Abstract
Cardiac hypertrophy is closely linked to impaired fatty acid oxidation, but the molecular basis of this link is unclear. Here, we investigated the loss of an obligate enzyme in mitochondrial long-chain fatty acid oxidation, carnitine palmitoyltransferase 2 (CPT2), on muscle and heart structure, function, and molecular signatures in a muscle- and heart-specific CPT2-deficient mouse (Cpt2M−/−) model. CPT2 loss in heart and muscle reduced complete oxidation of long-chain fatty acids by 87 and 69%, respectively, without altering body weight, energy expenditure, respiratory quotient, or adiposity. Cpt2M−/− mice developed cardiac hypertrophy and systolic dysfunction, evidenced by a 5-fold greater heart mass, 60–90% reduction in blood ejection fraction relative to control mice, and eventual lethality in the absence of cardiac fibrosis. The hypertrophy-inducing mammalian target of rapamycin complex 1 (mTORC1) pathway was activated in Cpt2M−/− hearts; however, daily rapamycin exposure failed to attenuate hypertrophy in Cpt2M−/− mice. Lysine acetylation was reduced by ∼50% in Cpt2M−/− hearts, but trichostatin A, a histone deacetylase inhibitor that improves cardiac remodeling, failed to attenuate Cpt2M−/− hypertrophy. Strikingly, a ketogenic diet increased lysine acetylation in Cpt2M−/− hearts 2.3-fold compared with littermate control mice fed a ketogenic diet, yet it did not improve cardiac hypertrophy. Together, these results suggest that a shift away from mitochondrial fatty acid oxidation initiates deleterious hypertrophic cardiac remodeling independent of fibrosis. The data also indicate that CPT2-deficient hearts are impervious to hypertrophy attenuators, that mitochondrial metabolism regulates cardiac acetylation, and that signals derived from alterations in mitochondrial metabolism are the key mediators of cardiac hypertrophic growth.
Keywords: acetylation, cardiac hypertrophy, cardiac metabolism, fatty acid oxidation, mammalian target of rapamycin (mTOR)
Introduction
The heart and muscle are continuously working organs with high energetic demands and limited capacity to store energetic reserves. The heart and muscle rely heavily on mitochondrial oxidative metabolism and particularly on fatty acids as substrate. Under circumstances when mitochondrial metabolism is reduced, the heart undergoes hypertrophic remodeling. For instance, mitochondrial metabolism is disrupted in cardiac hypertrophy induced by hypertension, obesity, diabetes, and aging (1–5). The cardiac hypertrophy that occurs in response to these stresses results in altered cardiac function, metabolism, and molecular signature. This hypertrophy is independently associated with increasing age, blood pressure, obesity, valve disease, and myocardial infarction and is a strong predictor of all-cause mortality (6–11). Despite the recognition of left ventricular remodeling as a significant risk factor for death, there is a lack of therapeutic options to treat hypertrophic remodeling beyond transplantation; thus, a better understanding of the molecular underpinnings of remodeling are needed.
To investigate how the complete loss of long-chain mitochondrial fatty acid oxidation affects heart and muscle function and health, we generated a heart and muscle carnitine palmitoyltransferase 2 (CPT2)-deficient2 mouse model. CPT2 is an enzyme required for mitochondrial long-chain fatty acid oxidation. CPT2 mediates the conversion of CPT1-generated acyl-carnitines to acyl-CoA within the matrix of the mitochondria. CPT2 is encoded by a single gene, and therefore no other enzymes can compensate. Thus, the loss of CPT2 incapacitates the acyl-carnitine-mediated mitochondrial oxidation of long-chain fatty acids. Here, we show that the loss of mitochondrial fatty acid oxidation caused by the genetic deletion of CPT2 causes deleterious cardiac hypertrophic remodeling and early lethality. We show that CPT2 deficiency–induced hypertrophy is devoid of fibrosis and progresses to dilation and expansion of the left ventricular chamber prior to death. CPT2-deficient hearts have activated mammalian target of rapamycin complex (mTORC1) and reduced protein acetylation, two molecular events shown to modulate hypertrophy. Unexpectedly, however, inhibition of mTOR and reversal of protein hypoacetylation failed to attenuate hypertrophy in the absence of CPT2. These data suggest that attenuation of hypertrophic remodeling is mechanistically dependent upon improving mitochondrial fatty acid oxidation, consistent with recent reports on the role of mTOR inhibition and acetylation improving cardiac function by increasing rates of fatty acid oxidation in aging hearts (12–15). Taken together, these results highlight the dependence of cardiac hypertrophy on mitochondrial fatty acid oxidation.
Results
Deletion of CPT2 in heart and skeletal muscle inhibits fatty acid oxidation without altering body weight
Genetically targeted Cre-recombinase-dependent floxed CPT2-deficient mice were generated as previously described (16). CPT2-targeted mice were crossed with muscle creatine kinase (MCK)-Cre mice to generate a model of cardiac and muscle CPT2 deficiency (Cpt2M−/−). The Cpt2M−/− mice were viable and born at the expected Mendelian ratios. In Cpt2M−/− mice, CPT2 protein was absent in the heart and muscle as indicated by Western blot (Fig. 1A). Unexpectedly, heterozygous Cpt2f/+ MCK-Cre (Cpt2M+/−) hearts had similar CPT2 protein compared with controls, whereas the skeletal muscle of Cpt2M+/− presented with intermediate CPT2 protein, suggesting a distinct regulatory mechanism of CPT2 protein abundance in a tissue-dependent manner. The loss of CPT2 in heart and gastrocnemius muscle resulted in an 87 and 69% reduction in complete fatty acid oxidation into CO2, respectively, and an 84 and 57% reduction in incomplete fatty acid oxidation into acid-soluble metabolites (ASMs), respectively (Fig. 1, B and C). The residual fatty acid oxidation is likely contributed by peroxisomal oxidation, with more peroxisomal oxidation in the Cpt2-deficient muscle, relative to the heart, suggesting an inability for peroxisomes to compensate for the loss of mitochondria long-chain fatty acid oxidation particularly in the heart. To confirm that impaired fatty acid oxidation in the heart and muscle causes the Cpt2M−/− mice to rely more on alternative substrates, 14C-2-deoxyglucose was injected retro-orbitally, and tissue-specific uptake of the 14C label was determined 30 min later. We show a 12-fold increase in deoxyglucose in Cpt2M−/− heart, relative to controls (Fig. 1D). These data suggest an increased reliance of Cpt2M−hearts on non-fatty acid substrates such as glucose.
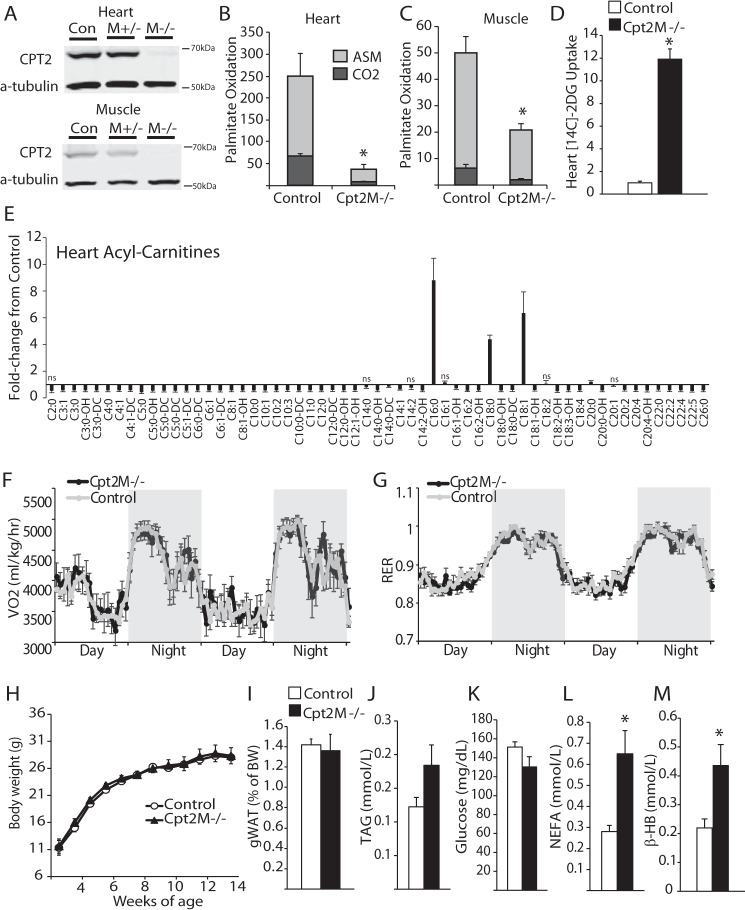
Figure 1.
Generation of cardiac and skeletal muscle CPT2 knock-out mouse. A, representative Western blot depicting the loss of CPT2 protein in heart and tibialis anterior muscle homogenates. Con, control. B and C, rate of [1-14C]palmitate oxidation to CO2 or ASMs in (B) heart or (C) gastrocnemius control or CPT2M−/− homogenates, expressed as nmol/mg protein/h (n = 5–6). D, 14C-2-deoxyglucose uptake into control and CPT2M−/− hearts (n = 3). E, relative acyl-carnitine profile of hearts from control and CPT2M−/− male mice at 8 weeks of age, all points significant unless marked with ns for non-significant (n = 7–9). F and G, whole-body oxygen consumption (VO2, F) and respiratory exchange ratio (RER, G) of control and CPT2M−/− male mice at 7 weeks of age (n = 7–9). H and I, body weight over time (H) and weight of gonadal white adipose tissue (gWAT, I) at 8 weeks of age from control and CPT2M−/− male mice (n = 6). J–M, plasma TAG (J), glucose (K), NEFAs (L), and βHB (M) from 8-week-old control and CPT2M−/− male mice (n = 6–8). *, significance by Student's t test (p ≤ 0.05).
To get a better understanding of oxidative flux in the heart, we performed relative quantification of cardiac acyl-carnitines by mass spectrometry. Acyl-carnitines are formed not only as intermediates of fatty acid oxidative flux but are also formed from CoA-containing mitochondrial metabolic intermediates by the exchange of CoA for carnitine. As expected, all medium-chain and several long-chain acyl-carnitine species were reduced by ∼50% in the Cpt2M−/− heart, relative to controls (Fig. 1E). Also as expected, several long-chain acyl-carnitines, the substrate for Cpt2, are increased in Cpt2M−/− hearts (Fig. 1E). We also observe reductions in all short-chain acyl-carnitine species (Fig. 1E). These data indicate reduced flux of fatty acid oxidation in Cpt2M−/− heart.
Because the heart and muscle are highly energetically demanding organs that rely on fatty acid oxidation for a majority of energy (17), we questioned whether a shift away from fatty acid oxidation in the Cpt2M−/− heart and muscle would result in altered whole-body substrate utilization. To determine substrate use, control and Cpt2M−/− mice were placed in metabolic chambers. Despite a loss of cardiac and muscle fatty acid oxidation, Cpt2M−/− mice showed similar whole-body oxygen consumption and respiratory exchange ratio relative to controls (Fig. 1, F and G). In agreement, we did not observe any change in body weight or adiposity between the control and Cpt2M−/− mice at 8 weeks of age (Fig. 1, H and I). Plasma triacylglycerol and glucose were similar between genotypes, whereas non-esterified fatty acids (NEFAs) and β-hydroxybutyrate (βHB) were increased 2.3- and 2-fold in Cpt2M−/− mice, respectively (Fig. 1, J–M).
Effect of CPT2 deficiency on mitochondria and lipid accumulation
To determine whether mitochondria were intact and functional in CPT2-deficient heart and muscle, the oxidation rates of octanoate, a medium-chain fatty acid that bypasses the CPT2-mediated acyl-carnitine shuttle, and pyruvate were determined. In the Cpt2M−/− gastrocnemius muscle, the rates of octanoate oxidation were similar to controls, yet the oxidation of pyruvate was increased 1.5-fold (Fig. 2, A and B). Upon examination of soleus muscle structure by TEM, mitochondrial area of Cpt2M−/− was not different from controls (Fig. 2, C and D). Total triacylglycerol composition in control and Cpt2M−/− soleus and gastrocnemius muscle were similar (Fig. 2E). These data suggest compensatory up-regulation of alternative mitochondrial substrate oxidative pathways in the muscle.
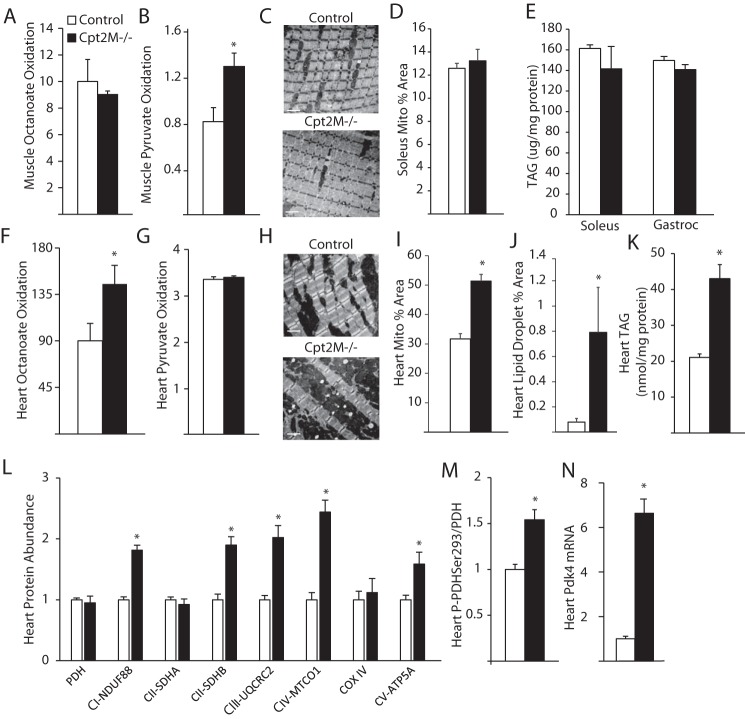
Figure 2.
Mitochondrial dysregulation in CPT2-deficient heart. A and B, gastrocnemius muscle homogenate oxidation rates for octanoate (A) and pyruvate (B) in control and Cpt2M−/− mice (n = 4–5). C and D, representative TEM images (C) and quantification of mitochondrial area from TEM images (D) of 12-week-old female control and CPT2M−/− soleus muscle (n = 3). Scale bars represent 2 μm. E, muscle TAG from control and CPT2M−/− 8-week-old male mice (n = 6). F and G, heart homogenate oxidation rates for octanoate (F) and pyruvate (G) in control and Cpt2M−/− mice, expressed as nmol/mg protein/h (n = 4–5). H–J, representative TEM images (H), and quantification of mitochondrial area (I) and lipid droplet area (J) from TEM images of 12-week-old female control and CPT2M−/− hearts (n = 3). Scale bars represent 2 μm. K, heart triacylglycerol from control and CPT2M−/− 8-week-old male mice (n = 6). L–N, heart protein abundance of mitochondrial proteins (L), quantification of phosphorylation of PDH (M), and mRNA abundance of Pdk4 (N) in 8-week-old male control and CPT2M−/− hearts (n = 6). *, significance by Student's t test (p ≤ 0.05).
In the heart, the rates of octanoate oxidation were increased 1.6-fold, but rates of pyruvate oxidation were unchanged in Cpt2M−/− relative to controls (Fig. 2, F and G). TEM analysis revealed increased mitochondrial and lipid droplet area in the hearts of Cpt2M−/− relative to controls (Fig. 2, H–J). The increase in lipid droplet was confirmed by a 2-fold increase in triacylglycerol (TAG) quantified from heart lipid extracts (Fig. 2K). In confirmation of enhanced mitochondria in the heart, we observed increased expression of several mitochondrial proteins (Fig. 2L). Specifically, the expression of mitochondrial electron transport protein subunits from complex I, II, III, and IV were significantly up-regulated; however, pyruvate dehydrogenase (PDH), complex II subunit A, and cytochrome oxidase IV were not increased in Cpt2M−/− relative to controls (Fig. 2L). To determine why pyruvate oxidation is not increased with increased mitochondrial content, the regulation of PDH by phosphorylation was determined to reveal a 1.5-fold increase in PDH phosphorylation at Ser-293 in Cpt2M−/− hearts (Fig. 2M). The phosphorylation of PDH at Ser-293 inhibits its activity and is regulated by pyruvate dehydrogenase kinases (Pdks). Indeed, the mRNA abundance of Pdk4 was 6.6-fold higher in Cpt2M−/− hearts (Fig. 2N). Together, these data suggest that the loss of CPT2 differentially regulates muscle and heart mitochondrial properties and that the heart maladaptively inhibits pyruvate oxidation but increases mitochondrial content.
Loss of CPT2 results in cardiac hypertrophy
Consistent with fatty acid oxidation deficiency causing cardiac hypertrophy, we observed a 3-fold increase in Cpt2M−/− heart mass at 10 weeks of age (Fig. 3A). To confirm that this effect was not dependent on developmental or skeletal muscle effects of CPT2 deficiency, the Cpt2f/f mice were bred to heart-specific tamoxifen-inducible Cre mice (Cpt2HT2−/−) and injected with tamoxifen at 8 weeks of age, and ∼6 months later a 3-fold increase in heart mass in the Cpt2HT2−/− mice was observed (Fig. 3A). To avoid confounding effects of the tamoxifen, Cpt2M−/− mice were used for subsequent experiments. Cardiomyocyte hypertrophy in Cpt2M−/− hearts was confirmed by determination of the cross-sectional area of cardiomyocytes on histological images, which revealed a 4-fold increase, relative to controls (Fig. 3B).
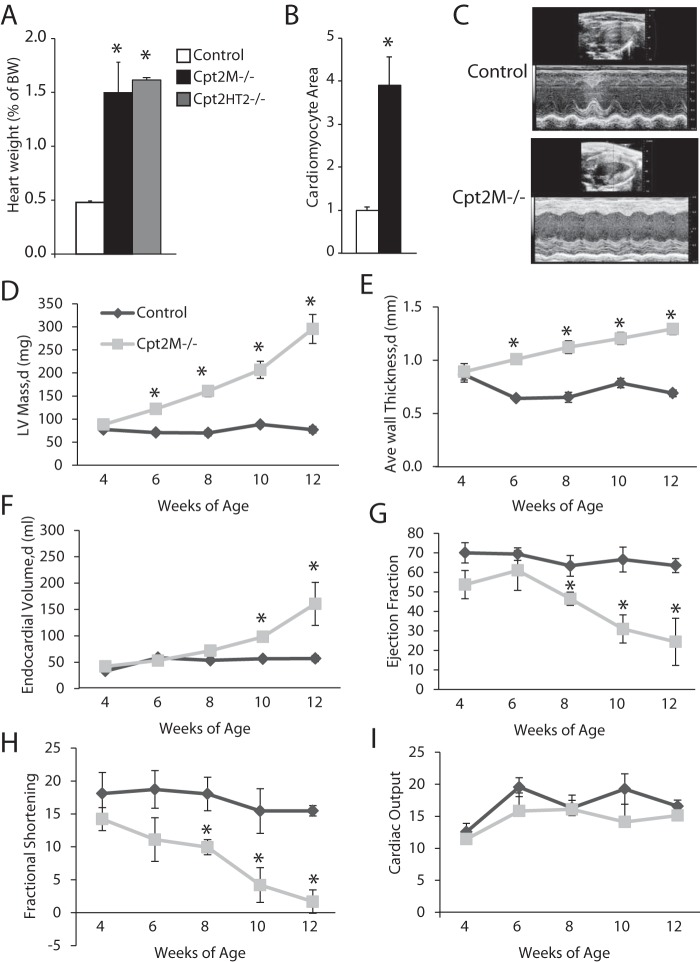
Figure 3.
Loss of CPT2 results in cardiac hypertrophy. A, heart weight relative to body weight (BW) of control, Cpt2M−/−, and Cpt2HERT2−/− mice (n = 3–6). B, relative cardiomyocyte cross-sectional area from control and Cpt2M−/− male mice at 8 weeks of age (n = 3). C, representative echocardiogram long-axis B-mode at diastole and M-mode images from control and Cpt2M−/− female mice at 12 weeks of age. D–I, echocardiogram-derived LV mass (D), LV wall thickness (E), LV end diastolic endocardial volume (F), ejection fraction (G), fractional shortening (H), and cardiac output (I) from control and Cpt2M−/− female mice from 4 to 12 weeks of age (n = 3–4). d, diastole. *, significance by Student's t test (p ≤ 0.05).
Echocardiograms were performed every 2 weeks in control and Cpt2M−/− to determine how cardiac remodeling progressed and altered cardiac function from 4 to 12 weeks of age. Representative echocardiograms are shown in long-axis B mode and M mode from 12-week-old control and Cpt2M−/− female mice (Fig. 3C). The echocardiogram image analysis revealed a gradual and linear-like increase in Cpt2M−/− left ventricular (LV) mass up to 3.4-fold greater than controls at 12 weeks of age (Fig. 3D). The increase in LV mass coincides with a 1.5-fold increase in thickening of the LV wall and a 3.7-fold increase in LV end-diastolic endocardial volume in Cpt2M−/− mice relative to age-matched littermate controls (Fig. 3, E and F). As the hypertrophy progressed in Cpt2M−/− mice from 4 to 12 weeks of age, a decline in ejection fraction from 54% to 22% and a decline in fractional shortening from 14% to 3.5% were observed (Fig. 3, G and H). The reduced ejection fraction indicates systolic dysfunction and reduced contractility in Cpt2M−/− mice. However, the increase in left ventricular mass and LV end diastolic endocardial volume coincided with reduced ejection fraction, resulting in maintained cardiac output in Cpt2M−/− mice to levels similar to controls (Fig. 3I). Thus, the loss of cardiac mitochondrial long-chain fatty acid oxidation caused by CPT2 deficiency resulted in the progressive development of severe cardiac hypertrophy and systolic dysfunction.
Because the Cpt2M−/− mice display systolic dysfunction, the question remained how long can Cpt2M−/− mice maintain sufficient heart function? To this end, we found that Cpt2M−/− hearts eventually failed, resulting in early lethality of Cpt2M−/− mice from 6 to 26 weeks of age in male and female mice (Fig. 4, A and B). At the end stage, prior to death, Cpt2M−/− mice show acute signs of illness characterized by lethargy, acute weight loss, kyphosis, and labored breathing. End stage echocardiogram analysis revealed dilated hypertrophy evidenced by a 5-fold increase in LV mass, a 7-fold increase in end-diastolic endocardial volume, reduced ejection fraction to 6.5%, and maintained cardiac output (Fig. 4, C–F).
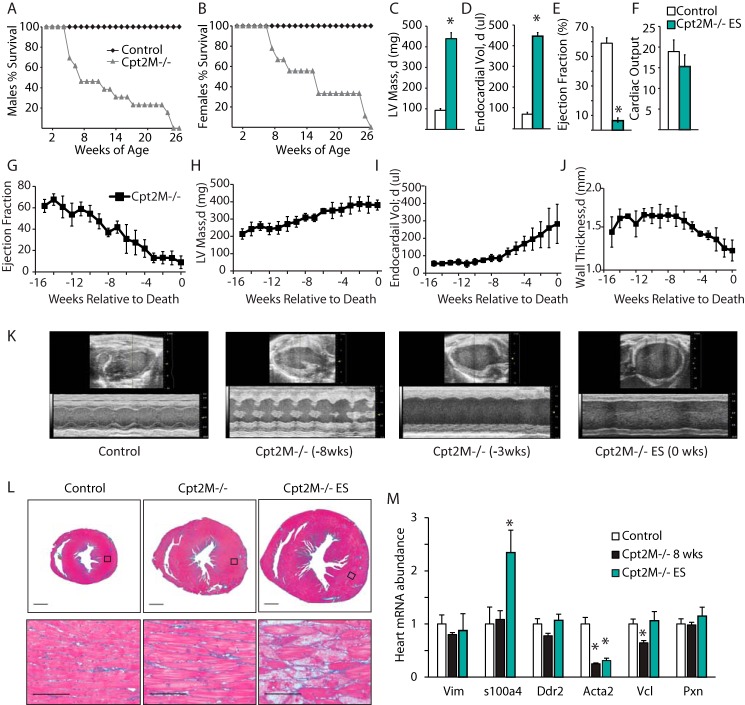
Figure 4.
Fibrosis-free dilated hypertrophic cardiac remodeling and early lethality in Cpt2M−/− mice. A and B, Kaplan–Meier plot of death in male (A) and female (B) control and Cpt2M−/− mice (n = 10–17). C–F, echocardiogram-derived LV mass (C), LV end diastolic endocardial volume (D), ejection fraction (E), and cardiac output (ml/min, F) in control and Cpt2M−/− mice at the end stage of disease (n = 4–5). G–J, weekly echocardiogram-derived ejection fraction (G), LV mass (H), LV end diastolic endocardial volume (I), and average LV wall thickness (J) in Cpt2M−/− female mice during the 16 weeks prior to death (n = 4). K, representative long-axis B-mode in diastole and M-mode images from a female control or Cpt2M−/− mouse 8, 3, or 0 weeks prior to death. L, representative short-axis Masson's Trichrome-stained heart histology from control or Cpt2M−/− male mice at mid- or end-stage hypertrophy as defined by LV end diastolic endocardial volume above 100 μl. M, relative mRNA abundance of cardiac fibrosis gene markers in hearts from control or Cpt2M−/− male 8-week-old or end-stage mice (n = 4–6). *, significance by Student's t test (p ≤ 0.05).
To determine how cardiac structure and function changed prior to end stage in Cpt2M−/− mice, weekly echocardiograms were performed until death occurred. Analysis of the echocardiogram images revealed a gradual decline in ejection fraction at end stage, indicating progressive decline in systolic function (Fig. 4G). This reduction in ejection fraction was accompanied by a cease in LV mass growth during the 3–4 weeks prior to the end stage, concomitant with a steady increase in end-diastolic endocardial volume and reduction in LV wall thickness (Fig. 4, H–J). The extreme remodeling and dilation are evident in the representative echocardiographic long-axis B-mode and M-mode images (Fig. 4K). These data suggest progressive dilation accompanied by attenuation in increasing heart mass that results in increased end-diastolic endocardial volume and reduction of ejection fraction preceding the end stage in Cpt2M−/− mice.
Cardiac fibrosis is believed to be a major contributor to mortality in cardiac hypertrophy because of its interference with electrical conduction. Despite the heart failure observed in the Cpt2M−/− mice, control and Cpt2M−/− female hearts did not show fibrosis indicated by Masson's Trichrome staining at the early stage when ejection fraction was 66% or at the end stage when ejection fraction was 18% (Fig. 4L). To confirm the lack of fibrosis, several gene markers were examined. Vimentin and fibroblast-specific protein 1 (s1004a) are gene markers for extracellular matrix remodeling from fibroblasts (18). Discoidin domain receptor (Ddr2), smooth muscle myosin heavy chain (Acta2), vinculin (Vcl), and paxillin (Pxn) are gene markers of extracellular matrix remodeling by cardiac fibroblasts and myofibroblasts (18). Consistent with the histological analysis, none of the fibrotic gene markers were increased in 8-week-old Cpt2M−/− mouse hearts, relative to controls (Fig. 4M). At the end stage, only one of the fibrotic gene markers was up-regulated, a 2.4-fold induction of s1004a, in Cpt2M−/− hearts (Fig. 4M). Together, these data suggest that CPT2 deficiency causes severe cardiac remodeling and heart failure independent of fibrosis.
Molecular underpinnings of CPT2 deficiency–induced cardiac hypertrophy
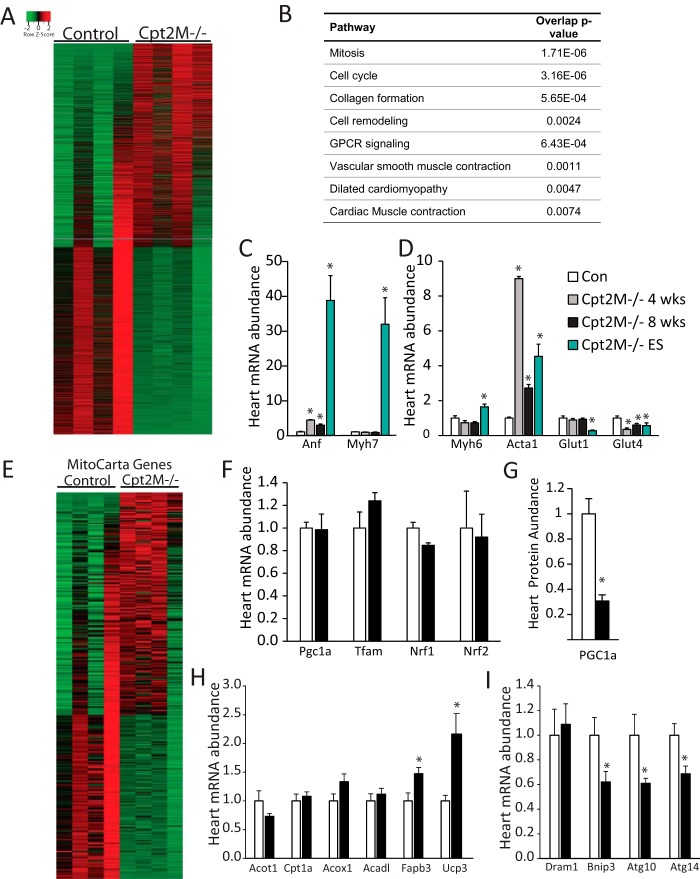
To further understand how loss of CPT2 regulates the molecular signature of the heart, RNA extracted from 8-week-old male control or Cpt2M−/− hearts was subjected to RNAseq analysis. Overall, 1646 genes were significantly up-regulated (adjusted p value < 0.05), and 2208 genes were significantly down-regulated (Fig. 5A and supplemental file). These differentially expressed genes were further analyzed for enriched pathways and biological processes using InnateDB and Ingenuity Pathway Analysis software. As expected, the most regulated pathways in our data set were related to cell cycle and mitosis, cellular remodeling, muscle contraction, and cardiomyopathy (Fig. 5B). Ingenuity Pathway Analysis software analysis for the most significantly regulated diseases and biofunctions were lipid metabolism and cardiovascular system development and function (supplemental Table 1). Intriguingly, although these functional analyses suggested changes in the cardiomyocyte and cellular remodeling genes, we did not observe the classical alterations in the gene markers of pathological hypertrophy and the fetal gene program: atrial natriuretic factor (Anf), β-myosin heavy chain (Myh7), skeletal α-actin (Acta1), α-myosin heavy chain (Myh6), glucose transporter 1 (Glut1), and glucose transporter 4 (Glut4). Specifically, Myh7 was not different between the genotypes; however, we observed a significant increase in Anf and Acta1 in 4- and 8-week-old Cpt2M−/− mice, relative to controls (Fig. 5, C and D). We next questioned whether fetal genes were expressed at the end stage of heart failure in Cpt2M−/− mice. The end-stage Cpt2M−/− heart showed increased Anf, Myh7, Myh6, and Acta1 by 39-, 32-, 1.6-, and 4.5-fold relative to controls (Fig. 5, C and D). Both Glut4 and Glut1 were significantly down-regulated (Fig. 5D). These data suggest a unique molecular signature of pathological hypertrophy wherein the fetal gene program was regulated atypically, not required for the initial phases of hypertrophy, and most pronounced at end-stage dilated hypertrophy.
Figure 5.
Transcriptional profile of Cpt2M−/− heart. A, heat map of all 3,854 genes significantly changed between 8-week-old male control and Cpt2M−/− hearts according to RNA sequencing (n = 4). B, biological function analysis of differentially expressed genes according to RNA sequencing showing the top eight enriched pathways in Cpt2M−/− hearts compared with control mice (n = 4). C and D, RT-PCR analysis of the fetal gene markers in control and Cpt2M−/− hearts from 4-week-old female, 8-week-old male, and end-stage heart failure female mice (n = 4–6). E, heat map of all 318 MitoCarta genes significantly changed between 8-week-old male control and Cpt2M−/− hearts according to RNA sequencing (n = 4). F and G, heart mRNA abundance of mitochondrial transcription factors (F) and protein abundance of PGC1α (G) in 8-week-old male control and Cpt2M−/− hearts (n = 6). H and I, heart mRNA abundance of PPARα target genes (H) and autophagy genes (I) in 8-week-old male control and Cpt2M−/− hearts (n = 6). *, significance by Student's t test (p ≤ 0.05).
Because mitochondrial protein abundance and area was increased, the genes categorized as mitochondrial by the Mouse MitoCarta2.0 data set (19, 20) were further assessed to reveal 318 of the 1158 nuclear and mtDNA genes found in the MitoCarta data set, of which 60% were increased and 40% decreased in Cpt2M−/− hearts (Fig. 5E). Transcription factors that drive mitochondrial biogenesis, Pgc1α, Tfam, Nrf1, and Nrf2, were not elevated in RNAseq data from Cpt2M−/− hearts and confirmed by RT-PCR (Fig. 5F). The lack of up-regulation of PGC1α was also confirmed at the protein level to reveal a surprising 70% reduction in Cpt2M−/− hearts (Fig. 5G). CPT2 deficiency in mouse adipose and liver up-regulates several PPAR-regulated genes (16, 22). Likewise several PPARα-driven genes were modestly but significantly up-regulated according to the RNAseq data; however, analysis of these genes by RT-PCR did not show significant elevations in Cpt2M−/− hearts (Fig. 5H). We also observed a significant 30–40% down-regulation of autophagy genes: BCL2-interacting protein 3 (Bnip3) and autophagy-related proteins 10 and 14 (Atg10 and Atg14), confirmed by RT-PCR (Fig. 5I). Together, these data suggest that the loss of cardiac CPT2 modulated nearly 4000 genes with major effects on many cellular functions and biological processes, including those related to hypertrophy and autophagy.
CPT2 deficiency–induced cardiac remodeling is not sensitive to rapamycin
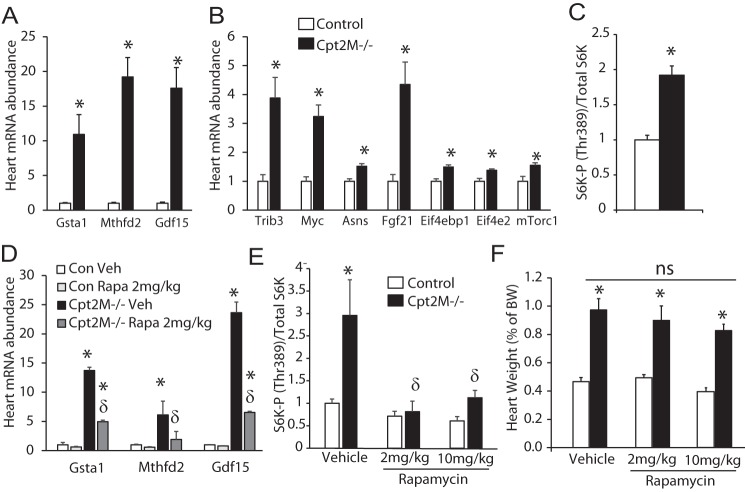
The target of rapamycin (mTOR) is a known activator of cell growth and an inhibitor of autophagy that is shown to play a role in cardiac hypertrophy (23–26). To determine whether mTOR was activated in Cpt2M−/− hearts, the expression of mTOR-regulated genes was measured. The mTOR-regulated genes (27) Gsta1, Mthfd2, Gdf15, Trib3, Myc, Asns, Fgf21, Eif4ebp1, Eif4e2, and mTorc1 were significantly up-regulated by1.4- to 19.2-fold in Cpt2M−/− hearts, relative to controls (Fig. 6, A and B). In further confirmation of increased mTOR signaling, the phosphorylation of ribosomal S6 kinase at Thr-389, the downstream mTORC1 target, was 2-fold greater in Cpt2M−/− hearts, relative to controls (Fig. 6C). These data, combined with reduced autophagy genes (Fig. 5I), suggest a role for the mTOR pathway in the Cpt2M−/− cardiac hypertrophy. To determine whether mTOR-mediated signaling was the major effector of cardiac hypertrophy in CPT2 deficiency, control and Cpt2M−/− mice were exposed to rapamycin, a potent inhibitor of mTORC1. Rapamycin at 2 or 10 mg/kg, or vehicle was injected i.p. into control or Cpt2M−/− mice once daily for 2 weeks, from 4 to 6 weeks of age. Following rapamycin treatment, we confirmed that rapamycin effectively inhibited mTOR signaling by reduced rapamycin-sensitive mTOR-regulated genes, reduced phosphorylation of S-6-kinase at Thr-389, and recovered autophagy genes (data not shown) in control and Cpt2M−/− rapamycin-treated hearts relative to vehicle-treated genotypic controls (Fig. 6, D and E). However, rapamycin failed to attenuate hypertrophy in the Cpt2M−/− mice at either the low or high dose (Fig. 6F). These data suggest that the mechanism through which rapamycin improves hypertrophic remodeling requires increased fatty acid oxidative flux through CPT2.
Figure 6.
Rapamycin-insensitive cardiac hypertrophy in Cpt2M−/− mice. A and B, the mRNA abundance of mTOR-regulated genes in control and Cpt2M−/− male 8-week-old hearts (n = 6). C, quantification of ribosomal S6 kinase phosphorylation at Thr-389 relative to total ribosomal S6 kinase in control and Cpt2M−/− male 8-week-old heart homogenates (n = 6). D and E, mRNA abundance of mTOR responsive genes (D) and phosphorylation of S-6-kinase at Thr-389 (E) in control and Cpt2M−/− hearts after 2 weeks of either vehicle or rapamycin treatment (n = 4–5). F, heart weight relative to body weight of control and Cpt2M−/− mice treated daily with either vehicle or rapamycin (2 or 10 mg/kg/day; n = 4–5). Significance by Student's t test between genotypes (*) or within genotype between treatments (δ) is shown (p ≤ 0.05).
Mitochondrial metabolism influences cardiac lysine acetylation but does not regulate hypertrophy
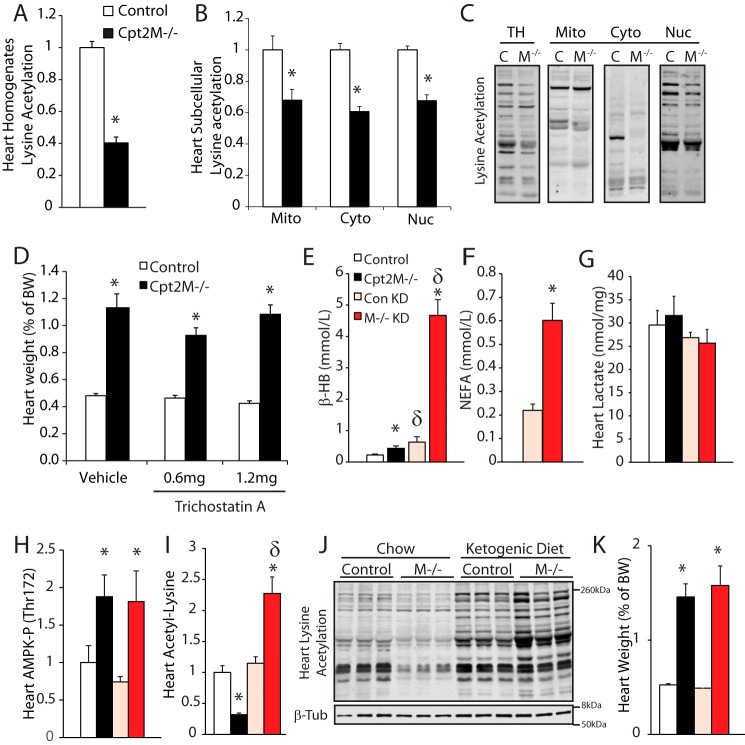
Protein acetylation is a post-translational modification known to influence many aspects of cardiomyocyte function. Acetyl-CoA is the substrate for protein acetylation and is the product of CPT2-mediated fatty acid oxidation. Accordingly, Western blot analysis of homogenates from control and Cpt2M−/− showed 60% reduced lysine acetylation (Fig. 7A). To determine whether this reduced acetylation was specific to the mitochondria, acetylation in subcellular fractions was determined to reveal 30–40% reduced lysine acetylation across the mitochondrial, cytosolic, and nuclear fractions (Fig. 7, B and C). These data suggest that fatty acid oxidation-derived acetyl-CoA is a critical substrate for protein acetylation in the heart.
Figure 7.
Modulation of protein acetylation does not regulate hypertrophy in CPT2-deficient hearts. A, quantification of lysine acetylation Western blot signal from control and Cpt2M−/− male 8-week-old heart homogenates (n = 6). B and C, quantification and representative images of acetyl-lysine Western blots of control (C) and Cpt2M−/− (M−/−) male 8–10-week-old heart homogenates (TH), mitochondrial (Mito), cytosolic (Cyto), and nuclear (Nuc) fractions (n = 3). D, heart weights of control and Cpt2M−/− female mice injected with vehicle or trichostatin A (n = 7). E–G, plasma βHB (E), NEFAs (F), and cardiac lactate (G) from control (Con) and Cpt2M−/− female mice fed either chow or ketogenic diet for 3 weeks (n = 7). H–K, heart AMPK phosphorylation (H), lysine acetylation (I and J), and weight (K) from control and Cpt2M−/− female mice fed either chow or ketogenic diet for 3 weeks (n = 7). Significance by Student's t test between genotypes (*) or within genotype between treatments (δ) is shown (p ≤ 0.05).
Because reduced protein acetylation is reported to promote cardiac hypertrophy, we next sought to investigate the role of protein acetylation in CPT2 deficiency–induced hypertrophy. The pan-HDAC inhibitor trichostatin A (TSA), a proven inhibitor of cardiac hypertrophy (28–31), was injected daily at a dose of 0.6 or 1.2 mg/kg into control or CPT2-deficient mice for 2 weeks from 6 to 8 weeks of age. TSA treatment at low or high dose failed to improve hypertrophy in CPT2-deficient mice (Fig. 7D). We next sought to modulate protein acetylation by placing control and Cpt2M−/− mice on a high-fat, low-carbohydrate ketogenic diet for 3 weeks from 7 to 10 weeks of age. Ketone bodies provide substrate for Cpt2-independent mitochondrial oxidative metabolism and acetylation substrates and act as inhibitors of deacetylase enzymes (32). The ketogenic diet effectively induced ketosis as indicated by increased plasma β-hydroxybutyrate in control Cpt2M−/− mice, compared with chow-fed mice (Fig. 7E). Strikingly, the ketogenic diet increased plasma free fatty acids and β-hydroxybutyrate 3- and 7.4-fold in Cpt2M−/− mice compared with controls (Fig. 7, E and F), suggesting either increased production and/or reduced utilization of ketone bodies in Cpt2M−/− mice. The lack of increased cardiac lactate in Cpt2-deficient mice compared with controls suggests ketone body utilization by the heart (Fig. 7G). Although ketone bodies provide an alternative substrate for mitochondrial metabolism in the CPT2-deficient heart, the phosphorylation of AMPK on Thr-172 remained elevated (Fig. 7H), suggesting that cardiac ATP-AMP balance was not fully restored by the ketogenic diet. However, the ketogenic diet did reverse the lysine-acetylation phenotype in Cpt2M−/−heart by increasing lysine acetylation by 2.3-fold in Cpt2M−/− heart, relative to ketogenic diet-fed controls (Fig. 7, I and J). Despite recovery of hypoacetylation, the ketogenic diet failed to attenuate cardiac hypertrophy in Cpt2M−/− mice (Fig. 7K). Together, these data suggest that protein acetylation in the heart is highly dependent on mitochondrial metabolic flux; however, acetylation-driven regulation of cell function and structure is not the predominant driver of hypertrophy in CPT2 deficiency.
Discussion
The classical concept of compensatory cardiac hypertrophy encompasses cardiomyocyte enlargement to reduce stress, as is the case in individuals with hypertension, vascular occlusion, or obesity. However, in the case of cardiomyocyte-specific fatty acid oxidation deficiency, there is no change in external stress on the heart; rather the stress is intracellular. Although in the face of physiological stress the heart is fully capable of switching between macromolecular substrates, mitochondrial fatty acid oxidation is obligate for proper cardiomyocyte structure and function. This is demonstrated by hypertrophic remodeling and cardiac failure in our model of cardiac CPT2-deficiency, as well as other animal models of cardiomyocyte-specific impairments in mitochondrial fatty acid oxidation (33–35). The observed hypertrophic remodeling increases in severity as the level of impairment in fatty acid oxidation increases. As such, blocking the acyl-carnitine shuttle required to initiate mitochondrial fatty acid oxidation in our Cpt2M−/− mice and the cardiac-CPT1b deficient mouse demonstrates severe cardiac remodeling and early lethality (33). In models of hypertension and pressure overload-induced hypertrophy, fibrosis is a hallmark of cardiomyopathy believed to be a key mediator of dysfunction and heart failure (18). The lack of fibrosis in cardiac-CPT2 deficiency–induced remodeling and heart failure suggests that initiation of fibrosis does not occur in direct consequence to reductions in fatty acid oxidation or to hypertrophic growth. The cause of the heart failure in Cpt2M−/− mice is likely due to cardiomyocyte energy insufficiency.
A hallmark of pathological cardiac hypertrophy is the activation of fetal cardiomyocyte genes believed to play a role in the pathogenesis of cardiac dysfunction. The fetal gene program is induced by biomechanical stress in both human and animal models. Several transcription factors have been identified that regulate these fetal genes, including nuclear factor of activated T-cells (NFAT), myocyte enhancer factor 2 (MEF2), and GATA4 (36). The activation of these transcription factors is multifaceted but is thought to be initiated by both external and internal stimuli (37). External stimuli include endothelin-1, angiotensin II, and catecholamines. Internal stimuli are less well-defined but are thought to be responsive to disruptions in mechanical stretch. In our model of limited cardiomyocyte fatty acid oxidative capacity, presumably in the absence of overload but rather internal metabolic stress, we do not see the classical early initiation of these fetal genes markers. Rather we show that the fetal gene markers are most pronounced at late-stage dilated cardiomyopathy. Intriguingly, reversion to the fetal gene program inherently increases glycolytic capacity and reduces fatty acid oxidative capacity, similar to that of the highly glycolytic fetal heart. Thus, the question remains: what is the role of the fetal gene program in cardiac hypertrophy and dysfunction? The fetal gene program, specifically shifts in β-MHC/Myh7 and α-MHC/Myh6 gene expression, is closely associated with a decline in contractile function in both animal models and humans (38). It has been argued that the fatty acid oxidation inhibitor etomoxir reverses these changes in fetal gene expression, and therefore fatty acid oxidation inhibitors should be considered therapy for heart failure (39–41). The lack of a classical induction of the fetal gene program in our model of fatty acid oxidation repression is consistent with the notion that reduced fatty acid oxidation may initially antagonize the fetal gene program. However, the fetal gene program is initiated in late stage remodeling of Cpt2M−/− mice concurrent with severe cardiac hypertrophy, dysfunction, and failure, thereby strongly arguing against the use of fatty acid oxidation inhibitors to treat pathological hypertrophy. Because the fetal gene reversion represses fatty acid oxidation and because inhibiting fatty acid oxidation independently elicits cardiac hypertrophy, these data suggest that the switch away from fatty acid oxidation is generating signals that initiate cardiac remodeling. Why would a metabolic shift elicit hypertrophy? One conceptual framework is that cardiomyocyte growth during fetal development is partially dependent on metabolically derived signals that arise from high-anaerobic, low-oxidative metabolism. Metabolic signals such as protein glycosylation and acetylation have emerged as potential mechanistic links between altered cardiac metabolism and cardiac hypertrophy (42–44). Thus, the loss of CPT2 may elicit its effects by disrupting regulation of post-transcriptional modifications such as acetylation, glycosylation, succinylation, and acylation that ultimately disrupt mitochondrial performance and structural stability of cardiomyocytes (42). Here, we investigated the role of protein acetylation in heart remodeling by CPT2 deficiency. We find that TSA exposure, shown to improve hypertrophy in both load and agonist-induced cardiac hypertrophy (28, 29), fails to improve hypertrophy in the absence of CPT2. Moreover, the ketogenic diet converted the CPT2-deficient heart from hypoacetylated to hyperacetylated without impacting hypertrophy. Our data suggest that protein acetylation is not the major driver of cardiac hypertrophy; thus, the role of protein acetylation in cardiomyocyte structure and function required further investigation.
A growing body of evidence supports mTOR activation in many models of cardiac hypertrophy (23). The mTORC1 inhibitor rapamycin attenuates cardiac hypertrophy in aortic banding, spontaneous hypertension, thyroid hormone, aging, and the fatty acid oxidation deficiency acyl-CoA synthetase 1 knock-out mouse (26, 46–50). Intriguingly, we do not observe improvement in cardiac hypertrophy by rapamycin treatment in Cpt2M−/− mice. These data suggest that either the mechanism inducing hypertrophy in our CPT2-deficient model is not dependent on the rapamycin-sensitive mTOR signaling pathway or that the mechanisms through which rapamycin improves hypertrophy, such as inhibiting protein synthesis and increasing autophagy (51), are limiting in CPT2 deficiency. Increased autophagy would not only reduce cell mass by increasing the breakdown of cellular proteins, but it would also result in the generation of free amino acids and fatty acids as mitochondrial metabolic substrates. In the case of the CPT2-deficient heart, it is possible that the loss of cell mass through autophagy is not rapid or substantial enough to overcome the rapid hypertrophic growth. Moreover, rapamycin is shown to improve mitochondrial fatty acid oxidation (12, 27); however, the absence of CPT2 would prevent improved fatty acid oxidation. It is possible that improvements in fatty acid oxidative flux are required for rapamycin-induced hypertrophy attenuation. Another distinct possibility for the lack of rapamycin-induced hypertrophic improvement is that the growth pathways responsible for hypertrophic growth in our Cpt2M−/− mouse are largely independent of mTOR signaling. Signaling pathways such as calcium-calmodulin, MAPK, and NF-κB are all implicated in cardiac hypertrophic remodeling (37).
In summary, the loss of CPT2 in the mouse heart causes severe cardiac hypertrophic remodeling, dilation, and heart failure. Although the hypertrophic-regulator mTORC1 is activated in Cpt2M−/− hearts, the inhibition of mTOR did not rescue the hypertrophy. Furthermore, hypoacetylation is an emerging causative agent in hypertrophy, yet hypertrophy was not attenuated by reversal of hypoacetylation using methods of chemical inhibition of deacetylase or metabolic modifications by ketogenic diet in Cpt2M−/− mice. These data suggest that mitochondrial fatty acid oxidative metabolic flux is the key mediator of cardiac hypertrophy; thus, the ability to improve mitochondrial metabolic flux stands as a primary target for therapeutic strategies to prevent or revert hypertrophy.
Experimental procedures
Animal
CPT2-deficient mice were generated as recently described (16); specifically C57BL/6 embryonic stem cells were used to target loxP recombination sites surrounding exon 4 of the Cpt2 gene. Exon 4 encompasses approximately one-third of the protein coding sequence including all of the critical catalytic residues (52), and this targeting strategy causes a frameshift in the remaining exons. CPT2-targeted mice were crossed with MCK-Cre or αMHC-MerCreMer (HERT2) mice (Jackson Laboratories stock no. 006475 or 005657). Littermates lacking the Cre gene (Cpt2flox/flox) were used as controls. The mice were given free access to water and standard chow (PicoLab 5053; Lab Diets, Richmond, IN) in pathogen-free barrier ventilated housing under 12-h light-dark cycles. All procedures were performed in accordance with Purdue Animal Care and Use Committee. Body weights were measured weekly or biweekly. Rates of O2 consumption and respiratory exchange ratios were determined in Oxymax chambers (Columbus Instruments) after mice were acclimated overnight. Rapamycin (LC Laboratories, Woburn, MA) was dissolved in ethanol at 10 mg/ml and diluted into 5% PEG-400, 5% Tween 80 in PBS and injected i.p. once daily for 2 weeks from 4 to 6 weeks of age at a dose of 2 mg/kg/day or for 3 weeks from 4 to 7 weeks of age at a dose of 10 mg/kg/day. Trichostatin A (Cayman Chemical, Ann Arbor, MI) was dissolved in pure DMSO at 20 mg/ml and further diluted into 10% DMSO in PBS (vehicle solution), injected via i.p. route once a day for 2 weeks at a dose of 0.6 or 1.2 mg/kg/day in 6–8-week-old mice. Customized ketogenic diet (F6689; fat:protein + carbohydrate, 4:1; BioServ, Flemington, NJ) was administered to mice ad libitum for 3 weeks from 7 to 10 weeks of age. Tissues were harvested after 3–4 h of food deprivation. Hearts were removed and rinsed in ice-cold PBS, and heart weight was determined.
Gene expression
Tissue RNA was isolated using TRIzol, and RNA was purified using PureLink RNA mini kit (Ambion, Life Technologies). RNAseq cDNA was synthesized using TruSeq stranded mRNA sample preparation (Illumina) kit, and two lanes of an Illumina Rapid Chemistry flow cell were clustered and sequenced in single-read mode for 51 base read on a HiSeq 2500 (Illumina) by the Purdue Genomics Core Facility. Sequence data quality was determined using FastQC software (version 0.11.2). Quality trimming was performed using FASTX Toolkit (version 0.0.13.2) to remove the bases with less than Phred33 score of 30, and the resulting reads of at least 25 base pairs were retained (which comprised >99% of total reads). For read-mapping, quality trimmed reads were mapped against the bowtie2-indexed mouse reference genome using TopHat package (version 2.1.0) with parameters library-type fr-firststrand, and number of allowed mismatches as 1. For counts, Tophat derived global mapping results and annotation (GTF/GFF) file for reference genome were fed to HTSeq package (version 0.6.1) to obtain the read counts for each gene feature for each replicate. Counts from all replicates were merged together using custom Perl scripts to generate a gene counts matrix for both samples (control and CPT2M). Genes with 0 counts across all replicates were discarded from the counts matrix. When genes have 0 count in one sample but not in others, the counts were converted from 0 to 1 to avoid having infinite values being calculated for fold change. Final combined counts matrix was utilized for further differential gene expression (DGE) analysis by DeSeq2 and edgeR packages. Additionally, DGE was calculated using the tuxedo protocol that directly utilizes the TopHat mapping files (bam) instead of count matrix. The tuxedo protocol uses fragments per kilobase of exon per million reads mapped, which is corrected (normalized) for the length of the gene and the library size to represent the gene expression as compared with raw counts in edgeR and DESeq2. Heat maps were generated using Heatmapper online tool (Wishart Research Group, University of Alberta, Edmonton, Canada) and functional analysis of the DGE was performed on Ingenuity Pathway Analysis software (Ingenuity Systems Inc., Redwood City, CA) and InnateDB online open source tool (53). Cutoffs were set for p value (p < 0.05) and fold change (−1.5 < fold change < 1.5). Enriched REACTOME and KEGG pathways, as well as Gene Ontology terms for biological processes, were calculated according to their corresponding p values for overrepresentation using hypergeometric test and corrected for multiple testing with a Benjamini–Hochberg test.
RT-PCR
cDNA was synthesized (Applied Biosystems High Capacity cDNA RT kit), and reactions were performed using SYBR Green (Bio-Rad) detection with specific primers (200 nm), equal amounts of cDNA (10 ng/reaction) and using an Applied Biosystems StepOnePlus Real-time PCR system with StepOne software (Applied Biosystems). The results were normalized to housekeeping gene and expressed as arbitrary units of 2−ΔCT.
Immunoblots and histology
Total homogenates were collected in sucrose media (10 mm Tris, 1 mm EDTA, 250 mm sucrose) with protease inhibitors. Lysates were collected in lysis buffer (50 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100) with protease inhibitors. Subcellular fractions were isolated as described (21). Briefly, hearts were collected, minced with scissors and homogenized with a Teflon pestle. Subsequent centrifugation and pellet and/or supernatant recovery steps were performed to obtain nuclear, cytosolic, and mitochondrial fractions. Homogenates, subcellular fractions, or lysates were equally loaded (15–30 μg) and electrophoresed on 8, 10, or 12% SDS-polyacrylamide gels; transferred to nitrocellulose membrane; blocked with 5% milk-TBST for 1 h; incubated with primary antibody (1:1,000–1:2,000) against CPT2 (Millipore, ABS85), HSP60 (Santa Cruz, sc13115), PDH (Cell Signaling, 3205), COXIV (Cell Signaling, 4850), SHDA (Cell Signaling, 11998; PGC1a (Cell Signaling, 2178), OXPHOS mixture (Abcam, MS604), S6K (Thr-389; Cell Signaling, 9234), S6K (Cell Signaling, 9202), acetylated lysine (Cell Signaling, 9441), AMPKα (Thr-172) (Cell Signaling, 4188), AMPKα (Cell Signaling, 5831), 4E-BP1 (Thr-37/46; Cell Signaling, 2855), or 4EBP1 (Cell Signaling, 9644); washed; and incubated with secondary anti-mouse or anti-rabbit antibodies conjugated to IR dye 800CW or 680LT (LiCor, Lincoln, NE). Protein was visualized with Odyssey CLx (LiCor) and quantified using Image Studio (LiCor). Hearts were collected for histology after total body perfusion with PBS followed by 2% paraformaldehyde + 2% gluteraldehyde in 0.1 m sodium cacodylate, pH 7.4. The hearts were paraffin embedded, sectioned, and stained for hematoxylin and eosin or Masson's trichrome stain by Purdue Histology Facility and Services. Tissue sections were scanned using Aperio Image Analysis (Leica, Buffalo Grove, IL), and images were captured using ImageScope software (Leica, Buffalo Grove, IL). Aperio-derived images were used to quantify the cardiomyocyte size in ImageJ software, and at least 60 round (cross-sectional) cells/mouse were averaged. The apex of the heart was processed, stained, and prepared for transmission electron microscope with the Life Sciences Microscope Facility at Purdue. Images were captured on a Philips CM-100 TEM. Mitochondrial and lipid droplet area was quantified using Image J software.
Lipid and plasma analysis
Heart total lipids were extracted with chloroform/methanol via Folch method (54), the chloroform phase was dried down by SpeedVac and resuspended in tert-butanol:methanol:Triton X-100 (3:1:1 ratio by volume), and lipids were quantified using colorimetric glycerol/triacylglycerol assay (Sigma). Blood glucose was determined by glucometer (NovaMax, Billerica, MA). Blood was collected in 5–10% 0.5 m EDTA and plasma NEFA (Wako, Richmond, VA), triacylglycerol (Sigma), and β-hydroxybutyrate (Cayman Chemical, Ann Arbor, MI) were determined by colorimetric assay per manufacturer's instruction. Levels of l-lactate in the heart were measured using a commercially available colorimetric kit (ab65330; Abcam, Cambridge, MA) prior deproteinization of the samples (ab204708; Abcam, Cambridge, MA). For acyl-carnitine profiling, samples (5–10 mg of tissue) were placed inside 2-ml vials with 1.4-mm ceramic (zirconium oxide) beads with 300 μl of water using Precellys 24 tissue homogenizer (Bertin Technologies, Rockville, MD). 200 μl of the homogenate was collected and submitted to lipid extraction using Bligh and Dyer method (55). By this protocol, phase separation is performed using CHCl3/MeOH/H2O (1:2:0.8), and the combined organic fractions are centrifuged; the bottom phase was transferred to a new tube and evaporated.
Dried lipid extracts were diluted in 200 μl of MeOH/H2O (stock solution). The stock solution was further diluted 40–70× (depending on the amount of tissue used for the lipid extraction) in 300 mm acetonitrile/methanol/ammonium acetate at 3:6.65:0.35 volume ratios, and the volume of 8 μl was directly delivered through a micro-autosampler (G1377A) to a QQQ6410 triple quadrupole mass spectrometer (Agilent Technologies, San Jose, CA) operated in the positive ion mode, equipped with Jet stream ESI ion source. The solvent pumped between injections was acetonitrile + 0.1% formic. For data analysis, relative amounts of ion abundances were used for the statistics. The values of ion intensities were normalized by total ion intensity. The multiple reaction monitoring in which the parent ion and the m/z 85 were monitored were used to plot the fold changes.
Oxidation assays and deoxyglucose uptake
Freshly isolated heart was minced and homogenized with a motor-driven Teflon pestle and glass mortar in ice-cold buffer oxidation buffer (100 mm KCl, 40 mm Tris·HCl, 10 mm Tris base, 5 mm MgCl2·6H2O, 1 mm EDTA, and 1 mm ATP, pH 7.4) at a 20-fold dilution (w/v), and the rate of [1-14C]palmitate, [1-14C]octanoate, or [U-14C]pyruvate was measured, as previously described (45). Briefly, carbon dioxide was trapped in a suspended well filled with sodium hydroxide-soaked filter paper. ASMs were detected from unprecipitated incubation materials exposed to sulfuric acid. For glucose uptake, anesthetized mice were injected retro-orbitally with 5 μCi of 2-deoxy[1-14C]glucose in saline (Moravek Biochemicals, Brea, CA) Tissues were harvested and flash frozen in liquid N2 30 min after injection. Radioactivity was measured in tissue homogenates by scintillation and normalized to the number of DPM present in 10 μl of whole blood obtained 5 min after injection.
Echocardiography
Echocardiograms were collected from isoflurane-anesthetized mice (1–3% in 1.5 liters/min medical grade air) by high frequency ultrasound (Vevo2100; FUJIFILM VisualSonics). M-mode, B-mode, and respiratory- and cardiac-gated images (electrocardiogram-gated Kilohertz Visualization) were collected for both medial short- and long-axis views. Echocardiogram-derived calculations were determined from the endocardial and epicardial area protocol (Vevo Lab; FUJIFILM VisualSonics). This protocol captures epicardial and endocardial heart length from long-axis images and LV wall circumference from short-axis images in both diastole and systole.
Author contributions
J. M. E. conceived and designed the experiments. A. S. P., L. Y. H., K. L. H., A. G. B., F. W. D., and J. M. E. performed the experiments. A. S. P., C. J. G., and J. M. E. analyzed the data. C. J. G. and J. M. E. contributed reagents/materials/analysis tools. A. S. P. and J. M. E. wrote the paper.
Supplementary Material
Acknowledgments
We thank Michael J. Wolfgang for generously gifting the CPT2 floxed mice. We thank Elizaveta J. Yurovich for collecting mouse body weight data and Aidan M. Hannon and Natalie M. Mudd for analyzing electron microscopy and histology images. We also thank Kolapo Ajuwon for providing access to Oxymax Chambers and the Metabolite Profiling Facility for the acyl-carnitine profiling.
This work was supported by Ralph W. and Grace M. Showalter Research Trust Fund Grant 208453 and McKinley Education Foundation 770520401. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental data and supplemental Table 1.
- CPT2
- carnitine palmitoyltransferase 2
- LV
- left ventricular
- mTOR
- mechanistic target of rapamycin
- mTORC
- mTOR complex
- TSA
- trichostatin A
- MCK
- muscle creatine kinase
- ASM
- acid-soluble metabolite
- NEFA
- non-esterified fatty acid
- βHB
- β-hyroxybutyrate
- PDH
- pyruvate dehydrogenase
- Pdk
- pyruvate dehydrogenase kinasel
- DGE
- differential gene expression
- ASM
- acid soluble metabolites
- TAG
- triacylglycerol
- TEM
- transmission electron microscopy.
References
- 1. Cho I. J., Chang H. J., Park H. B., Heo R., Shin S., Shim C. Y., Hong G. R., and Chung N. (2015) Aortic calcification is associated with arterial stiffening, left ventricular hypertrophy, and diastolic dysfunction in elderly male patients with hypertension. J. Hypertens. 33, 1633–1641 [DOI] [PubMed] [Google Scholar]
- 2. Brady T. M. (2016) The role of obesity in the development of left ventricular hypertrophy among children and adolescents. Curr. Hypertens. Rep. 18, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murdolo G., Angeli F., Reboldi G., Di Giacomo L., Aita A., Bartolini C., and Vedecchia P. (2015) Left ventricular hypertrophy and obesity: only a matter of fat? High Blood Press. Cardiovasc. Prev. 22, 29–41 [DOI] [PubMed] [Google Scholar]
- 4. Levelt E., Mahmod M., Piechnik S. K., Ariga R., Francis J. M., Rodgers C. T., Clarke W. T., Sabharwal N., Schneider J. E., Karamitsos T. D., Clarke K., Rider O. J., and Neubauer S. (2016) Relationship between left ventricular structural and metabolic remodeling in type 2 diabetes. Diabetes 65, 44–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McMullen J. R., and Jennings G. L. (2007) Differences between pathological and physiological cardiac hypertrophy: novel therapeutic strategies to treate heart failure. Clin. Exp. Pharmacol. Physiol. 34, 255–262 [DOI] [PubMed] [Google Scholar]
- 6. Levy D., Anderson K. M., Savage D. D., Kannel W. B., Christiansen J. C., and Castelli W. P. (1988) Echocardiographically detected left ventricular hypertrophy: revalence and risk factors: the Framingham Heart Study. Ann. Intern. Med. 108, 7–13 [DOI] [PubMed] [Google Scholar]
- 7. Bianco H. T., Izar M. C., Póvoa R. M., Bombig M. T., Fonseca H. A., Helfenstein T., Ferreira C. E., Nicolau J. C., Neto A. A., Feio C. M., Cerci M. S., and Fonseca F. A. (2014) Left ventricular hypertrophy and QTc dispersion are predictors of long-term mortality in subjects with type 2 diabetes. Int. J. Cardiol. 176, 1170–1172 [DOI] [PubMed] [Google Scholar]
- 8. Struthers A. D., and Morris A. D. (2002) Screening for and treating left-ventricular abnormalities in diabetes mellitus: a new way of reducing cardiac deaths. Lancet 359, 1430–1432 [DOI] [PubMed] [Google Scholar]
- 9. Levy D., Garrison R. J., Savage D. D., Kannel W. B., and Castelli W. P. (1989) Left ventricular mass and incidence of coronary heart disease in an elderly cohort: the Framingham Heart Study. Ann. Intern. Med. 110, 101–107 [DOI] [PubMed] [Google Scholar]
- 10. Benjamin E. J., and Levy D. (1999) Why is left ventricular hypertrophy so predictive of morbidity and mortality? Am. J. Med. Sci. 317, 168–175 [DOI] [PubMed] [Google Scholar]
- 11. Brown D. W., Giles W. H., and Croft J. B. (2000) Left ventricular hypertrophy as a predictor of coronary heart disease mortality and the effect of hypertension. Am. Heart J. 140, 848–856 [DOI] [PubMed] [Google Scholar]
- 12. Chiao Y. A., Kolwicz S. C., Basisty N., Gagnidze A., Zhang J., Gu H., Djukovic D., Beyer R. P., Raftery D., MacCoss M., Tian R., and Rabinovitch P. S. (2016) Rapamycin transiently induces mitochondrial remodeling to reprogram energy metabolism in old hearts. Aging 8, 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Anderson K. A., and Hirschey M. D. (2012) Mitochondrial protein acetylation regulates metabolism. Essays Biochem. 52, 23–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee C. F., and Tian R. (2015) Mitochondrion as a target for heart failure therapy: role of protein lysine acetylation. Circ. J. 79, 1863–1870 [DOI] [PubMed] [Google Scholar]
- 15. Sack M. N. (2012) The role of SIRT3 in mitochondrial homeostasis and cardiac adaptation to hypertrophy and aging. J. Mol. Cell Cardiol. 52, 520–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee J., Ellis J. M., and Wolfgang M. J. (2015) Adipose fatty acid oxidation is required for thermogenesis and potentiates oxidative stress-induced inflammation. Cell Rep. 10, 266–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lopaschuk G. D., Ussher J. R., Folmes C. D., Jaswal J. S., and Stanley W. C. (2010) Myocardial fatty acid metabolism in health and disease. Physiol. Rev. 90, 207–258 [DOI] [PubMed] [Google Scholar]
- 18. Fan D., Takawale A., Lee J., and Kassiri Z. (2012) Cardiac fibroblasts, fibrosis and extracellular matrix remodeling in heart disease. Fibrogenesis Tissue Repair 5, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Calvo S. E., Clauser K. R., and Mootha V. K. (2016) MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44, D1251–D1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pagliarini D. J., Calvo S. E., Chang B., Sheth S. A., Vafai S. B., Ong S. E., Walford G. A., Sugiana C., Boneh A., Chen W. K., Hill D. E., Vidal M., Evans J. G., Thorburn D. R., Carr S. A., et al. (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dimauro I., Pearson T., Caporossi D., and Jackson M. J. (2012) A simple protocol for the subcellular fractionation of skeletal muscle cells and tissue. BMC Res. Notes 5, 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee J., Choi J., Scafidi S., and Wolfgang M. J. (2016) Hepatic fatty acid oxidation restrains systemic catabolism during starvation. Cell Rep. 16, 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Balasubramanian S., Johnston R. K., Moschella P. C., Mani S. K., Tuxworth W. J. Jr., and Kuppuswamy D. (2009) mTOR in growth and protection of hypertrophying myocardium. Cardiovasc. Hematol. Agents Med. Chem. 7, 52–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee C.-H., Inoki K., and Guan K.-L. (2007) mTOR pathway as a target in tissue hypertrophy. Annu. Rev. Pharmacol. Toxicol. 47, 443–467 [DOI] [PubMed] [Google Scholar]
- 25. Kemi O. J., Ceci M., Wisloff U., Grimaldi S., Gallo P., Smith G. L., Condorelli G., and Ellingsen O. (2008) Activation or inactivation of cardiac Akt/mTOR signaling diverges physiological from pathological hypertrophy. J. Cell Physiol. 214, 316–321 [DOI] [PubMed] [Google Scholar]
- 26. Sciarretta S., Volpe M., and Sadoshima J. (2014) Mammalian target of rapamycin signaling in cardiac physiology and disease. Circ. Res. 114, 549–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schisler J. C., Grevengoed T. J., Pascual F., Cooper D. E., Ellis J. M., Paul D. S., Willis M. S., Patterson C., Jia W., and Coleman R. A. (2015) Cardiac energy dependence on glucose increases metabolites related to glutathione and activates metabolic genes controlled by mechanistic target of rapamycin. J. Am. Heart Assoc. 4, e001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cao D. J., Wang Z. V., Battiprolu P. K., Jiang N., Morales C. R., Kong Y., Rothermel B. A., Gillette T. G., and Hill J. A. (2011) Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc. Natl. Acad. Sci. U.S.A. 108, 4123–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ooi J. Y., Tuano N. K., Rafehi H., Gao X. M., Ziemann M., Du X. J., and El-Osta A. (2015) HDAC inhibition attenuates cardiac hypertrophy by acetylation and deacetylation of target genes. Epigenetics 10, 418–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kee H. J., Sohn I. S., Nam K. I., Park J. E., Qian Y. R., Yin Z., Ahn Y., Jeong M. H., Bang Y.-J., Kim N., Kim J.-K., Kim K. K., Epstein J. A., and Kook H. (2006) Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation 113, 51–59 [DOI] [PubMed] [Google Scholar]
- 31. Kook H., Lepore J. J., Gitler A. D., Lu M. M., Wing-Man Yung W., Mackay J., Zhou R., Ferrari V., Gruber P., and Epstein J. A. (2003) Cardiac hypertrophy and histone deacetylase–dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J. Clin. Invest. 112, 863–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Newman J. C., and Verdin E. (2014) β-Hydroxybutyrate: much more than a metabolite. Diabetes Res. Clin. Pract. 106, 173–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Haynie K. R., Vandanmagsar B., Wicks S. E., Zhang J., and Mynatt R. L. (2014) Inhibition of carnitine palymitoyltransferase1b induces cardiac hypertrophy and mortality in mice. Diabetes Obes. Metab. 16, 757–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cox K. B., Liu J., Tian L., Barnes S., Yang Q., and Wood P. A. (2009) Cardiac hypertrophy in mice with long-chain acyl-CoA dehydrogenase or very long-chain acyl-CoA dehydrogenase deficiency. Lab. Invest. 89, 1348–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ellis J. M., Mentock S. M., Depetrillo M. A., Koves T. R., Sen S., Watkins S. M., Muoio D. M., Cline G. W., Taegtmeyer H., Shulman G. I., Willis M. S., and Coleman R. A. (2011) Mouse cardiac acyl coenzyme A synthetase 1 deficiency impairs fatty acid oxidation and induces cardiac hypertrophy. Mol. Cell Biol. 31, 1252–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dirkx E., da Costa Martins P. A., and De Windt L. J. (2013) Regulation of fetal gene expression in heart failure. Biochim. Biophys. Acta 1832, 2414–2424 [DOI] [PubMed] [Google Scholar]
- 37. Heineke J., and Molkentin J. D. (2006) Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 7, 589–600 [DOI] [PubMed] [Google Scholar]
- 38. Palmer B. M. (2005) Thick filament proteins and performance in human heart failure. Heart Fail. Rev. 10, 187–197 [DOI] [PubMed] [Google Scholar]
- 39. Rupp H., Zarain-Herzberg A., and Maisch B. (2002) The use of partial fatty acid oxidation inhibitors for metabolic therapy of angina pectoris and heart failure. Herz 27, 621–636 [DOI] [PubMed] [Google Scholar]
- 40. Turcani M., and Rupp H. (1999) Modification of left ventricular hypertrophy by chronic etomoxir treatment. Br. J. Pharmacol. 126, 501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Turcani M., and Rupp H. (1997) Etomoxir improves left ventricular performance of pressure-overloaded rat heart. Circulation 96, 3681–3686 [DOI] [PubMed] [Google Scholar]
- 42. Carley A. N., Taegtmeyer H., and Lewandowski E. D. (2014) Matrix revisited: mechanisms linking energy substrate metabolism to the function of the heart. Circ. Res. 114, 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xie M., and Hill J. A. (2013) HDAC-dependent ventricular remodeling. Trends Cardiovasc. Med. 23, 229–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang J., Hu X., and Jiang H. (2016) HDAC inhibition: a novel therapeutic approach for attenuating heart failure by suppressing cardiac remodeling. Int. J. Cardiol. 214, 41–42 [DOI] [PubMed] [Google Scholar]
- 45. Ellis J. M., Li L. O., Wu P. C., Koves T. R., Ilkayeva O., Stevens R. D., Watkins S. M., Muoio D. M., and Coleman R. A. (2010) Adipose acyl-CoA synthetase-1 (ACSL1) directs fatty acids towards β-oxidation and is required for cold thermogenesis. Cell Metab. 12, 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Paul D. S., Grevengoed T. J., Pascual F., Ellis J. M., Willis M. S., and Coleman R. A. (2014) Deficiency of cardiac Acyl-CoA synthetase-1 induces diastolic dysfunction, but pathologic hypertrophy is reversed by rapamycin. Biochim. Biophys. Acta 1841, 880–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Soesanto W., Lin H.-Y., Hu E., Lefler S., Litwin S. E., Sena S., Abel E. D., Symons J. D., and Jalili T. (2009) Mammalian target of rapamycin is a critical regulator of cardiac hypertrophy in spontaneously hypertensive rats. Hypertension 54, 1321–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kuzman J. A., O'Connell T. D., and Gerdes A. M. (2007) Rapamycin prevents thyroid hormone-induced cardiac hypertrophy. Endocrinology 148, 3477–3484 [DOI] [PubMed] [Google Scholar]
- 49. McMullen J. R., Sherwood M. C., Tarnavski O., Zhang L., Dorfman A. L., Shioi T., and Izumo S. (2004) Inhibition of mTOR signaling with rapamycin regresses established cardiac hypertrophy induced by pressure overload. Circulation 109, 3050–3055 [DOI] [PubMed] [Google Scholar]
- 50. Dai D.-F., Karunadharma P. P., Chiao Y. A., Basisty N., Crispin D., Hsieh E. J., Chen T., Gu H., Djukovic D., Raftery D., Beyer R. P., MacCoss M. J., and Rabinovitch P. S. (2014) Altered proteome turnover and remodeling by short-term caloric restriction or rapamycin rejuvenate the aging heart. Aging Cell 13, 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gu J., Hu W., Song Z.-P., Chen Y.-G., Zhang D.-D., and Wang C.-Q. (2016) Rapamycin inhibits cardiac hypertrophy by promoting autophagy via the MEK/ERK/Beclin-1 pathway. Front. Physiol. 7, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hsiao Y. S., Jogl G., Esser V., and Tong L. (2006) Crystal structure of rat carnitine palmitoyltransferase II (CPT-II). Biochem. Biophys. Res. Commun. 346, 974–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lynn D. J., Winsor G. L., Chan C., Richard N., Laird M. R., Barsky A., Gardy J. L., Roche F. M., Chan T. H., Shah N., Lo R., Naseer M., Que J., Yau M., Acab M., et al. (2008) InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol. Syst. Biol. 4, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Folch J., Lees M., and Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 55. Bligh E. G., and Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.