Abstract
MicroRNA-155 (miR-155) regulates antitumor immune responses. However, its specific functions within distinct immune cell types have not been delineated in conditional KO mouse models. In this study, we investigated the role of miR-155 specifically within T cells during the immune response to syngeneic tumors. We found that miR-155 expression within T cells is required to limit syngeneic tumor growth and promote IFNγ production by T cells within the tumor microenvironment. Consequently, we found that miR-155 expression by T cells is necessary for proper tumor-associated macrophage expression of IFNγ-inducible genes. We also found that immune checkpoint–blocking (ICB) antibodies against programmed cell death protein 1/programmed death ligand 1 (PD-1/PD-L1) and cytotoxic T lymphocyte–associated protein 4 (CTLA-4) restored antitumor immunity in miR-155 T cell–conditional KO mice. We noted that these ICB antibodies rescued the levels of IFNγ-expressing T cells, expression of multiple activation and effector genes expressed by tumor-infiltrating CD8+ and CD4+ T cells, and tumor-associated macrophage activation. Moreover, the ICB approach partially restored expression of several derepressed miR-155 targets in tumor-infiltrating, miR-155–deficient CD8+ T cells, suggesting that miR-155 and ICB regulate overlapping pathways to promote antitumor immunity. Taken together, our findings highlight the multifaceted role of miR-155 in T cells, in which it promotes antitumor immunity. These results suggest that the augmentation of miR-155 expression could be used to improve anticancer immunotherapies.
Keywords: cellular immune response, leukocyte, microRNA (miRNA), tumor immunology, tumor microenvironment, T cell, miR-155
Introduction
MicroRNAs are a class of non-coding RNAs that function in immune cells by modulating gene expression posttranscriptionally via binding to the 3′ UTR of their target mRNA in a sequence-specific manner to repress the expression of these targets (1). MicroRNA-155 (miR-155) is a key regulator of the immune response in contexts ranging from autoimmunity to infection to immunization (2–4). miR-155 has more recently emerged as a regulator of tumor immunity because of its influence on the anti-tumor activity of a variety of immune cells, including T cells, natural killer cells, and myeloid populations (5–7). We previously reported a defect in the accumulation of IFNγ-expressing CD4+ and CD8+ T cells in tumors from mice genetically deficient in miR-155, and this correlated with increased growth of syngeneic B16f10 melanomas (8), an observation also made by an independent laboratory (9). Other studies found that overexpression of miR-155 in ovalbumin (OVA)-specific2 CD8+ T cells resulted in a robust antigen-specific immune response to B16 melanoma tumors that expressed OVA (5) and that miR-155 overexpression promoted the proliferation and effector function of CD8+ T cells in response to anti-tumor γc cytokines (10). A regulatory role for miR-155 in regulatory T cells was also reported in the setting of tumor immunity (11). Altogether, these studies provide evidence that miR-155 functions in various T cell types as they respond to solid tumors.
miR-155 is highly up-regulated in macrophages responding to LPS and other inflammatory stimuli (12). It is also expressed in tumor-associated macrophages (TAMs) (7) and is thought to be important in the polarization process of macrophages and monocytes to a pro-inflammatory anti-tumor phenotype (13). Ex vivo knockdown and overexpression of miR-155 in TAMs demonstrated that miR-155 expression in these cells promotes a pro-inflammatory M1 phenotype (14). This work, along with evidence showing that MMTV–PyMT mice develop spontaneous breast cancer at a higher rate when miR-155 is knocked down using a lentivirus-delivered inhibitory sponge in TAM populations (7), suggests that miR-155 expression within the macrophage compartment inhibits tumor growth by creating a pro-inflammatory tumor microenvironment. Additionally, there is evidence that miR-155 also regulates myeloid-derived suppressor cell responses in tumor-bearing mice (9, 15). Thus, in addition to T cells, miR-155 also appears to play important biological functions within the myeloid compartment during tumor immunity.
Despite this important progress, several unanswered questions about the role of miR-155 during antitumor immunity remain. The cell-intrinsic roles of miR-155 during T and myeloid cell responses to solid tumors have not been examined using miR-155–conditional knockout mice that do not require manipulations such as bone marrow reconstitution or adoptive transfers. Further, a potential role for miR-155 in regulating cross-talk between T cells and TAM populations within the tumor microenvironment has not been explored, nor has it been determined whether defective antitumor responses by miR-155−/− T cells can be reversed.
In this study, we employed miR-155–conditional knockout mice to test T cell- and macrophage-specific roles of miR-155 in response to a syngeneic B16f10 melanoma tumor. We found that miR-155 expression within the T cell compartment is required to promote optimal anti-tumor CD4+ and CD8+ T cell responses and reduce tumor growth. Additionally, miR-155 expression by T cells promoted the activation of TAMs through the induction of IFNγ-inducible genes, whereas its expression by LysM+ TAMs was not required for this response to occur. We also discovered that ICB therapy largely rescues anti-tumor immune responses in miR-155 T cell–conditional knockout (TCKO) mice and that it does so by restoring the levels of IFNγ-expressing T cells, TAM activation, and expression of several T cell activation and effector genes. Additionally, ICB also reduced the expression of several miR-155 target genes that were derepressed in T cells lacking miR-155. This indicates that miR-155 and ICB reagents regulate overlapping pathways. Our findings clearly demonstrate that T cell–expressed miR-155 plays a significant role in promoting the endogenous, multicellular immune response against solid tumors and that evaluation and/or augmentation of its expression may be a clinically relevant tool for immunotherapy.
Results
T cell–specific deletion of miR-155 reduces the levels of intratumor IFNγ-expressing T cells and promotes the growth of B16f10 tumors
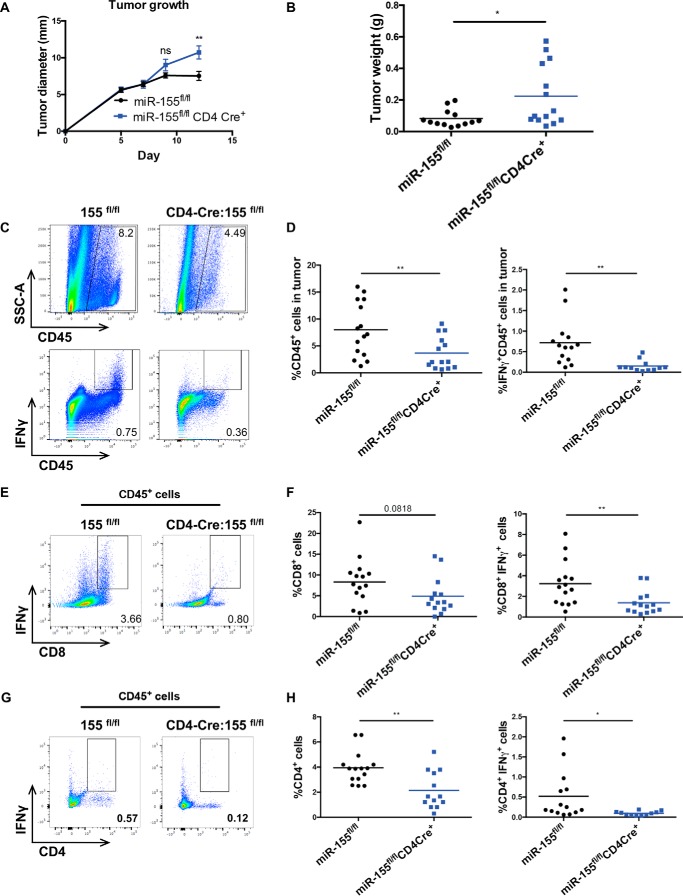
To assess the role of miR-155 expression within T cells following a solid tumor challenge, we injected syngeneic B16f10 melanoma cells into miR-155 TCKO mice in which miR-155 was conditionally deleted in CD4+ and CD8+ T cells via CD4-Cre (3). During the development of T cells in the thymus, all CD4+ and CD8+ T cells undergo a double-positive CD4+CD8+ stage in which they will express Cre under the control of CD4 and thus delete floxed genes in cells that will become either CD4+ or CD8+ T cells. On day 12 after injection, miR-155 TCKO mice exhibited modestly increased tumor sizes compared with 155fl/fl controls, as measured by diameter (Fig. 1A) and weight (Fig. 1B) of the transplanted tumors. These experiments were also performed using CD4Cre+ controls, and we obtained similar results (data not shown). We previously observed increased tumor growth in whole-body miR-155–deficient mice along with a defect in the numbers of intratumor IFNγ-expressing CD4+ and CD8+ T cells (8). Therefore, we assayed the production of this cytokine by tumor-infiltrating T cell populations from miR-155 TCKO mice and controls. We found an overall decrease in the percentage of CD45+ immune cells within tumors of these knockout mice, accompanied by a further reduction in the percentage of IFNγ+ cells within the CD45 compartment (Fig. 1, C and D). We also noted a trending decrease in the percentage of CD8+ T cells and a significant decrease in CD8+IFNγ+ T cells (Fig. 1, E and F) within the tumor microenvironment when miR-155 was specifically lacking from T cells. Additional analysis of the T cell compartments within the tumors also revealed a significant decrease in the percentages of CD4+ T cells and CD4+IFNγ+ T cells in our miR-155 TCKO mice compared with controls (Fig. 1, G and H).
Figure 1.
Reduced intratumor IFNγ-expressing T cells and increased tumor growth of B16f10 tumors in miR-155 TCKO mice. A, the largest diameter of tumors measured in millimeters during the time course of the tumor growth experiments. B, tumor weight in grams on day 12 after tumor injection at time of harvest. C, representative plot of CD45+ cells and IFNγ+CD45+ cells infiltrating tumors of miR-155fl/fl mice and miR-155 TCKO mice. Intracellular staining for IFNγ was completed by doing a 4-h stimulation with phorbol 12-myristate 13-acetate and ionomycin in the presence of Golgi Plug (BD Biosciences), followed by overnight fixation and staining for intracellular IFNγ the next day. D, percentages of cells shown in C. E, representative plots of CD8+IFNγ+ tumor-infiltrating T cells in miR-155fl/fl and miR-155 TCKO mice. F, percentages of cells shown in E. G, representative plots of CD4+IFNγ+ tumor-infiltrating T cells in miR-155fl/fl and miR-155 TCKO mice. H, percentages of cells shown in G. All plots represent data combined from two independently performed experiments. *, p < 0.05; **, p < 0.005; ns, not significant.
Reduced activation of TAMs in miR-155 TCKO tumor-bearing mice
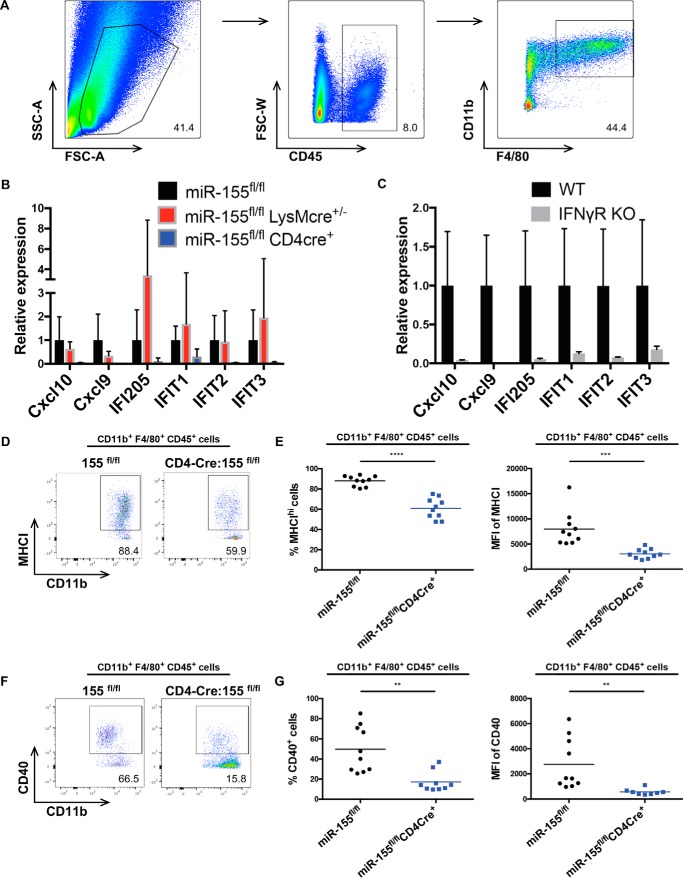
To further assess the effects of macrophage intrinsic versus extrinsic miR-155 expression on TAM phenotypes within the tumor microenvironment, we sorted macrophages from B16f10 tumors growing in miR-155 TCKO mice, miR-155 macrophage–conditional knockout mice (MCKO), and control mice on day 12 post-tumor administration based on their expression of CD45, CD11b, and F4/80 (Fig. 2A). Deletion of miR-155 from miR-155–floxed macrophages by LysM-Cre was verified by qPCR analysis of mature miR-155 transcript levels in bone marrow–derived macrophages (supplemental Fig. S1A). TAMs sorted from miR-155 TCKO mice had a marked decrease in mRNA levels encoding several IFNγ-inducible genes (Fig. 2B), consistent with reduced percentages of tumor-infiltrating, IFNγ-producing T cells in these mice, whereas TAMs from miR-155 MCKO mice expressed near WT levels of these genes. This particular gene subset was also confirmed to be IFNγ-inducible, as their expression was blocked in IFNγR−/− macrophages isolated from B16f10 tumors (Fig. 2C). Further, TAMs from miR-155 TCKO mice also had defective surface expression of MHCI and CD40, as determined by FACS, whereas miR-155 MCKO mice expressed WT levels of these cell surface proteins (Fig. 2, D–G, and data not shown). Also of note, miR-155 MCKO mice challenged with B16f10 tumors showed no difference in tumor growth or in the percentages of IFNγ-producing T cells in the tumor microenvironment (supplemental Fig. S1, B–D). Additionally, tumor growth in miR-155 MCKO mice given B16f10-OVA tumors was similar to control mice (supplemental Fig. S1E). Taken together, these results support a model whereby deletion of miR-155 expression in T cells results in decreased accumulation of IFNγ-producing T cells within the tumor microenvironment, and this results in decreased TAM expression of IFN response genes involved in their activation and antitumor responses.
Figure 2.
miR-155 TCKO mice have reduced TAM activation. A, sorting strategy to obtain CD45+CD11b+F4/80+ TAMs from B16f10 tumors on day 12 after tumor injection. (SSC-A represents side scatter, and FSC-A represents forward scatter.) B, qPCR performed on sorted TAMs from the indicated genotypes. C, qPCR on TAMs obtained from B16f10 tumor-bearing WT B6 mice and IFNγR−/− mice (The Jackson Laboratory). D, representative plot of CD11b+ F4/80+ MHCIhi TAMs from the indicated genotypes. E, percentages and mean fluorescence intensity (MFI) of cells shown in D. F, representative plot of CD11b+ F4/80+ CD40+ TAMs from the indicated genotypes. G, percentages and MFIs of cells shown in F. All data shown are representative of multiple independent experiments. Bar graphs represent the mean ± S.D. **, p < 0.005; ***, p < 0.0005; ****, p < 0.00005.
ICB therapy rescues defective anti-tumor immune responses by miR-155 TCKO mice
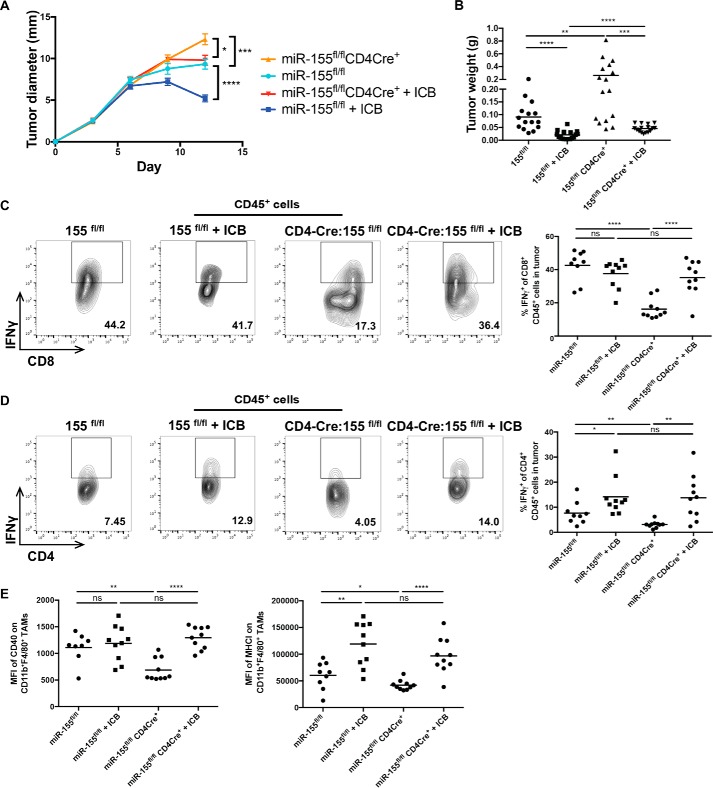
ICB of PD-1, PD-L1, and CTLA-4 has recently shown great promise in the clinic, as it promotes T cell responses to solid tumors by blocking inhibitory signals T cells receive from their environment (16–19). The utilization of combination therapy with these agents is effective for treating advanced cases of melanoma (20–22), and a previous study demonstrated that anti-PD1/PD-L1/CTLA-4 treatment extends the survival of mice challenged with the B16f10 melanoma cell line (23). Both miR-155 and ICB are critical drivers of T cell activation; thus, we were interested in understanding whether miR-155 is a downstream effector of ICB responsiveness. To determine whether ICB can reverse defective T cell–mediated tumor immunity observed in miR-155 TCKO mice, we administered a combination of αPD-1, αPD-L1, and αCTLA-4 antibodies to miR-155 TCKO and control mice with B16f10-OVA tumors. The results indicate that ICB significantly reduced tumor diameters and weights in miR-155 TCKO as well as control mice (Fig. 3, A and B). The percentage of CD8+ T cells producing IFNγ increased significantly within the tumors of treated miR-155 TCKO mice (Fig. 3C), and the percentage of CD4+ T cells producing IFNγ in miR-155 TCKO mice treated with ICB was rescued back to levels seen in control miR-155fl/fl mice treated with ICB (Fig. 3D). Additionally, defective CD40 and MHCI expression on TAMs from miR-155 TCKO mice returned to WT levels following ICB (Fig. 3E). These data indicate that ICB can rescue miR-155 tumor immunity phenotypes, where it reduces tumor growth, restores the levels of IFNγ-producing T cells, and promotes the activation of TAMs in miR-155 TCKO mice. This reveals that defective T cell antitumor immunity in the absence of T cell–expressed miR-155 is a defect that can be reversed.
Figure 3.
Restoration of tumor immunity following ICB therapy in miR-155 TCKO mice. A, B16f10-OVA tumor diameter growth measured over a 12-day time course. B, tumor weights of ICB treated and untreated miR-155fl/fl mice and miR-155 TCKO mice 12 days after tumor injection. C, representative flow plots of CD8+IFNγ+ T cells in tumors and percentages of IFNγ+ cells of CD8+ T cells in tumors from one representative experiment. D, representative flow plots of CD4+IFNγ+ T cells in tumors and percentages of IFNγ+ cells of CD4+ T cells in tumors from one representative experiment. E, MFI of CD40 and MHCI on CD45+CD11b+F4/80+ tumor-infiltrating macrophages of the indicated genotypes from one representative experiment. The plots represent data combined from multiple independent experiments unless otherwise indicated. *, p < 0.05; **, p < 0.005; ***, p < 0.0005; ****, p < 0.00005; ns, not significant.
Defective expression of effector and activation genes in miR-155−/− T cells is enhanced by ICB
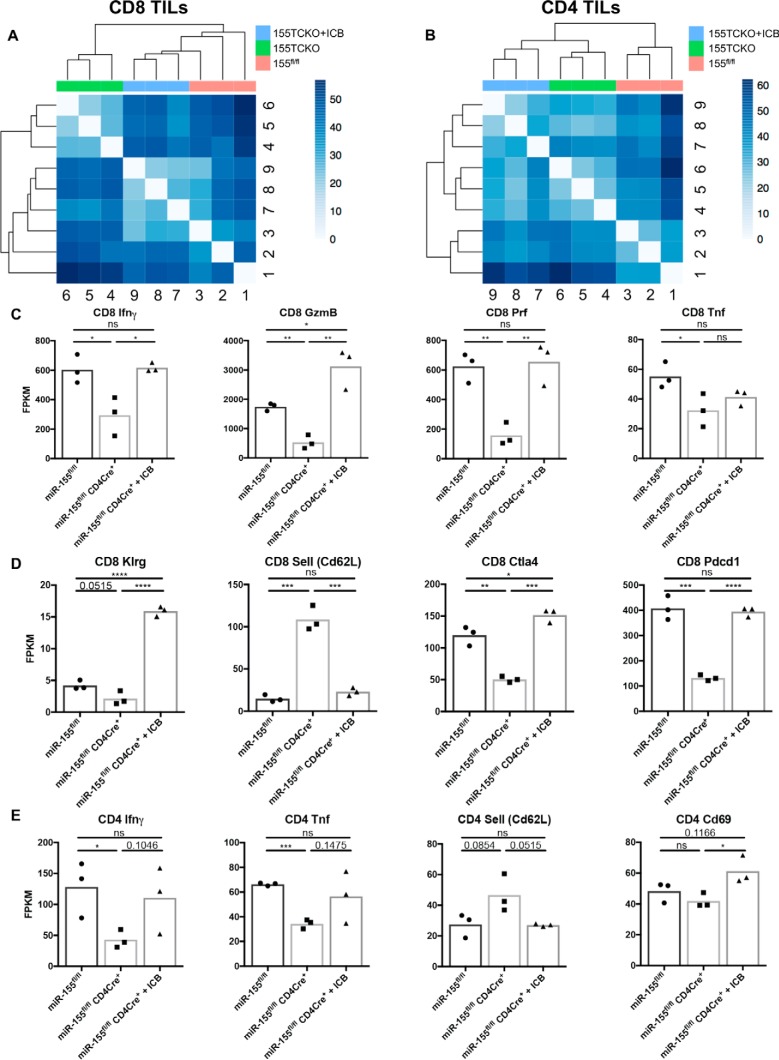
We next wanted to determine whether miR-155 expression within CD8+ and CD4+ T cells had an impact on additional genes that control effector function and/or activation of T cells within the tumor microenvironment, and if so, assess whether their expression is recovered following ICB. To accomplish this, we sorted both CD4+ and CD8+ T cell populations from miR-155fl/fl mice and miR-155 TCKO mice bearing B16f10-OVA tumors and miR-155 TCKO mice given ICB antibodies and performed RNA sequencing. The results indicate that CD8+ and CD4+ T cells each clustered according to genotype and treatment status (Fig. 4, A and B). The treatment effect occurring with ICB treated CD8+ T cells from miR-155 TCKO mice clustered more closely with CD8+ T cells from miR-155fl/fl mice than with untreated CD8+ T cells from TCKO mice (Fig. 4A), whereas ICB treated CD4+ T cells from miR-155 TCKO mice clustered more closely with untreated CD4+ T cells from TCKO mice than with CD4+ T cells from miR-155fl/fl mice (Fig. 4B), indicating a more robust recovery of the transcriptional profile of ICB treated miR-155 TCKO CD8+ T cells than miR-155 TCKO CD4+ T cells toward miR-155fl/fl controls. The transcription of CD8+ T cell effector genes, including IFNγ, granzyme B, perforin, and Tnf, were dependent on miR-155, all of which were rescued by ICB, with the exception of Tnf (Fig. 4C). The activation markers Klrg, Cd62l, Ctla4, and Pdcd1 all indicated that miR-155 TCKO CD8+ T cells were less activated than WT cells, but when treated with ICB reagents, these activation markers returned to WT levels (Fig. 4D). Finally, miR-155 expression in CD4+ T cells was required for the transcription of the effector genes IFNγ and Tnf in addition to the down-regulation of Cd62l in the absence of ICB (Fig. 4E). These genes, including the activation marker CD69, trended toward being restored to near WT levels with ICB. These results demonstrate an expanded role for miR-155 expression within tumor-infiltrating T cells, where its deletion reduces the expression of a subset of critical activation markers and effector genes and again shows that ICB can rescue most of these phenotypes.
Figure 4.
Tumor-infiltrating T cells from miR-155 TCKO mice have defective expression of activation and effector genes, and this is rescued by ICB. A, Euclidian distance plots showing the grouping of RNA sequencing data from CD8+ T cells sorted from B16f10-OVA tumors (12 days after tumor injection) of miR-155 TCKO mice treated with and without ICB and miR-155fl/fl mice. Higher numbers (darker colors) represent high dissimilarity, whereas lower numbers (lighter colors) represent high similarity. TIL, tumor-infiltrating lymphocyte. B, Euclidian distance plot for CD4+ T cells sorted from the mice in A. C, expression of CD8+ T cell effector molecules from mice with the indicated genotypes and treatment conditions. D, expression of CD8+ T cell activation markers from mice with the indicated genotypes and treatment conditions. E, expression of CD4+ T cell effector molecules and markers of activation from mice with the indicated genotypes and treatment conditions. *, p < 0.05; **, p < 0.005; ***, p < 0.0005; ****, p < 0.00005; ns, not significant.
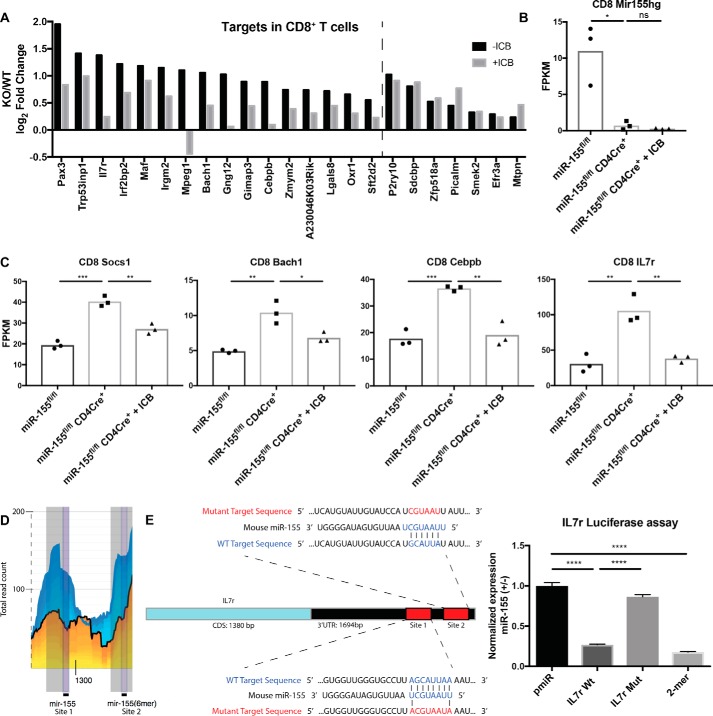
ICB reduces the expression of miR-155 targets in tumor-infiltrating CD8+ T cells from miR-155 TCKO mice, including the novel target IL7r
One potential mechanism whereby ICB may recover anti-tumor responses in miR-155 TCKO mice is by modulating the levels of direct miR-155 targets within T cells. To determine whether this was occurring, we utilized previously reported HITS-CLIP data generated with miR-155–deficient T cells (24) and found that, of the 191 predicted miR-155 targets from this dataset, 23 of these genes were significantly elevated in miR-155 TCKO CD8+ T cells compared with WT CD8+ T cells (Fig. 5A). 16 of these targets decreased in miR-155 TCKO CD8+ T cells from tumor-bearing mice treated with ICB. Seven targets were significantly increased in miR-155 TCKO CD4+ T cells, and four of these targets were reduced during ICB (supplemental Fig. S2A). miR-155 host gene expression was confirmed to be deleted in both CD8+ and CD4+ T cells (Fig. 5B and supplemental Fig. S2B). The targets Socs1, Bach1, and Cebpb, which are known targets of miR-155 during antitumor immunity and in other contexts (5, 25, 26), were all significantly elevated in miR-155 TCKO CD8+ tumor-infiltrating T cells and were rescued to near WT levels in ICB-treated miR-155 TCKO mice (Fig. 5C). IL7r is an important surface molecule whose expression is dynamically regulated throughout the development and activation of CD8+ T cells (27), and this putative target had a similar expression pattern as the targets in CD8+ T cells mentioned previously (Fig. 5C). Il7r expression was also decreased by ICB in CD4+ T cells (supplemental Fig. S2C). HITS-CLIP data reported previously (24) demonstrate decreased binding of the IL7r 3′ UTR by the RNA-induced silencing complex (RISC) in the absence of miR-155 (Fig. 5D), and we performed luciferase assays that confirmed functional miR-155 targeting of the IL7r 3′ UTR through a binding site–dependent mechanism (Fig. 5E). Together, these data support a model whereby miR-155 targets a subset of genes in T cells that regulate antitumor responses, with a large portion of these genes also being regulated by ICB. This indicates that miR-155 and ICB regulate overlapping pathways as they promote antitumor immunity mediated by T cells.
Figure 5.
Reduced expression of several miR-155 targets in tumor infiltrating T cells from miR-155 TCKO mice treated with ICB. A, log2-fold change of those miR-155 target genes, identified according to previously generated HITS-CLIP data from Loeb et al. (24), that were significantly derepressed in miR-155 TCKO CD8+ T cells compared with 155fl/fl control CD8+ T cells (−ICB). Expression of these genes was then compared between ICB-treated miR-155 TCKO mice and untreated control mice (+ICB) to determine the effect of ICB on the expression of predicted miR-155 targets in tumor-infiltrating CD8+ T cells. The dashed line delineates genes that were reduced with ICB from genes that were not. B, expression of the miR-155 host gene in CD8+ T cells from the indicated genotypes and treatment conditions. C, expression of the miR-155 targets Socs1, Bach1, Cebpb, and IL7r in CD8+ T cells of the indicated genotypes and treatment conditions. D, screenshot of the HITS-CLIP database from Loeb et al. (24), demonstrating the miR-155–dependent decrease in the binding of the IL7r 3′ UTR in miR-155 knockout CD4+ T cells (yellow area) compared with the binding demonstrated in WT CD4+ T cells (blue area). E, luciferase assay designed to test the functional binding of the miR-155 seed region to the 3′ UTR of IL7r. Utilizing GeneArt technology (Life Technologies), the indicated WT and mutant target sequences of the IL7r along with flanking regions were cloned into the pmiR reporter construct. 293T cells were transfected with either the WT or mutant pmiR reporter with and without miR-155 expression. After 24 h, luciferase expression was measured and normalized with Renilla expression for each sample. miR-155+ conditions were divided by miR-155− conditions within each group, and each group was normalized to the pmiR empty vector control, which was set at 1. Bar graphs represent the mean ± S.D. *, p < 0.05; **, p < 0.005; ***, p < 0.0005; ****, p < 0.00005; ns = not significant.
Discussion
The tumor microenvironment plays a key role in determining the type of immune response that is mounted against developing tumor cells. Recent successes in tumor immunotherapy, driven by the emergence of ICB therapies, demonstrate the utility of further understanding the molecular mechanisms that influence the anti-tumor immune response within the tumor microenvironment. miR-155 is one such regulator that is emerging as a critical modulator of the immune response to solid tumors. This study and previous work indicate that its expression within CD4+ and CD8+ T cells reduces the growth of syngeneic B16f10 tumors and increases the levels of IFNγ-expressing T cells within tumors. This study also found that miR-155 plays an expanded role in modulating the expression of effector molecules such as granzyme B, perforin, and Tnf in CD8+ T cells within the tumor microenvironment and that it regulates the expression of several activation genes in both CD8+ and CD4+ T cells in this environment. In addition to demonstrating that miR-155 functions in a cell-intrinsic manner within T cells to mediate effector gene expression, we show that a lack of miR-155 expression in T cells results in decreased IFN-inducible gene expression by tumor-associated macrophages, which is likely necessary for shaping a microenvironment that can reduce tumor growth. These are valuable insights into how miR-155 promotes cross-talk between T cells and macrophages within the tumor microenvironment. We previously observed no defects in T cell numbers in miR-155 TCKO mice under homeostatic conditions (3), and these data suggest that there is not a general decrease in T cell numbers in the periphery upon genetic deletion of miR-155. Thus, defects in the number of IFNγ-expressing T cells in tumors in miR-155 TCKO mice could involve defective initial activation in draining lymph nodes. It may also involve the inability of T cells that arrive at the tumor to become or remain activated and promote effector molecule production, resulting in reduced T cell numbers and defective antitumor responses within the tumor microenvironment. Finally, it is also possible that this can be explained by fewer activated T lymphocytes being able to home to and/or infiltrate into the tumor in the absence of miR-155. Future experiments will be required to carefully unravel the mechanistic basis for the defective levels of IFNγ-expressing T cells in tumor-bearing miR-155 TCKO mice.
The substantial rescue of CD8+ T cells by ICB, in combination with the partial rescue of CD4+ T cells, was sufficient to largely restore antitumor immunity in miR-155 TCKO mice. CD8+ T cells play critical roles in direct tumor killing. This may explain why a more complete rescue of miR-155−/− CD8+ T cells by ICB was necessary for controlling tumor growth in this model. However, even though miR-155−/− CD4+ T cells were rescued to a lesser extent by ICB than CD8+ T cells, the genes that trended toward being recovered include key effector genes such as IFNγ and Tnf, and these may be key to the role CD4+ T cells play in this context. Future studies will be needed to better understand why ICB has a larger impact on miR-155−/− CD8+ T cells versus miR-155−/− CD4+ T cells.
Macrophages are known to play vital roles in regulating the inflammatory microenvironment in a manner that can inhibit or promote tumor growth, depending on the type of macrophage response that occurs (28, 29). The secretion of IFNγ promotes a pro-inflammatory M1 macrophage response, but it is apparent that many other factors that are intrinsic or extrinsic to the macrophage compartment can shape this outcome (30). Previous studies utilizing whole-body knockout, overexpression, and other gene ablation techniques to study miR-155 expression within the myeloid compartment have demonstrated a role for miR-155 in maintaining M1 macrophage identity and function both in vitro and in vivo to prevent tumor metastasis and de novo tumor formation (7, 14, 31). However, our results, which are based on the commonly utilized LysM-Cre genetic deletion model, find that intrinsic expression of miR-155 within LysM-expressing macrophages is dispensable for the growth of B16f10 cells and TAM activation in this setting. Although the LysM-Cre model does have caveats (32, 33), our study indicates that the importance of macrophage-expressed miR-155 may be contextual and dependent on the tumor model and mode of miR-155 disruption being utilized. Further studies utilizing miR-155 MCKO mice and additional myeloid cell Cre driver strains will be necessary to understand the role of macrophage-expressed miR-155 within the context of different tumor types, including de novo and transplanted, and should provide additional understanding of where and when miR-155 regulates macrophage functions.
MicroRNAs fulfill many biological functions within immune cells by targeting the 3′ UTR of target mRNAs containing sites complementary to the six- to eight-nucleotide seed region of a given microRNA (1, 34, 35). The utility of this system allows a single microRNA to target multiple target genes that function within similar pathways to promote cellular effector functions. miR-155 is a canonical pro-inflammatory microRNA that allows immune cells, including those found within the tumor microenvironment, to fulfill their inflammatory effector functions in a variety of contexts (36). Targets such as Ship1, Socs1, and Ptpn2 are a few targets of miR-155 shown to individually promote specific functions within T cells (5, 8, 10), but it is likely that these, and many other targets, are functioning together to promote the effector function of these cells. Our data identify multiple derepressed miR-155 targets in tumor-infiltrating T cells by day 12 after tumor administration, including the target IL7r, which we show is a direct target of miR-155 in T cells. IL7 signaling regulates T cell survival, homeostasis, and proliferation during T cell development and homeostatic proliferation. Further, IL7r is down-regulated in T cells following T cell receptor (TCR) and/or IL2 stimulation (37), whereas miR-155 levels are known to increase during T cell activation (38). We show direct targeting of IL7r mRNA by miR-155, suggesting that miR-155 contributes to the repression of IL7r expression in response to activation signals. Thus, our data support a model whereby part of the role of miR-155 in promoting T cell activation involves repression of IL7r levels so that T cells can function properly upon activation. The down-regulation of IL7r levels in miR-155−/− T cells in response to ICB is an indicator of the increased level of activated miR-155−/− tumor-infiltrating T cells in response to this treatment. The observation that ICB partially restored IL7r and many other targets back toward WT levels in miR-155 TCKO mice suggests a possible mechanism whereby ICB is promoting T cell effector function by modulating direct targets of miR-155. Although the mechanistic basis for this is presently unclear, it suggests that miR-155 and ICB promote T cell antitumor responses through overlapping pathways.
It is clear from our study that miR-155 functions within the T cell compartment through a variety of mechanisms that include cross-talk with TAMs to constrain tumor growth in mice challenged with B16f10 tumors. This supports a growing literature indicating that miR-155 has significant translational potential as a promoter of antitumor responses (5, 10). Additionally, our study suggests that miR-155 and ICB therapy may function via overlapping pathways, as expression of miR-155 within T cells was largely dispensable for recovering the anti-tumor immune response by ICB, and ICB resulted in decreased expression of the targets derepressed in miR-155 TCKO tumor-infiltrating T cells. Previously, it has been observed that B16f10 tumors respond poorly to treatment with a single checkpoint-blocking antibody, such as CTLA-4, in the absence of other treatments (39). Our data indicate that antibody blocking of PD-1, PD-L1, and CTLA-4 further enhances T cell effector responses in the context of poorly immunogenic tumors and also suggest that increasing miR-155 expression in T cells may be a useful treatment alternative to ICB that may result in a similar outcome. The capacity of miR-155 to bolster the T cell compartment strongly suggests that overexpression of miR-155 within adoptively transferred, antigen-specific T cells or within chimeric antigen receptor T cells developed to specifically target tumors in cancer patients could improve responses by both cell types in a clinical setting. Further research in this area will hopefully lead to more treatment options for cancer patients and a greater ability to provide personalized treatment with successful outcomes.
Experimental procedures
Mice
C57BL/6 mice engineered to contain LoxP sites flanking miR-155 (The Jackson Laboratory) were crossed to mice expressing Cre under the control of the CD4 promoter (CD4-Cre, Taconic) as reported previously (3). miR-155 floxed mice were also crossed to mice expressing Cre under the control of the LysM promoter (LysM-Cre, The Jackson Laboratory) to restrict Cre-mediated deletion of miR-155 to myeloid cells. Groups of 5–10 sex-matched mice ranging from 6 to 12 weeks of age were used in each mouse experiment, and each experiment was repeated a minimum of two times. All experiments were performed in accordance with Institutional Animal Care and Use Committee (IACUC)-approved mouse protocols.
Cell culture and tumor experiments
B16f10 cells were acquired from the ATTC (catalog no. CRL-6475, lot no. 63048505) and grown in DMEM with 10% FBS, penicillin/streptomycin, and l-glutamine. B16f10-OVA cells were generated from B16f10 cells transfected with an OVA-expressing plasmid with resistance to G418 and grown similarly to B16f10 cells with addition of 0.1 mg/ml G418. Cells were cultured for at least 2 days, up to a confluency of 80–90%, and passaged once before injection into mice. 1 × 106 tumor cells were injected into the hind flank of each recipient in 100 μl of sterile PBS. Tumor growth was tracked by measuring the largest diameter of the tumor over a 12-day period. On day 12, mice were euthanized, and end point analysis of tumor and spleen tissues was performed.
Intracellular staining and flow cytometry
After mechanical disruption of tumor tissue, tumor cells were placed on an orbital shaker in Accumax (Innovative Cell Technologies) and incubated for 30 min at room temperature, followed by filtration through a 0.45-micron filter as performed previously (8). Spleen cells were also subjected to mechanical disruption and filtered through a 0.45-micron filter to obtain a single cell suspension following red blood cell lysis (Biolegend). Cells were stained with a combination of the following fluorophore-conjugated antibodies in Hanks' balanced salt solution supplemented with 10% BSA, pyruvate, EDTA, and HEPES: CD3 PB, CD3 Percp-cy5.5, CD4 PB, CD4 Percp-cy5.5, CD8 APC, CD45 PE-cy7, CD45 PB, Ly6c FITC, MHCII PB, Gr1 PE, CD40 FITC, CD86 Percp-cy5.5, F4/80 PE, F4/80 APC, CD11b Percp-cy5.5, CD11b APC, and IFNγ PE (eBioscience), all from Biolegend unless noted otherwise. After staining, the cells were washed, and flow cytometry was performed using a BD LSRFortessa (BD Biosciences), and data were subsequently analyzed using FlowJo software (Tree Star). Sorting of cells was completed on a FACSAria III (BD Biosciences) in the flow cytometry core facility at the University of Utah.
RNA isolation and qPCR
RNA was isolated from sorted cells with a miRNeasy mini/micro kit according to the instructions of the manufacturer (Qiagen). cDNA was made utilizing a qScript cDNA synthesis kit (Quanta Biosciences). Quantitative PCR was performed on a RocheLC480 utilizing qPCR master mix (Promega) with the following primer pairs: CXCL10, 5′GAAATCATCCCTGCGAGCCTATCC and 3′-GGGTCACCTACCGATCAGGATTAACG; CXCL9, 5′TTGGGCATCATCTTCCTGGAGCAG and 3′-GGTGATGTTTAGGGAGTTTCTGGAG; IFI205, 5′-AAGATCAAGGCATCTGGGAAAG and 3′-CCTCTGGGAATGTTCTGGTTC; IFIT1, 5′-CAGAAGCACACATTGAAGAA and 3′-TGTAAGTAGCCAGAGGAAGG; IFIT2, 5′-GGGAAAGCAGAGGAAATCAA and 3′-TGAAAGTTGCCATACAGAAG; IFIT3, 5′-GCCGTTACAGGGAAATACTGG and 3′-CCTCAACATCGGGGCTCT; L32, 5′-AAGCGAAACTGGCGGAAAC and 3′-TAACCGATGTTGGGCATCAG. Mature miR-155 expression was assessed by qPCR utilizing the miRCURY LNA Universal RT microRNA cDNA synthesis kit and the miRCURY LNA UniRT PCR primer for the miR-155 5p mature sequence (Exiqon).
ICB experiments
Mice of the indicated genotypes were given 1 × 106 B16f10-OVA cells injected subcutaneously into the hind flank. On days 3, 6, and 9 after tumor injection, each mouse in the treated group was given an intraperitoneal 250-μl injection of a mixture of αPD-1 (clone RMP1–14), αPD-L1 (clone 10F.9G2), and αCTLA-4 (clone 9H10) (BioXCell InVivoMAb) antibodies prepared at a concentration of 1 mg/ml, 1 mg/ml, and 0.4 mg/ml, respectively. Tumors and spleens from these mice were harvested and analyzed on day 12 after tumor injection.
RNA sequencing
Total RNA was isolated from flow-sorted CD45+CD3+CD4+ and CD45+CD3+CD8+ T cells from B16f10-OVA tumors with the miRNeasy micro kit (Qiagen). A cDNA library was prepared from RNA samples, and Illumina HiSeq 2500 sequencing was then performed at the University of Utah genomics core facility as described previously (3).
Luciferase assay
A 498-base pair sequence from the 3′ UTR of mouse IL7r (nucleotides 2537–3034 of IL7r mRNA with a flanking sequence to clone into the pmiR construct) that contains wild-type or mutant binding sites for miR-155 were synthesized by GeneArt Technology (Life Technology) and cloned into the pMiR reporter plasmid. 293T cells were utilized as described previously (40).
Statistical analysis
Student's t test was performed when comparing two groups, and significance was defined by a p value less than or equal to 0.05. Outlier data were identified using the ROUT method with a Q (false discovery rate) set to 1% in GraphPad Prism. The following denotes the meaning of statistical symbols: *, p < 0.05; **, p < 0.005; ***, p < 0.0005; ****, p < 0.00005.
Author contributions
T. B. H. coordinated and performed the experiments, analyzed the data, prepared the figures, and wrote the manuscript. S.-H. L. and W. W. T. provided technical assistance for the experiments and assisted with figure preparation. J. A. W., M. A., and M. C. R. provided technical assistance for the experiments. D. K. L., J. T., and A. G. R. managed the mouse colonies used in the experiments and provided technical assistance. W. P. V. advised on experiments and edited the manuscript. R. H. developed miR-155 TCKO mice. J. L. R. and M. A. W. advised on experiments. R. M. O. designed the research, advised on experiments, and edited the manuscript.
Supplementary Material
Acknowledgments
We thank the flow cytometry core facility as well as the genome sequencing and bioinformatics cores at the University of Utah. We also acknowledge Mingnan Chen at the University of Utah for providing B16f10-OVA tumor cells.
Funding for this research was provided by an American Cancer Society scholar award (to R. M. O.); NIA, National Institutes of Health Grant RO1AG047956 (to R. M. O.); and NCI, National Institutes of Health Grant F30CA189731 (to T. B. H.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. S1 and S2.
RNA sequencing data are available in the GEO database under accession number GSE101690.
- OVA
- ovalbumin
- TAM
- tumor-associated macrophage
- ICB
- immune checkpoint blocking
- TCKO
- T cell–conditional knockout
- MCKO
- macrophage–conditional knockout
- qPCR
- quantitative PCR
- PE
- phycoerythrin
- MFI
- mean fluorescence intensity
- MHCI
- major histocompatability complex I
- HITS-CLIP
- high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation
- PB
- Pacific Blue
- APC
- allophycocyanin.
References
- 1. O'Connell R. M., Rao D. S., Chaudhuri A. A., and Baltimore D. (2010) Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 10, 111–122 [DOI] [PubMed] [Google Scholar]
- 2. O'Connell R. M., Kahn D., Gibson W. S., Round J. L., Scholz R. L., Chaudhuri A. A., Kahn M. E., Rao D. S., and Baltimore D. (2010) MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 33, 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hu R., Kagele D. A., Huffaker T. B., Runtsch M. C., Alexander M., Liu J., Bake E., Su W., Williams M. A., Rao D. S., Möller T., Garden G. A., Round J. L., and O'Connell R. M. (2014) miR-155 promotes T follicular helper cell accumulation during chronic, low-grade inflammation. Immunity 41, 605–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vigorito E., Kohlhaas S., Lu D., and Leyland R. (2013) miR-155: an ancient regulator of the immune system. Immunol. Rev. 253, 146–157 [DOI] [PubMed] [Google Scholar]
- 5. Dudda J. C., Salaun B., Ji Y., Palmer D. C., Monnot G. C., Merck E., Boudousquie C., Utzschneider D. T., Escobar T. M., Perret R., Muljo S. A., Hebeisen M., Rufer N., Zehn D., Donda A., et al. (2013) MicroRNA-155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity 38, 742–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Trotta R., Chen L., Ciarlariello D., Josyula S., Mao C., Costinean S., Yu L., Butchar J. P., Tridandapani S., Croce C. M., and Caligiuri M. A. (2012) miR-155 regulates IFN-γ production in natural killer cells. Blood 119, 3478–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zonari E., Pucci F., Saini M., Mazzieri R., Politi L. S., Gentner B., and Naldini L. (2013) A role for miR-155 in enabling tumor-infiltrating innate immune cells to mount effective antitumor responses in mice. Blood 122, 243–252 [DOI] [PubMed] [Google Scholar]
- 8. Huffaker T. B., Hu R., Runtsch M. C., Bake E., Chen X., Zhao J., Round J. L., Baltimore D., and O'Connell R. M. (2012) Epistasis between microRNAs 155 and 146a during T cell-mediated antitumor immunity. Cell Rep. 2, 1697–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang J., Yu F., Jia X., Iwanowycz S., Wang Y., Huang S., Ai W., and Fan D. (2015) MicroRNA-155 deficiency enhances the recruitment and functions of myeloid-derived suppressor cells in tumor microenvironment and promotes solid tumor growth. Int. J. Cancer 136, E602–E613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ji Y., Wrzesinski C., Yu Z., Hu J., Gautam S., Hawk N. V., Telford W. G., Palmer D. C., Franco Z., Sukumar M., Roychoudhuri R., Clever D., Klebanoff C. A., Surh C. D., Waldmann T. A., et al. (2015) miR-155 augments CD8+ T-cell antitumor activity in lymphoreplete hosts by enhancing responsiveness to homeostatic γc cytokines. Proc. Natl. Acad. Sci. U.S.A. 112, 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen S., Wang L., Fan J., Ye C., Dominguez D., Zhang Y., Curiel T. J., Fang D., Kuzel T. M., and Zhang B. (2015) Host miR155 promotes tumor growth through a myeloid-derived suppressor cell-dependent mechanism. Cancer Res. 75, 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O'Connell R. M., Taganov K. D., Boldin M. P., Cheng G., and Baltimore D. (2007) MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 104, 1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He M., Xu Z., Ding T., Kuang D. M., and Zheng L. (2009) MicroRNA-155 regulates inflammatory cytokine production in tumor-associated macrophages via targeting C/EBPβ. Cell Mol. Immunol. 6, 343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cai X., Yin Y., Li N., Zhu D., Zhang J., Zhang C. Y., and Zen K. (2012) Re-polarization of tumor-associated macrophages to pro-inflammatory M1 macrophages by microRNA-155. J. Mol. Cell. Biol. 4, 341–343 [DOI] [PubMed] [Google Scholar]
- 15. Li L., Zhang J., Diao W., Wang D., Wei Y., Zhang C. Y., and Zen K. (2014) MicroRNA-155 and microRNA-21 promote the expansion of functional myeloid-derived suppressor cells. J. Immunol. 192, 1034–1043 [DOI] [PubMed] [Google Scholar]
- 16. Topalian S. L., Drake C. G., and Pardoll D. M. (2015) Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 27, 450–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hodi F. S., O'Day S. J., McDermott D. F., Weber R. W., Sosman J. A., Haanen J. B., Gonzalez R., Robert C., Schadendorf D., Hassel J. C., Akerley W., van den Eertwegh A. J., Lutzky J., Lorigan P., Vaubel J. M., et al. (2010) Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363, 711–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Topalian S. L., Hodi F. S., Brahmer J. R., Gettinger S. N., Smith D. C., McDermott D. F., Powderly J. D., Carvajal R. D., Sosman J. A., Atkins M. B., Leming P. D., Spigel D. R., Antonia S. J., Horn L., Drake C. G., et al. (2012) Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brahmer J. R., Tykodi S. S., Chow L. Q., Hwu W. J., Topalian S. L., Hwu P., Drake C. G., Camacho L. H., Kauh J., Odunsi K., Pitot H. C., Hamid O., Bhatia S., Martins R., Eaton K., et al. (2012) Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 366, 2455–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wolchok J. D., Kluger H., Callahan M. K., Postow M. A., Rizvi N. A., Lesokhin A. M., Segal N. H., Ariyan C. E., Gordon R. A., Reed K., Burke M. M., Caldwell A., Kronenberg S. A., Agunwamba B. U., Zhang X., et al. (2013) Nivolumab plus ipilimumab in advanced melanoma. N. Engl. J. Med. 369, 122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Larkin J., Chiarion-Sileni V., Gonzalez R., Grob J. J., Cowey C. L., Lao C. D., Schadendorf D., Dummer R., Smylie M., Rutkowski P., Ferrucci P. F., Hill A., Wagstaff J., Carlino M. S., Haanen J. B., et al. (2015) Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 373, 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Postow M. A., Chesney J., Pavlick A. C., Robert C., Grossmann K., McDermott D., Linette G. P., Meyer N., Giguere J. K., Agarwala S. S., Shaheen M., Ernstoff M. S., Minor D., Salama A. K., Taylor M., et al. (2015) Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N. Engl. J. Med. 372, 2006–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Curran M. A., Montalvo W., Yagita H., and Allison J. P. (2010) PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc. Natl. Acad. Sci. U.S.A. 107, 4275–4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Loeb G. B., Khan A. A., Canner D., Hiatt J. B., Shendure J., Darnell R. B., Leslie C. S., and Rudensky A. Y. (2012) Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol. Cell 48, 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gottwein E., Mukherjee N., Sachse C., Frenzel C., Majoros W. H., Chi J. T., Braich R., Manoharan M., Soutschek J., Ohler U., and Cullen B. R. (2007) A viral microRNA functions as an orthologue of cellular miR-155. Nature 450, 1096–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. O'Connell R. M., Rao D. S., Chaudhuri A. A., Boldin M. P., Taganov K. D., Nicoll J., Paquette R. L., and Baltimore D. (2008) Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 205, 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mazzucchelli R., and Durum S. K. (2007) Interleukin-7 receptor expression: intelligent design. Nat. Rev. Immunol. 7, 144–154 [DOI] [PubMed] [Google Scholar]
- 28. Noy R., and Pollard J. W. (2014) Tumor-associated macrophages: from mechanisms to therapy. Immunity 41, 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chanmee T., Ontong P., Konno K., and Itano N. (2014) Tumor-associated macrophages as major players in the tumor microenvironment. Cancers 6, 1670–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shiao S. L., Ganesan A. P., Rugo H. S., and Coussens L. M. (2011) Immune microenvironments in solid tumors: new targets for therapy. Genes Dev. 25, 2559–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu F., Jia X., Du F., Wang J., Wang Y., Ai W., and Fan D. (2013) miR-155-deficient bone marrow promotes tumor metastasis. Mol. Cancer Res. 11, 923–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ye M., Iwasaki H., Laiosa C. V., Stadtfeld M., Xie H., Heck S., Clausen B., Akashi K., and Graf T. (2003) Hematopoietic stem cells expressing the myeloid lysozyme gene retain long-term, multilineage repopulation potential. Immunity 19, 689–699 [DOI] [PubMed] [Google Scholar]
- 33. Bell J. J., and Bhandoola A. (2008) The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature 452, 764–767 [DOI] [PubMed] [Google Scholar]
- 34. Krol J., Loedige I., and Filipowicz W. (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610 [DOI] [PubMed] [Google Scholar]
- 35. Mehta A., and Baltimore D. (2016) MicroRNAs as regulatory elements in immune system logic. Nat. Rev. Immunol. 16, 279–294 [DOI] [PubMed] [Google Scholar]
- 36. Seddiki N., Brezar V., Ruffin N., Lévy Y., and Swaminathan S. (2014) Role of miR-155 in the regulation of lymphocyte immune function and disease. Immunology 142, 32–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carrette F., and Surh C. D. (2012) IL-7 signaling and CD127 receptor regulation in the control of T cell homeostasis. Semin. Immunol. 24, 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Haasch D., Chen Y. W., Reilly R. M., Chiou X. G., Koterski S., Smith M. L., Kroeger P., McWeeny K., Halbert D. N., Mollison K. W., Djuric S. W., and Trevillyan J. M. (2002) T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell. Immunol. 217, 78–86 [DOI] [PubMed] [Google Scholar]
- 39. van Elsas A., Hurwitz A. A., and Allison J. P. (1999) Combination immunotherapy of B16 melanoma using anti-cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) and granulocyte/macrophage colony-stimulating factor (GM-CSF)-producing vaccines induces rejection of subcutaneous and metastatic tumors accompanied by autoimmune depigmentation. J. Exp. Med. 190, 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hu R., Huffaker T. B., Kagele D. A., Runtsch M. C., Bake E., Chaudhuri A. A., Round J. L., and O'Connell R. M. (2013) MicroRNA-155 confers encephalogenic potential to Th17 cells by promoting effector gene expression. J. Immunol. 190, 5972–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.