Abstract
Breast cancer is the most common malignancy diagnosed in women, and the incidence of breast cancer is increasing every year. Obesity has been identified as one of the major risk factors for breast cancer progression. The mechanisms by which obesity contributes to breast cancer development is not yet understood; however, there are a few mechanisms counted as potential producers of breast cancer in obesity, including insulin resistance, chronic inflammation and inflammatory cytokines, adipokines, and sex hormones. Recent emerging evidence suggests that alterations in microRNA (miRNA) expressions are found in several diseases, including breast cancer and obesity; however, miRNA roles in obesity-linked breast cancer are beginning to unravel. miRNAs are thought to be potential noninvasive biomarkers for diagnosis and prognosis of cancer patients with comorbid conditions of obesity as well as therapeutic targets. Recent studies have evidenced that nutrients and other dietary factors protect against cancer and obesity through modulation of miRNA expressions. Herein, we summarize a comprehensive overview of up-to-date information related to miRNAs and their molecular targets involved in obesity-associated breast cancer. We also address the mechanisms by which dietary factors modulate miRNA expression and its protective roles in obesity-associated breast cancer. It is hoped that this review would provide new therapeutic strategies for the treatment of obesity-associated breast cancer to reduce the burden of breast cancer.
Keywords: obesity, breast cancer, adipocyte, microRNA, dietary components
Introduction
Breast cancer is the most commonly diagnosed cancer (1.7 million cases, 11.9%) among women in 140 of 184 countries worldwide and the second leading cause of cancer deaths among women worldwide (522,000 deaths, 6.4% in 2012). According to GLOBOCAN 2012 from WHO, breast cancer incidence has increased by >20%, and mortality has increased by 14% compared with 2008 (1). There are several risk factors associated with the development of breast cancer, including potentially modifiable and nonmodifiable factors (2). The modifiable risk factors include overweight or obesity (postmenopausal breast cancer), use of menopausal hormone therapy, less physical activity, shift work (particularly at night), cigarette smoking (particularly starting smoking before the first pregnancy), and consumption of alcohol and high-calorie diets (3, 4). The nonmodifiable risk factors include an inherited mutation of breast cancer–susceptibility genes, such as breast cancer type 1/2 susceptibility protein, E-cadherin 1, and phosphatase and tensin homolog (PTEN); family history of breast cancer; high breast tissue density; certain benign breast conditions; type 2 diabetes (independent of obesity); and reproductive factors, such as nulliparity, recent use of oral contraceptives, long menstrual history, and increased amounts of sex hormones (5, 6).
Among the risk factors, obesity has been identified as one of the major risk factors for breast cancer development. Meta-analysis studies have shown an ~30% increased risk of breast cancer recurrence or death in obese women compared with normal-weight women (7, 8). Numerous studies have shown the occurrence of obesity and cancer at the biochemical, physiological, pathologic, and epidemiologic levels, but only a small part of the molecular mechanisms of obesity-associated breast cancer has been studied (9). Recently, the association of obesity and cancer with microRNAs (miRNAs) has been proposed. Studies suggest that up- or downregulation of some specific miRNAs is the common biological factor between obesity and cancer (10). Hence, recent updates and exploration of these miRNAs are important because they may serve as potential targets and novel biomarkers in obesity-linked cancer therapies.
Surgery, radiotherapy, chemotherapy, and hormonal therapy are among the major treatments for breast cancer with different stages; however, acquisition of drug resistance and toxicities remain and limit the overall response and survival of breast cancer patients (11). Therefore, identification of safer chemopreventive agents from natural sources is necessary to improve breast cancer patients’ survival and quality of life by overcoming drug resistance and decreasing drug-induced toxicities. Recently, the use of natural agents for the treatment of obesity and obesity-related breast cancer has been substantially increased not only because of their minimal toxicity but because they target several signaling pathways. Studies have reported on the ability of essential nutrients, phytochemicals, and other bioactive functional foods to modulate the expression of miRNA that regulates diverse biological processes, including adipogenesis, insulin resistance, adipokines production, cell proliferation, apoptosis, migration, and invasion (12, 13). Therefore, this review emphasizes recent knowledge about the regulation of miRNAs and their molecular targets involved in obesity-associated breast cancer. Further, we describe the mechanisms by which dietary factors modulate miRNA expression and its protective roles in obesity-associated breast cancer.
Obesity-Associated Breast Cancer
Obesity is a chronic medical condition resulting from increased fat mass and energy storage in adipose tissue, genetic predisposition and environment, increased calorie uptake, and decreased physical activity, which leads to an adverse effect on health (14). Obesity is clinically defined as based on the BMI (in kg/m2) of ≥30; BMI is defined as body weight (in kilograms) divided by the squared body height (in meters). The WHO-recommended BMI cutoffs for the classification of weight are summarized in Table 1.
TABLE 1.
The BMI weight cutoffs
| BMI, kg/m2 | Classification (description) |
| <18.5 | Underweight (thin) |
| 18.5–24.9 | Normal weight (healthy and acceptable weight) |
| 25.0–29.9 | Overweight (pre-obese) |
| 30.0–34.9 | Class I obesity (mild obesity) |
| 35.0–39.9 | Class II obesity (moderate obesity) |
| ≥40.0 | Class III obesity (severe or morbid obesity) |
Obesity is strongly associated with changes in the physiological function of adipose tissue, leading to adipocyte differentiation, insulin resistance, altered expression of hormones, growth factors, inflammatory cytokines, and altered secretion of adipokines. The increased circulating concentrations of acute-phase proteins and inflammatory cytokines are caused by the increased production of multiple proteins by adipose tissue and thus result in the maintenance of low-grade inflammation, which leads to insulin resistance and metabolic syndrome (15). All these factors are involved in the development of several pathologic conditions, such as cardiovascular disease, type 2 diabetes mellitus, and several types of cancers (16). Among the pathological conditions, breast cancer is one of the major diseases affected by obesity risk (17). Hence, in the following sections, we have provided an overview of mechanisms of actions including 1) insulin resistance and deregulation of insulin signaling, 2) chronic inflammation and inflammatory cytokines, 3) adipokines, and 4) sex steroids that lead to cell growth, proliferation, metastasis, and inhibited apoptosis.
Mechanisms underlying obesity-related breast cancer
Insulin resistance and deregulation of insulin signaling.
Insulin resistance is one of the major mechanisms explaining the link between obesity and breast cancer (18). The increased serum insulin is positively correlated with BMI, and this commonly results in hyperinsulinemia. Hyperinsulinemia contributes to carcinogenesis by the action of insulin along with their binding proteins (19). The rise in insulin and insulin growth factor plays a major role in creating a procarcinogenic environment. The binding of these ligands to their cognate receptors, namely the insulin receptor and insulin-like growth factor I receptor, triggers several signaling networks, including rat sarcoma (RAS)/rapidly accelerated fibrosarcoma/MAPK and phosphatidylinositol 3-kinase/mammalian target of rapamycin systems. Finally, these effects alter the expression of genes involved in cell proliferation, cell cycle progression, survival, angiogenesis, and invasion, which leads to neovascularization and metastasis and promotes breast tumorigenesis (18, 20).
Chronic inflammation and inflammatory cytokines.
Obesity is correlated with a chronic inflammatory response, a key feature of adipose tissue dysfunction and thought to be a major contributing factor for obesity-associated breast cancer. Adipocyte dysfunction results in an increased production of cytokines and activation of pro-inflammatory signaling pathways such as TNF-α, IL-1β, and NF-κB, which are believed to be involved in carcinogenesis (21). Studies have shown that cytokines, including TNF-α, IL-6, and IFNs, are present in the tumor microenvironment and metastatic sites, suggesting the strong association of these factors with breast cancer development (22).
Obesity is correlated with increased levels of circulating FFAs, which increases insulin resistance by the activation of pro-inflammatory pathways (23). Toll-like receptor (TLR) signaling and the activity of NF-κB are enhanced by SFA in macrophages (24). Moreover, TLR4 signaling, expression of myeloid differentiation primary response gene 88 (MyD88), and the activity of NF-κB with the secretion of IL-6 and TNF-α are substantially increased in the adipose tissue explants and adipocytes by the treatment of SFAs (25). TLR2 agonists (tripalmitoylated CysSerLys4) and TLR4 agonists (LPS) increase TLR2/4 expression in macrophages, adipocytes, and adipose tissues, which results in the activation of several inflammatory biomediators (26), suggesting that TLRs play an important role in obesity-induced inflammation.
Oxidative stress induced by chronic inflammation could create a microenvironment favorable to cancer progression in obesity (27). Obesity-induced inflammation involves other inflammatory components including matrix metalloproteinases that are involved in breast cancer invasion and metastasis (28). The increased amounts of matrix metalloproteinases in obesity and their role in the process of mature adipocytes might represent a potential molecular association between obesity and breast cancer (29).
Adipokines.
Adipose tissue produces several types of hormones and cytokines, called adipokines. These adipokines are mainly secreted from adipocytes and stromal cells. Among the several adipokines, leptin and adiponectin are the major contributing factors for obesity-associated breast cancer development (30).
Leptin plays a crucial role in regulating energy balance by decreasing appetite and increasing metabolism (31). The increased concentrations of leptin are strongly correlated with obese individuals because of its increased release from adipocytes (32). The low concentrations of plasma leptin decrease the risk of breast cancer in premenopausal women; however, high levels of leptin are positively correlated with breast cancer risk in postmenopausal women (33). Leptin and its receptor are highly expressed in breast tumors in association with distant metastasis (34, 35). Leptin receptors activate multiple signaling networks, such as phosphatidylinositol 3-kinase, MAPK, and signal transducer and activator of transcription (STAT) systems (36). Several studies have reported that leptin amplifies estrogen signaling by increasing aromatase activity (37) or by the transactivation of estrogen receptor α (ERα) via extracellular signal-regulated kinase and STAT3 signaling pathways in cancer cells (38). Our recent study showed that leptin increases the expression of human telomerase reverse transcriptase (hTERT) via the transactivation of ERα in breast and ovarian cancer cells (12). Collectively, leptin acts as proliferative, anti-apoptotic self-renewal and angiogenesis and is a survival factor in obesity-linked breast cancer.
Adiponectin is mainly secreted by adipocytes and has important anti-inflammatory and insulin-sensitizing effects (39). The serum concentrations of adiponectin are inversely related to obesity and the risk of breast cancer. However, a few studies suggested that this inverse correlation occurs only in postmenopausal, and not in premenopausal, women, and this might be because of the changes in the concentrations of female sex hormones, especially estrogen (40, 41). Adiponectin has anticancer effects from increased insulin sensitivity and the activation of AMP-activated protein kinase and inactivation of MAPK pathways (42). Adiponectin-mediated activation of AMP-activated protein kinase inhibits phosphorylation of protein kinase B (AKT) through protein phosphatase-2 activity, which leads to a reduction in the invasion of breast cancer cells (43). Thus, adiponectin is known to be an antiproliferative apoptosis inducer and an inhibitor of angiogenesis and metastasis in obesity-associated breast cancer.
Sex hormones.
A high concentration of sex steroids produced by fat tissue is a well-known mechanism for the progression of cancers, including breast, ovarian, and endometrium, that are linked to hormones. Studies have shown that increased circulating concentrations of dehydroepiandrosterone, Δ4-androstenedione, testosterone, and estradiol and decreased concentrations of sex hormone-binding globulin increase the postmenopausal risk of breast cancer (44). Particularly, the increased production of estradiol from androgenic precursors by aromatase activity in adipose tissue is the predominant mechanism that explains the increased risk of breast cancer from obesity in postmenopausal women. This association was strongly linked with luminal subtypes (ER/progesterone receptor–positive) of breast cancer (45). In addition, the rise in circulating concentrations of androgen is also associated with an increased risk of pre- and postmenopausal breast cancer, suggesting the possible role of androgen in obesity-associated breast cancer (46). Likewise, the development and progression of breast cancer regulation by sex steroids are well established.
Role of miRNAs in Obesity-Associated Breast Cancer
miRNA function
Two decades ago, the central dogma of molecular biology was that genetic information is transferred from DNA to protein by mRNA. However, a new class of RNA regulatory genes, called miRNAs, recently has been found to have structural, catalytic, and/or regulatory functions, including genetic regulation in the cells (47). The miRNA genes or introns are transcribed by RNA polymerase II to yield primary miRNAs in the nucleus and further processed by Drosha to release pre-miRNAs. The pre-miRNAs are transported into the cytoplasm by exportin-5, which leads to the liberation of mature miRNAs. miRNAs are able to regulate genes at the posttranscriptional level by binding to the 3′-untranslated region (3′UTR) of target mRNAs. This results in the cleavage of target mRNA degradation by the argonaute-containing RNA-induced silencing complex or a decreased translation rate (48). Currently, >2500 miRNAs have been identified in humans (miRBase v21, September 2016), and ∼60% of the human genomes have been predicted to be miRNA targets (49). Through the regulation of mRNA expression, miRNAs have been found to regulate normal and pathologic cellular processes including cancer, obesity, diabetes, and drug resistance (10). Interestingly, chronic diseases that have different pathological etiologies may share similarities in their molecular processes. In this context, it is now evident that some specific miRNA could be a mutual factor between obesity and cancer (10).
Although most of the miRNAs are found in the cellular microenvironment, a number of circulating or extracellular miRNAs (EC miRNAs) have been detected in extracellular environments, such as biological fluids and cell culture medias, which are considered real-time “liquid biopsies” (50). Studies have shown that miRNAs are found in blood and biological fluids, such as breast milk, saliva, urine, colostrum, and bronchial lavage, peritoneal, cerebrospinal, and seminal fluids (51). EC miRNAs are more stable and long lasting under harsh conditions compared with cellular miRNAs, which are easily degraded in the extracellular environment. Thus, it is clear that EC miRNAs are resistant to ribonuclease activity by some protective mechanisms in the extracellular environment. Several mechanisms have been proposed to demonstrate how miRNAs are released and protected from the endogenous ribonuclease activity in circulation. One of the mechanisms suggested that miRNAs are encapsulated in exosomes, membrane vesicles, and microvesicles along with mRNAs and proteins. Further, exosomes play important roles in cell-to-cell communication (52). miRNAs present in microvesicles are released from secreting cells, whereas miRNAs as free oligonucleotides without exosomes are released from other cell types (53). Physiological and pathological processes, including pregnancy and tumors, may be altered the level of EC miRNAs. The ability of EC miRNAs to be transferred from one cell to another cell suggests that EC miRNAs act as hormones and should have a receptor with which miRNAs interact. Several EC miRNAs receptors have been proposed; among them, the TLR family is most important and interesting. According to specific miRNA patterns, tumor cells also release miRNAs and alter the normal concentrations in the biological fluids (52). Hence, these EC miRNAs could serve as cancer biomarkers. In the following sections, we update the most recently documented evidence of the regulatory role of miRNAs in obesity, breast cancer, and obesity-associated breast cancer (Figure 1).
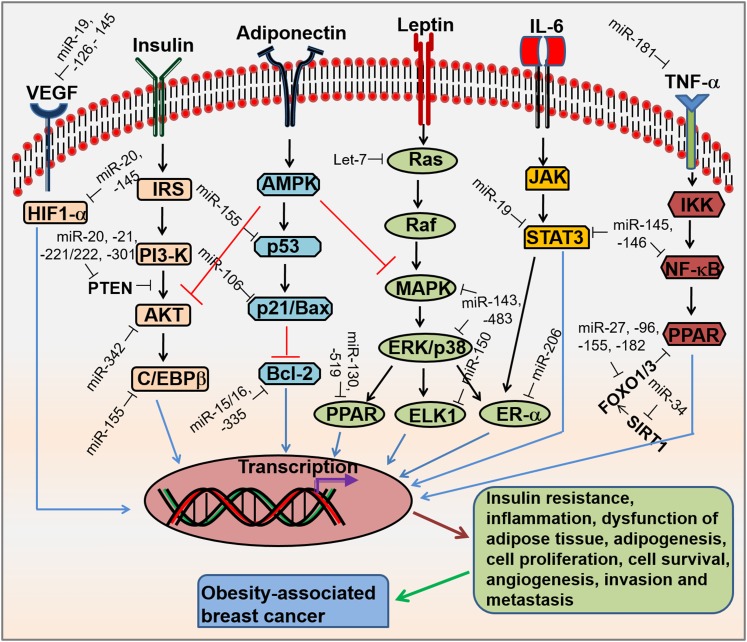
FIGURE 1.
Signaling pathways and miRNAs involved in obesity-associated breast cancer. Obesity increases the concentrations of VEGF, insulin, leptin, and inflammatory cytokines (IL-6 and TNF-α), which results in binding to its cognate cell surface receptors and activates the receptors. This activation leads to the regulation of several signaling pathways such as HIF1-α, PI3K/AKT, Ras/Raf/MAPK, JAK/STAT3, and IKK/NF-κβ. The low concentration of adiponectin abolished adiponectin signaling, leading to activation of AKT and MAPK pathways. In addition, individual or multiple miRNAs affect signaling pathways by targeting any single gene or multiple genes. As a result, this increases insulin resistance, inflammation, dysfunctional adipose tissue, adipogenesis, cell proliferation, cell survival, angiogenesis, invasion, and and metastasis, which ultimately induces the progression of breast cancer. AKT, protein kinase B; AMPK, AMP-activated protein kinase; Bcl-2, B-cell lymphoma 2; C/EBPβ, CCAAT/enhancer-binding family of proteins β ELK1, Ets-like transcription factor 1; ERK, extracellular signal-regulated kinase; ER-α, estrogen receptor alpha; FOXO1/3, forkhead box protein O1/3; HIF-1α, hypoxia-inducible factor-1α IKK, IκB kinase; IRS, insulin receptor substrate; JAK, janus family of protein kinase; miRNA, microRNA; PI3-K, phosphatidylinositol 3-kinase; PTEN, phosphatase and tensin homolog; Raf, rapidly accelerated fibrosarcoma; Ras, rat sarcoma; SIRT, Sirtuin; STAT3, signal transducer and activator of transcription 3; VEGF, vascular endothelial growth factor.
Role of miRNAs in breast cancer
miRNAs are known to be dysregulated in all types of cancer, including breast cancer, and play a major role in cell differentiation, proliferation, apoptosis, tumorigenesis, metastasis, and drug resistance (54). Based on the expression and function of miRNA in breast cancer progression, they can be classified as a tumor suppressor miRNAs (tsmiRs) and oncogenic miRNAs (oncomiRs). tsmiRs are located in fragile sites caused by genomic deletion, mutation, and epigenetic silencing, which leads to the loss of function of tsmiRs and results in upregulation of their target oncogenes (55). oncomiRs are located in chromosomal regions that are amplified and translocated in a gene, which leads to upregulation of oncomiRs and results in suppression of their target tumor-suppressor genes in cancer cells (56). The oncomiRs and tsmiRs with their most important targets and functions that are involved in the pathogenesis of breast cancer are summarized in Table 2. In this section, we focus on the impact of miRNA regulation on critical regulators of breast cancer, including tumor initiation, proliferation, metastasis, apoptosis, and drug resistance.
TABLE 2.
miRNAs regulated in breast cancer1
| miRNA | Up-/Downregulation | Functions | Target genes | References |
| Let-7 | Down | Inhibits proliferation and metastasis | HMGA2, H-RAS, K-RAS, c-MYC, CCND2, PBX3 | 57 |
| miR-1 | Down | Inhibits proliferation and migration | FRIZZLED 7, TANKYRASE 2 | 58 |
| miR-7 | Down | Promotes drug sensitivity | MRP2 | 59 |
| miR-9a | Up | Promotes metastasis | CDH1 | 57 |
| miR-10a | Down | Inhibits proliferation | RARβ, THRα | 60 |
| miR-10b | Up | Promotes migration, invasion, and metastasis | HOXD10, SYNDECAN 1 | 57 |
| miR-15a | Down | Inhibits metastasis and induces apoptosis | BCL-2, E2F | 57 |
| miR-15b | Up | Promotes migration | MTSS1 | 61 |
| miR-16a | Down | Inhibits metastasis and induces apoptosis | WIP1, BCL-2, E2F, CDK6, CCND1 | 57 |
| miR-17–5p | Down | Inhibits proliferation | AIB1 | 62 |
| miR-19a-3p | Down | Inhibits proliferation, metastasis, and angiogenesis | FRA1, STAT3, VEGF | 57 |
| miR-20a/b | Up | Promotes proliferation, angiogenesis, and metastasis | CCND1, PTEN, HIF-1α | 57, 63 |
| miR-21 | Up | Promotes proliferation, metastasis, and EMT | PDCD4, PTEN, CDC25, MSH2, MESPIN | 57 |
| miR-22 | Down | Inhibits metastasis | CDK6, SIRT1, SP1 | 57 |
| miR-26a | Down | Inhibits proliferation and metastasis | GREB1, MTDH, CCND2, CCNE2 | 57 |
| miR-27a | Up | Promotes cell viability and cell cycle | ZBTB4, ST4, MYT1, FOXO1 | 64 |
| miR-29b | Down | Inhibits metastasis and angiogenesis | ITGβ1, MMP2, TIAM1 | 65 |
| miR-30a | Down | Inhibits proliferation and metastasis | MTDH | 66 |
| miR-30c | Down | Promotes drug sensitivity | TWF1, IL-11 | 67, 68 |
| miR-31 | Down | Promotes drug sensitivity and inhibits metastasis | RHOA, RADIXIN, IGA5, PRKCE | 65, 69 |
| miR-34(a,b,c) | Down | Induces apoptosis and inhibits proliferation | CCND1, FRA1, c-MYC, NOTCH 1, EYA2, CDK4/6, CCND1, SIRT1, AXL | 57 |
| miR-93 | Up | Promotes angiogenesis and metastasis | LATS2 | 70 |
| miR-96 | Up | Promotes cell viability and cell cycle | FOXO1 | 64 |
| miR-103/107 | Up | Promotes migration and global miRNA biogenesis | DICER, DAPK4, KLF4 | 71 |
| miR-106b | Up | Promotes invasion and metastasis | p21, AIB1, pRB, BRMS1, CDKN1A | 72, 73 |
| miR-124 | Down | Inhibits metastasis | SLUG, EZH2, ROCK2, CDK4 | 57 |
| miR-125a-5p | Down | Inhibits proliferation | HDAC4/5, HER3, HUR | 57 |
| miR-125b | Down | Inhibits cell proliferation, invasion, and metastasis | EPOR, ENPEP, CK-α, HER2 | 57 |
| miR-126 | Down | Inhibits cell proliferation, invasion, and metastasis | IGFBP2, PITPNC1, MERTK, VEGF | 57 |
| miR-128 | Down | Inhibits self-renewal | BMI-1, ABCC5 | 74 |
| miR-142 | Up | Promotes self-renewal | APC | 75 |
| miR-142-3p | Down | Inhibits proliferation, invasion, and metastasis | WASL, ITGα, RAC1, COFLIN2 | 76 |
| miR-143 | Down | Inhibits proliferation and metastasis | HER3, DNMT3 | 57, 77 |
| miR-145 | Down | Inhibits proliferation, angiogenesis, and metastasis | EGFR, c-MYC, VEGF, N-CDH, HIF-2α, MUCIN 1, HER3, ROCK1 | 57 |
| miR-146a/b | Down | Inhibits proliferation and metastasis | ICAM1, NF-κB, STAT3 | 57 |
| miR-155 | Up | Promotes proliferation, metastasis, and telomere length | RHOA, CXCR4, SOX1, p53, FOXO3 | 57 |
| miR-181a | Up | Inhibits apoptosis | ATM | 57 |
| miR-181b-3p | Up | Promotes invasion and metastasis | SMAD3, YWHAG | 78 |
| miR-182 | Up | Invasion and metastasis | BRCA1, FOXO1 | 57, 64 |
| miR-185 | Down | Inhibits proliferation | DNMT1 | 79 |
| miR-194 | Down | Inhibits metastasis | TALIN 2 | 57 |
| miR-199b-5p | Down | Inhibits proliferation | HER2 | 57 |
| miR-200a | Down | Inhibits migration | SLUG, BMI-1, ZEB1/2, EPH2 | 57, 80 |
| miR-200b | Down | Inhibits cell proliferation and induces apoptosis | SP1 | 81 |
| miR-200c | Down | Inhibits self-renewal and metastasis | BMI-1, ZEB1/2 | 57 |
| miR-205 | Down | Inhibits proliferation and metastasis | HER3, p53 | 57 |
| miR-206 | Down | Inhibits cell growth, metastasis, and drug sensitivity | ERα, CCND2 | 57 |
| miR-216b | Down | Inhibits cell growth and metastasis | P2 × 7, SDCBP | 57, 82 |
| miR-221/222 | Up | Promotes drug resistance and cell proliferation and is antiapoptotic | ERα, PTEN, p57, p27 | 57, 83 |
| miR-301a | Up | Promotes proliferation and metastasis | PTEN | 57 |
| miR-326 | Down | Promotes drug sensitivity | ABCC1 | 84 |
| miR-328 | Down | Promotes drug sensitivity | ABCG2 | 85 |
| miR-335 | Down | Induces apoptosis and inhibits metastasis | SOX4, TENASCIN c, SP1, BCL-2 | 57 |
| miR-345 | Down | Promotes drug sensitivity | MRP2 | 59 |
| miR-339-5p | Down | Induces apoptosis | BCL-6 | 57 |
| miR-342 | Down | Inhibits proliferation | EGFR, HER2, AKT, PKC | 57 |
| miR-373/520c | Up | Promotes invasion and metastasis | CD44 | 57 |
| miR-429 | Down | Inhibits cell proliferation and metastasis | ZEB1, TUBB2A, CRKL | 57 |
| miR-451 | Down | Promotes drug sensitivity | MDR1 | 86 |
| miR-487a | Down | Promotes drug sensitivity | ABCG2 | 87 |
| miR-489 | Down | Promotes drug sensitivity | SMAD3 | 88 |
| miR-491-5p | Down | Promotes proliferation | JMJD2B, EGFR, HER2 | 57, 89 |
| miR-495 | Up | Promotes proliferation and hypoxia resistance | CDH1, REDD1 | 90 |
| miR-498 | Down | Induces apoptosis and inhibits proliferation and metastasis | hTERT, HER2 | 13, 91 |
| miR-708 | Down | Inhibits metastasis | NEURONATIN | 57 |
| miR-888 | Up | Promotes metastasis | CDH1, ACTγ1, CDC42 | 57 |
ABCC, ATP-binding cassette subfamily C member; ABCG2, ATP-binding cassette sub-family G member 2; ACTγ1, actin gamma 1; AIB1, amplified in breast cancer 1; AKT, protein kinase B; APC, activated protein C; ATM, ataxia-telangiectasia mutated; AXL, axillary lymphoscintigraphy; BCL, B-cell lymphoma; BMI-1, B lymphoma Moloney murine leukemia virus insertion region 1 homolog; BRCA1, breast cancer type 1 susceptibility protein; BRMS1, breast cancer metastasis-suppressor 1; CCND, cyclin D; CCNE2, G1/S-specific cyclin-E2; CDC, cell division cycle; CDH1, cadherin-1; CDK, cyclin-dependent kinase; CDKN1A, cyclin dependent kinase inhibitor 1A; CD44, cluster of differentiation 44; CK-α, cytokeratin-α c-MYC, avian myeloblastosis; CRKL, crk-like protein; CXCR4, C-X-C chemokine receptor type 4; DAPK4, death-associated protein kinase 4; DNMT, DNA (cytosine-5)-methyltransferase; EGFR, epidermal growth factor receptor; EMT, epithelial mesenchymal transition; ENPEP, glutamyl aminopeptidase; EPH2, erythropoietin-producing human hepatocellular receptors 2; EPOR, erythropoietin receptor; ERα, estrogen receptor alpha; EYA2, eyes absent 2; EZH2, enhancer of zeste homolog 2; FOX, forkhead box protein; FRA1, fos-related antigen-1; GREB1, gene regulated by estrogen in breast cancer-1; HDAC4/5, histone deacetylase 4/5; HER, human epidermal growth factor receptor; HIF, hypoxia-inducible factor; HMGA, high-mobility group AT-hook; HOXD10, homeobox 10; H-RAS, Harvey rat sarcoma; hTERT, human telomerase reverse transcriptase; HUR, human antigen R; ICAM1, intercellular adhesion molecule 1; IGA5, immunoglobulin A5; IGFBP2, insulin-like growth factor–binding protein 2; ITG, integrin; JMJD2B, jumonji domain-containing protein 2B; KLF4, kruppel-like factor 4; K-RAS, kirsten rat sarcoma; LATS2, large tumor suppressor kinase 2; MDR1, multidrug resistance protein 1; MERTK, c-Mer tyrosine kinase; miRNA, microRNA; MMP, matrix metalloproteinase; MRP2, multidrug resistance protein; MSH2, mutS protein homolog 2; MTDH, metadherin; MTSS1, metastasis suppressor 1; MYT1, myelin transcription factor 1; N-CDH, N-Cadherin; P2 × 7, P2 × purinoceptor 7; PBX3, pre B cell leukemia transcription factor 3; PDCD4, programmed cell death 4; PITPNC1, phosphatidylinositol transfer protein cytoplasmic 1; PKC, protein kinase C; pRB, retinoblastoma protein; PRKCE, protein kinase C epsilon; PTEN, phosphatase and tensin homolog; RAC1, ras-related C3 botulinum toxin substrate 1; RARβ, retinoic acid receptor β REDD1, regulated in development and DNA damage response 1; RHOA, ras homolog gene family, member A; ROCK, ρ-associated protein kinase; SDCBP, syndecan binding protein; SIRT1, Sirtuin1; SMAD3, mothers against decapentaplegic homolog 3; SOX, sex determining region Y box; SP, specificity protein; STAT3, signal transducer and activator of transcription 3; ST4, suppression of tumorigenicity 4; THRα, thyroid hormone receptor α TIAM1, T-lymphoma invasion and metastasis 1; TUBB2A, tubulin beta-2A; TWF1, twinfilin actin-binding protein homolog 1; VEGF, vascular endothelial growth factor; WASL, Wiskott-Aldrich syndrome-like; WIP1, wt-p53-induced phosphatase; YWHAG, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase; ZBTB4, zinc finger BTB broad complex/tramtrack/bric-a-brac domain protein 4; ZEB1/2, zinc finger E-box-binding homeobox 1/2.
Cancer-initiating cells or cancer stem-like cells (CSCs) that have deregulated biological properties of normal stem cells are considered responsible for tumor initiation, development, and progression. The breast CSCs were the first CSCs identified in human solid breast tumors (92). CSCs have been described as displaying a specific miRNA profile. Some miRNAs have been associated with the inhibition of CSCs, including miR-200c-141, miR-200b-200a-429, and miR-183-96-182 (93). In addition, Harvey-RAS (H-Ras) and high-mobility group AT-hook 2 (HMGA2) are targeted by let-7, which leads to the suppression of breast CSC self-renewal and differentiation (94). Wnt/β-catenin signaling is targeted by miR-1, which results in the inhibition of breast CSC proliferation and migration (58). Collectively, these miRNAs play important roles in the regulation of breast CSC tumor seeding, self-renewal capacity, and metastasis.
Metastasis causes 90% of cancer deaths that are involved in the multistep process for cancer aggressiveness (95). Epithelial mesenchymal transition (EMT) has been shown to contribute to breast metastatic tumor progression, particularly at specific stages (i.e., invasion and migration) where tumor cells disassemble and migrate to tissue and organ sites distant from the primary tumors (96). It is critical to understand the genes involved in the progression of metastasis from primary tumor sites. In this context, several miRNAs have been found to regulate breast cancer metastasis. For instance, miR-10b and miR-21 have been associated with the promotion of breast cancer cell migration and invasion through the regulation of EMT (97–100). In contrast, several miRNAs may act as breast cancer metastasis-suppressive miRNAs, including miR-29b, miR-34a, miR-126, miR-200, miR-206, and miR-335, through the inhibition of metastatic cell invasion and migration (101, 102). The miR-200 family inhibits the EMT phenotype by targeting cadherin-1 (CDH1) transcriptional repressors zinc finger E-box binding homeobox 1 and 2 (ZEB1 and 2) (103). miR-34a targets p53 mRNA, which leads to the inhibition of invasive breast cancer cells through the repression of SNAIL and EMT (104). Therefore, these miRNAs play a major role in the regulation of metastasis and EMT in breast cancer cells.
Surgery in combination with adjuvant therapy, such as cytotoxic anticancer drugs, hormonal therapy, and targeted drugs, is the major treatment of breast cancer patients. There are several mechanisms that have been proposed for drug resistance to chemotherapeutic agents, such as the alteration of ATP-binding cassette drug transporters that efflux anticancer agents, perturbations in epigenetic modifications, the induction of cell-survival and antiapoptotic pathways, and changes in the availability of drug targets (105–107). In addition to these well-known mechanisms of drug resistance, multiple miRNAs have been recently identified as critical regulators of the acquisition of drug resistance in breast cancer. For instance, miR-451 increases the doxorubicin sensitivity via direct targeting of the multidrug resistance 1 (mdr1) gene, an essential factor in drug resistance (86). miR-7 and miR-345 directly target 3′-UTR of the multidrug resistance–associated protein-2 (MRP2), resulting in the increased sensitivity of cisplatin in Michigan cancer foundation (MCF) 7 cells (59). Hence, the miRNA-regulated reversing of drug resistance through drug transporters and drug metabolic enzymes is very important for establishing effective chemotherapeutic agents.
Studies have shown that miRNAs have a role in regulating TLR signaling that modulates immune response and inflammation in breast cancer. For example, polyinosinic:polycytidylic acid–mediated activation of TLR3 induces upregulation of miR-29b, miR-29c, miR-148b, and miR-152 in breast cancer cells, leading to demethylation of retinoic acid receptor β (RARβ) and increasing breast cancer sensitivity to retinoic acid (108).
Serological biomarkers, such as carcinoembryonic antigen, a soluble form of mucin 1 protein (cancer antigens 15–3 and 27.29), and circulating cytokeratin fragments (tissue-type plasminogen activator, trehalose 6-phosphate synthase, and cytokeratin-19 fragment), are considered potential cancer biomarkers; however, they have low specificity and sensitivity as alternatives to biopsy and imaging (109). Hence, there is a need to develop low-cost and noninvasive cancer biomarkers to enhance therapy and provide information on chemoresistance and the risk of relapses. A liquid biopsy is a noninvasive concept of using body fluids, such as blood, saliva, urine, and more recently milk, as sources of circulating cells or cell components to provide information on cancer target tissues. It can be used for early diagnosis, tumor staging, analysis of the risk of metastasis, and real-time monitoring of therapies. Although liquid biopsy used to be the study of circulating cells, now it has been extended to other cell components, such as circulating DNA, miRNA, microvesicles, and exosomes (110). miRNAs produced from breast cancer tumors are circulated into the blood and milk, but most of the studies have been focused only on blood and few on urine. For instance, miR-1, miR-16, miR-21, and miR-103 were isolated in blood and used as diagnostic, staging, and prognostic biomarkers for breast cancer. miR-21, miR-125b, miR-155, and miR-451 were found in the urine of breast cancer patients (53, 111). A recent study showed evidence that milk contains high concentrations of miRNAs, and this could be used to evaluate the health status of mammary glands during lactation and breast cancer progression (51). The serological expression of miR-21, miR-210, and miR-373 were increased in breast cancer patients by trastuzumab with neoadjuvant chemotherapy (112), suggesting that miRNAs can be used to monitor therapy.
Role of miRNAs in obesity
Adipose tissue is a major contributor to the pathophysiology of obesity. The progression of adipogenesis has 2 phases, determination and maturation. During the determination phase, adipocyte precursor cells are produced from embryonic stem cells or mesenchymal stem cells, which leads to the differentiation of adipocyte precursor cells into preadipocytes. The maturation phase is the terminal differentiation of preadipocytes, in which preadipocytes are converted into mature adipocytes and subsequently generate new small fat cells and lipid content (113). These complex processes are regulated by several transcription factors, including PPARγ, members of CCAAT/enhancer-binding family of proteins (C/EBP), adipocyte determination and differentiation-dependent factor 1 (ADD1), sterol regulatory element-binding protein 1 (SREBP1), and extracellular hormones (113, 114). In recent years, there has been substantial attention paid to the role of miRNAs in regulating adipogenesis and obesity (115) (Table 3). miRNAs can enhance or suppress adipogenic differentiation of mesenchymal stem cells (MSCs) and mature adipocyte differentiation by regulating transcription factors and signaling pathways related to adipogenesis. Knockdown of enzymes involved in miRNAs biogenesis, such as Drosha and Dicer, inhibited the adipocyte differentiation in human MSCs (133), demonstrating the role of miRNAs in adipocyte development (119). Inhibition of Dicer in 3T3-L1 cells (mouse preadipocytes) resulted in the suppression of adipogenesis through the downregulation of adipocyte markers, including Pparγ, Tnf-α, and fatty acid–binding protein 4 (FabpP4) (136). Overexpression of miR-155 and miR-221/222 inhibits adipogenesis in human MSCs through the targeting of PPARγ, C/EBPα, and p27 transcripts (137).
TABLE 3.
miRNAs regulated in obesity1
| miRNA | Up-/Downregulation | Functions | Target genes | References |
| Let-7 | Down | Inhibits adipose tissue inflammation and adipogenesis | Hmga2, CDC34 | 115, 116 |
| miR-8 | Up | Promotes adipogenesis and antagonizes Wnt signaling | TCF | 117 |
| miR-14 | Down | Inhibits adipogenesis and regulator of intracellular TG and diglyceride content | p38MAPK | 117 |
| miR-15a | Up | Reduces cell number but increases pre-adipocyte size | Dlk1 | 118 |
| miR-17/92 | Up | Promotes adipogenesis | Rb2/p130 | 119 |
| miR-21 | Up | Promotes adipogenesis | TGFBR2, Ap1 | 116, 120 |
| miR-26 | Up | Promotes adipogenesis | ADAM17 | 120 |
| miR-27a/b | Down | Inhibits adipocyte differentiation | Pparγ, C/EBPα | 116 |
| miR-29 | Up | Promotes adipogenesis and insulin resistance | Akt | 117 |
| miR-30a/d | Up | Promotes adipogenesis | RUNX2 | 120 |
| miR-30c | Up | Promotes adipogenesis | PAI-1, ALK2 | 120 |
| miR-31 | Down | Inhibits adipogenesis | C/ebpα | 116 |
| miR-93 | Up | Promotes fat mass and insulin resistance | Sirt7, TBX3 | 121 |
| miR-103 | Up | Promotes adipogenesis | PDK1, Wnt3a | 116 |
| miR-124 | Up | Promotes adipogenesis | CREB | 117 |
| miR-125a-3p | Up | Promotes adipogenesis | RhoA | 122 |
| miR-125b-3p | Up | Promotes adipogenesis | MMP11 | 123 |
| miR-130 | Down | Inhibits adipogenesis | Pparγ | 124 |
| miR-132 | Up | Promotes adipocyte differentiation | SIRT1 | 125 |
| miR-138 | Down | Inhibits adipocyte differentiation | EID-1 | 126 |
| miR-141 | Up | Promotes insulin resistance | YWHAG | 127 |
| miR-143 | Down | Inhibits adipogenesis | Erk5, Mapk7 | 116, 120 |
| miR-146b-5p | Down | Inhibits insulin signaling | Sirt1, IRAK1, TRAF6 | 128 |
| miR-150 | Down | Promotes adipose tissue inflammation and insulin resistance | Elk1, Etf1, Myb | 129 |
| miR-155 | Down | Inhibits adipocyte differentiation and brown adipocyte-like phenotype | C/EBPβ | 130 |
| miR-181a | Up | Promotes adipocyte differentiation | Tnf-α | 131 |
| miR-193b | Up | Promotes adipocyte differentiation | RUNX1 | 132 |
| miR-199 | Up | Promotes adipocyte differentiation | LIF | 133 |
| miR-204 | Up | Promotes adipocyte differentiation | Runx2 | 120 |
| miR-210 | Up | Promotes adipogenesis | TCF7L2 | 120 |
| miR-211 | Up | Promotes adipogenesis and blocks osteogenesis in MSCs | Runx2 | 117 |
| miR-221/222 | Down | Inhibits adipogenesis | CDKN1B | 120 |
| miR-223 | Down | Inhibits insulin resistance and inflammation | Pknox1 | 134 |
| miR-320 | Up | Promotes adipogenesis | Runx2 | 120 |
| miR-326 | Down | Inhibits adipogenesis | C/ebpα | 117 |
| miR-335 | Up | Promotes adipogenesis | RUNX2 | 120 |
| miR-346 | Up | Promotes adipocytes and differentiation | LIF | 133 |
| miR-369-5p | Down | Inhibits adipogenesis | FABP4 | 135 |
| miR-375 | Up | Promotes adipogenesis | Erk1/2 | 117 |
| miR-378/378* | Up | Promotes adipogenesis | Ago2, Klf15, Fabp4, Fasn, Scd-1, Resistin | 120 |
| miR-448 | Down | Inhibits adipogenesis | Klf5 | 120 |
| miR-483-5p | Up | Promotes adipogenesis | ERK1 | 122 |
| miR-519d | Up | Promotes adipogenesis | PPARγ | 120 |
| miR-637 | Up | Promotes adipogenesis | OSTERIX | 120 |
ADAM17, disintegrin and metalloproteinase domain 17; Ago2, argonaute 2; Akt, protein kinase B; ALK2, activin receptor-like kinase 2; Ap1, activator protein 1; C/EBP, CCAAT-enhancer-binding protein; CDC34, cell division cycle 34; CDKN1B, cyclin-dependent kinase inhibitor 1b; CREB, cAMP responsive element binding protein; Dlk1, delta like noncanonical notch ligand 1; EID-1, e1a-like inhibitor of differentiation 1; Elk1, Ets-like transcription factor 1; ERK, extracellular signal-regulated kinase; Etf1, electron transfer flavoprotein 1; FABP4, fatty acid–binding protein 4; Fasn, fatty acid synthase; Hmga2, high-mobility group AT-hook 2; IRAK1, IL-1 receptor-associated kinase 1; Klf, kruppel-like factor; LIF, leukemia inhibitory factor; miRNA, microRNA; MMP11, matrix metalloproteinase 11; MSC, mesenchymal stem cell; Myb, myeloblastosis; PAI-1, plasminogen activator inhibitor-1; PDK1, pyruvate dehydrogenase kinase 1; Pknox1, PBX/knotted 1 homeobox 1; Rb2/p130, retinoblastoma-like protein 2; RhoA, ras homolog gene family; RUNX, runt-related transcription factor; Scd-1, stearoyl-CoA desaturase-1; SIRT, Sirtuin; TBX3, T-box transcription factor 3; TCF, T cell factor; TCF7L2, transcription factor 7-like 2; TGFBR2, TGF beta receptor 2; TRAF6, TNF receptor-associated factor 6; Wnt3a, wnt family member 3A; YWHAG, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase.
A large panel of miRNA screening studies in human preadipocyte cells with the use of antisense oligonucleotides demonstrated that the knockdown of miR-9 and miR-143 suppresses adipogenic markers such as glucose transporter type 4 (GLUT4), hormone-sensitive lipase (HSL), FABP adipocyte 2 (FABPAP2), PPARγ2, and TG accumulation (138). Interestingly, some miRNAs have a dual role in adipogenesis. For example, during the terminal differentiation of adipocytes, ectopic expression of miR-143 enhances adipogenesis by targeting MAPKK5-MAPK7 signaling cascades, whereas miR-143 inhibits adipocyte differentiation during clonal expansion (139). miR-369-5p inhibits the expression of adiponectin C1Q and collagen domain-containing (ADIPOQ) during adipogenesis by direct targeting of FABP4 (135). The miR-30 family targets SREBP1, activin receptor-like kinase 2 (ALK2), and runt-related transcription factor 2 (RUNX2) transcripts, which leads to enhancing adipocyte differentiation (140). miR-27 and miR-130 suppress mouse and human adipocyte differentiation by directly inhibiting PPARγ (141, 124).
Several miRNAs have been associated with insulin resistance. For instance, miR-221 is upregulated by targeting TNF-α and adiponectin receptor 1 (ADIPOR1) in obesity, which contributes to the development of insulin resistance (142). The development of obesity-associated insulin resistance is impaired in miR-143 knockout mice. Insulin-induced AKT kinase activation and glucose homeostasis are reduced in transgenic mice with miR-143. The expression of miR-143 is increased in the liver of genetically modified obese mouse models (143). The expression of miR-93 is negatively correlated with insulin sensitivity in women with an insulin-resistant condition (121). Multiple miRNAs, such as miR-27a and miR-222, are upregulated in 3T3-L1 adipocyte cells incubated with extracellular glucose, which leads to increased insulin resistance (144). In insulin-resistant 3T3-L1 adipocytes cells, inhibition of miR-320 modulates the expression of p85 and the phosphorylation of AKT and GLUT4, which leads to increasing insulin sensitivity and glucose uptake (145).
Adipose tissue dysfunction due to chronic inflammation is a hallmark feature of obesity. The increased infiltration of macrophages and release of inflammatory cytokines, including chemokine (C-C motif) ligand 2, TNF-α, and IL-6, impair insulin signaling and endothelial dysfunction, resulting in insulin resistance (146, 147). Several individual miRNAs have been demonstrated to have important roles in inflammation. In response to inflammatory cytokines TNF-α and IL-6, the expression of miR-146b was significantly increased in human differentiated adipocytes (148). In contrast, miR-221 expression was decreased in response to either leptin or TNF-α in human preadipocytes (142). Interestingly, the expression of miR-132 is downregulated in white adipose tissue from patients with obesity (149). The mRNA and protein concentrations of adiponectin were increased by ectopic expression of miR-21 in mouse adipocyte cells (150). In response to adiponectin, the expression of miR-155 was markedly increased by jun nuclear kinase (JNK)–NF-κB–dependent mechanisms in mouse macrophage cells (151). Thus, miRNAs could regulate adipocyte differentiation, insulin sensitivity, and inflammation by targeting genes, including adipogenic transcription factors, activating macrophages, and signaling cascades.
Role of miRNAs in obesity-associated breast cancer
Although insulin resistance, chronic inflammation, adipokines, and sex hormones have been proposed as mechanisms of action for obesity-induced breast cancer, miRNAs represent another molecular mechanism for the regulation of breast cancer by obesity-related mechanisms. However, the role of miRNAs in the hallmark of cancer processes, such as cell proliferation, angiogenesis, and EMT, are beginning to be associated with obesity. The activity of Dicer, a miR-processing machinery gene, has been influenced by the regulation of obesity (152) and breast cancer (153). Multiple specific miRNAs that regulate obesity have also been involved in breast cancer (Table 4). As described in the previous section, adipokines play an important role in the pathogenesis of obesity by increased concentrations of leptin and decreased concentrations of adiponectin. This process has been associated with an altered expression of miRNAs, including let-7, miR-27, and miR-143, which links to both obesity (115) and cancer (154). Further, these miRNAs have been shown to regulate PPARγ, which is known as a negative regulator of carcinogenesis, suggesting that these miRNAs play a vital role in obesity and cancer. miR-31 is known for inhibiting cell proliferation and metastasis in breast cancer (155), and it also inhibits adipogenesis by directly targeting C/EBPα (156).
TABLE 4.
Uniquely regulated miRNAs in breast cancer, obesity, and obesity-associated breast cancer1
| Up-/Downregulation of miRNAs |
|||
| miRNA | Breast cancer | Obesity | Obesity-associated breast cancer |
| let-7 | Down | Down | — |
| miR-21 | Up | Up | — |
| miR-30c | Down | Up | — |
| miR-31 | Down | Down | — |
| miR-93 | Up | Up | — |
| miR-124 | Down | Up | — |
| miR-143 | Down | Down | — |
| miR-155 | Up | Down | — |
| miR-181a | Up | Up | — |
| miR-221/222 | Up | Down | — |
| miR-302b | Up | — | Up |
| miR-326 | Down | Down | — |
| miR-335 | Down | Up | — |
| miR-498 | Down | — | Down |
miRNA, microRNA.
Interestingly, studies showed that miR-138 targets e1a-like inhibitor of differentiation 1 (EID-1), which is thought to be involved in reentry into the cell cycle with the transcriptional activation of genes responsible for cell differentiation and negative regulation of adipogenesis, suggesting the potential role of miR-138 in obesity and cancer (126). The decreased expression of miR-143 during adipogenesis mimics the effect of TNF-α treatment of differentiated adipocytes (138). miR-143 has also been linked to breast cancer by targeting human epidermal growth factor receptor 3 (HER3), which results in inhibition of cell proliferation and metastasis (157). Similarly, miR-335 has been implicated in insulin secretion, which leads to differential regulation of glucose metabolism in the nonobese diabetic model (158), and is also involved in the cell cycle proliferation and metastasis of breast cancer cells (101, 159). Another well-characterized miRNA, miR-9, plays a role in insulin production and trafficking (160) and cancer metastasis (161). Our recent investigation has suggested that a mechanistic involvement of miR-498 suppresses leptin-induced in vitro and in vivo tumor growth (12). A recent study demonstrated that breast cancer cells that coculture with immature adipocytes or cytokines upregulate miR-302b via activation of SRC, sex determining region Y box-2 (SOX2), and avian myeloblastosis (c-MYC), which leads to an increase in the tumor-initiating cell abundance and metastatic progression (162).
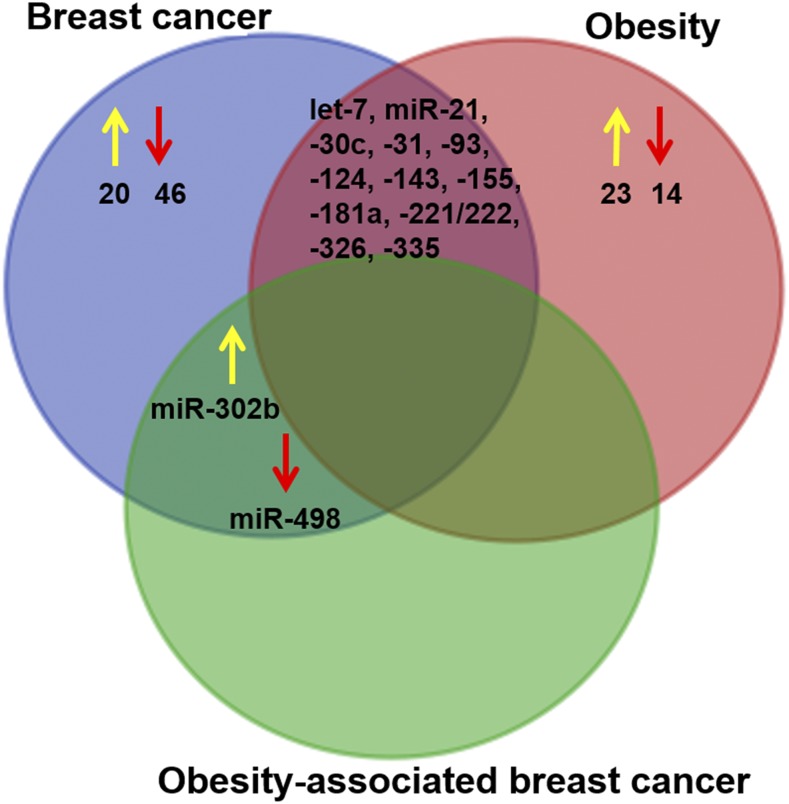
Uniquely expressed and shared miRNAs between breast cancer and obesity are presented in a Venn diagram (Figure 2). The results showed that 13 of 103 miRNAs (let-7, miR-21, miR-30c, miR-31, miR-93, miR-124, miR-143, miR-155, miR-181a, miR-326, and miR-335) were commonly regulated in both breast cancer and obesity. In addition, there are 2 miRNAs, miR-302b and miR-498, that were commonly regulated in breast cancer and obesity-associated breast cancer but not in obesity alone. The above studies have suggested molecular evidence for the critical role of miRNAs in the chronic diseases of obesity and breast cancer; however, the functional consequences of miRNAs in obesity-associated breast cancer are totally unknown. It is hoped that miRNAs will serve as novel biomarkers and molecular targets for obesity-associated breast cancer therapy.
FIGURE 2.
The Venn diagram shows the number of uniquely regulated microRNAs between breast cancer, obesity, and obesity-associated breast cancer. The yellow arrow indicates upregulation, and the red arrow indicates downregulation.
Nutritional Modulation of miRNAs in Obesity-Associated Breast Cancer
Nutrition
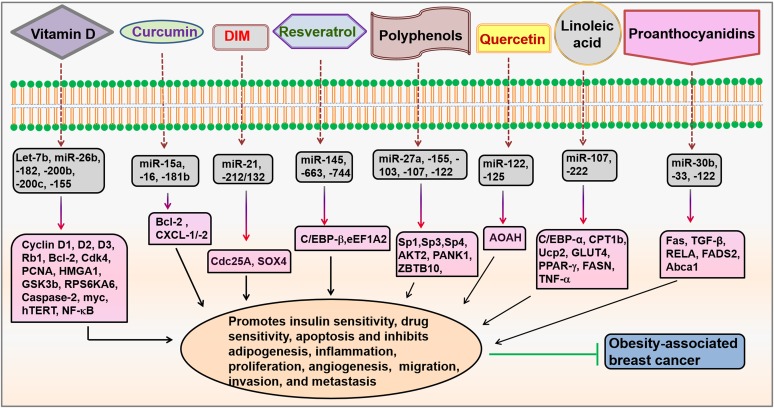
The metabolic syndrome and its chronic diseases are known to be caused by unhealthy nutrients and dietary components, imbalanced dietary energy intake, and expenditure on genetic factors. The healthy diet and lifestyle are strongly associated with multimodal disease prevention strategy. Recent studies have shown the impact of nutrients, phytochemicals, and other bioactive functional foods on epigenetic processes that have an important role in the regulation of many target genes through modulation of miRNA expression. Dietary components that alter endogenous miRNA expression and function through epigenetic factors, which leads to the regulation of several biological processes, including cell proliferation, apoptosis, migration, invasion, angiogenesis, insulin resistance, and adipokine synthesis, are the basis of nutrigenomics science and are empowered to reduce disease mortality (163). In addition to endogenous miRNA, recent studies emphasize the role of exogenous miRNA from dietary sources that are bioavailable and affect gene expression in humans and mice (164). Nutrigenomics is a promising approach to the prevention of obesity-linked cancer because epigenetic changes occur frequently in obesity and cancer. However, the studies examining the impact of various dietary components on miRNA expression and functions are little known. In this section, we provide existing and the most recent scientific proof that is related to the impact of nutrients and dietary components on the modulation of miRNA expression (Figure 3) in breast cancer (Table 5) and obesity (Table 6).
FIGURE 3.
Modulation of miRNAs by dietary agents. Dietary agents, such as vitamin D, curcumin, DIM, resveratrol, polyphenols, quercetin, linoleic acid, and proanthocyanidins, modulate miRNAs that regulate different signaling molecules involved in obesity-associated breast cancer. Abca1, ATP-binding cassette transporter 1; AKT, protein kinase B; AOAH, acyloxyacyl hydrolase; Bcl-2, B-cell lymphoma 2; C/EBP, CCAAT/enhancer-binding family of proteins; Cdc25A, cell division cycle 25A; Cdk4, cyclin-dependent kinase 4; CPT1b, carnitine palmitoyltransferase 1b; CXCL-1/-2, chemokine (C-X-C motif) ligands 1/2; DIM, 3,3′-diindolylmethane; eEF1A2, eukaryotic translation elongation factor 1α 2; FADS2, fatty acid desaturase 2; Fas, fatty acid synthase; FASN, fatty acid synthase; GLUT4, glucose transporter type 4; GSK3b, glycogen synthase kinase-3 β HMGA1, high-mobility group AT-hook 1; hTERT, human telomerase reverse transcriptase; miRNA, microRNA; myc, avian myeloblastosis; PANK1, pantothenate kinase 1; PCNA, proliferating-cell nuclear antigen; Rb1, retinoblastoma-associated protein 1; RELA, v-rel reticuloendotheliosis viral oncogene homolog A; RPS6KA6, ribosomal protein S6 kinase; SOX4, sex determining region Y box-4; Sp, specificity protein; Ucp2, uncoupling protein 2; ZBTB10, zinc finger and broad complex/tramtrack/bric-a-brac domain containing 10.
TABLE 5.
Nutritional modulation of miRNAs in breast cancer1
| Dietary component | miRNA | Up-/Downregulation | Cell type | Experimental conditions | Target genes | Functions | References |
| 1,25 (OH)2D3 | Let-7b | Up | MCF12F cells | 250 nM, 24 h | CCND1/2/3, RB1, MYC, CDK4 | Inhibits cell proliferation and induces apoptosis | 165 |
| Curcumin | miR-15a | Up | MCF-7 cells | 60 μM, 24 h | BCL-2 | Inhibits cell proliferation and induces apoptosis | 166 |
| Curcumin | miR-16 | Up | MCF-7 cells | 60 μM, 24 h | BCL-2 | Inhibits cell proliferation and induces apoptosis | 166 |
| Diindolylmethane | miR-21 | Up | MCF-7 and MDA-MB-468 cells and in vivo xenograft tumor model | 30–60 μM, 24–96 h (in vitro), 5 mg/kg body weight 7 wk (in vivo) | CDC25A | Inhibits cell proliferation, colony formation, and in vivo tumor growth and induces cell cycle arrest | 167 |
| Retinoic acid | miR-21 | Up | T47D, MCF-7, and MDA-MB-231 cells | 1 μM, 6 h | MASPIN, IL-1β, ICAM-1, PLAT | Inhibits cell growth and motility | 168 |
| 1,25 (OH)2D3 | miR-26b | Down | MCF12F cells | 250 nM, 24 h | RPS6KA6, PCNA, HMGA1, GSK3-β | Inhibits cell proliferation and induces apoptosis | 165 |
| Pomegranate polyphenols | miR-27a | Down | BT474 and MDA-MB-231 cells and in vivo xenograft tumor model | 2.5–10 μg/mL, 24 h (in vitro), 0.8 mg/kg body weight for 35 d | SP1/3/4, ZBTB10 | Inhibits cell proliferation and in vivo tumor growth and induces apoptosis | 169 |
| Butyrate | miR-31 | Up | MDA-MB-231 cells | 4 mM, 4 h | BMI-1 | Induces cellular senescence | 170 |
| Vitamin C | miR-93 | Down | MCF-10A, T47D cells, and rat mammary gland | Rat fed with vitamin C in 1% drinking water for 240 d, 1 mM, 48 h | Nrf2, Nqo1, Sod3 | Inhibits clonalibility, mammosphere formation, migration, and mammary tumor growth damage | 171 |
| Resveratrol | miR-145 | Up | MDA-MB-231, MCF-7, BT549 cells | 30 μM, 24 h | C/EBP-β | Suppresses cell proliferation | 172 |
| Pomegranate polyphenols | miR-155 | Down | BT474 and MDA-MB-231 cells and in vivo xenograft tumor model | 2.5–10 μg/mL, 24 h (in vitro), 0.8 mg/kg body weight for 35 d (in vivo) | AKT2 | Inhibits cell proliferation and in vivo tumor growth and induces apoptosis | 169 |
| Curcumin | miR-181b | Up | MDA-MB-231 cells and in vivo xenograft tumor model | 25 μM, 6 h (in vitro), 0.1% for 35 d (in vivo) | CXCL1/2 | Inhibits cell proliferation and in vivo tumor growth and induces apoptosis | 173 |
| 1,25 (OH)2D3 | miR-182 | Down | MCF12F cells | 250 nM, 24 h | CCND, BCL-2, CASPASE-2 | Cell proliferation, apoptosis | 165 |
| 1,25 (OH)2D3 | miR-200b | Down | MCF12F cells | 250 nM, 24 h | CCND2, MYC | Cell proliferation, apoptosis | 165 |
| 1,25 (OH)2D3 | miR-200c | Down | MCF12F cells | 250 nM, 24 h | MYC | Cell proliferation, apoptosis | 165 |
| DIM | miR-212/132 | Up | MDA-MB-231, T47D | 25 μM, 48 h | SOX4 | Inhibits proliferation, migration and invasion | 174 |
| 1,25 (OH)2D3 | miR-498 | Up | MCF-7 cells | 100 nM, 24 h | hTERT | Inhibits cell proliferation and metastasis and induces apoptosis | 13 |
| Resveratrol | miR-663 | Up | MCF-7 cells | 100 μM, 24 h | eEF1A2 | Inhibits cell proliferation | 175 |
| Resveratrol | miR-744 | Up | MCF-7 cells | 100 μM, 24 h | eEF1A2 | Inhibits cell proliferation | 175 |
AKT2, protein kinase B2; BCL-2, B-cell lymphoma 2; BMI-1, B lymphoma Moloney murine leukemia virus insertion region 1 homolog; C/EBPβ, CCAAT-enhancer-binding protein β CCND, cyclin D; CDC25A, cell division cycle 25A; CDK4, cyclin-dependent kinase 4; CXCL1/2, chemokine (C-X-C motif) ligand 1/2; DIM, 3,3′-diindolylmethane; eEF1A2, elongation factor 1α 2; GSK3-β, glycogen synthase kinase-3 β HMGA1, high-mobility group AT-hook 1; hTERT, human telomerase reverse transcriptase; ICAM-1, intercellular adhesion molecule 1; MDA-MB-231, MD Anderson-metastatic breast-231; miRNA, microRNA; MYC, avian myeloblastosis; Nqo1, NAD(P)H dehydrogenase [quinone] 1; Nrf, nuclear factor erythroid; PCNA, proliferating-cell nuclear antigen; PLAT, plasminogen activator tissue type; RB1, Retinoblastoma-associated protein 1; RPS6KA6, ribosomal protein S6 kinase; Sod3, superoxide dismutase 3; SOX4, sex determining region Y box-4; SP1/3/4, specificity protein 1/3/4; ZBTB10, zinc finger and broad complex/tramtrack/bric-a-brac domain containing 10; 1,25 (OH)2D3, 1,25-dihydroxycholecalciferol.
TABLE 6.
Nutritional modulation of miRNAs in obesity1
| Dietary component | miRNA | Up-/Downregulation | Cell type | Experimental conditions | Target genes | Functions | References |
| Epigallocatechin gallate | Let-7a | Up | HepG2 cells | 100 μM, 24 h | K-RAS | Glucose metabolism and insulin sensitivity | 176 |
| Grape extract with Resveratrol | miR-21 | Up | Human blood sample | 139 mg phenolics and 8 mg resveratrol/d, 1 y | TNFα, IL-1β | Inflammatory response | 177 |
| Cocoa proanthocyanidins and grape seed proanthocyanidins | miR-30b | Down | HepG2 cells | 100 mg/L, 5 h | TGFβ, RELA, FADS2 | Inflammation, NF-ҝB and PPAR signaling | 178 |
| Grape seed proanthocyanidins | miR-33 | Down | FAO cells and mouse liver | 25 mg/L, 1 h (in vitro), 250 mg/kg body weight (in vivo) | Abca1 | Inflammatory response | 179 |
| Polyphenol extract from Hibiscus sabdariffa | miR-103 | Up | Hyperlipidemic mice liver | 28.6 mg ⋅ kg−1 ⋅ d−1, 10 wk | Pank1 | TG storage, acetyl-CoA metabolism | 180 |
| CLA | miR-107 | Down | Mouse adipose tissue | 3 or 10 mg/d, 37 d | C/ebpα, Cpt1b, Ucp2 | FA metabolism | 181 |
| Polyphenol extract from H. sabdariffa | miR-107 | Up | Liver of hyperlipidemic mice | 28.6 mg ⋅ kg−1 ⋅ d−1, 10 wk | Pank1 | TG storage, acetyl-CoA metabolism | 180 |
| Coffee polyphenols | miR-122 | Up | Hepa 1–6 cells and mouse liver | 2.5 μg/mL, 24 h (in vitro), 0.5–1% for 2–15 wk (in vivo) | Srebp, Fasn | FA synthesis | 182 |
| Grape seed proanthocyanidins | miR-122 | Down | FAO cells and mouse liver | 25 mg/L, 1 h (in vitro), 28.6 mg ⋅ kg−1 ⋅ d−1, 10 wk (in vivo) | Fasn | FA synthesis | 183 |
| Polyphenol extract from H. sabdariffa | miR-122 | Up | Hyperlipidemic mice liver | 28.6 mg ⋅ kg−1 ⋅ d−1, 10 wk (in vivo) | Fasn, Srebp | FA synthesis | 180 |
| Quercetin | miR-122 | Up | Mouse liver | 2 mg/g diet, 6 wk | Aoah | Lipid metabolism | 184 |
| Quercetin | miR-125 | Up | Mouse liver | 2 mg/g diet, 6 wk | Aoah | Inflammation | 184 |
| Vitamin E–deficient diet | miR-125 | Down | Rat liver | 6 mo | Tnfα | Inflammation | 185 |
| Allyl-isothiocyanate | miR-155 | Down | RAW 264.7 cells and mouse liver | 1–10 μM, 6 h (in vitro), 15 mg/kg body weight, 7 d (in vivo) | Nrf2, p65 | Inflammation | 186 |
| Grape extract with resveratrol | miR-155 | Down | Human blood sample | 139 mg phenolics and 8 mg resveratrol/d, 1 y | TNFα | Inflammation | 177 |
| 1,25 (OH)2D3 | miR-155 | Down | RAW264.7 cells | 20 nM, 24 h | NF-κB | Inflammation and innate immunity | 187 |
| Grape extract with resveratrol | miR-181 | Up | Human blood sample | 139 mg phenolics and 8 mg resveratrol/d, 1 y | TNFα, IL-1β | Inflammation | 177 |
| CLA | miR-222 | Down | Mouse adipose tissue | 3 or 10 mg/d, 37 d | Glut4, Pparγ, Fasn, Ucp2, Tnf-α | Adipogenesis | 181 |
| Western-type diet | miR-302a | Down | Mouse liver | LDL knockout mice, 2 wk | Abca1, Elovl6 | FA utilization and insulin resistance | 188 |
| Fisetin | miR-378 | Down | High-fat diet, 10 wk | 0.05% w/w fisetin with 20% fat diet, 10 wk | Srebp-1, Scd1, Fasn, Nrf1 | FA oxidation, lipogenesis | 189 |
| High-fat diet | miR-467b | Down | Mouse liver tissue | High-fat diet, 8 wk | Lpl | Insulin resistance | 190 |
Abca1, ATP-binding cassette transporter 1; Aoah, acyloxyacyl hydrolase; C/ebpα, CCAAT-enhancer-binding protein α CLA, conjugated linoleic acid; Cpt1b, carnitine palmitoyltransferase 1b; Elovl6, elongation of very long chain fatty acid protein 6; FADS2, fatty acid desaturase 2; Fasn, fatty acid synthase; Glut4, glucose transporter type 4; K-RAS, kirsten rat sarcoma; Lpl, lipoprotein lipase; miRNA, microRNA; Nrf, nuclear factor erythroid 2-related factor; Pank1, pantothenate kinase 1; RELA, v-rel reticuloendotheliosis viral oncogene homolog A; Scd1, stearoyl-CoA desaturase-1; Srebp, sterol regulatory element-binding protein; Ucp2, uncoupling protein 2; 1,25 (OH)2D3, 1,25-dihydroxycholecalciferol.
Nutritional modulation of miRNAs in breast cancer
Vitamins are one of the essential nutrients involved in the suppression of breast cancer through the regulation of miRNA expression. Vitamin C induces the upregulation of nuclear factor erythroid 2–related factor 2 (Nrf2) and its related genes (superoxide dismutase and NAD(P)H:quinone oxidoreductase) by suppressing miR-93 concentrations in MCF-10A and T47D cells. In response to vitamin C, Nrf2 expression increased through downregulation of miR-93 in an August Copenhagen Irish rat model of 17-β estradiol-induced mammary cancer (171). The active metabolite of vitamin D (1,25-dihydroxycholecalciferol [1,25 (OH)2D3]) modulates miR-182 expression by targeting p53 and proliferating-cell nuclear antigen (PCNA) expression levels, resulting in protection of breast epithelial cells against cellular stress (165). Our previous study demonstrated that 1,25 (OH)2D3–induced upregulation of miR-498 binds to 3′UTR of hTERT, resulting in a decrease of hTERT mRNA stability, indicating that miR-498 is an immediate-response gene and mediates 1,25 (OH)2D3 anti-cancer activity (13). Ectopic expression of miR-125b inhibits the anticancer effect of 1,25 (OH)2D3 by suppressing vitamin D receptor expression in MCF-7 cells (191). These findings suggest that vitamins may influence breast cancer prevention and treatment by miRNA modulation.
FAs are another dietary component involved in the modulation of miRNA expression in breast cancer cells. For instance, miR-31 is upregulated by butyrate, an SCFA and known histone deacetylase inhibitor, resulting in downregulation of the polycomb group protein B lymphoma Moloney murine leukemia virus insertion region 1 homolog and induces cellular senescence (170). Another important dietary factor involved in the prevention of breast cancer is DHA, an omega-3 FA, which is directly obtained from maternal milk, fish oil, or algae oil or chemically synthesized from α-linolenic acid. DHA has been to shown modulate miRNA expression, which regulates the growth and metastatic genes, such as colony-stimulating factor (CSF) 1. Treatment of breast cancer cells with DHA downregulates the expression of miR-21, which is highly expressed in breast cancer cells, resulting in inhibition of CSF-1 transcription by increasing PTEN expression. Further, ectopic expression of miR-21 decreased the ability of DHA to downregulate CSF mRNA expression in breast cancer cells (192).
Curcumin isolated from the rhizomes of turmeric (Curcuma longa) has been shown to regulate cancer signaling pathways through miRNA expression in breast cancer. For instance, the curcumin-induced upregulation of miR-15a and miR-16 directly targets B-cell lymphoma 2 (BCL-2) mRNA, a key anti-apoptotic protein, resulting in the induction of apoptosis and inhibition of cell proliferation in MCF-7 breast cancer cells. The ability of curcumin to decrease BCL-2 expression was compromised when miR-15a and miR-16 were silenced by oligonucleotides (166). Another well-studied curcumin-regulated miRNA in breast cancer is miR-21, which mediates the anti-cancer effects of curcumin by the suppression of PTEN/phosphatidylinositol 3-kinase/AKT, programmed cell death protein 4, and NF-κB signaling pathways (193). Curcumin upregulates miR-181b, thereby inhibiting inflammatory cytokines such as chemokine (C-X-C motif) ligand 1/2, resulting in the suppression of in vitro and in vivo breast cancer invasion (173).
Resveratrol (3,4′,5-trihydroxystillbene) is a bioactive ingredient present in several plants including mulberries, grapes, plums, and peanuts, and it possesses anticancer, antioxidant, and anti-inflammatory properties. Several studies have shown that the anticancer effects of resveratrol are mediated through miRNA expression. miR-663 is upregulated in response to resveratrol, which is able to inhibit the elongation factor 1α 1/2 (eEF1A2) mRNA level, leading to the inhibition of MCF-7 breast cancer cell proliferation (175). Resveratrol treatment increased the expression of tumor suppressor miRNAs, including miR-141 and miR-200c, which inhibit breast CSCs-like characteristics of MD Anderson-metastatic breast-231 (MDA-MB-231) luc-D3H2LN derived from MDA-MB-231 cells. In hormone mammary tumors, miR-21, miR-129, miR-204, and miR-489 were upregulated, and DNA methyltransferase 3b expression was downregulated on resveratrol treatment (194).
The indole-3-carbinol, a glucosinolate found in cabbage, kale, radish, cauliflower, and Brussels sprouts, undergoes a rapid condensation reaction in the stomach, leading to the formation of 3,3′-diindolylmethane (DIM). DIM has been found to modulate genes involved in cell proliferation and the cell cycle through miRNAs in breast cancer. The DIM-activated aryl hydrocarbon receptor induces a highly conserved miRNA cluster, named miR-212/132, which directly targets the premetastatic SOX4 resulting in a decrease of SOX4 mRNA expression in breast cancer cells. Interestingly, ectopic expression of miR-212/132 inhibits the migration and invasion of breast cancer cells, suggesting that an anticancer effect of DIM is mediated through miR-212/132 to control metastasis in breast cancer patients (174). DIM increased the efficacy of herceptin in Sloan-Kettering Breast cancer 3 and MDA-MB-231 cells by inducing apoptosis and inhibiting cell growth and colony formation through upregulation of miR-200. Moreover, the ectopic expression of miR-200 in combination with DIM and herceptin downregulates Forkhead box protein M1, which results in the suppression of breast cancer cells (195).
Recently, food-borne or dietary miRNAs have been shown to be very stable and to circulate in the blood, which leads to the regulation of gene expression in multiple tissues and plays an important role in cancer progression (196). Studies showed that bovine miRNAs in cow milk and avian miRNAs in chicken eggs are bioavailable through peripheral blood mononuclear cells and apparently other peripheral tissues (164, 197). The physiological concentrations of these miRNAs affect gene expression in in vitro and in vivo systems. The plasma concentrations of miR-29b and miR-200c were decreased by 61% when mice fed a milk-miRNA–deficient diet for 4 wk, demonstrating that endogenous miRNAs do not compensate for dietary miRNA deficiency. The study reported that dietary miRNAs bind to TLRs or by the surface antigen-mediated delivery of exosome, which leads to exhibit its biological effects on the cells (198). A recent study suggested that cow milk contains >245 miRNAs, and it has been implicated in all aspects of health and disease (197). For instance, enrichment of miR-22 derived from mammary secretory cells inhibits estrogen signaling by targeting ERα, which leads to the inhibition of breast cancer progression (199). The plant-derived miRNAs have a therapeutic potential role in breast cancer. Study has shown that women’s serum containing plant miR-159 and its concentration are inversely correlated with the morbidity and progression of breast cancer (200). miR-159 was identified mostly in extracellular vesicles. Interestingly, synthetic miR-159 directly targets the 3′UTR of transcription factor 7 (TCF7), which leads to the suppression of in vitro and in vivo breast cancer cells (200).
Nutritional modulation of miRNAs in obesity
Only a few studies have investigated the effects of dietary components and their derivatives on the expression of miRNAs that are involved in obesity and obesity-related metabolic alterations in different experimental models.
Dietary polyphenols have been shown to improve dyslipidemia and insulin resistance by the modulation of specific miRNAs. Proanthocyanidins, a rich polyphenol in diets, increased the expression of ATP-binding cassette transporter 1 (ABCA1) by downregulating miR-33, which leads to increasing the hepatic cholesterol efflux and the production of new HDL cholesterol in hepatocytes of obese rats. In addition, proanthocyanidins also altered the expression of miR-122 and its target gene, fatty acid synthase (Fasn), which resulted in reduced lipogenesis in obese rats (183). Chronic treatment with grape seed proanthocyanidin extract can increase the tolerance to lipid overload and postprandial lipemia in a dose-dependent manner by altering miR-33a and miR-122 and their target genes (183). Polyphenols extracts from the Hibiscus sabdariffa plant reduced fatty liver disease by regulating lipid and glucose metabolism through the modulation of miR-122, miR-103, and miR-107 in diet-induced hyperlipidemic mice (180), suggesting that miR-122, miR-103, and miR-107 may be considered therapeutic targets against obesity and its related metabolic alterations.
A high-fat diet has been known to cause obesity and obesity-related metabolic diseases that can modulate the miRNA expression. The downregulation of miR-122 and upregulation of hepatic IκB kinase and β-oxidation–related genes are seen in the offspring with the consumption of a high-fat diet during pregnancy and lactation, resulting in the disturbances of hepatic lipid metabolism in offspring and in their childhood (201). Nonalcoholic fatty liver disease (NAFLD) is characterized by excessive fat deposited (steatosis) in the liver, which leads to insulin resistance, hyperlipidemia, and obesity. A high-fat diet upregulates miR-103 and miR-107 in a mouse model of high-fat, diet-induced NAFLD (202). Quercetin and coffee induced the expression of miR-122, which prevents diet-induced liver steatosis in mice (184, 203). Dietary lycopene downregulates Fabp7 by the upregulation of miR-21 expression, which leads to the reduction of intracellular lipid accumulation in the liver (204). Experimental animal consumption of a methionine-choline-deficient diet downregulates miR-122, resulting in diet-induced NAFLD and liver steatosis (205)
CLA with dietary supplementation has been shown to reduce body fat stores in abdominal white adipose tissue and increase lean body mass by enhancing lipolysis, fat oxidation, and fat cell apoptosis and reducing the size of fat cells, the uptake and storage of FAs, and the inhibition of enzymes involved in lipid metabolism (206). CLA treatment in mice significantly increased insulin sensitivity and decreased insulin resistance by downregulating miR-103 and miR-107 in mice fed a standard-fat diet, suggesting that miRNAs are regulated by dietary FAs in obesity (181). Fisetin, a flavonol found in vegetables and fruits, has been shown to protect fat accumulation by the downregulation of miR-378 in mice fed a high-fat diet, indicating the protective role of dietary fisetin mediated through miRNA molecules on the consequences of obesity (189).
Vitamin D modulates the intracellular mechanisms of insulin action mediated by vitamin D receptor and insulin receptor substrate-1 in type 2 diabetic mice, indicating the protective effects of vitamin D against obesity and its related metabolic disorders. 1,25 (OH)2D3 suppresses the differentiation of preadipocytes by decreasing the expression of adipogenesis-related genes in a dose-dependent manner, suggesting that 1,25 (OH)2D3 could regulate the posttranslational expression of genes involved in lipid and glucose metabolism. 1,25 (OH)2D3 co-administered with testosterone upregulates the expression of miR-29a/b, which target PPARα, leading to the alteration of lipid metabolism (207). In addition, our recent study demonstrated that the miR-498–mediated hTERT downregulation in response to 1,25 (OH)2D3 is a key event in mediating the inhibition of leptin signaling and high-fat-diet–induced tumor growth, suggesting the role of 1,25 (OH)2D3 in obesity-linked cancer (12).
Vitamin E consists of 2 groups of compounds, including tocopherols and tocotrienols, that can regulate miRNA expression. Rats fed a vitamin E–deficient diet decreased the expression of miR-122a and miR-125 compared with rats fed a vitamin E–sufficient diet, which results in the alteration of lipid metabolism and inflammation (185). Thus, these studies suggested that vitamins are important regulators of lipid metabolism and obesity, and they may exert these properties through miRNA expression.
Nutritional modulation of miRNAs in obesity and breast cancer
Because numerous studies have reported the biological effects of essential nutrients, phytochemicals, and other bioactive functional foods on breast cancer and/or obesity, only a few studies have been conducted on the effects of nutritional factors that target the key pathways underlying obesity-linked breast cancer.
Curcumin has been shown to have chemopreventive properties and reverse hyperlipidemia, hyperglycemia, and other symptoms related to obesity by targeting NF-κB, STAT3, cyclooxygenase-2 (COX-2), AKT, and mammalian target of rapamycin signaling cascades, resulting in inhibition of obesity-linked cancer growth. Studies documented that resveratrol has been shown to inhibit breast cancer growth and maintain glycemic control in diabetic patients and inhibit inflammatory signaling by TNF-α, IL-6, C-reactive protein, and NF-κB pathways. Ursolic acid is a naturally occurring triterpenoid derived from apples, rosemary, and other fruits and vegetables. Ursolic acid has been found to inhibit oncogene promoter-induced inflammation, hyperplasia, and tumor growth in a mouse model of postmenopausal breast cancer. In addition, ursolic acid is a strong insulin-sensitizing and anti-inflammatory agent that regulates NF-κB, COX-2, and AKT pathways against the effects of obesity (208).
Therefore, like curcumin, resveratrol and ursolic acid are believed to have anticancer and antiobesity properties. However, our recent study demonstrated the effect of 1,25 (OH)2D3 on obesity-associated breast cancer through miRNA regulation, and to our knowledge, no studies have been conducted related to the effects of nutrients and nutritional factors on the regulation of miRNA in obesity-linked breast cancer. Hence, further studies are warranted for identifying possible miRNA regulation by nutrients.
Conclusions and Future Perspectives
Obesity is a complex and multifaceted condition that increases body weight and the amount of body visceral fat, which are responsible for 9% of breast cancer progression. Hence, it is critical to understand the molecular mechanisms and how obesity influences the carcinogenic processes in the breast. The known molecular mechanisms involved in obesity-associated breast cancer are mediated through insulin resistance, chronic inflammation and inflammatory cytokines, adipokines, and sex hormones. Over the past decade, growing bodies of literature have explored the discovery of miRNAs and their molecular targets involved in the pathogenesis of many diseases, including cancer, obesity, and diabetes. Given the deregulation of miRNAs during several pathogenic processes, we predict that miRNAs act as novel biomarkers and therapeutic targets for obesity-associated breast cancer.
In this review, we summarized the potential role of miRNAs as therapeutic targets in linking breast cancer and obesity. In breast cancer, miRNAs have been shown to regulate common hallmarks, such as tumor initiation, cell proliferation, apoptosis, migration, and drug resistance (Table 2). miRNAs have also been shown to regulate adipocyte differentiation, insulin sensitivity, and inflammation by targeting genes, including adipogenic transcription factors, activating macrophages, and signaling cascades (Table 3). It is exciting to note that specific miRNAs could regulate identical targets in breast cancer as well as obesity models (Table 4), which might provide new insights into identifying the connection between obesity and cancer. Furthermore, because of high stability, reproducibility, and easy detection of miRNAs in body fluids, miRNAs can represent a novel class of potential biomarkers for diagnosis and prognosis of cancer patients with comorbid conditions of obesity as well as therapeutic targets. However, the challenge remains to determine the functional consequences of these miRNAs and how they are regulated in obesity-associated breast cancer. For example, the causative role of miRNAs, which regulates both adipose and breast cancer tissues, must be understood. Further, the actions of adipose miRNA in regulating extracellular factors, such as hormones and adipokines that are involved in breast cancer progression, remain largely unknown. Novel methodologies with more mechanistic studies are warranted to develop customized therapies for obesity-linked breast cancer patients by either overexpression or knockdown of miRNAs.
This review also summarized the nutritional modulation of miRNA expressions and their mRNA targets in obesity-linked breast cancer. Several dietary components and phytochemicals, including vitamins, FAs, curcumin, resveratrol, indole-3-carbinol, garcinol, dietary polyphenols, and matrine, have been shown to modulate miRNA expression and target multiple genes in breast cancer (Table 5) and obesity (Table 6). In recent years, there has been more attention on studying the effect of dietary components and their derivatives as important epigenetic factors involved in posttranscriptional regulation of apoptosis, cell proliferation, metastasis, adipogenesis, lipids, and glucose metabolism genes. It has been suggested that dietary components can be used to decrease the expression of oncogenic miRNAs and increase the expression of tumor-suppressor miRNAs to affect drug-resistance mechanisms associated with standard chemotherapy. miRNA profiling can be a useful method for the assessment of nutritional status, development of a suitable diet, and the design of future therapeutic approaches in obesity-linked breast cancer. However, more research is needed on the efficacy of dietary factors against obesity-associated breast cancer. The modulation of miRNA profiles by dietary agents during breast cancer progression would provide new insights into early biomarkers for obesity-associated breast cancer prevention. Most of the studies that were highlighted in this review of the modulation of miRNAs by dietary components focused on cell lines or experimental animals treated with dietary components, resulting in the change of miRNA and its target gene expressions. Hence, there is a need for experimental functional studies, including the use of transgenic mice with a gain or loss of miRNA genes, to provide a better understanding of the complex regulation of miRNAs by dietary factors involved in these processes. It is hoped that this review will encourage the exploration of the promising role of miRNAs as effector molecules, which will provide new therapeutic strategies for the treatment of obesity-associated breast cancer to reduce the burden of breast cancer.
Acknowledgments
We thank Professor Ram Rajasekharan, Director, CSIR-Central Food Technological Research Institute, for his support and encouragement. Both authors read and approved the final manuscript.
Footnotes
Abbreviations used: AKT, protein kinase B; C/EBP, CCAAT/enhancer-binding family of proteins; CSC, cancer stem-like cell; CSF, colony-stimulating factor; DIM, 3,3′-diindolylmethane; EC miRNA, extracellular miRNA; EMT, epithelial mesenchymal transition; ERα, estrogen receptor α FABP, fatty acid–binding protein; hTERT, human telomerase reverse transcriptase; MCF, Michigan Cancer Foundation; MDA-MB-231, MD Anderson-metastatic breast-231; miRNA, microRNA; MSC, mesenchymal stem cell; NAFLD, nonalcoholic fatty liver disease; onconmiR, oncogenic microRNA; PTEN, phosphatase and tensin homolog; RAS, rat sarcoma; SOX, sex determining region Y box; STAT, signal transducer and activator of transcription; TLR, toll-like receptor; tsmiR, tumor suppressor microRNA; 3′UTR, three prime untranslated region; 1,25 (OH)2D3, 1,25-dihydroxycholecalciferol.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359–86. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society. Cancer facts and figures. Boston (MA): American Cancer Society; 2016. [Google Scholar]
- 3.Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Bouvard V, Altieri A, Cogliano V; WHO International Agency for Research on Cancer Monograph Working Group. Carcinogenicity of alcoholic beverages. Lancet Oncol 2007;8:292–3. [DOI] [PubMed] [Google Scholar]
- 4.Key J, Hodgson S, Omar RZ, Jensen TK, Thompson SG, Boobis AR, Davies DS, Elliott P. Meta-analysis of studies of alcohol and breast cancer with consideration of the methodological issues. Cancer Causes Control 2006;17:759–70. [DOI] [PubMed] [Google Scholar]
- 5.Economopoulou P, Dimitriadis G, Psyrri A. Beyond BRCA: new hereditary breast cancer susceptibility genes. Cancer Treat Rev 2015;41:1–8. [DOI] [PubMed] [Google Scholar]
- 6.Hulka BS, Moorman PG. Breast cancer: hormones and other risk factors. Maturitas 2001;38:103–13–. [DOI] [PubMed] [Google Scholar]
- 7.Chan DS, Vieira AR, Aune D, Bandera EV, Greenwood DC, McTiernan A, Navarro Rosenblatt D, Thune I, Vieira R, Norat T. Body mass index and survival in women with breast cancer-systematic literature review and meta-analysis of 82 follow-up studies. Ann Oncol 2014;25:1901–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Protani M, Coory M, Martin JH. Effect of obesity on survival of women with breast cancer: systematic review and meta-analysis. Breast Cancer Res Treat 2010;123:627–35. [DOI] [PubMed] [Google Scholar]
- 9.De Pergola G, Silvestris F. Obesity as a major risk factor for cancer. J Obes 2013;2013:291546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ali AS, Ali S, Ahmad A, Bao B, Philip PA, Sarkar FH. Expression of microRNAs: potential molecular link between obesity, diabetes and cancer. Obes Rev 2011;12:1050–62. [DOI] [PubMed] [Google Scholar]
- 11.Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med 2002;53:615–27. [DOI] [PubMed] [Google Scholar]
- 12.Kasiappan R, Sun Y, Lungchukiet P, Quarni W, Zhang X, Bai W. Vitamin D suppresses leptin stimulation of cancer growth through microRNA. Cancer Res 2014;74:6194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasiappan R, Shen Z, Tse AK, Jinwal U, Tang J, Lungchukiet P, Sun Y, Kruk P, Nicosia SV, Zhang X, et al. . 1,25-Dihydroxyvitamin D3 suppresses telomerase expression and human cancer growth through microRNA-498. J Biol Chem 2012;287:41297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung WW, Mao P. Recent advances in obesity: genetics and beyond. ISRN Endocrinol 2012;2012:536905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trayhurn P, Wood IS. Adipokines: inflammation and the pleiotropic role of white adipose tissue. Br J Nutr 2004;92:347–55. [DOI] [PubMed] [Google Scholar]
- 16.Kanasaki K, Koya D.. Biology of obesity: lessons from animal models of obesity. J Biomed Biotechnol 2011;2011:197636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthews SB, Thompson HJ. The obesity-breast cancer conundrum: an analysis of the issues. Int J Mol Sci 2016;17:989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arcidiacono B, Iiritano S, Nocera A, Possidente K, Nevolo MT, Ventura V, Foti D, Chiefari E, Brunetti A. Insulin resistance and cancer risk: an overview of the pathogenetic mechanisms. Exp Diabetes Res 2012;2012:789174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giovannucci E. Insulin-like growth factor-I and binding protein-3 and risk of cancer. Horm Res 1999;51 Suppl 3:34–41. [DOI] [PubMed] [Google Scholar]
- 20.Rose DP, Vona-Davis L. The cellular and molecular mechanisms by which insulin influences breast cancer risk and progression. Endocr Relat Cancer 2012;19:R225–41. [DOI] [PubMed] [Google Scholar]
- 21.Coussens LM, Werb Z. Inflammation and cancer. Nature 2002;420:860–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicolini A, Carpi A, Rossi G. Cytokines in breast cancer. Cytokine Growth Factor Rev 2006;17:325–37. [DOI] [PubMed] [Google Scholar]
- 23.Boden G. Obesity and free fatty acids. Endocrinol Metab Clin North Am 2008;37:635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suganami T, Mieda T, Itoh M, Shimoda Y, Kamei Y, Ogawa Y. Attenuation of obesity-induced adipose tissue inflammation in C3H/HeJ mice carrying a Toll-like receptor 4 mutation. Biochem Biophys Res Commun 2007;354:45–9. [DOI] [PubMed] [Google Scholar]
- 25.Youssef-Elabd EM, McGee KC, Tripathi G, Aldaghri N, Abdalla MS, Sharada HM, Ashour E, Amin AI, Ceriello A, O’Hare JP, et al. . Acute and chronic saturated fatty acid treatment as a key instigator of the TLR-mediated inflammatory response in human adipose tissue, in vitro. J Nutr Biochem 2012;23:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fresno M, Alvarez R, Cuesta N. Toll-like receptors, inflammation, metabolism and obesity. Arch Physiol Biochem 2011;117:151–64. [DOI] [PubMed] [Google Scholar]
- 27.Baglietto L, English DR, Hopper JL, MacInnis RJ, Morris HA, Tilley WD, Krishnan K, Giles GG. Circulating steroid hormone concentrations in postmenopausal women in relation to body size and composition. Breast Cancer Res Treat 2009;115:171–9. [DOI] [PubMed] [Google Scholar]
- 28.Chavey C, Mari B, Monthouel MN, Bonnafous S, Anglard P, Van Obberghen E, Tartare-Deckert S. Matrix metalloproteinases are differentially expressed in adipose tissue during obesity and modulate adipocyte differentiation. J Biol Chem 2003;278:11888–96. [DOI] [PubMed] [Google Scholar]
- 29.Katiyar SK, Meeran SM. Obesity increases the risk of UV radiation-induced oxidative stress and activation of MAPK and NF-kappaB signaling. Free Radic Biol Med 2007;42:299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vona-Davis L, Rose DP. Adipokines as endocrine, paracrine, and autocrine factors in breast cancer risk and progression. Endocr Relat Cancer 2007;14:189–206. [DOI] [PubMed] [Google Scholar]
- 31.Vendrell J, Broch M, Vilarrasa N, Molina A, Gomez JM, Gutierrez C, Simon I, Soler J, Richart C. Resistin, adiponectin, ghrelin, leptin, and proinflammatory cytokines: relationships in obesity. Obes Res 2004;12:962–71. [DOI] [PubMed] [Google Scholar]
- 32.Hamilton BS, Paglia D, Kwan AY, Deitel M. Increased obese mRNA expression in omental fat cells from massively obese humans. Nat Med 1995;1:953–6. [DOI] [PubMed] [Google Scholar]
- 33.Harris HR, Tworoger SS, Hankinson SE, Rosner BA, Michels KB. Plasma leptin levels and risk of breast cancer in premenopausal women. Cancer Prev Res (Phila) 2011;4:1449–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garofalo C, Koda M, Cascio S, Sulkowska M, Kanczuga-Koda L, Golaszewska J, Russo A, Sulkowski S, Surmacz E. Increased expression of leptin and the leptin receptor as a marker of breast cancer progression: possible role of obesity-related stimuli. Clin Cancer Res 2006;12:1447–53. [DOI] [PubMed] [Google Scholar]
- 35.Ishikawa M, Kitayama J, Nagawa H. Enhanced expression of leptin and leptin receptor (OB-R) in human breast cancer. Clin Cancer Res 2004;10:4325–31. [DOI] [PubMed] [Google Scholar]
- 36.Dieudonne MN, Machinal-Quelin F, Serazin-Leroy V, Leneveu MC, Pecquery R, Giudicelli Y. Leptin mediates a proliferative response in human MCF7 breast cancer cells. Biochem Biophys Res Commun 2002;293:622–8. [DOI] [PubMed] [Google Scholar]
- 37.Catalano S, Marsico S, Giordano C, Mauro L, Rizza P, Panno ML, Ando S. Leptin enhances, via AP-1, expression of aromatase in the MCF-7 cell line. J Biol Chem 2003;278:28668–76. [DOI] [PubMed] [Google Scholar]
- 38.Catalano S, Mauro L, Marsico S, Giordano C, Rizza P, Rago V, Montanaro D, Maggiolini M, Panno ML, Ando S. Leptin induces, via ERK1/ERK2 signal, functional activation of estrogen receptor alpha in MCF-7 cells. J Biol Chem 2004;279:19908–15. [DOI] [PubMed] [Google Scholar]
- 39.Cnop M, Havel PJ, Utzschneider KM, Carr DB, Sinha MK, Boyko EJ, Retzlaff BM, Knopp RH, Brunzell JD, Kahn SE. Relationship of adiponectin to body fat distribution, insulin sensitivity and plasma lipoproteins: evidence for independent roles of age and sex. Diabetologia 2003;46:459–69. [DOI] [PubMed] [Google Scholar]
- 40.Gross AL, Newschaffer CJ, Hoffman-Bolton J, Rifai N, Visvanathan K. Adipocytokines, inflammation, and breast cancer risk in postmenopausal women: a prospective study. Cancer Epidemiol Biomarkers Prev 2013;22:1319–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macis D, Guerrieri-Gonzaga A, Gandini S. Circulating adiponectin and breast cancer risk: a systematic review and meta-analysis. Int J Epidemiol 2014;43:1226–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dieudonne MN, Bussiere M, Dos Santos E, Leneveu MC, Giudicelli Y, Pecquery R. Adiponectin mediates antiproliferative and apoptotic responses in human MCF7 breast cancer cells. Biochem Biophys Res Commun 2006;345:271–9. [DOI] [PubMed] [Google Scholar]
- 43.Kim KY, Baek A, Hwang JE, Choi YA, Jeong J, Lee MS, Cho DH, Lim JS, Kim KI, Yang Y. Adiponectin-activated AMPK stimulates dephosphorylation of AKT through protein phosphatase 2A activation. Cancer Res 2009;69:4018–26. [DOI] [PubMed] [Google Scholar]
- 44.Brown KA, Simpson ER. Obesity and breast cancer: progress to understanding the relationship. Cancer Res 2010;70:4–7. [DOI] [PubMed] [Google Scholar]
- 45.Renehan AG, Zwahlen M, Egger M. Adiposity and cancer risk: new mechanistic insights from epidemiology. Nat Rev Cancer 2015;15:484–98. [DOI] [PubMed] [Google Scholar]
- 46.Kaaks R, Berrino F, Key T, Rinaldi S, Dossus L, Biessy C, Secreto G, Amiano P, Bingham S, Boeing H, et al. . Serum sex steroids in premenopausal women and breast cancer risk within the European Prospective Investigation into Cancer and Nutrition (EPIC). J Natl Cancer Inst 2005;97:755–65. [DOI] [PubMed] [Google Scholar]
- 47.Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet 2001;2:919–29. [DOI] [PubMed] [Google Scholar]
- 48.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet 2016;17:719–32. [DOI] [PubMed] [Google Scholar]
- 50.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654–9. [DOI] [PubMed] [Google Scholar]
- 51.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem 2010;56:1733–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huan J, Hornick NI, Shurtleff MJ, Skinner AM, Goloviznina NA, Roberts CT Jr., Kurre P. RNA trafficking by acute myelogenous leukemia exosomes. Cancer Res 2013;73:918–29. [DOI] [PubMed] [Google Scholar]
- 53.Izzotti A, Carozzo S, Pulliero A, Zhabayeva D, Ravetti JL, Bersimbaev R. Extracellular MicroRNA in liquid biopsy: applicability in cancer diagnosis and prevention. Am J Cancer Res 2016;6:1461–93. [PMC free article] [PubMed] [Google Scholar]
- 54.Ambros V. The functions of animal microRNAs. Nature 2004;431:350–5. [DOI] [PubMed] [Google Scholar]
- 55.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. . Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 2004;101:2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. . A microRNA polycistron as a potential human oncogene. Nature 2005;435:828–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaboli PJ, Rahmat A, Ismail P, Ling KH. MicroRNA-based therapy and breast cancer: a comprehensive review of novel therapeutic strategies from diagnosis to treatment. Pharmacol Res 2015;97:104–21. [DOI] [PubMed] [Google Scholar]
- 58.Liu T, Hu K, Zhao Z, Chen G, Ou X, Zhang H, Zhang X, Wei X, Wang D, Cui M, et al. . MicroRNA-1 down-regulates proliferation and migration of breast cancer stem cells by inhibiting the Wnt/beta-catenin pathway. Oncotarget 2015;6:41638–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pogribny IP, Filkowski JN, Tryndyak VP, Golubov A, Shpyleva SI, Kovalchuk O. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int J Cancer 2010;127:1785–94. [DOI] [PubMed] [Google Scholar]
- 60.Khan S, Wall D, Curran C, Newell J, Kerin MJ, Dwyer RM. MicroRNA-10a is reduced in breast cancer and regulated in part through retinoic acid. BMC Cancer 2015;15:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kedmi M, Sas-Chen A, Yarden Y. MicroRNAs and growth factors: an alliance propelling tumor progression. J Clin Med 2015;4:1578–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol 2006;26:8191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kayani M, Kayani MA, Malik FA, Faryal R. Role of miRNAs in breast cancer. Asian Pac J Cancer Prev 2011;12:3175–80. [PubMed] [Google Scholar]
- 64.Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem 2009;284:23204–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bouyssou JM, Manier S, Huynh D, Issa S, Roccaro AM, Ghobrial IM. Regulation of microRNAs in cancer metastasis. Biochim Biophys Acta 2014;1845:255–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang N, Wang X, Huo Q, Sun M, Cai C, Liu Z, Hu G, Yang Q. MicroRNA-30a suppresses breast tumor growth and metastasis by targeting metadherin. Oncogene 2014;33:3119–28. [DOI] [PubMed] [Google Scholar]
- 67.Bockhorn J, Dalton R, Nwachukwu C, Huang S, Prat A, Yee K, Chang YF, Huo D, Wen Y, Swanson KE, et al. . MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat Commun 2013;4:1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bockhorn J, Yee K, Chang YF, Prat A, Huo D, Nwachukwu C, Dalton R, Huang S, Swanson KE, Perou CM, et al. . MicroRNA-30c targets cytoskeleton genes involved in breast cancer cell invasion. Breast Cancer Res Treat 2013;137:373–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Körner C, Keklikoglou I, Bender C, Wörner A, Münstermann E, Wiemann S. MicroRNA-31 sensitizes human breast cells to apoptosis by direct targeting of protein kinase C epsilon (PKCepsilon). J Biol Chem 2013;288:8750–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fang L, Du WW, Yang W, Rutnam ZJ, Peng C, Li H, O’Malley YQ, Askeland RW, Sugg S, Liu M, et al. . MiR-93 enhances angiogenesis and metastasis by targeting LATS2. Cell Cycle 2012;11:4352–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T, et al. . A MicroRNA targeting dicer for metastasis control. Cell 2010;141:1195–207. [DOI] [PubMed] [Google Scholar]
- 72.Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson AL, et al. . MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol 2008;28:2167–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Asghari F, Haghnavaz N, Baradaran B, Hemmatzadeh M, Kazemi T. Tumor suppressor microRNAs: targeted molecules and signaling pathways in breast cancer. Biomed Pharmacother 2016;81:305–17. [DOI] [PubMed] [Google Scholar]
- 74.Li M, Fu W, Wo L, Shu X, Liu F, Li C. miR-128 and its target genes in tumorigenesis and metastasis. Exp Cell Res 2013;319:3059–64. [DOI] [PubMed] [Google Scholar]
- 75.Isobe T, Hisamori S, Hogan DJ, Zabala M, Hendrickson DG, Dalerba P, Cai S, Scheeren F, Kuo AH, Sikandar SS, et al. . miR-142 regulates the tumorigenicity of human breast cancer stem cells through the canonical WNT signaling pathway. ELife 2014;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schwickert A, Weghake E, Bruggemann K, Engbers A, Brinkmann BF, Kemper B, Seggewiss J, Stock C, Ebnet K, Kiesel L, et al. . microRNA miR-142–3p inhibits breast cancer cell invasiveness by synchronous targeting of WASL, integrin alpha V, and additional cytoskeletal elements. PLoS One 2015;10:e0143993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ng EK, Li R, Shin VY, Siu JM, Ma ES, Kwong A. MicroRNA-143 is downregulated in breast cancer and regulates DNA methyltransferases 3A in breast cancer cells. Tumour Biol 2014;35:2591–8. [DOI] [PubMed] [Google Scholar]
- 78.Yoo JO, Kwak SY, An HJ, Bae IH, Park MJ, Han YH. miR-181b-3p promotes epithelial-mesenchymal transition in breast cancer cells through Snail stabilization by directly targeting YWHAG. Biochim Biophys Acta 2016;1863:1601–11. [DOI] [PubMed] [Google Scholar]
- 79.Tang H, Liu P, Yang L, Xie X, Ye F, Wu M, Liu X, Chen B, Zhang L, Xie X. miR-185 suppresses tumor proliferation by directly targeting E2F6 and DNMT1 and indirectly upregulating BRCA1 in triple-negative breast cancer. Mol Cancer Ther 2014;13:3185–97. [DOI] [PubMed] [Google Scholar]
- 80.Tsouko E, Wang J, Frigo DE, Aydogdu E, Williams C. miR-200a inhibits migration of triple-negative breast cancer cells through direct repression of the EPHA2 oncogene. Carcinogenesis 2015;36:1051–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yao Y, Hu J, Shen Z, Yao R, Liu S, Li Y, Cong H, Wang X, Qiu W, Yue L. MiR-200b expression in breast cancer: a prognostic marker and act on cell proliferation and apoptosis by targeting Sp1. J Cell Mol Med 2015;19:760–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jana S, Sengupta S, Biswas S, Chatterjee A, Roy H, Bhattacharyya A. miR-216b suppresses breast cancer growth and metastasis by targeting SDCBP. Biochem Biophys Res Commun 2017;482:126–33. [DOI] [PubMed] [Google Scholar]
- 83.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem 2008;283:29897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liang Z, Wu H, Xia J, Li Y, Zhang Y, Huang K, Wagar N, Yoon Y, Cho HT, Scala S, et al. . Involvement of miR-326 in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1. Biochem Pharmacol 2010;79:817–24. [DOI] [PubMed] [Google Scholar]
- 85.Pan YZ, Morris ME, Yu AM. MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol 2009;75:1374–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF, Pogribny IP. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther 2008;7:2152–9. [DOI] [PubMed] [Google Scholar]
- 87.Ma MT, He M, Wang Y, Jiao XY, Zhao L, Bai XF, Yu ZJ, Wu HZ, Sun ML, Song ZG, et al. . MiR-487a resensitizes mitoxantrone (MX)-resistant breast cancer cells (MCF-7/MX) to MX by targeting breast cancer resistance protein (BCRP/ABCG2). Cancer Lett 2013;339:107–15. [DOI] [PubMed] [Google Scholar]
- 88.Jiang L, He D, Yang D, Chen Z, Pan Q, Mao A, Cai Y, Li X, Xing H, Shi M, et al. . MiR-489 regulates chemoresistance in breast cancer via epithelial mesenchymal transition pathway. FEBS Lett 2014;588:2009–15. [DOI] [PubMed] [Google Scholar]
- 89.Hui Z, Yiling C, Wenting Y, XuQun H, ChuanYi Z, Hui L. miR-491-5p functions as a tumor suppressor by targeting JMJD2B in ERalpha-positive breast cancer. FEBS Lett 2015;589:812–21. [DOI] [PubMed] [Google Scholar]
- 90.Hwang-Verslues WW, Chang PH, Wei PC, Yang CY, Huang CK, Kuo WH, Shew JY, Chang KJ, Lee EY, Lee WH. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene 2011;30:2463–74. [DOI] [PubMed] [Google Scholar]
- 91.Leivonen SK, Sahlberg KK, Makela R, Due EU, Kallioniemi O, Borresen-Dale AL, Perala M. High-throughput screens identify microRNAs essential for HER2 positive breast cancer cell growth. Mol Oncol 2014;8:93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA 2003;100:3983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al. . Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 2009;138:592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, et al. . let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007;131:1109–23. [DOI] [PubMed] [Google Scholar]
- 95.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer 2003;3:453–8. [DOI] [PubMed] [Google Scholar]
- 96.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell 2006;127:679–95. [DOI] [PubMed] [Google Scholar]
- 97.Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 2008;27:4373–9. [DOI] [PubMed] [Google Scholar]
- 98.Qi L, Bart J, Tan LP, Platteel I, Sluis T, Huitema S, Harms G, Fu L, Hollema H, Berg A. Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer 2009;9:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Song B, Wang C, Liu J, Wang X, Lv L, Wei L, Xie L, Zheng Y, Song X. MicroRNA-21 regulates breast cancer invasion partly by targeting tissue inhibitor of metalloproteinase 3 expression. J Exp Clin Cancer Res 2010;29:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007;449:682–8. [DOI] [PubMed] [Google Scholar]
- 101.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008;451:147–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang S, Kim K, Jin UH, Pfent C, Cao H, Amendt B, Liu X, Wilson-Robles H, Safe S. Aryl hydrocarbon receptor agonists induce microRNA-335 expression and inhibit lung metastasis of estrogen receptor negative breast cancer cells. Mol Cancer Ther 2012;11:108–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008;10:593–601. [DOI] [PubMed] [Google Scholar]
- 104.Kim NH, Kim HS, Li XY, Lee I, Choi HS, Kang SE, Cha SY, Ryu JK, Yoon D, Fearon ER, et al. . A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J Cell Biol 2011;195:417–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Baker EK, El-Osta A. The rise of DNA methylation and the importance of chromatin on multidrug resistance in cancer. Exp Cell Res 2003;290:177–94. [DOI] [PubMed] [Google Scholar]
- 106.Glasspool RM, Teodoridis JM, Brown R. Epigenetics as a mechanism driving polygenic clinical drug resistance. Br J Cancer 2006;94:1087–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee AJ, Endesfelder D, Rowan AJ, Walther A, Birkbak NJ, Futreal PA, Downward J, Szallasi Z, Tomlinson IP, Howell M, et al. . Chromosomal instability confers intrinsic multidrug resistance. Cancer Res 2011;71:1858–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Galli R, Paone A, Fabbri M, Zanesi N, Calore F, Cascione L, Acunzo M, Stoppacciaro A, Tubaro A, Lovat F, et al. . Toll-like receptor 3 (TLR3) activation induces microRNA-dependent reexpression of functional RARbeta and tumor regression. Proc Natl Acad Sci USA 2013;110:9812–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kazarian A, Blyuss O, Metodieva G, Gentry-Maharaj A, Ryan A, Kiseleva EM, Prytomanova OM, Jacobs IJ, Widschwendter M, Menon U, et al. . Testing breast cancer serum biomarkers for early detection and prognosis in pre-diagnosis samples. Br J Cancer 2017;116:501–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem 2013;59:110–8. [DOI] [PubMed] [Google Scholar]
- 111.Zhu J, Zheng Z, Wang J, Sun J, Wang P, Cheng X, Fu L, Zhang L, Wang Z, Li Z. Different miRNA expression profiles between human breast cancer tumors and serum. Front Genet 2014;5:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Müller V, Gade S, Steinbach B, Loibl S, von Minckwitz G, Untch M, Schwedler K, Lubbe K, Schem C, Fasching PA, et al. . Changes in serum levels of miR-21, miR-210, and miR-373 in HER2-positive breast cancer patients undergoing neoadjuvant therapy: a translational research project within the Geparquinto trial. Breast Cancer Res Treat 2014;147:61–8. [DOI] [PubMed] [Google Scholar]
- 113.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 2006;7:885–96. [DOI] [PubMed] [Google Scholar]
- 114.Rosen ED, Spiegelman BM. Molecular regulation of adipogenesis. Annu Rev Cell Dev Biol 2000;16:145–71. [DOI] [PubMed] [Google Scholar]
- 115.Arner P, Kulyte A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat Rev Endocrinol 2015;11:276–88. [DOI] [PubMed] [Google Scholar]
- 116.McGregor RA, Choi MS. microRNAs in the regulation of adipogenesis and obesity. Curr Mol Med 2011;11:304–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Son YH, Ka S, Kim AY, Kim JB. Regulation of adipocyte differentiation via microRNAs. Endocrinol Metab (Seoul) 2014;29:122–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Andersen DC, Jensen CH, Schneider M, Nossent AY, Eskildsen T, Hansen JL, Teisner B, Sheikh SP. MicroRNA-15a fine-tunes the level of Delta-like 1 homolog (DLK1) in proliferating 3T3–L1 preadipocytes. Exp Cell Res 2010;316:1681–91. [DOI] [PubMed] [Google Scholar]
- 119.Wang Q, Li YC, Wang J, Kong J, Qi Y, Quigg RJ, Li X. miR-17–92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Natl Acad Sci USA 2008;105:2889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hamam D, Ali D, Kassem M, Aldahmash A, Alajez NM. microRNAs as regulators of adipogenic differentiation of mesenchymal stem cells. Stem Cells Dev 2015;24:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cioffi M, Vallespinos-Serrano M, Trabulo SM, Fernandez-Marcos PJ, Firment AN, Vazquez BN, Vieira CR, Mulero F, Camara JA, Cronin UP, et al. . MiR-93 controls adiposity via inhibition of Sirt7 and Tbx3. Cell Reports 2015;12:1594–605. [DOI] [PubMed] [Google Scholar]
- 122.Chen K, He H, Xie Y, Zhao L, Zhao S, Wan X, Yang W, Mo Z. miR-125a-3p and miR-483-5p promote adipogenesis via suppressing the RhoA/ROCK1/ERK1/2 pathway in multiple symmetric lipomatosis. Sci Rep 2015;5:11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rockstroh D, Loffler D, Kiess W, Landgraf K, Korner A. Regulation of human adipogenesis by miR125b-5p. Adipocyte 2016;5:283–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lee EK, Lee MJ, Abdelmohsen K, Kim W, Kim MM, Srikantan S, Martindale JL, Hutchison ER, Kim HH, Marasa BS, et al. . miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol 2011;31:626–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Strum JC, Johnson JH, Ward J, Xie H, Feild J, Hester A, Alford A, Waters KM. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol Endocrinol 2009;23:1876–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang Z, Bian C, Zhou H, Huang S, Wang S, Liao L, Zhao RC. MicroRNA hsa-miR-138 inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells through adenovirus EID-1. Stem Cells Dev 2011;20:259–67. [DOI] [PubMed] [Google Scholar]
- 127.Capobianco V, Nardelli C, Ferrigno M, Iaffaldano L, Pilone V, Forestieri P, Zambrano N, Sacchetti L. miRNA and protein expression profiles of visceral adipose tissue reveal miR-141/YWHAG and miR-520e/RAB11A as two potential miRNA/protein target pairs associated with severe obesity. J Proteome Res 2012;11:3358–69. [DOI] [PubMed] [Google Scholar]
- 128.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA 2006;103:12481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ying W, Tseng A, Chang RC, Wang H, Lin YL, Kanameni S, Brehm T, Morin A, Jones B, Splawn T, et al. . miR-150 regulates obesity-associated insulin resistance by controlling B cell functions. Sci Rep 2016;6:20176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen Y, Siegel F, Kipschull S, Haas B, Frohlich H, Meister G, Pfeifer A. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat Commun 2013;4:1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li H, Chen X, Guan L, Qi Q, Shu G, Jiang Q, Yuan L, Xi Q, Zhang Y. MiRNA-181a regulates adipogenesis by targeting tumor necrosis factor-alpha (TNF-alpha) in the porcine model. PLoS One 2013;8:e71568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sun L, Xie H, Mori MA, Alexander R, Yuan B, Hattangadi SM, Liu Q, Kahn CR, Lodish HF. Mir193b-365 is essential for brown fat differentiation. Nat Cell Biol 2011;13:958–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Oskowitz AZ, Lu J, Penfornis P, Ylostalo J, McBride J, Flemington EK, Prockop DJ, Pochampally R. Human multipotent stromal cells from bone marrow and microRNA: regulation of differentiation and leukemia inhibitory factor expression. Proc Natl Acad Sci USA 2008;105:18372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhuang G, Meng C, Guo X, Cheruku PS, Shi L, Xu H, Li H, Wang G, Evans AR, Safe S, et al. . A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation 2012;125:2892–903. [DOI] [PubMed] [Google Scholar]
- 135.Bork S, Horn P, Castoldi M, Hellwig I, Ho AD, Wagner W. Adipogenic differentiation of human mesenchymal stromal cells is down-regulated by microRNA-369-5p and up-regulated by microRNA-371. J Cell Physiol 2011;226:2226–34. [DOI] [PubMed] [Google Scholar]
- 136.Mudhasani R, Imbalzano AN, Jones SN. An essential role for Dicer in adipocyte differentiation. J Cell Biochem 2010;110:812–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Skårn M, Namlos HM, Noordhuis P, Wang MY, Meza-Zepeda LA, Myklebost O. Adipocyte differentiation of human bone marrow-derived stromal cells is modulated by microRNA-155, microRNA-221, and microRNA-222. Stem Cells Dev 2012;21:873–83. [DOI] [PubMed] [Google Scholar]
- 138.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, et al. . MicroRNA-143 regulates adipocyte differentiation. J Biol Chem 2004;279:52361–5. [DOI] [PubMed] [Google Scholar]
- 139.Chen L, Wu P, Guo XH, Hu Y, Li YL, Shi J, Wang KZ, Chu WY, Zhang JS. miR-143: a novel regulator of MyoD expression in fast and slow muscles of Siniperca chuatsi. Curr Mol Med 2014;14:370–5. [DOI] [PubMed] [Google Scholar]
- 140.Zaragosi LE, Wdziekonski B, Brigand KL, Villageois P, Mari B, Waldmann R, Dani C, Barbry P. Small RNA sequencing reveals miR-642a-3p as a novel adipocyte-specific microRNA and miR-30 as a key regulator of human adipogenesis. Genome Biol 2011;12:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Karbiener M, Fischer C, Nowitsch S, Opriessnig P, Papak C, Ailhaud G, Dani C, Amri EZ, Scheideler M. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem Biophys Res Commun 2009;390:247–51. [DOI] [PubMed] [Google Scholar]
- 142.Chou WW, Wang YT, Liao YC, Chuang SC, Wang SN, Juo SH. Decreased microRNA-221 is associated with high levels of TNF-alpha in human adipose tissue-derived mesenchymal stem cells from obese woman. Cell Physiol Biochem 2013;32:127–37. [DOI] [PubMed] [Google Scholar]
- 143.Jordan SD, Kruger M, Willmes DM, Redemann N, Wunderlich FT, Bronneke HS, Merkwirth C, Kashkar H, Olkkonen VM, Bottger T, et al. . Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol 2011;13:434–46. [DOI] [PubMed] [Google Scholar]
- 144.Herrera BM, Lockstone HE, Taylor JM, Ria M, Barrett A, Collins S, Kaisaki P, Argoud K, Fernandez C, Travers ME, et al. . Global microRNA expression profiles in insulin target tissues in a spontaneous rat model of type 2 diabetes. Diabetologia 2010;53:1099–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ling HY, Ou HS, Feng SD, Zhang XY, Tuo QH, Chen LX, Zhu BY, Gao ZP, Tang CK, Yin WD, et al. . CHANGES IN microRNA (miR) profile and effects of miR-320 in insulin-resistant 3T3-L1 adipocytes. Clin Exp Pharmacol Physiol 2009;36:e32–9. [DOI] [PubMed] [Google Scholar]
- 146.Maury E, Brichard SM. Adipokine dysregulation, adipose tissue inflammation and metabolic syndrome. Mol Cell Endocrinol 2010;314:1–16. [DOI] [PubMed] [Google Scholar]
- 147.Hotamisligil GS. Inflammation and metabolic disorders. Nature 2006;444:860–7. [DOI] [PubMed] [Google Scholar]
- 148.Shi C, Zhu L, Chen X, Gu N, Chen L, Zhu L, Yang L, Pang L, Guo X, Ji C, et al. . IL-6 and TNF-alpha induced obesity-related inflammatory response through transcriptional regulation of miR-146b. J Interferon Cytokine Res 2014;34:342–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Heneghan HM, Miller N, McAnena OJ, O’Brien T, Kerin MJ. Differential miRNA expression in omental adipose tissue and in the circulation of obese patients identifies novel metabolic biomarkers. J Clin Endocrinol Metab 2011;96:E846–50. [DOI] [PubMed] [Google Scholar]
- 150.Kang M, Yan LM, Zhang WY, Li YM, Tang AZ, Ou HS. Role of microRNA-21 in regulating 3T3–L1 adipocyte differentiation and adiponectin expression. Mol Biol Rep 2013;40:5027–34. [DOI] [PubMed] [Google Scholar]
- 151.Subedi A, Park PH. Autocrine and paracrine modulation of microRNA-155 expression by globular adiponectin in RAW 264.7 macrophages: involvement of MAPK/NF-kappaB pathway. Cytokine 2013;64:638–41. [DOI] [PubMed] [Google Scholar]
- 152.Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes 2009;58:1050–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO, et al. . MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol 2007;8:R214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.van Schooneveld E, Wildiers H, Vergote I, Vermeulen PB, Dirix LY, Van Laere SJ. Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast Cancer Res 2015;17:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 2009;137:1032–46. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 156.Tang YF, Zhang Y, Li XY, Li C, Tian W, Liu L. Expression of miR-31, miR-125b-5p, and miR-326 in the adipogenic differentiation process of adipose-derived stem cells. OMICS 2009;13:331–6. [DOI] [PubMed] [Google Scholar]
- 157.Yan X, Chen X, Liang H, Deng T, Chen W, Zhang S, Liu M, Gao X, Liu Y, Zhao C, et al. . miR-143 and miR-145 synergistically regulate ERBB3 to suppress cell proliferation and invasion in breast cancer. Mol Cancer 2014;13:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Esguerra JL, Bolmeson C, Cilio CM, Eliasson L. Differential glucose-regulation of microRNAs in pancreatic islets of non-obese type 2 diabetes model Goto-Kakizaki rat. PLoS One 2011;6:e18613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Png KJ, Yoshida M, Zhang XH, Shu W, Lee H, Rimner A, Chan TA, Comen E, Andrade VP, Kim SW, et al. . MicroRNA-335 inhibits tumor reinitiation and is silenced through genetic and epigenetic mechanisms in human breast cancer. Genes Dev 2011;25:226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Joglekar MV, Joglekar VM, Hardikar AA. Expression of islet-specific microRNAs during human pancreatic development. Gene Expr Patterns 2009;9:109–13. [DOI] [PubMed] [Google Scholar]
- 161.Guo LM, Pu Y, Han Z, Liu T, Li YX, Liu M, Li X, Tang H. MicroRNA-9 inhibits ovarian cancer cell growth through regulation of NF-kappaB1. FEBS J 2009;276:5537–46. [DOI] [PubMed] [Google Scholar]
- 162.Picon-Ruiz M, Pan C, Drews-Elger K, Jang K, Besser AH, Zhao D, Morata-Tarifa C, Kim M, Ince TA, Azzam DJ, et al. . Interactions between adipocytes and breast cancer cells stimulate cytokine production and drive Src/Sox2/miR-302b-mediated malignant progression. Cancer Res 2016;76:491–504. [DOI] [PubMed] [Google Scholar]
- 163.Ross SA, Davis CD. The emerging role of microRNAs and nutrition in modulating health and disease. Annu Rev Nutr 2014;34:305–36. [DOI] [PubMed] [Google Scholar]
- 164.Zempleni J, Baier SR, Howard KM, Cui J. Gene regulation by dietary microRNAs. Can J Physiol Pharmacol 2015;93:1097–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Peng X, Vaishnav A, Murillo G, Alimirah F, Torres KE, Mehta RG. Protection against cellular stress by 25-hydroxyvitamin D3 in breast epithelial cells. J Cell Biochem 2010;110:1324–33. [DOI] [PubMed] [Google Scholar]
- 166.Yang J, Cao Y, Sun J, Zhang Y. Curcumin reduces the expression of Bcl-2 by upregulating miR-15a and miR-16 in MCF-7 cells. Med Oncol 2010;27:1114–8. [DOI] [PubMed] [Google Scholar]
- 167.Jin Y. 3,3′-Diindolylmethane inhibits breast cancer cell growth via miR-21-mediated Cdc25A degradation. Mol Cell Biochem 2011;358:345–54. [DOI] [PubMed] [Google Scholar]
- 168.Terao M, Fratelli M, Kurosaki M, Zanetti A, Guarnaccia V, Paroni G, Tsykin A, Lupi M, Gianni M, Goodall GJ, et al. . Induction of miR-21 by retinoic acid in estrogen receptor-positive breast carcinoma cells: biological correlates and molecular targets. J Biol Chem 2011;286:4027–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Banerjee N, Talcott S, Safe S, Mertens-Talcott SU. Cytotoxicity of pomegranate polyphenolics in breast cancer cells in vitro and vivo: potential role of miRNA-27a and miRNA-155 in cell survival and inflammation. Breast Cancer Res Treat 2012;136:21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cho JH, Dimri M, Dimri GP. MicroRNA-31 is a transcriptional target of histone deacetylase inhibitors and a regulator of cellular senescence. J Biol Chem 2015;290:10555–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Singh B, Ronghe AM, Chatterjee A, Bhat NK, Bhat HK. MicroRNA-93 regulates NRF2 expression and is associated with breast carcinogenesis. Carcinogenesis 2013;34:1165–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sachdeva M, Liu Q, Cao J, Lu Z, Mo YY. Negative regulation of miR-145 by C/EBP-beta through the Akt pathway in cancer cells. Nucleic Acids Res 2012;40:6683–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kronski E, Fiori ME, Barbieri O, Astigiano S, Mirisola V, Killian PH, Bruno A, Pagani A, Rovera F, Pfeffer U, et al. . miR181b is induced by the chemopreventive polyphenol curcumin and inhibits breast cancer metastasis via down-regulation of the inflammatory cytokines CXCL1 and -2. Mol Oncol 2014;8:581–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hanieh H. Aryl hydrocarbon receptor-microRNA-212/132 axis in human breast cancer suppresses metastasis by targeting SOX4. Mol Cancer 2015;14:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Vislovukh A, Kratassiouk G, Porto E, Gralievska N, Beldiman C, Pinna G, El’skaya A, Harel-Bellan A, Negrutskii B, Groisman I. Proto-oncogenic isoform A2 of eukaryotic translation elongation factor eEF1 is a target of miR-663 and miR-744. Br J Cancer 2013;108:2304–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tsang WP, Kwok TT. Epigallocatechin gallate up-regulation of miR-16 and induction of apoptosis in human cancer cells. J Nutr Biochem 2010;21:140–6. [DOI] [PubMed] [Google Scholar]
- 177.Tomé-Carneiro J, Larrosa M, Yañez-Gascon MJ, Dávalos A, Gil-Zamorano J, Gonzálvez M, García-Almagro FJ, Ruiz Ros JA, Tomas-Barberán FA, Espín JC, et al. . One-year supplementation with a grape extract containing resveratrol modulates inflammatory-related microRNAs and cytokines expression in peripheral blood mononuclear cells of type 2 diabetes and hypertensive patients with coronary artery disease. Pharmacol Res 2013;72:69–82. [DOI] [PubMed] [Google Scholar]
- 178.Arola-Arnal A, Blade C. Proanthocyanidins modulate microRNA expression in human HepG2 cells. PLoS One 2011;6:e25982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.La VD, Bergeron C, Gafner S, Grenier D. Grape seed extract suppresses lipopolysaccharide-induced matrix metalloproteinase (MMP) secretion by macrophages and inhibits human MMP-1 and -9 activities. J Periodontol 2009;80:1875–82. [DOI] [PubMed] [Google Scholar]
- 180.Joven J, Espinel E, Rull A, Aragones G, Rodriguez-Gallego E, Camps J, Micol V, Herranz-Lopez M, Menendez JA, Borras I, et al. . Plant-derived polyphenols regulate expression of miRNA paralogs miR-103/107 and miR-122 and prevent diet-induced fatty liver disease in hyperlipidemic mice. Biochim Biophys Acta 2012;1820:894–9. [DOI] [PubMed] [Google Scholar]
- 181.Parra P, Serra F, Palou A. Expression of adipose microRNAs is sensitive to dietary conjugated linoleic acid treatment in mice. PLoS One 2010;5:e13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Noratto GD, Kim Y, Talcott ST, Mertens-Talcott SU. Flavonol-rich fractions of yaupon holly leaves (Ilex vomitoria, Aquifoliaceae) induce microRNA-146a and have anti-inflammatory and chemopreventive effects in intestinal myofibroblast CCD-18Co cells. Fitoterapia 2011;82:557–69. [DOI] [PubMed] [Google Scholar]
- 183.Baselga-Escudero L, Blade C, Ribas-Latre A, Casanova E, Salvado MJ, Arola L, Arola-Arnal A. Grape seed proanthocyanidins repress the hepatic lipid regulators miR-33 and miR-122 in rats. Mol Nutr Food Res 2012;56:1636–46. [DOI] [PubMed] [Google Scholar]
- 184.Boesch-Saadatmandi C, Wagner AE, Wolffram S, Rimbach G. Effect of quercetin on inflammatory gene expression in mice liver in vivo - role of redox factor 1, miRNA-122 and miRNA-125b. Pharmacol Res 2012;65:523–30. [DOI] [PubMed] [Google Scholar]
- 185.Gaedicke S, Zhang X, Schmelzer C, Lou Y, Doering F, Frank J, Rimbach G. Vitamin E dependent microRNA regulation in rat liver. FEBS Lett 2008;582:3542–6. [DOI] [PubMed] [Google Scholar]
- 186.Wagner AE, Boesch-Saadatmandi C, Dose J, Schultheiss G, Rimbach G. Anti-inflammatory potential of allyl-isothiocyanate–role of Nrf2, NF-(kappa) B and microRNA-155. J Cell Mol Med 2012;16:836–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chen Y, Liu W, Sun T, Huang Y, Wang Y, Deb DK, Yoon D, Kong J, Thadhani R, Li YC. 1,25-Dihydroxyvitamin D promotes negative feedback regulation of TLR signaling via targeting microRNA-155-SOCS1 in macrophages. J Immunol 2013;190:3687–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hoekstra M, van der Sluis RJ, Kuiper J, Van Berkel TJ. Nonalcoholic fatty liver disease is associated with an altered hepatocyte microRNA profile in LDL receptor knockout mice. J Nutr Biochem 2012;23:622–8. [DOI] [PubMed] [Google Scholar]
- 189.Jeon TI, Park JW, Ahn J, Jung CH, Ha TY. Fisetin protects against hepatosteatosis in mice by inhibiting miR-378. Mol Nutr Food Res 2013;57:1931–7. [DOI] [PubMed] [Google Scholar]
- 190.Ahn J, Lee H, Chung CH, Ha T. High fat diet induced downregulation of microRNA-467b increased lipoprotein lipase in hepatic steatosis. Biochem Biophys Res Commun 2011;414:664–9. [DOI] [PubMed] [Google Scholar]
- 191.Mohri T, Nakajima M, Takagi S, Komagata S, Yokoi T. MicroRNA regulates human vitamin D receptor. Int J Cancer 2009;125:1328–33. [DOI] [PubMed] [Google Scholar]
- 192.Mandal CC, Ghosh-Choudhury T, Dey N, Choudhury GG, Ghosh-Choudhury N. miR-21 is targeted by omega-3 polyunsaturated fatty acid to regulate breast tumor CSF-1 expression. Carcinogenesis 2012;33:1897–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chen J, Xu T, Chen C. The critical roles of miR-21 in anti-cancer effects of curcumin. Ann Transl Med 2015;3:330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Qin W, Zhang K, Clarke K, Weiland T, Sauter ER. Methylation and miRNA effects of resveratrol on mammary tumors vs. normal tissue. Nutr Cancer 2014;66:270–7. [DOI] [PubMed] [Google Scholar]
- 195.Ahmad A, Ali S, Ahmed A, Ali AS, Raz A, Sakr WA, Rahman KM. 3, 3′-Diindolylmethane enhances the effectiveness of herceptin against HER-2/neu-expressing breast cancer cells. PLoS One 2013;8:e54657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Cui J, Zhou B, Ross SA, Zempleni J. Nutrition, microRNAs, and human health. Adv Nutr 2017;8:105–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Baier SR, Nguyen C, Xie F, Wood JR, Zempleni J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J Nutr 2014;144:1495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, et al. . MicroRNAs bind to toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA 2012;109:E2110–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Pandey DP, Picard D. miR-22 inhibits estrogen signaling by directly targeting the estrogen receptor alpha mRNA. Mol Cell Biol 2009;29:3783–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chin AR, Fong MY, Somlo G, Wu J, Swiderski P, Wu X, Wang SE. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res 2016;26:217–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Benatti RO, Melo AM, Borges FO, Ignacio-Souza LM, Simino LA, Milanski M, Velloso LA, Torsoni MA, Torsoni AS. Maternal high-fat diet consumption modulates hepatic lipid metabolism and microRNA-122 (miR-122) and microRNA-370 (miR-370) expression in offspring. Br J Nutr 2014;111:2112–22. [DOI] [PubMed] [Google Scholar]
- 202.Alisi A, Da Sacco L, Bruscalupi G, Piemonte F, Panera N, De Vito R, Leoni S, Bottazzo GF, Masotti A, Nobili V. Mirnome analysis reveals novel molecular determinants in the pathogenesis of diet-induced nonalcoholic fatty liver disease. Lab Invest 2011;91:283–93. [DOI] [PubMed] [Google Scholar]
- 203.Murase T, Misawa K, Minegishi Y, Aoki M, Ominami H, Suzuki Y, Shibuya Y, Hase T. Coffee polyphenols suppress diet-induced body fat accumulation by downregulating SREBP-1c and related molecules in C57BL/6J mice. Am J Physiol Endocrinol Metab 2011;300:E122–33. [DOI] [PubMed] [Google Scholar]
- 204.Ahn J, Lee H, Jung CH, Ha T. Lycopene inhibits hepatic steatosis via microRNA-21-induced downregulation of fatty acid-binding protein 7 in mice fed a high-fat diet. Mol Nutr Food Res 2012;56:1665–74. [DOI] [PubMed] [Google Scholar]
- 205.Dolganiuc A, Petrasek J, Kodys K, Catalano D, Mandrekar P, Velayudham A, Szabo G. MicroRNA expression profile in Lieber-DeCarli diet-induced alcoholic and methionine choline deficient diet-induced nonalcoholic steatohepatitis models in mice. Alcohol Clin Exp Res 2009;33:1704–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Fischer-Posovszky P, Kukulus V, Zulet MA, Debatin KM, Wabitsch M. Conjugated linoleic acids promote human fat cell apoptosis. Horm Metab Res 2007;39:186–91. [DOI] [PubMed] [Google Scholar]
- 207.Wang WL, Chatterjee N, Chittur SV, Welsh J, Tenniswood MP. Effects of 1alpha,25 dihydroxyvitamin D3 and testosterone on miRNA and mRNA expression in LNCaP cells. Mol Cancer 2011;10:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Woźniak Ł, Sk ą, pska S, Marsza ł, ek K. Ursolic acid–a pentacyclic triterpenoid with a wide spectrum of pharmacological activities. Molecules 2015;20:20614–41. [DOI] [PMC free article] [PubMed] [Google Scholar]