CFPK cells. The dose rate was ∼2 J/m2/s. One minute after light exposure began, dishes were placed on ice water, and cells were harvested and processed as described previously (23), with exceptions noted below. Timing of the irradiations was such that 10 plates were successively irradiated in a batch, with the irradiation, repair, and placement of plates on ice taking 12 min per batch. By this approach, all cells were irradiated at an OD of ∼0.4. Experiments utilized a total of either 600 ml or 300 ml of cells irradiated and repaired in this manner, with the exception of uvrD− cells, which generate sufficient excision product (23) that only 30 ml of cells were needed for XR-seq experiments.
In repair experiments with CF-10237 derivatives, dilutions of unirradiated cells at OD ≈ 0.4 were plated on LB and M9 to determine whether substantial numbers of RNA polymerase revertants had arisen, as suggested (33). Only revertants are able to grow on M9, and revertants were present at less than 0.1% in the populations subsequently irradiated.
The XR-seq procedure was modified slightly from prior experiments (23). In an early step of XR-seq, the partially purified product of excision is ligated on both ends to adapters. In these experiments, we used a modified 3′-adapter called “A3b,” which includes an 8-base random sequence. The purpose of this sequence was to allow the identification and elimination of duplicate reads introduced in the PCR step. A3b is made by annealing the oligonucleotides A3Fb: SpC3-3′-GGAACCGTGGGCTCTTAAGGTNNNNNNNNGCCGTGAGTATGCTTGACTCGG(P)-5′ and A3Rb: 5′-CGGCACTCATACGAACTGAGCCNNNNN-SpC3-3′. These oligonucleotides were purchased from Integrated DNA Technologies, and are phosphorylated (P), possess random A,G,C or T (N), or are blocked by spacer-C3 (SpC3-3) as indicated. The 5′-adapter was as described in Hu et al. (22).
We used cutadapt (34) to trim adapter sequences with the following parameter: a, GGCTCAGTTCGTATGAGTGCCG. We analyzed the reads of 13-mer length having dithymines at the expected damage site (8th and 9th positions of the 13-mer reads). The reads were aligned to the E. coli reference genome retrieved from the National Center for Biotechnology Information (NCBI, accession number NC_000913.2) using bowtie (35) with the following parameters: q; nomaqround; phred33-quals; S; seed 123. Samtools (36) and bedtools (37) were used to process the alignment. Strands were separated by a custom script. The genomic coordinates were normalized and converted to bedGraph and BigWig files using bedtools (37) and ucsctools (38), respectively. Per-gene repair signals were computed using bedtools and normalized to reads per kilobase per million mapped reads with custom scripts. Publicly available RNA sequencing (RNA-seq) data sets (27) were analyzed as we reported before (23).
The time-course subtractive analysis of the UvrD− XR-seq datasets was performed with custom scripts. Random sampling was applied to retrieve 12 million reads from each sample. Each unique genomic location was counted, and the counts of a sample of a certain time point were subtracted from the subsequent one (time points were 1, 3, 10, 30, and 90 min).
Author contributions
O. A., A. S., and C. P. S. designed research, analyzed data, and wrote the paper. O. A. and C. P. S. performed research. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
This work was supported by National Institutes of Health Grants GM118102 and ES027255. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Table S1.
- NTS
- nontranscribed strand
- TS
- transcribed strand
- TCR
- transcription-coupled repair
- TRCF
- transcription–repair coupling factor
- XR-seq
- excision repair–sequencing
- RNAP
- RNA polymerase
- ppGpp
- guanosine pentaphosphate
- IPTG
- isopropyl-1-thio-β-d-galactopyranoside
- OD
- optical density.
References
- 1. Wood R. D. (1997) Nucleotide excision repair in mammalian cells. J. Biol. Chem. 272, 23465–23468 [DOI] [PubMed] [Google Scholar]
- 2. Sancar A. (2016) Mechanisms of DNA Repair by Photolyase and Excision Nuclease (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 55, 8502–8527 [DOI] [PubMed] [Google Scholar]
- 3. Mellon I., Spivak G., and Hanawalt P. C. (1987) Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51, 241–249 [DOI] [PubMed] [Google Scholar]
- 4. Mellon I., and Hanawalt P. C. (1989) Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature 342, 95–98 [DOI] [PubMed] [Google Scholar]
- 5. Hanawalt P. C., and Spivak G. (2008) Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9, 958–970 [DOI] [PubMed] [Google Scholar]
- 6. Selby C. P., and Sancar A. (1990) Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. J. Biol. Chem. 265, 21330–21336 [PubMed] [Google Scholar]
- 7. Selby C. P., and Sancar A. (1991) Gene- and strand-specific repair in vitro: partial purification of a transcription-repair coupling factor. Proc. Natl. Acad. Sci. U.S.A. 88, 8232–8236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Selby C. P., Witkin E. M., and Sancar A. (1991) Escherichia coli mfd mutant deficient in “mutation frequency decline” lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc. Natl. Acad. Sci. U.S.A. 88, 11574–11578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Witkin E. M. (1966) Radiation-induced mutations and their repair. Science 152, 1345–1353 [DOI] [PubMed] [Google Scholar]
- 10. Bockrath R. C., and Palmer J. E. (1977) Differential repair of premutational UV-lesions at tRNA genes in E. coli. Mol. Gen. Genet. 156, 133–140 [DOI] [PubMed] [Google Scholar]
- 11. Selby C. P., and Sancar A. (1993) Molecular mechanism of transcription-repair coupling. Science 260, 53–58 [DOI] [PubMed] [Google Scholar]
- 12. Selby C. P. (2017) Mfd protein and transcription-repair coupling in Escherichia coli. Photochem. Photobiol. 93, 280–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Selby C. P., and Sancar A. (1995) Structure and function of transcription-repair coupling factor. I. Structural domains and binding properties. J. Biol. Chem. 270, 4882–4889 [DOI] [PubMed] [Google Scholar]
- 14. Selby C. P., and Sancar A. (1995) Structure and function of transcription-repair coupling factor. II. Catalytic properties. J. Biol. Chem. 270, 4890–4895 [DOI] [PubMed] [Google Scholar]
- 15. Park J. S., Marr M. T., and Roberts J. W. (2002) E. coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell 109, 757–767 [DOI] [PubMed] [Google Scholar]
- 16. Manelyte L., Kim Y. I., Smith A. J., Smith R. M., and Savery N. J. (2010) Regulation and rate enhancement during transcription-coupled DNA repair. Mol. Cell 40, 714–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deaconescu A. M., Chambers A. L., Smith A. J., Nickels B. E., Hochschild A., Savery N. J., and Darst S. A. (2006) Structural basis for bacterial transcription-coupled DNA repair. Cell 124, 507–520 [DOI] [PubMed] [Google Scholar]
- 18. Fan J., Leroux-Coyau M., Savery N. J., and Strick T. R. (2016) Reconstruction of bacterial transcription-coupled repair at single-molecule resolution. Nature 536, 234–237 [DOI] [PubMed] [Google Scholar]
- 19. Kamarthapu V., Epshtein V., Benjamin B., Proshkin S., Mironov A., Cashel M., and Nudler E. (2016) ppGpp couples transcription to DNA repair in E. coli. Science 352, 993–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Magnusson L. U., Farewell A., and Nyström T. (2005) ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 13, 236–242 [DOI] [PubMed] [Google Scholar]
- 21. Ahn B. (2000) A physical interaction of UvrD with nucleotide excision repair protein UvrB. Mol. Cells 10, 592–597 [DOI] [PubMed] [Google Scholar]
- 22. Hu J., Adar S., Selby C. P., Lieb J. D., and Sancar A. (2015) Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 29, 948–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Adebali O., Chiou Y. Y., Hu J., Sancar A., and Selby C. P. (2017) Genome-wide transcription-coupled repair in Escherichia coli is mediated by the Mfd translocase. Proc. Natl. Acad. Sci. U.S.A. 114, E2116–E2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Husain I., Van Houten B., Thomas D. C., Abdel-Monem M., and Sancar A. (1985) Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proc. Natl. Acad. Sci. U.S.A. 82, 6774–6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Orren D. K., Selby C. P., Hearst J. E., and Sancar A. (1992) Post-incision steps of nucleotide excision repair in Escherichia coli: disassembly of the UvrBC-DNA complex by helicase II and DNA polymerase I. J. Biol. Chem. 267, 780–788 [PubMed] [Google Scholar]
- 26. Wade J. T., and Grainger D. C. (2014) Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat. Rev. Microbiol. 12, 647–653 [DOI] [PubMed] [Google Scholar]
- 27. Thomason M. K., Bischler T., Eisenbart S. K., Förstner K. U., Zhang A., Herbig A., Nieselt K., Sharma C. M., and Storz G. (2015) Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J. Bacteriol. 197, 18–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lloréns-Rico V., Cano J., Kamminga T., Gil R., Latorre A., Chen W. H., Bork P., Glass J. I., Serrano L., and Lluch-Senar M. (2016) Bacterial antisense RNAs are mainly the product of transcriptional noise. Sci. Adv. 2, e1501363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oller A. R., Fijalkowska I. J., Dunn R. L., and Schaaper R. M. (1992) Transcription-repair coupling determines the strandedness of ultraviolet mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 89, 11036–11040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kunala S., and Brash D. E. (1995) Intragenic domains of strand-specific repair in Escherichia coli. J. Mol. Biol. 246, 264–272 [DOI] [PubMed] [Google Scholar]
- 31. Mellon I., and Champe G. N. (1996) Products of DNA mismatch repair genes mutS and mutL are required for transcription-coupled nucleotide-excision repair of the lactose operon in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 93, 1292–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miller J. H. (1972) Assay of the lac operon enzymes. in Experiments in Molecular Genetics , pp. 349–376, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33. Vinella D., Potrykus K., Murphy H., and Cashel M. (2012) Effects on growth by changes of the balance between GreA, GreB, and DksA suggest mutual competition and functional redundancy in Escherichia coli. J. Bacteriol. 194, 261–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12, 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 35. Langmead B., Trapnell C., Pop M., and Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., and 1000 Genome Project Data Processing Subgroup (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Quinlan A. R. (2014) BEDTools: the Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinformatics 47, 11.12.1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kent W. J., Zweig A. S., Barber G., Hinrichs A. S., and Karolchik D. (2010) BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26, 2204–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.