Abstract
N3-Methyladenine (3-MeA) is formed in DNA by reaction with S-adenosylmethionine, the reactive methyl donor, and by reaction with alkylating agents. 3-MeA protrudes into the DNA minor groove and strongly blocks synthesis by replicative DNA polymerases (Pols). However, the mechanisms for replicating through this lesion in human cells remain unidentified. Here we analyzed the roles of translesion synthesis (TLS) Pols in the replication of 3-MeA-damaged DNA in human cells. Because 3-MeA has a short half-life in vitro, we used the stable 3-deaza analog, 3-deaza-3-methyladenine (3-dMeA), which blocks the DNA minor groove similarly to 3-MeA. We found that replication through the 3-dMeA adduct is mediated via three different pathways, dependent upon Polι/Polκ, Polθ, and Polζ. As inferred from biochemical studies, in the Polι/Polκ pathway, Polι inserts a nucleotide (nt) opposite 3-dMeA and Polκ extends synthesis from the inserted nt. In the Polθ pathway, Polθ carries out both the insertion and extension steps of TLS opposite 3-dMeA, and in the Polζ pathway, Polζ extends synthesis following nt insertion by an as yet unidentified Pol. Steady-state kinetic analyses indicated that Polι and Polθ insert the correct nt T opposite 3-dMeA with a much reduced catalytic efficiency and that both Pols exhibit a high propensity for inserting a wrong nt opposite this adduct. However, despite their low fidelity of synthesis opposite 3-dMeA, TLS opposite this lesion replicates DNA in a highly error-free manner in human cells. We discuss the implications of these observations for TLS mechanisms in human cells.
Keywords: DNA damage; DNA damage response; DNA enzyme; DNA polymerase; DNA repair; N3-methyladenine, translesion synthesis in human cells
Introduction
N3-Methyladenine (3-MeA)2 is formed in DNA by reaction with alkylating agents and by reaction with S-adenosylmethionine, the reactive methyl donor in most cellular reactions (1, 2). It has been estimated that the action of S-adenosylmethionine on DNA generates ∼600 3-MeA adducts in a mammalian cell per day and the half-life of 3-MeA is between 4 and 24 h (3). 3-MeA is highly cytotoxic because it blocks DNA synthesis by the replicative DNA polymerases (4). Structural studies with the catalytic subunit of DNA polymerase (Pol) δ, the major eukaryotic replicative Pol, have shown that a number of residues in its active site interact with bases in the DNA minor groove, such that a mispaired base can be sensed up to 4 bases away from the primer terminus (5). Thus, an infringement of the minor groove at the N3 position of a purine would be highly blocking to replication by Polδ, necessitating the requirement of translesion synthesis (TLS) DNA Pols for inserting a nucleotide (nt) opposite 3-MeA and then extending synthesis from the inserted nt for a distance of at least 4–5 nts before Polδ could take over.
Among the human TLS Pols, Pols η, ι, κ, and Rev1, Polι is most suited to incorporate a nt opposite lesions that protrude into the minor groove. The active site of Polι is much narrower than that of other TLS Pols; consequently, the purine template A or G is pushed into the syn conformation by the incoming nt, and two hydrogen bonds are formed between the Hoogsteen edge of the purine template and the Watson-Crick edge of the incoming nt (6–8). Because the adoption of a syn conformation by 3-MeA would move this minor groove lesion into the more spacious major groove, Polι could incorporate the correct nt T opposite 3-MeA by Hoogsteen base pairing. Structural studies with Polκ have indicated that its active site can accommodate lesions that protrude into the minor groove at the template-primer junction, whereby it could proficiently extend synthesis from the 3-MeA·T base pair at the template-primer junction (9). Thus, proficient and error-free replication through the 3-MeA lesion could be performed by the sequential action of Pols ι and κ.
To analyze the genetic control of TLS through 3-MeA in human cells, we used the stable 3-deaza analog of this adduct, 3-dMeA. We show that TLS through this adduct occurs via three different pathways. As expected from structural studies, we provide evidence that Pols ι and κ function together in mediating TLS through this lesion. In addition, we find that Polθ and Polζ are also required and they mediate replication through this lesion via two independent pathways. Even though Pols ι and θ incorporate nts opposite 3-dMeA with a low fidelity, replication through the lesion occurs in an error-free manner in human cells. We discuss the implications of these and other observations for the role of TLS in promoting proficient and predominantly error-free replication through DNA lesions in human cells.
Results
Genetic control of replication through the 3-dMeA lesion in human cells
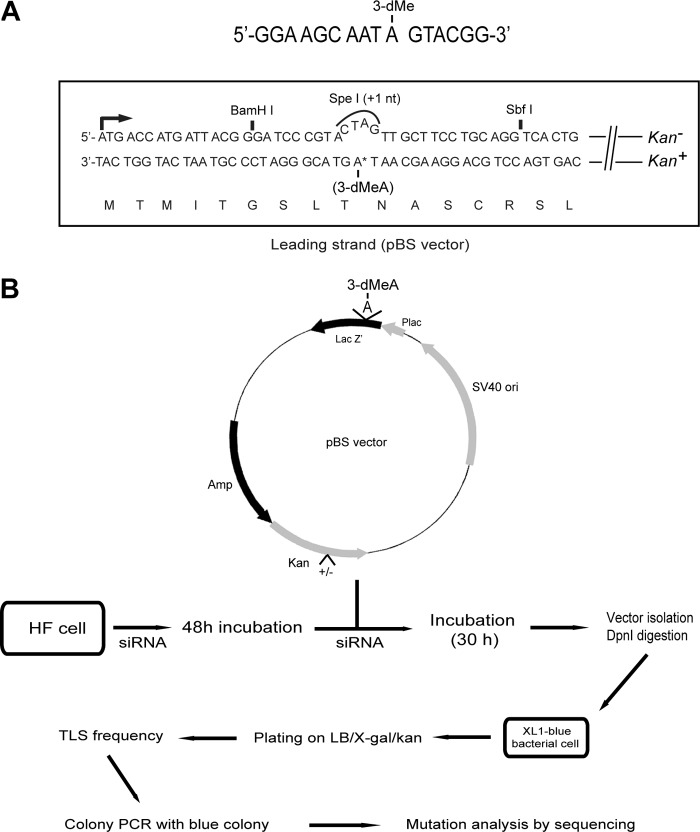
3-dMeA was incorporated in the lacZ target sequence in the leading strand (Fig. 1A) and the schematic of siRNA depletions and the various steps for the analyses of TLS frequency and for mutation analyses is shown in Fig. 1B. In the plasmid system, TLS through the lesion generates Kan+ blue colonies and the frequency of Kan+ blue colonies among the total Kan+ colonies gives a very reliable and highly repeatable estimate of TLS frequency.
Figure 1.
Schematic for TLS assays opposite 3-dMeA in human cells. A, the target 16-mer sequence containing 3-dMeA is shown on the top. The lacz′ sequence in the leading strand in the pBS vector, containing the 3-dMeA adduct, is shown below. The lesion containing the DNA strand is in-frame and it carries the Kan+ gene. TLS through the adduct generates Kan+ blue colonies. B, strategy for TLS assays and mutation analysis. The sequence of siRNA depletions and other steps for analyzing TLS and mutational products are outlined.
To identify the TLS pols required for replicating through the 3-dMeA lesion, we examined the effects of siRNA depletion of various TLS Pols, including Pols ι, κ, θ, and ζ. For all the TLS Pols analyzed, we ascertained that siRNA treatment led to a highly efficient depletion of the intended protein similar to that shown previously (10, 11). The TLS data we obtained from independent TLS assays in human cells are highly reproducible as evidenced from the high repeatability of data for TLS opposite UV-induced lesions, cis-syn TT dimer, and the (6–4)-TT photoproduct reported in different studies (10, 12, 13).
TLS opposite 3-dMeA in normal human fibroblasts treated with control (NC) siRNA occurs with a frequency of ∼46% (Table 1), and the TLS frequency remains the same in Polη-depleted cells indicating that Polη is not required. By contrast, TLS frequency is reduced to ∼27% in Polι- or Polκ-depleted cells. To determine whether Pols ι and κ function together or independently, we examined the effects of their simultaneous depletion on TLS frequency. Our observation that their simultaneous depletion causes no further reduction in TLS frequency than that conferred by their individual depletion indicates that they function together in mediating replication through the 3-dMeA lesion. Depletion of Polθ or the Rev3 catalytic or Rev7 accessory subunit of Polζ also led to a reduction in TLS frequency to ∼25%. To determine whether Polθ and Polζ function together in one TLS pathway or whether they constitute independent pathways, we examined the effects of their co-depletion on TLS frequency. Our results that TLS frequency is reduced to ∼13% upon the simultaneous depletion of Pols θ and ζ indicate that these Pols act independently.
Table 1.
Effects of siRNA knockdowns of TLS polymerases on the replicative bypass of 3-dMeA carried on the leading strand template in human fibroblasts
| siRNA | No. Kan+ colonies | No. blue colonies among Kan+ | TLS |
|---|---|---|---|
| % | |||
| NC | 446 | 206 | 46.2 |
| Polη | 358 | 161 | 45.0 |
| Polι | 326 | 90 | 27.6 |
| Polκ | 268 | 72 | 26.9 |
| Polι + Polκ | 385 | 94 | 24.4 |
| Polθ | 415 | 105 | 25.3 |
| Rev3 | 427 | 108 | 25.3 |
| Rev7 | 305 | 78 | 25.6 |
| Polθ + Rev3 | 405 | 52 | 12.8 |
| Polι + Polθ | 306 | 43 | 14.1 |
| Polκ + Polθ | 348 | 47 | 13.5 |
| Polι + Rev3 | 350 | 48 | 13.7 |
| Polι + Rev7 | 295 | 42 | 14.2 |
| Polκ + Rev3 | 408 | 52 | 12.7 |
Our genetic results suggested that replication through 3-dMeA is mediated via three independent pathways in which Pols ι and κ function together in one pathway and Polθ and Polζ function in two additional pathways. To verify this inference, we determined whether simultaneous depletion of Polι or Polκ together with Polθ or with Polζ leads to a further reduction in TLS frequency. In accord with this inference, co-depletion of Polι or Polκ with Polθ confers a reduction in TLS frequency to ∼14%, and a similar reduction in TLS frequency occurs upon the simultaneous depletion of Polι or Polκ with Polζ (Table 1). Thus, TLS opposite 3-dMeA occurs via three independent Polι/κ, Polθ, and Polζ pathways (Fig. 2), and each of these pathways contributes about equally to the replication of 3-deMeA-damaged DNA.
Figure 2.
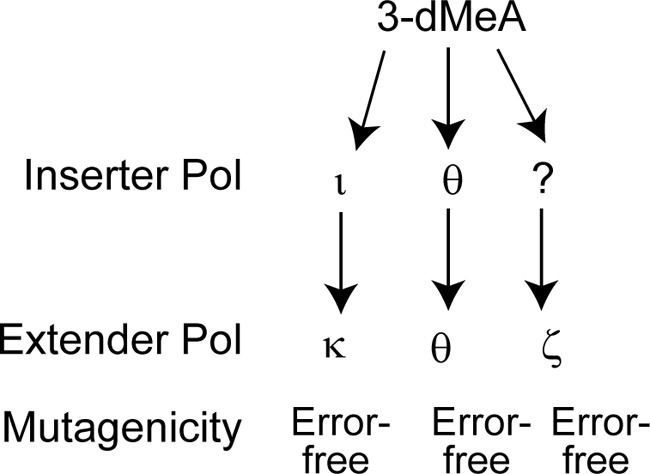
TLS pathways for replicating through 3-dMeA in human cells. Replication through 3-dMeA occurs via 3 different Polι/Polκ-, Polθ-, and Polζ-dependent pathways and all three pathways mediate error-free TLS through the lesion. The roles of Pols in inserting a nt opposite 3-dMeA and in extension of synthesis from the inserted nt are indicated.
Non-catalytic role of Rev1 in the Polι/Polκ pathway
Rev1 specifically incorporates a C opposite template G (14). In the Rev1 active site, G does not form a base pair with the incoming C; instead, template G is pushed out into a solvent-filled cavity and a conserved Arg residue in Rev1 then pairs with the incoming C (15, 16). Thus, Rev1 DNA polymerase activity is specifically adapted to insert a C opposite N2-dG DNA adducts and would play no role in TLS opposite 3-dMeA. However, in mammalian cells, Rev1 also plays an indispensable role as a scaffolding component of Y-family Pols η, ι, and κ (10). Hence, even though the Rev1 catalytic activity would play no role in TLS opposite 3-dMeA, its scaffolding role would be required for TLS mediated by the Polι/Polκ pathway but not for TLS mediated by the Polθ or Polζ pathways. To confirm this, we examined the effects of co-depletion of Rev1 with Polι, Polκ, Polθ, or Polζ. As shown in Table 2, Rev1 depletion reduces the TLS frequency to ∼23%, and this frequency remained the same upon co-depletion of Rev1 with Polι or Polκ. By contrast, simultaneous depletion of Rev1 with Polθ or Rev3 reduced TLS frequency to ∼12%. The epistasis of Rev1 with Polι or Polκ and the reduction in TLS frequency upon co-depletion of Rev1 with Polθ or Polζ is consistent with the requirement of Rev1 for Polι/Polκ-dependent TLS but not for TLS mediated by Polθ or Polζ.
Table 2.
Effect of siRNA knockdowns of Rev1 in combination with other TLS Pols on the replicative bypass of 3-dMeA carried on the leading strand template in human fibroblasts
| siRNA | No. Kan+ colonies | No. blue colonies among Kan+ | TLS |
|---|---|---|---|
| % | |||
| NC | 412 | 190 | 46.1 |
| Rev1 | 408 | 96 | 23.5 |
| Rev1 + Poι | 371 | 90 | 24.3 |
| Rev1 + Polκ | 415 | 95 | 22.9 |
| Rev1 + Polθ | 402 | 51 | 12.7 |
| Rev1 + Rev3 | 390 | 48 | 12.3 |
Error-free replication through 3-dMeA in human cells
Sequence analyses of TLS products from cells treated with control siRNA revealed no mutational events, and we also found no evidence of mutational events among the TLS products obtained from Polι-, Rev1-, Polθ-, or Polζ-depleted cells (Table 3). Thus, TLS opposite 3-dMeA occurs in a highly error-free manner in human cells.
Table 3.
Effects of siRNA knockdown of TLS polymerases on mutation frequencies and nucleotide inserted opposite 3-dMeA on the leading strand DNA template in human fibroblasts
| siRNA | No. of Kan+ blue colonies sequenced | Nucleotide inserted |
Mutation frequency | |||
|---|---|---|---|---|---|---|
| A | G | C | T | |||
| % | ||||||
| NC | 336 | 0 | 0 | 0 | 336 | 0 |
| Polι | 96 | 0 | 0 | 0 | 96 | 0 |
| Rev1 | 96 | 0 | 0 | 0 | 96 | 0 |
| Rev3 | 96 | 0 | 0 | 0 | 96 | 0 |
| Polθ | 96 | 0 | 0 | 0 | 96 | 0 |
Catalytic efficiency and fidelity of TLS Pols for DNA synthesis opposite 3-dMeA
Polι primarily inserts a T opposite 3-dMeA, and in the presence of 4 dNTPs, it inserts a T opposite the lesion but does not extend synthesis from the inserted nt (Fig. 3). Using steady-state kinetic analyses, we determined the catalytic efficiency and fidelity of Polι for nt insertion opposite undamaged A versus 3-dMeA. As shown in Table 4, Polι inserts a T nt opposite an undamaged A with over 1000-fold higher catalytic efficiency than an incorrect nt. However, compared with the incorporation of a T opposite undamaged A, T is incorporated opposite 3-dMeA with an ∼8-fold lower efficiency. Moreover, compared with the efficiency for T incorporation opposite 3-dMeA, the efficiency for incorporating the incorrect nt A or G opposite 3-dMeA is reduced by only ∼25–30-fold, and the efficiency for incorporating a C is reduced by ∼90-fold. Thus, the catalytic efficiency of Polι for incorporating the correct nt opposite 3-dMeA is reduced and the efficiency for incorporating an incorrect nt is enhanced. As determined by steady-state kinetic analyses, Polκ extends synthesis from the 3-dMeA:T base pair by incorporating dATP opposite the next 5′ template base with an ∼10-fold reduced efficiency compared with that for the undamaged A:T base pair (Table 5).
Figure 3.
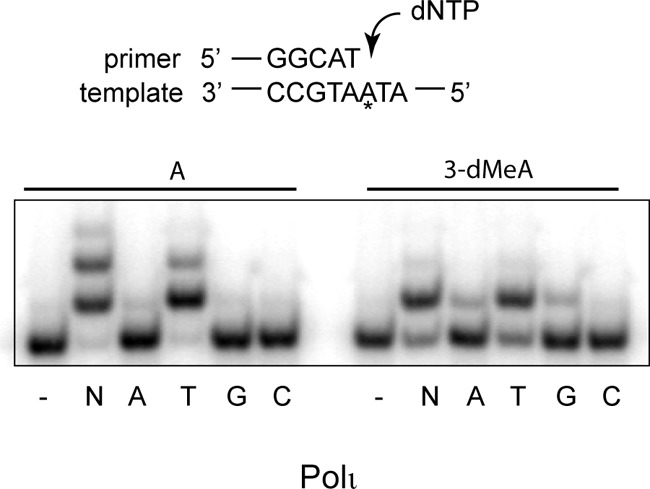
Deoxynucleotide incorporation by Polι(1–452) opposite undamaged A and 3-dMeA. Polι (0.2 nm) was incubated with DNA (10 nm) and with 50 μm of one of the 4 dNTPS (A, T, G, or C) or with 50 μm of each of 4 dNTPS (N) for 10 min at 37 °C. The DNA substrate is shown above the gel; the asterisk indicates the site of an A or 3-dMeA.
Table 4.
Steady-state kinetic analyses of nucleotide incorporation opposite undamaged A or 3-dMeA by human Polι
Polι (0.02–0.2 nm) was incubated with primer:template DNA substrate (10 nm) and increasing concentrations of dNTPs for 10 min at 37 °C. The nucleotide incorporation rate was plotted against dNTP concentration and the data were fit to the Michaelis-Menten equation. Apparent Km and kcat values were obtained from the fit and used to calculate the efficiency of deoxynucleotide incorporation (kcat/Km).
| Template nucleotide | Incoming nucleotide | kcat | Km | kcat/Km | Catalytic efficiency relative to T |
|---|---|---|---|---|---|
| min−1 | μm | ||||
| T | 10.7 ± 0.6 | 5.3 ± 0.9 | 2 | 1 | |
| A | A | 0.54 ± 0.02 | 220 ± 38.4 | 0.002 | 1 × 10−3 (↓ 1000 × ) |
| G | 0.48 ± 0.04 | 332 ± 70 | 0.0014 | 7 × 10−4 (↓ 1428 × ) | |
| C | ND | ND | |||
| T | 3.2 ± 0.09 | 12.4 ± 1.2 | 0.25 | 1 | |
| 3-dMeA | A | 1.5 ± 0.22 | 192 ± 96 | 0.0078 | 3.1 × 10−2 (↓ 32 × ) |
| G | 0.62 ± 0.02 | 59 ± 12.0 | 0.01 | 4 × 10−2 (↓ 25 × ) | |
| C | 0.37 ± 0.02 | 131.6 ± 29 | 0.0028 | 1.1 × 10−2 (↓ 89 × ) |
Table 5.
Kinetic parameters of extension reaction catalyzed by human Polκ from A:T or 3-dMeA:T base pair
Polκ (0.02–0.07 nm) was incubated with primer:template DNA substrate (10 nm) and increasing concentration of dNTPs for 5–10 min at 37 °C. The nucleotide incorporation rate was plotted against dNTP concentration and the data were fit to the Michaelis-Menten equation. Apparent Km and kcat values were obtained from the fit and used to calculate the efficiency of deoxynucleotide incorporation (kcat/Km).
| Primer:template base pair | dNTP added | kcat | Km | kcat/Km | Relative catalytic efficiency |
|---|---|---|---|---|---|
| min−1 | μm | ||||
| A:T | dATP | 56.8 ± 2.4 | 0.5 ± 0.02 | 113.6 | 1 |
| 3-dMeA:T | dATP | 14 ± 0.5 | 1.46 ± 0.1 | 10.3 | 11 ↓ |
Polθ primarily incorporates a T opposite 3-dMeA; however, it incorporates an A also, albeit not as well, and in the presence of 4 dNTPs, Polθ replicates DNA through the 3-dMeA lesion (Fig. 4). Steady-state kinetic analyses indicate that opposite undamaged A, Polθ incorporates a T with an over 103 higher catalytic efficiency than any of the incorrect nts; however, compared with that opposite undamaged A, the catalytic efficiency of Polθ for T incorporation opposite 3-dMeA is reduced almost 100-fold (Table 6). Moreover, compared with the efficiency for T incorporation opposite 3-dMeA, Polθ incorporates an A opposite this lesion with only an ∼10-fold reduction in efficiency, a G with an ∼30-fold reduction in efficiency, and a C with an ∼120-fold reduction in efficiency (Table 6). Thus, the catalytic efficiency and fidelity of Polθ for incorporating the correct nt opposite 3-dMeA is greatly reduced. The efficiency of Polθ for incorporating dATP opposite the next 5′ T template base is reduced ∼20-fold compared with the efficiency for extending synthesis from the undamaged A:T base pair (Table 7).
Figure 4.

Deoxynucleotide incorporation by Polθ opposite undamaged A and 3-dMeA. Polθ (1 nm) was incubated with DNA (10 nm) and with 25 μm of one of the 4 dNTPS (A, T, G, or C) or with 25 μm of each of 4 dNTPs (N) for 10 min at 37 °C. The DNA substrate is shown above the gel; the asterisk indicates the site of an A or 3-dMeA.
Table 6.
Steady-state kinetic analyses of nucleotide incorporation opposite undamaged A or 3-dMeA by human Polθ
Polθ (1 nm) was incubated with primer:template DNA substrate (10 nm) and increasing concentration of dNTPs for 10 min, at 37 °C. The nucleotide incorporation rate was plotted against dNTP concentration and the data were fit to the Michaelis-Menten equation. Apparent Km and kcat values were obtained from the fit and used to calculate the efficiency of deoxynucleotide incorporation (kcat/Km).
| Template nucleotide | Incoming nucleotide | kcat | Km | kcat/Km | Catalytic efficiency relative to T |
|---|---|---|---|---|---|
| min−1 | μm | ||||
| T | 0.35 ± 0.02 | 0.0078 ± 0.0018 | 45.2 | 1 | |
| A | A | 0.62 ± 0.06 | 52.6 ± 10.8 | 0.011 | 2.4 × 10−4 (↓ 4113 × ) |
| G | 0.59 ± 0.05 | 79.5 ± 15.6 | 0.007 | 1.5 × 10−4 (↓ 6464 × ) | |
| C | 0.79 ± 0.05 | 35.5 ± 5.5 | 0.022 | 4.8 × 10−4 (↓ 2056 × ) | |
| T | 0.6 ± 0.02 | 1.3 ± 0.11 | 0.46 | 1 | |
| 3-dMeA | A | 0.64 ± 0.03 | 13.5 ± 2.0 | 0.047 | 0.102 (↓ 10 ×) |
| G | 0.21 ± 0.008 | 14.4 ± 1.8 | 0.0146 | 3.2 × 10−2 (↓ 32 × ) | |
| C | 0.47 ± 0.02 | 123 ± 23 | 0.0038 | 8.3 × 10−3 (↓ 120 × ) |
Table 7.
Kinetic parameters of extension reaction catalyzed by human Polθ from A:T or 3-dMeA:T base pair
Polθ (1 nm) was incubated with primer:template DNA substrate (10 nm) and increasing concentration of dNTPs for 10 min, at 37 °C. The nucleotide incorporation rate was plotted against dNTP concentration and the data were fit to the Michaelis-Menten equation. Apparent Km and kcat values were obtained from the fit and used to calculate the efficiency of deoxynucleotide incorporation (kcat/Km).
| Primer:template base pair | dNTP added | kcat | Km | kcat/Km | Relative catalytic efficiency |
|---|---|---|---|---|---|
| min−1 | μm | ||||
| A:T | dATP | 0.84 ± 0.02 | 0.10 ± 0.009 | 8.4 | 1 |
| 3-dMeA:T | dATP | 0.55 ± 0.3 | 1.3 ± 0.18 | 0.42 | 20↓ |
Polζ is a proficient extender of synthesis from nts incorporated opposite DNA lesions by another TLS Pol (17–21). We find that the proficiency of Polζ for extension of synthesis from the 3-dMeA:T base pair is reduced ∼30-fold compared with that for extending the undamaged A:T base pair (Table 8). Because Polζ exhibits poor proficiency for inserting nts opposite DNA lesions, we presume that another as yet unidentified TLS Pol inserts nts opposite 3-dMeA from which Polζ then extends synthesis.
Table 8.
Kinetic parameters of extension reaction catalyzed by yeast Polζ from A:T or 3-dMeA:T base pair
Polζ (0.2 nm) was incubated with primer:template DNA substrate (10 nm) and increasing concentration of dNTPs for 10 min, at 37 °C. The nucleotide incorporation rate was plotted against dNTP concentration and the data were fit to the Michaelis-Menten equation. Apparent Km and kcat values were obtained from the fit and used to calculate the efficiency of deoxynucleotide incorporation (kcat/Km).
| Primer:template base pair | dNTP added | kcat | Km | kcat/Km | Relative catalytic efficiency |
|---|---|---|---|---|---|
| min−1 | μm | ||||
| A:T | dATP | 0.5 ± 0.012 | 0.04 ± 0.005 | 12.5 | 1 |
| 3-dMeA:T | dATP | 0.7 ± 0.07 | 1.7 ± 0.64 | 0.41 | 33 ↓ |
Discussion
A major role of TLS in promoting replication through the 3-dMeA lesion
Our results indicating that TLS in normal human fibroblasts accounts for almost 50% lesion bypass strongly suggest that TLS provides a primary means for replicating through this DNA lesion. Because the 3-dMeA lesion could have been removed from the plasmid by the 3-alkyladenine DNA glycosylase (22), or by nucleotide excision repair, and because removal of the lesion by these repair processes will generate Kan+ white colonies, the proportion of Kan+ blue colonies, indicative of TLS, will be correspondingly reduced. Thus, the TLS frequency in excision repair proficient cells underestimates the frequency with which TLS actually occurs. The high frequency of TLS in repair proficient cells supports the premise that TLS provides the primary mechanism for replicating through the 3-dMeA lesion and that alternative lesion bypass mechanisms such as filling-in of the gap opposite the lesion site by template switching play a much less significant and subsidiary role. TLS also plays a prominent role in promoting replication through other DNA lesions such as UV induced cis-syn TT dimer (12) and (6–4)-TT photoproduct (13), thymine glycol (11, 23) generated by the reaction of oxygen-free radicals with thymine; and N1-methyladenine (24) generated by reaction with naturally occurring methyl halides and by reaction with environmental methylating agents.
Action mechanisms of TLS Pols in DNA synthesis opposite 3-dMeA
The ability of Polι to push the purine template into a syn conformation and to form a Hoogsteen base pair with the incoming pyrimidine nt provides a mechanism by which Polι could insert a T opposite 3-dMeA (7, 8), and the ability of Polκ to accommodate the 3-dMeA:T base pair at the primer terminus (9) would allow it to extend synthesis from this base pair. In the absence of structural information, it is unclear whether Polθ pushes the 3-dMeA lesion into a syn conformation for forming a Hoogsteen base pair with the incoming dTTP and then it extends synthesis from the 3-dMeA:T Hoogsteen base pair; or whether the minor groove disruption by 3-dMeA presents no steric hindrance to the Polθ active site; consequently, it forms a 3-dMeA:T Watson-Crick (W-C) base pair and then extends synthesis from the W-C base pair. For the Polζ-dependent pathway, we expect Polζ to extend synthesis from the T nt inserted opposite 3-dMeA by a DNA polymerase whose identity remains to be determined. However, the possibility that human Polζ can insert a nt opposite 3-dMeA as well as extend synthesis from the inserted nt cannot be entirely excluded at this point.
Highly error-free replication through the 3-dMeA lesion
Sequence analysis of TLS products has indicated that all three pathways dependent upon Polι/Polκ, Polθ, and Polζ, respectively, conduct TLS opposite 3-dMeA in an error-free manner. In striking contrast to the lack of mutational TLS products in human cells, in vitro biochemical studies have indicated that TLS Pols synthesize DNA opposite 3-dMeA with a low fidelity. Thus, whereas Polι incorporates a T opposite undamaged A with a 1000-fold or even higher catalytic efficiency than an incorrect nt, it incorporates a T opposite 3-dMeA with a reduced efficiency than opposite undamaged A and the catalytic efficiency for inserting the incorrect nt opposite 3-dMeA is enhanced. For Polθ, the proficiency for incorporating a T opposite 3-dMeA is greatly reduced compared with that opposite undamaged A, and the catalytic efficiency of Polθ for inserting an incorrect nt opposite 3-dMeA is increased. The striking discrepancy between the low fidelity of DNA synthesis by Polι or Polθ opposite 3-dMeA observed in in vitro biochemical studies versus the high fidelity of TLS opposite this lesion observed in human cells strongly suggests that the fidelity of TLS Pols is tightly regulated during replication through this DNA lesion in human cells. We have previously noted that TLS opposite DNA lesions such as cis-syn TT dimer (12), (6–4)-TT photoproduct (13), thymine glycol (11, 23), or an N1-methyladenine (24) also occurs in a much more error-free manner in human cells than biochemical studies would have predicted. This accumulating evidence reinforces the notion that the catalytic efficiency and fidelity of TLS Pols opposite DNA lesions are regulated such that replication through DNA lesions occurs in a predominantly error-free manner in human cells.
Experimental procedures
Construction of plasmid vectors containing 3-dMeA
Since the half-life of 3-MeA in vitro has been established to be between 12 and 24 h (3), that precludes the construction of oligos containing this DNA adduct. To circumvent this problem, a stable 3-deaza analog of the nucleoside 3′-methyl-2′-deoxyadenosine was incorporated into oligos as 3-dMeA. 3-dMeA projects into the DNA minor groove and blocks synthesis by replicative Pols. Hence, for all the genetic and biochemical studies, we used 3-dMeA lesion-containing DNAs.
3-Deaza-3-methyldeoxyadenosine oligonucleotides were synthesized on a Model 8909 Expedite DNA synthesizer using standard DNA synthesis chemistry. The 3-deaza-3-methyl-deoxyadenosine was incorporated using an offline coupling mode for incorporation of the 3-deaza-3-Me-dA-cyanoethyl phosphoramidite (purchased from Berry and Associates, Inc., Dexter, MI 48130). The oligos were deprotected using standard concentrated ammonia deprotection, and purified and analyzed by reverse phase HPLC on a Beckman System Gold HPLC. Oligonucleotides were additionally analyzed and confirmed by MALDI-MS on a Bruker Autoflex MALDI mass spectrophotometer.
The in-frame target sequence of the lacz′ gene containing 3-dMeA is shown in Fig. 1A. Because the lacz′ sequence in the 3-dMeA-containing DNA strand is in-frame, it encodes functional β-galactosidase (β-gal); the opposite DNA strand harbors an SpeI restriction site containing a +1 frameshift, which makes it non-functional for β-gal. The 3-dMeA-containing strand carries the kanamycin gene (Kan+), whereas the other DNA strand has the kan− gene (Fig. 1A). The detailed methods for the construction of lesion-containing SV40-based duplex plasmids have been published previously (12, 13).
Assays for translesion synthesis and mutation analyses of TLS products in human cells
The schematic of TLS assay is shown in Fig. 1B and the detailed methods for TLS assays have been published previously (12, 13). Briefly, human fibroblast GM637 cells were transfected with the particular siRNA and after 48 h of incubation, the target vector DNA and siRNA (second transfection) were co-transfected. After 30 h of incubation, plasmid DNA was transfected into Escherichia coli XL1-Blue super competent cells (Stratagene) and cells plated on LB/kan plates containing isopropyl 1-thio-β-d-galactopyranoside (GenDEPOT) and 100 μg/ml of X-gal (GenDEPOT). TLS frequency is determined from the number of blue colonies among total colonies growing on LB/Kan plates and mutation frequencies and mutational changes were analyzed by DNA sequencing.
DNA polymerase assays
DNA substrates consisted of a radiolabeled oligonucleotide primer annealed to a 75-nt oligonucleotide DNA template by heating a mixture of primer/template at a 1:1.5 molar ratio to 95 °C and allowing it to cool to room temperature for several hours. The template 75-mer oligonucleotide contained the sequence 5′-AGC AAG TCA CCA ATG TCT AAG AGT TCG TAT AAT GCC TAC ACT GGA GTA CCG GAG CAT CGT CGT GAC TGG GAA AAC-3′ and it contained an undamaged A or 3-dMeA at the underlined position. For examining the incorporation of dATP, dTTP, dCTP, or dGTP nucleotides individually, or of all 4 dNTPs, a 44-mer primer 5′-GTT TTC CCA GTC ACG ACG ATG CTC CGG TAC TCC AGT GTA GGC AT-3′ was annealed to the above 75-mer template.
The standard DNA polymerase reaction (5 μl) contained 25 mm Tris·HCl (pH 7.5), 5 mm MgCl2, 1 mm dithiothreitol, 100 μg/ml of BSA, 10% glycerol, 10 nm DNA substrate, and 0.2 nm Polι or 1 nm Polθ. For nucleotide incorporation assays, 50 or 25 μm dATP, dTTP, dCTP, or dGTP (Roche Biochemicals) were used for Polι and Polθ, respectively, and for examining synthesis through the 3-dMeA lesion, 50 or 25 μm each of all 4 dNTPs were used for Polι and Polθ, respectively. Reactions were carried out for 10 min at 37 °C. Reaction products were resolved on a 12% polyacrylamide gel containing 8 m urea and analyzed by a PhosphorImager.
Steady-state kinetic analysis
Steady-state kinetic analyses for deoxynucleotide incorporation were performed as described (25). Gel band intensities of the substrate and products of the deoxynucleotide incorporation reactions were quantified using a PhosphorImager and ImageQuant software (Molecular Dynamics). The observed rate of deoxynucleotide incorporation, vobs, was determined by dividing the amount of product formed by the reaction time and protein concentration. The vobs was graphed as a function of the deoxynucleotide concentration, and the data were fit to the Michaelis-Menten equation describing a hyperbola: vobs = (kcat[E] × [dNTP])/(Km + [dNTP]). From the best fit curve, the apparent Km and kcat steady-state kinetics parameter were obtained for the incorporation of dATP, dGTP, dCTP, and dTTP and the efficiencies of the nucleotide incorporation (kcat/Km) were determined.
Author contributions
J. H. Y. performed and analyzed the experiments on the genetic control of TLS and mutagenicity, J. R. C. performed the biochemical experiments and analyzed the data, J. P. contributed to the genetic experiments, S. P. and L. P. designed and coordinated the study and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgment
We thank Richard Hodge, Director, UTMB Synthetic Organic Chemistry Core for construction of the 3-deaza-3-methyl-deoxyadenosine oligonucleotides.
This work was supported by National Institutes of Health Grants ES021452, ES022948, and ES020833, and in part by National Institutes of Health NIEHS Center Grant P30 ES06676. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- 3-MeA
- N3-methyladenine
- 3-dMeA
- 3-deaza-3-methyladenine
- Pol
- polymerase
- TLS
- translesion synthesis
- nt
- nucleotide
- Kan
- kanamycin.
References
- 1. Lindahl T., and Barnes D. E. (2000) Repair of endogenous DNA damage. Cold Spring Harbor Symp. Quant. Biol. 65, 127–133 [DOI] [PubMed] [Google Scholar]
- 2. Rydberg B., and Lindahl T. (1982) Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-l-methionine is a potentially mutagenic reaction. EMBO J. 1, 211–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Singer B., and Brent T. P. (1981) Human lymphoblasts contain DNA glycosylase activity excising N-3 and N-7-methyl and ethyl purines but not O6-alkylguanines. Proc. Natl. Acad. Sci. U.S.A. 78, 856–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sedgwick B., Bates P. A., Paik J., Jacobs S. C., and Lindahl T. (2007) Repair of alkylated DNA: recent advances. DNA Repair 6, 429–442 [DOI] [PubMed] [Google Scholar]
- 5. Swan M. K., Johnson R. E., Prakash L., Prakash S., and Aggarwal A. K. (2009) Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase δ. Nat. Struct. Mol. Biol. 16, 979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nair D. T., Johnson R. E., Prakash L., Prakash S., and Aggarwal A. K. (2005) Human DNA polymerase ι incorporates dCTP opposite template G via a G. C+ Hoogsteen base pair. Structure 13, 1569–1577 [DOI] [PubMed] [Google Scholar]
- 7. Nair D. T., Johnson R. E., Prakash S., Prakash L., and Aggarwal A. K. (2004) Replication by human DNA polymerase ι occurs via Hoogsteen base-pairing. Nature 430, 377–380 [DOI] [PubMed] [Google Scholar]
- 8. Nair D. T., Johnson R. E., Prakash L., Prakash S., and Aggarwal A. K. (2006) An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-ι active site. Structure 14, 749–755 [DOI] [PubMed] [Google Scholar]
- 9. Lone S., Townson S. A., Uljon S. N., Johnson R. E., Brahma A., Nair D. T., Prakash L., Prakash S., and Aggarwal A. K. (2007) Human DNA polymerase κ encircles DNA: implications for mismatch extension and lesion bypass. Mol. Cell 25, 601–614 [DOI] [PubMed] [Google Scholar]
- 10. Yoon J. H., Park J., Conde J., Wakamiya M., Prakash L., and Prakash S. (2015) Rev1 promotes replication through UV lesions in conjunction with DNA polymerases η, ι, and κ but not DNA polymerase ζ. Genes Dev. 29, 2588–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoon J. H., Roy Choudhury J., Park J., Prakash S., and Prakash L. (2014) A role for DNA polymerase θ in promoting replication through oxidative DNA lesion, thymine glycol, in human cells. J. Biol. Chem. 289, 13177–13185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoon J.-H., Prakash L., and Prakash S. (2009) Highly error-free role of DNA polymerase η in the replicative bypass of UV induced pyrimidine dimers in mouse and human cells. Proc. Natl. Acad. Sci. U.S.A. 106, 18219–18224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoon J.-H., Prakash L., and Prakash S. (2010) Error-free replicative bypass of (6–4) photoproducts by DNA polymerase ζ in mouse and human cells. Genes Dev. 24, 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prakash S., Johnson R. E., and Prakash L. (2005) Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74, 317–353 [DOI] [PubMed] [Google Scholar]
- 15. Nair D. T., Johnson R. E., Prakash L., Prakash S., and Aggarwal A. K. (2005) Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science 309, 2219–2222 [DOI] [PubMed] [Google Scholar]
- 16. Swan M. K., Johnson R. E., Prakash L., Prakash S., and Aggarwal A. K. (2009) Structure of the human REV1-DNA-dNTP ternary complex. J. Mol. Biol. 390, 699–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haracska L., Unk I., Johnson R. E., Johansson E., Burgers P. M., Prakash S., and Prakash L. (2001) Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev. 15, 945–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnson R. E., Haracska L., Prakash S., and Prakash L. (2001) Role of DNA polymerase η in the bypass of a (6–4) TT photoproduct. Mol. Cell. Biol. 21, 3558–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnson R. E., Washington M. T., Haracska L., Prakash S., and Prakash L. (2000) Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406, 1015–1019 [DOI] [PubMed] [Google Scholar]
- 20. Nair D. T., Johnson R. E., Prakash L., Prakash S., and Aggarwal A. K. (2006) Hoogsteen base pair formation promotes synthesis opposite the 1, N6-ethenodeoxyadenosine lesion by human DNA polymerase ι. Nat. Struct. Mol. Biol. 13, 619–625 [DOI] [PubMed] [Google Scholar]
- 21. Nair D. T., Johnson R. E., Prakash L., Prakash S., and Aggarwal A. K. (2008) Protein-template directed synthesis across an acrolein-derived DNA adduct by yeast Rev1 DNA polymerase. Structure 16, 239–245 [DOI] [PubMed] [Google Scholar]
- 22. Shrivastav N., Li D., and Essigmann J. M. (2010) Chemical biology of mutagenesis and DNA repair: cellular responses to DNA alkyaltion. Carcinogenesis 31, 59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yoon J.-H., Bhatia G., Prakash S., and Prakash L. (2010) Error-free replicative bypass of thymine glycol by the combined action of DNA polymerases κ and ζ in human cells. Proc. Natl. Acad. Sci. U.S.A. 107, 14116–14121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Conde J., Yoon J. H., Roy Choudhury J., Prakash L., and Prakash S. (2015) Genetic control of replication through N1-methyladenine in human cells. J. Biol. Chem. 290, 29794–29800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson R. E., Prakash L., and Prakash S. (2006) Yeast and human translesion DNA synthesis polymerases: expression, purification, and biochemical characterization. Methods Enzymol. 408, 390–407 [DOI] [PubMed] [Google Scholar]