Abstract
A high proliferation rate has been observed to facilitate somatic cell reprogramming, but the pathways that connect proliferation and reprogramming have not been reported. DNA methyltransferase 1 (DNMT1) methylates hemimethylated CpG sites produced during S phase and maintains stable inheritance of DNA methylation. Impairing this process results in passive DNA demethylation. In this study, we show that the cell proliferation rate positively correlated with the expression of Dnmt1 in G1 phase. In addition, as determined by whole-genome bisulfate sequencing and high-performance liquid chromatography, global DNA methylation of mouse embryonic fibroblasts was significantly higher in G1 phase than in G2/M phase. Thus, we suspected that high cellular proliferation requires more Dnmt1 expression in G1 phase to prevent passive DNA demethylation. The methylation differences of individual CpG sites between G1 and G2/M phase were related to the methylation status and the positions of their surrounding CpG sites. In addition, larger methylation differences were observed on the promoters of pluripotency-related genes; for example, Oct4, Nanog, Sox2, Esrrb, Cdh1, and Epcam. When such methylation differences or passive DNA demethylation accumulated with Dnmt1 suppression and proliferation acceleration, DNA methylation on pluripotency-related genes was decreased, and their expression was up-regulated, which subsequently promoted pluripotency and mesenchymal–epithelial transition, a necessary step for reprogramming. We infer that high cellular proliferation rates promote generation of induced pluripotent stem cells at least partially by inducing passive DNA demethylation and up-regulating pluripotency-related genes. Therefore, these results uncover a connection between cell reprogramming and DNA methylation.
Keywords: cell proliferation, DNA methylation, DNA methyltransferase, p53, reprogramming
Introduction
Several mechanisms associated with the generation of induced pluripotent stem cells (iPSCs)3 have been reported. Among these mechanisms, a high proliferation rate is beneficial for the reprogramming of mouse embryonic fibroblasts (MEFs) (1, 2). In addition, it has been suggested that the generation efficiency of iPSCs correlates with the number of cell cycles that occur rather than with the actual time elapsed (2). Thus, certain biological events that occur during the cell cycle may stochastically allow cells to enter pathways that lead to pluripotency. Our first hypothesis was that accelerated proliferation leads to particular epigenetic modulations and subsequently facilitates reprogramming. Therefore, the relationship between cell proliferation and the expression of genes related to epigenetic modulations was examined.
DNA methylation is subject to complex regulation during reprogramming. Ten-eleven translocation methylcytosine dioxygenase 1 (Tet1) mediates active DNA demethylation and replaces Oct4 to promote reprogramming, which is also modulated by vitamin C (Vc) (3–5). In addition, during DNA replication, the newly synthesized DNA strand has no cytosine methylation. The stable inheritance of DNA methylation during proliferation relies on DNA methyltransferase 1 (DNMT1), which methylates hemimethylated CpGs not only during S phase but also during G2/M phase (6–8). Normally, global DNA methylation is stable during proliferation. However, inhibition of such DNMT1-mediated methylation by suppressing Dnmt1 expression or by promoting cell proliferation accumulates the hemimethylated CpGs along with the cell cycle progress, gradually reduces global DNA methylation, and results in “passive” DNA demethylation (9).
During iPSCs generation, an both increase in proliferation rate and a decrease in global DNA methylation are observed. It is reasonable to suggest that a high proliferation rate might lead to passive DNA demethylation, regulate the expression of certain genes, and facilitate reprogramming. Thus, in this study, a connection between passive DNA demethylation and proliferation was established and studied during reprogramming.
Results
Dnmt1 expression in G1 phase correlates with proliferation rates
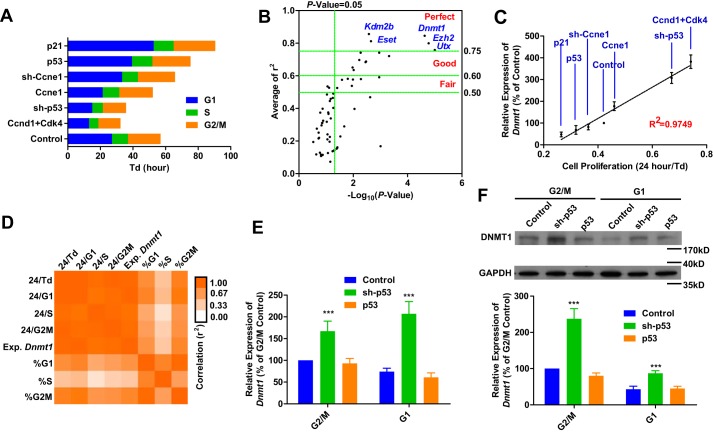
To explore the potential connection between proliferation rate and the expression of genes related to epigenetic regulation, like histone modification and DNA methylation, the cell proliferation rate, especially the length of G1 phase, was modulated by regulating the expression of p53, p21, Ccne1, Ccnd1, and Cdk4 in MEFs (Fig. 1A). The expression of 102 genes related to DNA methylation, histone methylation, or other epigenetic modulations was determined by qPCR (Table S1). The expression of Dnmt1, Eset, Ezh2, Kdm2b, and Utx had the most significant correlation with proliferation rate (Fig. 1, B and C), suggesting that high expression of these genes is required in cells with a high proliferation rate.
Figure 1.
Dnmt1 expression correlates with proliferation rate. A–D, the proliferation and cell cycle of MEFs were modulated by retrovirus-delivered gene expression (A). MEFs infected with retroviruses encoding Flag and sh-Luc were used as controls. The correlation between cell proliferation (Td) and gene expression was determined by qPCR (B). The average r2 values are shown on the y axis, whereas the p values for the correlation efficiencies with baseline (0.5000) are shown on the x axis The correlation between cell proliferation (Td) and Dnmt1 expression is listed in C. The correlations among Dnmt1 expression, the respective lengths of different phases of the cell cycle, and percent occupancy of different phases of the cell cycle are summarized in D. E and F, MEFs were infected with retroviruses encoding sh-p53 and p53. MEFs infected with retroviruses encoding Flag and sh-Luc were used as controls. The expression of Dnmt1 was determined at the mRNA (E) and protein levels (F, with representative images from five independent experiments). One-way ANOVA with Dunnett post hoc test was used for comparisons between control and other groups. ***, p < 0.001.
Among the five identified genes, Dnmt1 was selected for further investigation because of the connection between reprogramming and DNA methylation (4, 5). Because the expression of Dnmt1 is relatively high during S phase (10, 11), the correlation described above might result from an increased percentage of cells in S phase. This possibility was partially excluded by the higher correlation of Dnmt1 expression with G1 phase length or doubling time (Td) than with the percentage of cells in S phase (Fig. 1D). In addition, the acceleration of cell proliferation with sh-p53 up-regulated Dnmt1 expression, both at the mRNA and protein levels, in G1 phase (Fig. 1, E and F). Because the stable inheritance of DNA methylation during proliferation relies on DNMT1-mediated methylation on hemimethylated CpGs not only in S phase but also in G2/M phase (6–8), we proposed that DNMT1 in G1 phase might also contribute to the stable inheritance of DNA methylation.
Dnmt1 up-regulation compensates for the accelerated proliferation
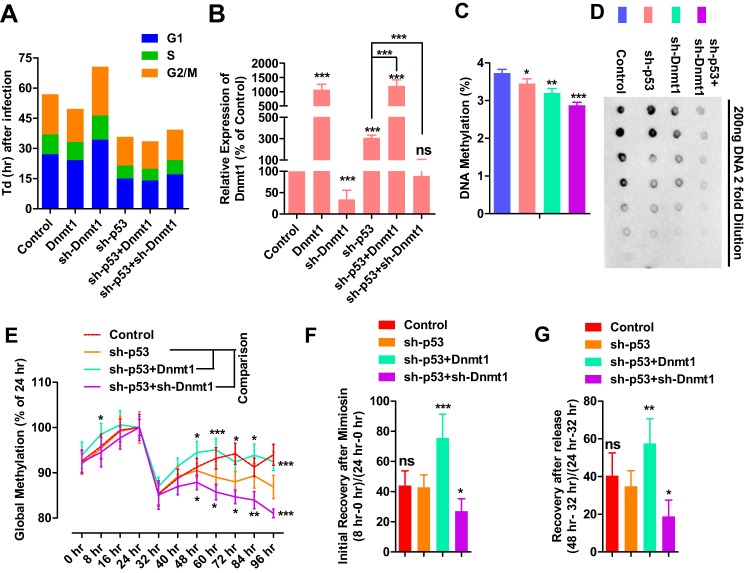
The hypothesis above was supported by the ability of sh-p53 to shorten G1 phase and up-regulate Dnmt1 expression (Fig. 1, A, E, and F), which suggested that more DNMT1 was required to maintain DNA methylation in a shorter cell cycle. In addition, sh-Dnmt1 decreased the proliferation rate and induced a longer G1 phase (Fig. 2A), suggesting that less DNMT1 required a longer cell cycle to maintain DNA methylation. When sh-p53 was combined with sh-Dnmt1, both sh-p53-induced Dnmt1 up-regulation and sh-Dnmt1-induced cell cycle arrest were attenuated (Fig. 2, A and B).
Figure 2.
Dnmt1 expression is required for stable DNA methylation inheritance. A and B, MEFs were infected with different retroviruses encoding Flag+sh-Luc (control), sh-Luc+Dnmt1 (Dnmt1), Flag+sh-Dnmt1 (sh-Dnmt1), Flag+sh-p53 (sh-p53) or sh-p53+Dnmt1, or sh-p53+sh-Dnmt1. Cell cycle information was collected 3 days after infection by counting cells and performing PI staining (A). Dnmt1 expression was determined at the same time by qPCR (B). C and D, MEFs were infected with different retroviruses encoding Flag+sh-Luc, Flag+shp53, Flag+sh-Dnmt1, or sh-p53+sh-Dnmt1. Global DNA methylation levels were determined 3 days later with HPLC (C) and dot-blot (D). E–G, MEFs were infected with different retroviruses encoding Flag+sh-Luc, Flag+sh-p53, sh-p53+Dnmt1, or sh-p53+sh-Dnmt1 at hour −48. Two days after infection (hour 0), 0.5 μm mimosine was used to treat cells for an additional 24 h. After mimosine withdrawal, cells were further cultured for 72 h (hours 24–96). DNA methylation levels were determined by HPLC and are summarized in (E) after normalizing to the methylation levels at hour 24. Percentage recoveries of DNA methylation during (F) or after (G) mimosine treatment were compared as indicated. Two-way ANOVA with Bonferroni post hoc test was used for comparisons between the sh-p53 group and the other two groups with sh-p53 in (E). One-way ANOVA with Dunnett post hoc test was used for comparisons between control and other groups in B and C or the sh-p53 group and other groups in F and G. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
Although the data here could not serve as direct evidence for the function of DNMT1 in G1 phase, it confirmed the balance between proliferation rate and Dnmt1 expression. Cells with different proliferation rates require different amounts of DNMT1 to maintain stable DNA methylation during proliferation. A shorter cell cycle requires a larger amount of DNMT1 whereas a longer cell cycle requires less.
sh-p53 induced cell proliferation, shortened G1 phase, and made cells require more DNMT1. Dnmt1 up-regulation induced by sh-p53 was a kind of compensative effect for the higher proliferation rate. However, such Dnmt1 up-regulation did not fully compensate for the accelerated proliferation induced by sh-p53 or fully rescued the inheritance of DNA methylation, which led to significant DNA demethylation, as indicated by the dot-blot and HPLC in Fig. 2, C and D.
On the other hand, overexpression of sh-Dnmt1 not only decreased Dnmt1 expression but also impaired cell proliferation, which compensated for Dnmt1 down-regulation (Fig. 2A). However, the compensation was not strong enough to prevent DNA demethylation (Fig. 2, C and D). When sh-p53 was co-expressed with sh-Dnmt1, the two shRNAs inhibited the compensative effects, Dnmt1 up-regulation and proliferation arrest, induced by each other (Fig. 2, A and B) and resulted in even larger DNA demethylation (Fig. 2, C and D).
We also found that a larger change on Dnmt1 expression was induced by sh-p53 than that induced by p53. We propose that higher proliferation requires more DNMT1 for stable inheritance DNA methylation, and the cells are forced to express more Dnmt1. Although lower proliferation requires less DNMT1, cells have no way to reduce Dnmt1 expression immediately. In addition, a larger change in cell cycle was induced by sh-Dnmt1 than that induced by Dnmt1 (Fig. 2A). Less DNMT1 forces cells to have a longer cell cycle for stable DNA methylation, whereas sufficient DNMT1 does not guarantee high proliferation. In summary, enough DNMT1 is necessary but not sufficient for high proliferation.
To further confirm the balance mentioned above, mimosine was used to synchronized the cells by G1/S arrest, and DNA methylation was monitored after mimosine treatment or mimosine withdrawal. As indicated in supplemental Fig. S1A, 0.5 μm mimosine arrested cells in G1 phase, and the majority of cells moved from G1 phase to S phase and then to G2/M after mimosine withdrawal. Although infection with different retroviruses, like those encoding Flag, sh-Luc, sh-p53, Dnmt1, and sh-Dnmt1, affected the cell cycle differently (Fig. 2A), 24-h mimosine treatment forced the cells into similar G1/S arrest (supplemental Fig. S1B).
Because DNA methylation is not completely inherited after S phase, G1/S arrest by mimosine provided additional time for DNMT1 to function, which subsequently resulted in an increase in global DNA methylation after mimosine treatment (hours 0–24, Fig. 2E). In addition, cells moved forward from G1/S arrest to S phase and induced a dramatic decrease in global DNA methylation during the first 8 h after mimosine withdrawal (hours 24–32, Fig. 2E). After S phase, global DNA methylation gradually returned to the basal level, possibly because of the function of DNMT1 (hours 32–48, Fig. 2E). Thus, the dynamic regulation of DNA methylation confirmed the ability of DNMT1 to maintain DNA methylation outside of S phase.
The percentage increase in DNA methylation during the first 8 h of mimosine treatment (0–8 h versus 0–24 h) and the percentage of recovery during the first 8 h after mimosine withdrawal (24–32 h versus 24–48 h) were used to measure the ability of cells to complete DNA demethylation after S phase. Proliferation acceleration with sh-p53 did not affect the two percentages (Fig. 2, F and G). However, the two percentages were increased by Dnmt1 overexpression and decreased by sh-Dnmt1 overexpression (Fig. 2, F and G), suggesting an ability of Dnmt1 to affect the inheritance of DNA methylation.
DNA methylation of MEFs is higher in G1 than in G2/M phase
We have proposed that DNMT1 in G1 phase also contributes to the stable inheritance of DNA methylation. However, the data above only confirmed the function of DNMT1 outside of S phase, but did not directly support the functions of DNMT1 in G1 phase. To provide direct evidence for this hypothesis, DNA methylation in G1 phase and G2/M phase was compared.
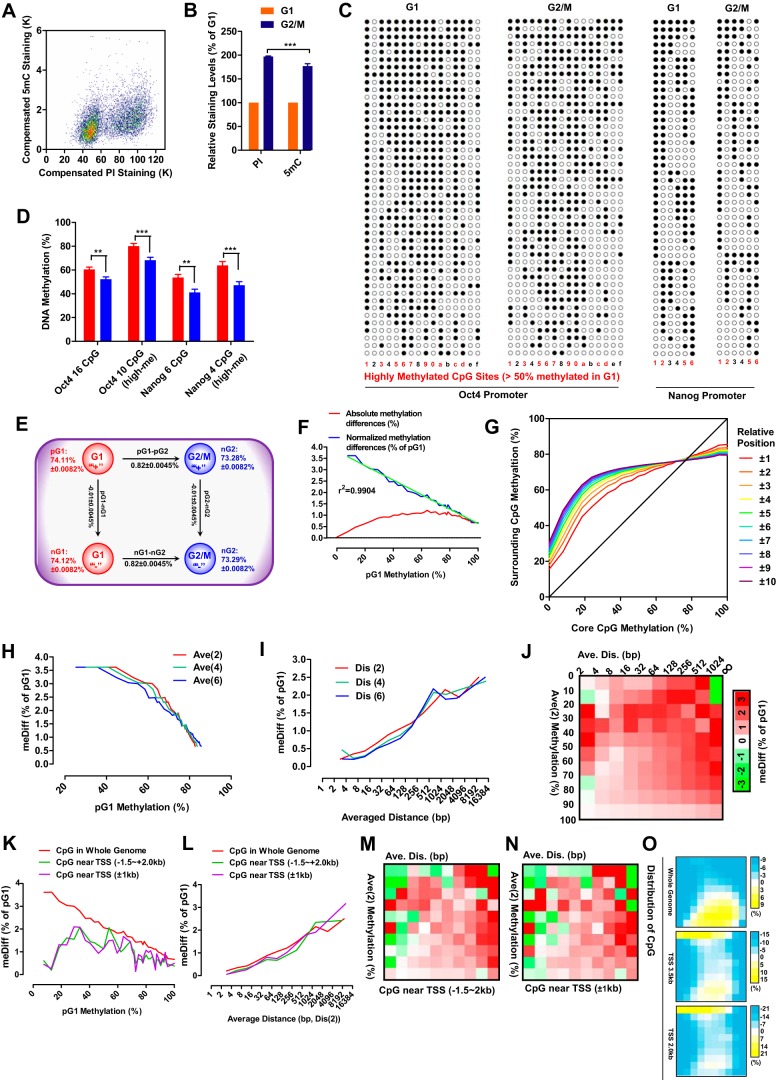
DNA and 5-methylcytosine were stained with propidium iodide (PI) and specific antibodies, respectively, as shown in Fig. 3A. The amount of DNA in G2/M cells was 196% ± 3% (n = 5) of that in G1 cells, whereas the amount of methylated cytosine in G2/M cells was only 181% ± 5% (n = 5) of that in G1 cells (Fig. 3B). To confirm this, the levels of methylation of the Oct4 and Nanog promoters were determined in both G1 and G2/M MEFs (Fig. 3, C and D). The average methylation level of 16 CpGs on the Oct4 promoter was higher in G1 than in G2/M MEFs. When 10 highly methylated CpGs (methylation over 50% in G1 phase) were selected for additional analysis, the difference in methylation was larger. A similar phenomenon was observed on the Nanog promoter. Thus, DNA methylation of MEFs is higher in G1 than in G2/M phase
Figure 3.
The DNA methylation is higher in G1 than in G2/M phase. A and B, MEFs were stained with PI and an antibody against 5mC before FACS analysis (A). The relative levels of DNA (PI staining) and 5mC (antibody staining) of MEFs in G2/M and G1 phase are shown in B. C and D, promoter methylation patterns of Oct4 and Nanog in G2/M and G1 MEFs (C). The methylation levels of all CpG sites and highly methylated CpG sites (>50% methylation in G1) were compared between G2/M and G1 phases (D). E, schematic of methylation differences. F, absolute methylation differences between G1 and G2/M phase and normalized methylation differences (against corresponding pG1, meDiff) were plotted as a function of pG1 methylation. G, the average methylation of surrounding CpGs was plotted as a function of core CpG methylation. H, methylation differences were plotted as a function of the average methylation of the two (Ave(2)), four (Ave(4)), and six (Ave(6)) closest surrounding CpGs. I, methylation differences were plotted as a function of the average distance between core CpGs and the two (Dis(2)), four (Dis(4)) and six (Dis(6)) closest surrounding CpGs. J, CpGs were grouped according to methylations level and the positions of the two closest surrounding CpGs. The average methylation differences of each group are represented as a heatmap. K–O, CpGs within −1.5 to 2.0 kb or −1.0 to +1.0 kb of TSSs were selected. Similar analyses as in F, I, and J were performed in K–N. The distributions of the three types of CpGs along the methylation levels and positions of the two closest surrounding CpGs are shown in O. Two-way ANOVA with Bonferroni post hoc test and two-tailed t test were used for comparisons between the indicated groups in B and D, respectively. **, p < 0.01; ***, p < 0.001.
DNA methylation was then further analyzed with whole-genome bisulfate sequencing (WGBS) to confirm the higher methylation level in G1 phase. WGBS covered the mouse genome ∼30 times in both samples (MEFs in G1 and G2/M phase) and displayed no preference for different regions of the genome (supplemental Fig. S1, A–H). Because WGBS distinguished the positive (the top strand annotated in the genome) and negative strand (the bottom strand annotated in the genome), each qualified CpG site had four reads, methylation levels on the positive strand in G1 phase (pG1), on the negative strand in G1 phase (nG1), and on two strands in G2/M phase (pG2 and nG2) (Fig. 3E). When the four reads of all qualified CpGs (over 11 million) were analyzed, no significant differences were found between positive and negative strands. However, methylation was significantly higher in G1 phase (74.12% ± 0.0082%) than in G2/M phase (73.28% ± 0.0082%, p < 1 × 10−6) (Fig. 3E). Our results differ from the constant DNA methylation during replication and cell cycle arrest suggested by a previous report (12). However, the constant DNA methylation observed in that study might result from the low sequencing depth (5.3–8.6×) and the inclusion of all CpGs with a sequencing depth of greater than 1 in the analysis (12).
Absolute methylation differences (nG1-pG2/2-nG2/2) of CpGs between G1 and G2/M phase were plotted against their pG1 methylation levels (Fig. 3F). The highest differences were discovered in CpGs with pG1 methylation at around 70%, whereas the lowest differences were found in CpGs with nearly no pG1 methylation. Because CpGs on the template strand can only be recognized as methylated or unmethylated, absolute methylation differences were then normalized against the corresponding pG1 methylation levels, and the normalized results indicated the probability that the methylation of particular CpGs was not inherited after G2/M phase or required to be inherited in G1 phase. The normalized methylation differences were negatively correlated with pG1 methylation levels (Fig. 3F). Hereafter, methylation differences refer to the normalized methylation differences, and the abbreviation meDiff is used in the figures.
As the cellular machinery involved in inheritance of DNA methylation only senses the methylation of CpGs in a binary model, and the methylation difference indicates how DNMT1 works during G1 phase, the mechanisms underlying the correlation between methylation differences and pG1 methylation were investigated. We considered each individual CpG as a core CpG and the CpGs close to the core CpG as their surrounding CpGs. The methylation levels of core CpGs were found to be similar to those of their surrounding CpGs (Fig. 3G), which was consistent with a previous report on the similar methylation status of adjacent CpGs (13). In addition, negative correlations were observed between methylation differences and the average methylation levels of the two, four, or six closest surrounding CpGs (Fig. 3H). The distance between the core CpGs and their surrounding CpGs should also be critical, not only because of the limited size of the DNA methylation inheritance machinery but also because of the connection between methylation levels and the positions of surrounding CpGs (supplemental Fig. S3, A and B). The average distance between the core and the surrounding CpGs was positively correlated with methylation differences (Fig. 3I). Therefore, the methylation differences of core CpGs may be determined by the methylation status and the positions of their surrounding CpGs. Methylation differences were then assessed after grouping CpGs according to the average methylation of the two closest CpGs and the average distance between the core and the two closest CpGs (Fig. 3J). The largest methylation differences were observed with CpGs whose surrounding CpGs had very close positions and medium methylation levels or faraway positions and extremely low methylation levels. These results suggest that the methylation differences are enriched in the genome with particular CpG density and methylation status.
We next analyzed the methylation differences of CpGs near transcription start sites (TSSs, −1.5 to +2.0 kb or −1.0 to +1.0 kb). The methylation and the distance from the surrounding CpGs of these two groups of CpGs were different from those of CpGs in the whole genome (supplemental Fig. S3, C–E), which resulted in lower methylation differences of CpGs with low pG1 methylation (Fig. 3K); but how methylation differences were affected by the distance between the core CpG and two closest CpGs was not influenced. (Fig. 3L). As indicated in Fig. 3, M and N, the patterns of methylation differences of these CpGs were similar to those shown in Fig. 3F, suggesting that DNMT1 does not function differentially in regions close to TSSs compared with the whole genome. The difference shown in Fig. 3K is likely due to the different distributions of CpGs when being grouped according to methylation levels and the positions of the two closest surrounding CpGs (Fig. 3O).
Promoters of pluripotency-related genes have higher methylation differences
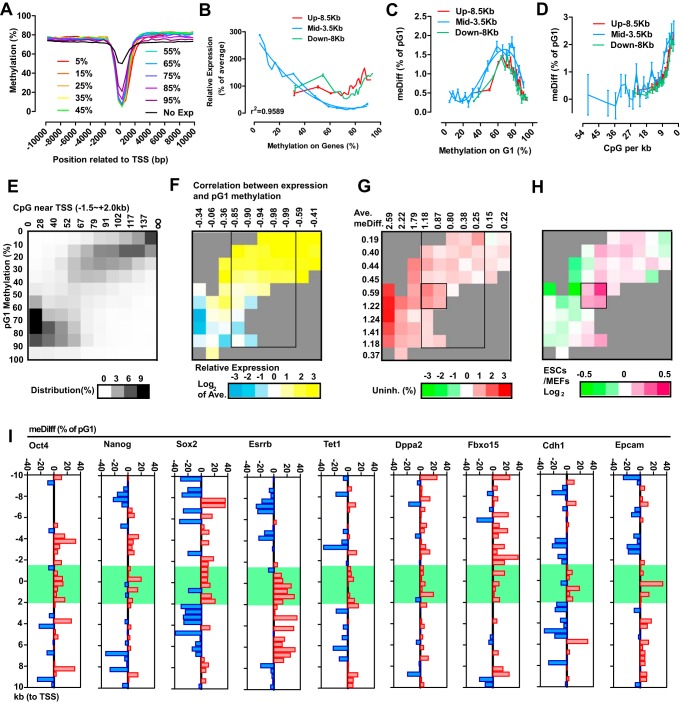
The expression levels of protein-coding genes were obtained from previously reported RNA-seq (supplemental Table S2). Based on the ranking of their expression (from the top 5% to 95%), protein-coding genes with detectable expression were equally divided into 10 groups, and the average methylation levels of these groups of genes were plotted, along with the relative distances to TSSs (Fig. 4A). Negative correlation between gene expression and methylation levels was only observed in a region from −1.5 to +2.0 kb (Fig. 4A). The methylation levels of particular genes were then determined as the average methylation levels of CpGs in three different regions, −10 to −1.5 kb (Up, 8.5 kb), −1.5 to +2.0 kb (Mid, 3.5 kb), and +2.0 to +10 kb (Down, 8 kb) (Fig. 4B). The distributions of methylation and CpG numbers in the Mid region were different from those in the Up and Down regions (supplemental Fig. S3, F and G). In addition, only methylation in the Mid region correlated negatively with gene expression, especially when the methylation level was relatively low (below 70%) (Fig. 4B). Methylation differences of particular genes were calculated similarly as for individual CpGs and were also linked to the pG1 methylation level and CpG number in the Mid region of the gene (Fig. 4, C and D).
Figure 4.
Pluripotency-related genes were preferentially demethylated. A, depending on their expression ranking, genes with detectable expression were divided into 10 groups from top 5% to 95%. Methylation levels were calculated every 500 bp in a 20-kb region around the TSS. Genes whose expression were not detected in current RNA-sequencing were analyzed together as the No Exp group. B–D, correlations between gene methylation and normalized expression are plotted in B. Methylation differences were plotted against gene methylation (C) and CpG density (D). E–H, genes were grouped according to CpG density and pG1 methylation. The distribution of genes (E), normalized expression (F), methylation differences (G), and differences in gene expression between MEFs and ES cells/iPSCs (H) were summarized as heat maps. The parameters used for grouping are labeled in E. The correlation efficiencies between pG1 methylation and gene expression in each column (with a certain number of CpGs in the Mid regions) are listed just above in F. The average methylation differences of genes in each column and row are provided in G. I, methylation differences were calculated every 500 bp in a 20-kb region around the TSS for the indicated pluripotency-related genes.
Genes were then grouped according to their pG1 methylation levels and the number of CpGs in their Mid regions (Fig. 4E). Genes with high pG1 methylation and a few number of CpGs as well as genes with low pG1 methylation and a large number of CpGs accounted for the majority of the analyzed genes. When analyzing gene expression in these groups, a better correlation between pG1 methylation and gene expression was observed with genes that had moderate numbers of CpGs (52∼117) in their Mid regions, as indicated in Fig. 4F. Within groups of genes that had a moderate number of CpGs, four gene groups with prominent methylation differences are further highlighted in Fig. 4G. Because the final four gene groups displayed significantly more up-regulation during the conversion from MEFs to iPSCs (Fig. 4H), methylation differences or higher methylation in G1 phase might be preferentially enriched in pluripotency-related genes. To confirm the hypothesis, average methylation levels of genes were determined every 500 bp in a region from −10 to +10 kb. As indicated in Fig. 4I, Oct4, Nanog, Sox2, Esrrb, Tet1, Dppa2, Fbxo15, Cdh1, and Epcam have significant higher methylation in their Mid region in G1 phase. Therefore, if these methylation differences are not fully rescued in G1 phase, then the accumulated demethylation on these gene might up-regulate these genes and promote pluripotency.
Passive demethylation facilitates reprogramming
Cells normally maintain constant global methylation levels during proliferation, relying on DNMT1 not only in S phase but also in G2/M and G1 phase. Thus, under conditions in which proliferation is accelerated and Dnmt1 expression is suppressed, DNA methylation might not be inherited completely and result in passive DNA demethylation. Thus, passive DNA demethylation can be considered as unrescued and accumulated methylation differences as mentioned above. To confirm the preference of passive DNA demethylation for pluripotency-related genes, sh-Dnmt1- and sh-p53-induced passive DNA demethylation was investigated during reprogramming.
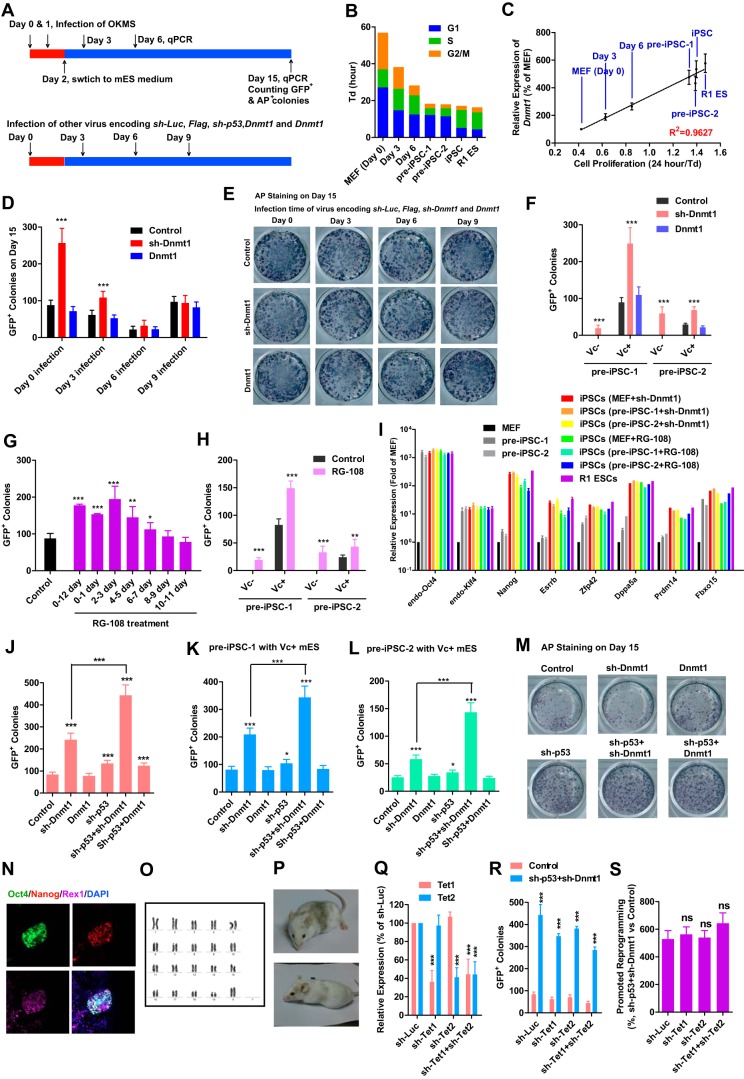
Retroviruses encoding Oct4, Klf4, c-Myc, and Sox2 were introduced to MEFs twice, on day 0 and day 1 respectively. 10% FBS–containing medium was switched to MES medium with or without vitamin C (Fig. 5A). qPCR samples were collected on days 3, 6, 9, and 15. Alkaline phosphatase-positive (AP+) and GFP+ colonies were always counted and collected on day 15.
Figure 5.
Passive DNA demethylation facilitates reprogramming. A, schematic of the reprogramming timeline. B and C, correlation of cell proliferation with Dnmt1 expression in MEFs (day 0), pre-iPSCs, and R1 ES cells and during reprogramming (days 3 and 6). D and E, retroviruses encoding Flag, sh-Luc, sh-p53, sh-Dnmt1, and Dnmt1 were introduced into MEFs on days 0, 3, 6, or 9 during reprogramming. GFP+ colonies were counted on day 15 just before AP staining. F, retroviruses encoding Flag, sh-Luc, sh-p53, sh-Dnmt1, and Dnmt1 were introduced into two pre-iPSC lines. MES medium with or without Vc was used to induce the conversion from pre-iPSCs to iPSCs. GFP+ colonies were counted 7 days after infection. G and H, 10 μm RG-108 was used to treat cells in different periods during reprogramming (G) or during the conversion from pre-iPSCs to iPSCs (H). GFP+ colonies were counted. I, expression of pluripotent genes was determined in iPSCs generated in D–H by qPCR. J–P, sh-Dnmt1 and sh-p53 were used alone or together to induce passive DNA demethylation during reprograming or conversion from pre-iPSCs to iPSCs. Reprogramming efficiencies of MEFs (J) and pre-iPSCs (K and L) to GFP+ iPSCs are shown. One colony generated from MEFs with sh-p53 and sh-Dnmt1 was further characterized using immunofluorescence (N), karyotyping (O), and chimera formation (P). Q–S, retroviruses encoding sh-Tet1 and sh-Tet2 were delivered into MEFs alone or together. Their abilities to affect the expression of Tet1 and Tet2 were determined 3 days after infection (Q). These two shRNAs were used during reprogramming together with Flag+sh-Luc or sh-p53+sh-Dnmt1. GFP+ colonies were counted on day 15 (R). GFP+ colonies in sh-p53+sh-Dnmt1 groups were normalized to control (Flag+sh-Luc) groups and subjected to additional comparisons (S). One-way ANOVA with Dunnett post hoc test was used for comparisons between control and other groups or the indicated groups. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
The expression of Dnmt1 was correlated with proliferation during MEF reprogramming (Fig. 5, B and C). To determine the function of Dnmt1 suppression during different stages of reprogramming, retroviruses encoding Flag, sh-Luc, sh-p53, sh-Dnmt1, and Dnmt1 were introduced to cells on days 0, 3, 6, or 9. sh-Dnmt1–promoted reprogramming was only observed when retroviruses were delivered at early stages of reprogramming, on day 0 or day 3 (Fig. 5D).
Although sh-Dnmt1 overexpression increased the number of GFP+ colonies, it did not affected or slightly decreased the number of AP+ colonies (Fig. 5, D and E). AP+ colonies normally included iPSC and pre-iPSC colonies, whereas GFP+ colonies only included iPSC colonies. sh-Dnmt1 overexpression might promote reprogramming by facilitating the conversion from pre-iPSCs to iPSCs. Two pre-iPSC lines established in a previous report were used (14). Pre-iPSC-1 (pre-3a-2 in the previous report) is sensitive to Vc whereas pre-iPSC-1 (pre-2-1 in the previous report) is not. sh-Dnmt1 but not Dnmt1 overexpression facilitated the conversion from pre-iPSCs to iPSCs (Fig. 5F).
RG-108, an inhibitor of DNMT1 (15), also promoted the conversion of MEFs and pre-iPSCs to iPSCs (Fig. 5, G and H). The iPSCs generated above were isolated, cultured, and determined to express endogenous pluripotency markers, like Nanog, Esrrb, and so on, at levels close to those in R1 ES cells, suggesting their pluripotency (Fig. 5I).
Because sh-Dnmt1 overexpression slightly inhibited cell proliferation (Fig. 2A), which does not favor passive DNA demethylation, cell proliferation was accelerated with sh-p53 in the presence of sh-Dnmt1. The reprogramming efficiency of both MEFs and pre-iPSCs was further enhanced when sh-p53 and sh-Dnmt1 were used together (Fig. 5, J–M). Selected GFP+ colonies generated with sh-p53 and sh-Dnmt1 overexpression displayed up-regulated expression of pluripotent genes and normal karyotypes and were able to give rise to chimeric mice (Fig. 5, N–P). Therefore, passive DNA demethylation and the methylation differences mentioned above facilitate reprogramming.
To get rid of the influences from active DNA demethylation, retroviruses encoding sh-Tet1 and sh-Tet2 were used alone or together. The two shRNAs decreased the expression of Tet1 and Tet2, respectively, by about 60% (Fig. 5Q). When these two shRNAs were used during reprogramming, the numbers of GFP+ colonies were counted and compared between the Flag+sh-Luc and sh-p53+sh-Dnmt1 groups. Although reducing the expression of either sh-Tet1 or sh-Tet2 impaired the overall reprogramming efficiency (Fig. 5R), it did not affect the ability of passive DNA demethylation induced by sh-p53+sh-Dnmt1 to promote reprogramming (Fig. 5S). Thus, passive DNA demethylation affects reprogramming independent of active DNA demethylation. However, the two kinds of DNA demethylation may still connect with each other during reprogramming, which requires additional investigation.
Passive DNA demethylation up-regulates pluripotency-related genes
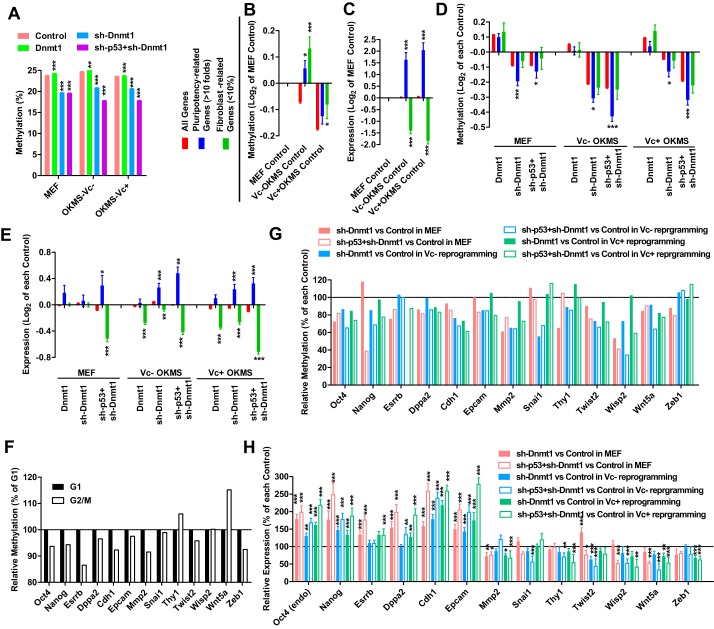
To provide a basis for understanding the differences between active and passive DNA demethylation and how these differences affect reprogramming, control vectors (Flag and sh-Luc), Dnmt1, sh-Dnmt1, or sh-Dnmt1+sh-p53 were overexpressed in three experimental systems, MEFs, Vc−, and Vc+ reprogramming. Gene expression was measured using RNA-seq (GSE93416), and DNA methylation was determined by reduced representation bisulfite sequencing (RRBS, GSE93058) (supplemental Table S3). Passive DNA demethylation induced by both sh-Dnmt1 and sh-Dnmt1+sh-p53 was observed (Fig. 6A). In addition, the methylation levels of protein-coding genes determined in WGBS positively correlated with that determined in RRBS (supplemental Fig. S3H).
Figure 6.
Passive DNA demethylation up-regulates pluripotent and mesenchymal genes. A retrovirus was used to introduce Flag+sh-Luc (control), Dnmt1, sh-Dnmt1, and sh-Dnmt1+sh-p53 into MEFs and into MEFs during OKMS-induced reprogramming without Vc (Vc−) or with Vc (Vc+). A, global DNA methylation were determined 5 days after infection by RRBS. B–E, fibroblast- and pluripotency-related genes were selected according to their expression in MEFs and iPSCs. The changes in methylation and expression of these two groups of genes were compared with global changes of all genes. Changes in two control groups during reprogramming were first normalized to the control group in MEFs before comparisons (B and C). Changes in groups with Dnmt1, sh-Dnmt1, and/or sh-p53 overexpression were first normalized to the corresponding control group in three experimental systems (D and E). F–H, pluripotent genes (Oct4, Nanog, Esrrb, and Dppa2), epithelial genes (Cdh1 and Epcam), and mesenchymal genes (Mmp2, Snai1, Thy1, Twist2, Wisp2,Wnt5a, and Zeb1) were analyzed. Their methylation levels in G1 and G2/M MEFs were generated from WGBS (F). Their methylation and expression levels during overexpression with Dnmt1, sh-Dnmt1, and/or sh-p53 were generated from RRBS (G) and qPCR (H) respectively. One-way ANOVA with Dunnett post hoc test was used for comparisons between the indicated control groups and other groups. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Based on the gene expression profiles of MEFs, iPSCs, and ES cells (16, 17), two groups of genes were selected for further investigation. Genes whose expression in ES cells and iPSCs was over 10-fold or below 10% of that in MEFs were analyzed as pluripotency- or fibroblast-related genes. Gene methylation and expression in control groups of MEFs were used for normalization (Fig. 6, B and C). The methylation levels of fibroblast-related genes were higher than those of pluripotency-related genes during reprogramming, especially during reprograming with Vc (Fig. 6B). In addition, up-regulation of pluripotency-related genes and down-regulation of fibroblast-related genes were observed during reprogramming (Fig. 6C), which was consistent with our normal understanding of reprogramming.
The ability of passive DNA demethylation to affect gene methylation and expression was determined by normalizing gene methylation and expression in the Dnmt1, sh-Dnmt1, and sh-Dnmt1+sh-p53 groups to those in their corresponding control (Flag+sh-Luc) groups within the same experimental system (Fig. 6, D and E). For Fig. 6, B and C, we used control group MEFs for normalization to indicate the difference between MEFs and MEFs during reprogramming. For Fig. 6, D and E, we used control groups in different experimental systems for normalization to indicate the influence of passive DNA demethylation.
In all three experimental systems, sh-Dnmt1 and sh-Dnmt1+sh-p53 induced greater demethylation of pluripotency-related genes than of fibroblast-related genes, resulting in up-regulation of pluripotency-related genes and down-regulation of fibroblast-related genes (Fig. 6, D and E), which was consistent with its ability to promote reprogramming (Fig. 5).
Further analyses were performed on representatives of several groups of genes: pluripotent, mesenchymal, and epithelial genes. The methylation differences of these genes between G1 and G2/M phases were generated from current WGBS (Fig. 6G). The methylation changes of these genes during passive DNA demethylation induced by sh-Dnmt1 with or without sh-p53 were generated from the current RRBS (Fig. 6H). Gene expression was determined by qPCR (Fig. 6I). Pluripotent genes, like Oct4, Nanog, Esrrb, and Dppa2, had lower methylation in G2/M than in G1 phase. Their methylation and expression were significantly modulated by passive DNA demethylation (Fig. 6, G–I).
Mesenchymal–epithelial transition (MET) is a necessary step at the early stage of reprogramming (18). Inducing an opposite epithelial–mesenchymal transition (EMT) inhibits reprogramming, whereas inducing MET promotes reprogramming. MEFs are mesenchymal cells, whereas iPSCs and ES cells are epithelial cells. Genes that are significantly up-regulated during reprogramming (pluripotency-related genes; Fig. 6, B–E) are enriched with epithelial genes, whereas genes that are significantly down-regulated during reprogramming (fibroblast-related genes; Fig. 6, B–E) are enriched with mesenchymal genes. When analyzing epithelial genes like Epcam and Cdh1, their methylation and expression were regulated similarly as those of the pluripotent genes mentioned above (Fig. 6, G–I). The up-regulation of epithelial genes was consistent with the down-regulation of mesenchymal genes, like Mmp2, Snai1, Thy1, Twist2, Wisp2, Wnt5a, and Zeb1, during passive DNA demethylation, which suggests the progress of MET. Therefore, passive DNA demethylation preferentially affects a set of genes including several pluripotent genes and epithelial genes and subsequently facilitates pluripotency generation and MET.
Discussion
DNA methylation can be stably passed on from parent to daughter cells; however, DNA methylation is also flexible, as it is affected by multiple factors. Given that this is the case, how do cells achieve a balance between the stability and flexibility of DNA methylation? This study indicates that inheritance of DNA methylation is not completed during DNA replication or in S phase. The uninherited DNA methylation appears as hemimethylated CpGs and can be remethylated by DNMT1 during G2/M and G1 phase. Therefore, by modulating the balance between Dnmt1 expression and proliferation rate, inheritance of DNA methylation may be regulated. In addition, if the uninherited DNA methylation after S phase is enriched in particular regions or on promoters of a special subset of genes, as demonstrated in this study, inheritance of DNA methylation may be involved in gene expression regulation and favor or prohibit certain cell fates.
The ability of DNMT1 to methylate hemimethylated CpGs has been reported previously (6–8). The direct evidence for its function in G1 phase is the higher DNA methylation in G1 than in G2/M phase. Basing on WGBS, the difference in global DNA demethylation levels of G1 and G2/M MEFs is significant but relatively small, about 0.8% when considering absolute methylation difference and about 1.1% after normalization (Fig. 3E). However, the methylation decrease after S phase in the mimosine experiments shown in Fig. 2E is close to 15%. Such a large difference should be due to two facts. First, G1 MEFs included cells from different stages of G1 phase, during which DNMT1 constantly functions to complete inheritance of DNA methylation, and so did MEFs in G2/M phase. Thus, the significant decrease in methylation shown in Fig. 2E is the difference between the highest methylation in G1 and the lowest methylation in G2 and does not represent the actual methylation difference between G1 and G2/M phase. Based on the data shown in Fig. 2E, global DNA methylation in G1 and G2/M MEFs was at about 94% and 91% of the highest methylation level, which results in a difference of around 2–3%. In addition, the inclusion of MEFs in S phase in the two samples might explain the remaining gap.
Based on the results shown in Fig. 3, H and I, we propose that methylation of surrounding CpGs is beneficial for inheritance of DNA methylation, possibly because the methylated surrounding CpGs keep the DNMT1 machinery in a conformation that favors following methylation on core CpGs. We also proposed that inheritance of DNA methylation was impaired by the distance between core and surrounding CpGs, possibly because a large distance results in the exclusion of surrounding CpGs from the DNMT1 machinery. Thus, CpGs whose surrounding CpGs have faraway positions and extremely low methylation levels were observed to have large methylation differences (Fig. 3J). However, CpGs whose surrounding CpGs have very close positions and medium methylation levels also have large methylation differences (Fig. 3J). A possible explanation is as follows. If the distance between CpGs is too small (<4 bp) for surrounding CpGs to be excluded from the DNMT1 activation site, inheritance of DNA methylation is impaired, especially when the methylation of the surrounding CpGs is around 20–70%. However, additional investigation is required to prove this.
The mesenchymal and epithelial states have been recognized as two major states of cells (19). Expression changes induced by passive DNA demethylation (sh-Dnmt1+sh-p53) facilitated MET (Fig. 6I), whereas those by active DNA demethylation and Tet1 facilitated EMT, especially in Vc-containing systems (5). Thus, it is reasonable for us to suggest that the two models of induced DNA demethylation yielded almost opposite expression changes in mesenchymal and epithelial genes in the presence of Vc, especially when considering that Tet1-induced demethylation employs 5-hydroxymethylcytosine and 5-formylcytosine as intermediates. Therefore, the influence of passive DNA demethylation by sh-Dnmt1 and active DNA demethylation by Tet1 on EMT or MET and on different cell fate conversions should be studied.
Experimental procedures
Materials
Information related to the materials, the assay kits, and the deposited data used in this study is listed in supplemental Table S2. All procedures related to animal studies were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (Publication 80-23) and were approved by the Institutional Review Board of the Guangzhou Institutes of Biomedicine and Health. All efforts were made to minimize the number of animals used and their suffering.
Generation of iPSCs
MEFs were maintained in high-glucose DMEM supplemented with 10% FBS, non-essential amino acids, and GlutaMAX. MEFs were subjected to a mycoplasma test (MycoAlertTM, Lonza, Allendale, NJ) to ensure that they were free of mycoplasma before use. The retrovirus was produced using Plat-E cells and pMXs-based retroviral vectors as described previously, except that a calcium phosphate transfection protocol was used (20).
Within two passages, the MEFs were split into 12-well plates (1.5 × 104 cells/well). After addition of Polybrene to 4 μg/ml, the viral supernatant was used for infection of cells. Oct4, Klf4, c-Myc, and Sox2 were introduced into cells twice, on day 0 and day 1, and MES or MES-Vc (high-glucose DMEM, non-essential amino acids, GlutaMAX, leukemia inhibitory factor (LIF), 2-mercaptoethanol, and 10% FBS with or without Vc) was used on day 2. The medium was replaced daily with freshly prepared medium. GFP+ and AP+ colonies were counted on day 14.
Pre-iPSCs were treated in MES with or without Vc. A retrovirus was used to induce exogenous expression on day 0. GFP+ colonies were counted 7 days later.
AP staining and TUNEL staining
Cells were fixed with 4% paraformaldehyde in PBS, incubated at room temperature for 2 min, and then washed twice with 0.5 ml of TBST (20 mm Tris-HCl, 500 mm NaCl, 0.05% Tween 20, pH 7.5). Freshly prepared 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium color development substrate (Promega; 0.33 mg/ml 5-bromo-4-chloro-3-indolyl phosphate, 0.167 mg/ml nitro blue tetrazolium, 100 mm Tris-HCl (pH 9.0), 150 mm NaCl, and 1 mm MgCl2) was added. The plates were incubated in the dark at room temperature for 15 min and then rinsed with PBS. Apoptotic cell death was analyzed using the TUNEL assay with an in situ cell death detection kit (TMR Red, Roche, 12156792910).
Cell line characterization
Characterization of iPSCs was performed as reported previously (20). Primary antibodies against NANOG and REX1 were used together with the appropriate Alexa-conjugated secondary antibodies. Immunofluorescence results were obtained using a Zeiss LSM 800 confocal microscope.
For karyotype analysis, demecolcine (50 μg/ml, Sigma) was added to the cells for 1 h. The cells were trypsinized, pelleted, resuspended in 0.075 m KCl, and incubated for 20 min at 37 °C. The cells were then fixed with acetic acid:methanol (1:3) for 10 min at 37 °C. The cells were collected by centrifugation, resuspended in fixative solution, dropped on a cold slide, and incubated at 75 °C for 3 h. The slides were treated with trypsin and colorant, and metaphase chromosomes were analyzed using an Olympus BX51 microscope.
For immunoblotting, antibodies against DNMT1 and GAPDH were used together with the appropriate HRP-conjugated secondary antibodies. The gels used for immunoblotting with different antibodies were run under the same experimental conditions. Representative images were cropped from the original images with no modification of the relative intensities.
qPCR
Total RNA was extracted from cells using TRIzol (Invitrogen), and 5 μg of RNA was used to synthesize cDNA with ReverTra Ace® (Toyobo) and oligo(dT) (Takara) according to the instructions of the manufacturer. The transcript levels of genes were determined using SYBR Premix Ex TaqII (Tli RNaseH Plus, Takara) and a CFX-96 real-time system (Bio-Rad). The primers used are listed in supplemental Table S1.
During the investigation of correlations between cell proliferation and the expression of genes related to epigenetic modulations, the expression of several G1/S arrest-related genes, p53, p21, Ccne1, Ccnd1, and Cdk4, was regulated using a retroviral system. The expression of 102 genes related to DNA methylation, histone methylation, or other epigenetic modulations was determined by qPCR 4 days after infection. Td was calculated by dividing the 96-h log2 increase in cell number over a 4-day period. The correlation between cell proliferation (24/Td) and gene expression (normalized to the control group) was calculated for each gene using a linear regression model in GraphPad Prism 5.0. The experiments were repeated at least five times. The average correlation efficiencies (r2) are shown on the y axis in Fig. 1B. The p values between the correlation efficiencies and baseline (multiple r2 at 0.5000) are provided on the x axis in Fig. 1B.
FACS
Cells were dissociated with trypsin/EDTA, fixed with 4% paraformaldehyde, and permeabilized with 0.1% Triton X-100. Antibodies against 5-methylcytosine were used in conjunction with the appropriate secondary antibodies to stain the cells. The cells were then used for FACS analysis using a BD FACSAria II flow cytometer.
DNA methylation
DNA was extracted using the Wizard genomic DNA purification kit (Promega, A1125) according to the instructions of the manufacturer. DNA methylation levels were determined using various methods.
For Bisulfite sequencing, genomic DNA (700 ng) obtained from various samples was exposed overnight to a mixture of 40.5% sodium bisulfite and 10 mm hydroquinone. Subsequently, regions of the Oct4 and Nanog promoters were amplified by PCR using primers described previously (20). The PCR products were cloned into the pMD18-T vector (Takara), propagated in DH5α, and sequenced.
For HPLC, purified DNA was digested with DNA Degradase Plus (Zymo Research, E2021) at 37 °C for 24 h to obtain nucleosides for subsequent HPLC analysis (the 50-μl reaction volume contained 3 μl of DNA degradase, 5 μl of enzyme buffer, and 5 μg of DNA). The resulting DNA digestion solution was analyzed by electrophoresis on a 1% agarose gel to verify complete digestion.
HPLC was performed according to a method reported previously. The DNA digestion solution was diluted 4- to 5-fold, filtered through a 0.2-μm nylon membrane (Agilent, 5190-5106), and transferred to an HPLC sample vial (Agilent, 5188-6591). Each sample (10-μl volume) was loaded and analyzed on an Agilent 1260 BIO machine with a C18 reverse-phase column (2.1 × 50 mm, 1.8 μm, Agilent ZORBAX Eclipse Plus C18). The mobile phase was 7 mm ammonium acetate (pH 6.7)/5% methanol (v/v), the flow rate was 0.3 ml per min, and the detector was set at 280 nm.
Calibration curves were generated using 2′-deoxycytidine (dC; Sigma, D3897) and 5-methyl-2′-deoxycytidine (5mdC; Chemcruz, sc278256). The concentrations of dC and 5mdC in the samples were calculated by interpolation from the calibration curves. The DNA methylation level was calculated as 5mdC / (dC + 5mdC) × 100%.
For WGBS, MEFs were digested and fixed with 70% ethanol overnight. The fixed cells were treated with 0.25 mg/ml RNase A at 37 °C for 30 min to remove RNA and stained with 50 μg/ml propidium iodide (Sigma, P4170) for 30 min. The resulting cell suspension was used to enrich G1/S and S/G2 phase cells by FACS on a BD FACSAria II flow cytometer. Approximately 3 × 106 cells in each phase of the cell cycle were sorted by FACS, and DNA was extracted from the sorted cells as described above. The purity of the DNA was determined using a K5500 spectrophotometer, and DNA quantification was performed using a Qubit® 3.0 fluorometer. The purified DNA was shipped on dry ice to Annoroad Gene Technology Co. Inc. (Beijing, China) for WGBS. Clean reads were mapped to the mouse genome (GRCm38/mm10) using methods and software reported previously (21). The raw sequencing data, clean reads, and methylation information of each cytosine that was sequenced at least once during WGBS were provided to the authors for further analysis.
For RRBS, MEFs were infected with a retrovirus carrying the indicated genes twice: once on day 0 and once on day 1. The cells used in the MEF experimental system were cultured as normal MEFs for an additional 5 days. The cells used in the Vc−/Vc+ OKMS reprogramming experimental systems (Vc−/Vc+) were also infected with a retrovirus carrying the four Yamanaka factors during the first 2 days and were subjected to further reprogramming over the next 5 days.
Five days after infection, ∼3 × 106 cells in each group were harvested by digestion with 0.25% trypsin. DNA was extracted from the cells, purified, and quantified as described above. The purified DNA was shipped on dry ice to Annoroad Gene Technology Co. Inc. for RRBS. RRBS was performed according to protocols reported previously (22).
RNA-seq
RNA was extracted from cells using TRIzol reagent (Invitrogen). Illumina mRNA-seq libraries were prepared for each RNA sample using the TruSeq RNA Sample Preparation Kit v2; the mRNA-seq libraries were then sequenced on an Illumina NextSeq 500 instrument with the NextSeq 500 Mid Output Kit v2.
The obtained RNA-seq reads were processed by RNA-seq by expectation maximization to estimate transcript abundances. The reads were aligned to the (Ensembl v67) transcriptome, and the number of reads associated with a given transcript was used to estimate the abundance of that transcript in transcripts per million.
Statistical analysis
All experiments were repeated at least five times (n ≥ 5), with the exception of sequencing. The data were analyzed and compared using a two-tailed t test, one-way ANOVA with Dunnett's post-hoc test, or two-way ANOVA with Bonferroni post hoc test in GraphPad Prism 5.0. Error bars and n represent the standard deviation (standard error when indicated) and the number of independent experiments, respectively. Asterisks represent significant differences (*, p < 0.05; **, p < 0.01; ***, p < 0.001) from the indicated control groups. Detailed statistical information is listed in supplemental Table S4.
When groups of CpGs in WGBS or RRBS were analyzed, the standard error was too small to be plotted because the number of CpGs was always large. However, when the methylation and expression results for particular genes were analyzed, the standard error is provided. Logarithmic differences were calculated as log2. Logarithmic differences in expression and methylation are shown with standard errors, but the p values were generated using the original values.
Accession numbers
GSE10871 and GSE14012 were used to provide gene expression profiles of ES cells and neural stem cells relative to that of MEFs (16, 17).
Author contributions
H. Z. and D. P. conceived and supervised the study and wrote the manuscript. H. Z. and S. H. designed the experiments and analyzed the data. S. H., H. S., L. Lin, Y. Z., J. C., L. Liang, Y. L., M. Z., X. Y., X. W., F. W., F. Z., J. C., and H. Z. performed the experiments.
Supplementary Material
Acknowledgment
We thank the Guangzhou Branch of the Supercomputing Center of the Chinese Academy of Sciences for support.
This work was supported by National Natural Science Foundation of China Grants 91519305, 31422032, 31671475, U1601228, and 31421004; Special Research Project for Frontier Science of the Chinese Academy of Sciences Grant QYZDB-SSW-SMC031; Guangdong Natural Science Foundation Grant 2014A030308002; Guangzhou Science and Technology Program Grant 201607010239; Guangzhou Health Care Collaborative Innovation Program Grant 201508020250; and the Guangdong Special Support Program Grant 2014TQ01R157. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S3 and Tables S1–S4.
- iPSC
- induced pluripotent stem cell
- MEF
- mouse embryonic fibroblast
- Vc
- vitamin c
- Td
- doubling time
- PI
- propidium iodide
- WGBS
- whole-genome bisulfate sequencing
- TSS
- transcription start site
- RNA-seq
- RNA sequencing
- RRBS
- reduced representation bisulfite sequencing
- MET
- mesenchymal–epithelial transition
- EMT
- epithelial–mesenchymal transition
- ANOVA
- analysis of variance
- meDiff.
- normalized methylation differences between G1 and G2/M phase
- AP
- alkaline phosphatase
- OKMS
- MEF reprogramming induced by overexpression of Oct4, Klf4, c-Myc, and Sox2
- dC
- 2′-deoxycytidine
- 5mdC
- 5-methyl-2′-deoxycytidine
- 5mC
- 5-methylcytosine.
References
- 1. Ruiz S., Panopoulos A. D., Herrerías A., Bissig K. D., Lutz M., Berggren W. T., Verma I. M., and Izpisua Belmonte J. C. (2011) A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr. Biol. 21, 45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hanna J., Saha K., Pando B., van Zon J., Lengner C. J., Creyghton M. P., van Oudenaarden A., and Jaenisch R. (2009) Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462, 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Costa Y., Ding J., Theunissen T. W., Faiola F., Hore T. A., Shliaha P. V., Fidalgo M., Saunders A., Lawrence M., Dietmann S., Das S., Levasseur D. N., Li Z., Xu M., Reik W., et al. (2013) NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 495, 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gao Y., Chen J., Li K., Wu T., Huang B., Liu W., Kou X., Zhang Y., Huang H., Jiang Y., Yao C., Liu X., Lu Z., Xu Z., Kang L., et al. (2013) Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell 12, 453–469 [DOI] [PubMed] [Google Scholar]
- 5. Chen J., Guo L., Zhang L., Wu H., Yang J., Liu H., Wang X., Hu X., Gu T., Zhou Z., Liu J., Liu J., Wu H., Mao S. Q., Mo K., et al. (2013) Vitamin C modulates TET1 function during somatic cell reprogramming. Nat. Genet. 45, 1504–1509 [DOI] [PubMed] [Google Scholar]
- 6. Probst A. V., Dunleavy E., and Almouzni G. (2009) Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 10, 192–206 [DOI] [PubMed] [Google Scholar]
- 7. Schermelleh L., Haemmer A., Spada F., Rösing N., Meilinger D., Rothbauer U., Cardoso M. C., and Leonhardt H. (2007) Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 35, 4301–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Easwaran H. P., Schermelleh L., Leonhardt H., and Cardoso M. C. (2004) Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep. 5, 1181–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang J., Hevi S., Kurash J. K., Lei H., Gay F., Bajko J., Su H., Sun W., Chang H., Xu G., Gaudet F., Li E., and Chen T. (2009) The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 41, 125–129 [DOI] [PubMed] [Google Scholar]
- 10. Bou Kheir T., and Lund A. H. (2010) Epigenetic dynamics across the cell cycle. Essays Biochem. 48, 107–120 [DOI] [PubMed] [Google Scholar]
- 11. Hervouet E., Nadaradjane A., Gueguen M., Vallette F. M., and Cartron P. F. (2012) Kinetics of DNA methylation inheritance by the Dnmt1-including complexes during the cell cycle. Cell Div. 7, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vandiver A. R., Idrizi A., Rizzardi L., Feinberg A. P., and Hansen K. D. (2015) DNA methylation is stable during replication and cell cycle arrest. Sci. Rep. 5, 17911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Landau D. A., Clement K., Ziller M. J., Boyle P., Fan J., Gu H., Stevenson K., Sougnez C., Wang L., Li S., Kotliar D., Zhang W., Ghandi M., Garraway L., Fernandes S. M., et al. (2014) Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell 26, 813–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen J., Liu H., Liu J., Qi J., Wei B., Yang J., Liang H., Chen Y., Chen J., Wu Y., Guo L., Zhu J., Zhao X., Peng T., Zhang Y., et al. (2013) H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 45, 34–42 [DOI] [PubMed] [Google Scholar]
- 15. Brueckner B., Garcia Boy R., Siedlecki P., Musch T., Kliem H. C., Zielenkiewicz P., Suhai S., Wiessler M., and Lyko F. (2005) Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 65, 6305–6311 [DOI] [PubMed] [Google Scholar]
- 16. Sridharan R., Tchieu J., Mason M. J., Yachechko R., Kuoy E., Horvath S., Zhou Q., and Plath K. (2009) Role of the murine reprogramming factors in the induction of pluripotency. Cell 136, 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mikkelsen T. S., Hanna J., Zhang X., Ku M., Wernig M., Schorderet P., Bernstein B. E., Jaenisch R., Lander E. S., and Meissner A. (2008) Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q., Qin B., Xu J., Li W., Yang J., Gan Y., et al. (2010) A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7, 51–63 [DOI] [PubMed] [Google Scholar]
- 19. Zheng H., Hutchins A. P., Pan G., Li Y., Pei D., and Pei G. (2014) Where cell fate conversions meet Chinese philosophy. Cell Res. 24, 1162–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu X., Sun H., Qi J., Wang L., He S., Liu J., Feng C., Chen C., Li W., Guo Y., Qin D., Pan G., Chen J., Pei D., and Zheng H. (2013) Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT-MET mechanism for optimal reprogramming. Nat. Cell Biol. 15, 829–838 [DOI] [PubMed] [Google Scholar]
- 21. Krueger F., and Andrews S. R. (2011) Bismark: a flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics 27, 1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lister R., Pelizzola M., Dowen R. H., Hawkins R. D., Hon G., Tonti-Filippini J., Nery J. R., Lee L., Ye Z., Ngo Q. M., Edsall L., Antosiewicz-Bourget J., Stewart R., Ruotti V., Millar A. H., et al. (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.