PhyH-DI, a β-propeller phytase from Bacillus sp. HJB17 that interacts with PhyH-DII and other typical β-propeller phytases, can increase the efficiency of phytases in hydrolyzing phytate. Native and selenomethionine-substituted PhyH-DI were expressed, purified and crystallized.
Keywords: β-propeller phytases, crystallization, X-ray diffraction, selenomethionine-substituted protein
Abstract
Phytases are phosphatases that hydrolyze phytates to less phosphorylated myo-inositol derivatives and inorganic phosphate. β-Propeller phytases, which are very diverse phytases with improved thermostability that are active at neutral and alkaline pH and have absolute substrate specificity, are ideal substitutes for other commercial phytases. PhyH-DI, a β-propeller phytase from Bacillus sp. HJB17, was found to act synergistically with other single-domain phytases and can increase their efficiency in the hydrolysis of phytate. Crystals of native and selenomethionine-substituted PhyH-DI were obtained using the vapour-diffusion method in a condition consisting of 0.2 M sodium chloride, 0.1 M Tris pH 8.5, 25%(w/v) PEG 3350 at 289 K. X-ray diffraction data were collected to 3.00 and 2.70 Å resolution, respectively, at 100 K. Native PhyH-DI crystals belonged to space group C121, with unit-cell parameters a = 156.84, b = 45.54, c = 97.64 Å, α = 90.00, β = 125.86, γ = 90.00°. The asymmetric unit contained two molecules of PhyH-DI, with a corresponding Matthews coefficient of 2.17 Å3 Da−1 and a solvent content of 43.26%. Crystals of selenomethionine-substituted PhyH-DI belonged to space group C2221, with unit-cell parameters a = 94.71, b = 97.03, c = 69.16 Å, α = β = γ = 90.00°. The asymmetric unit contained one molecule of the protein, with a corresponding Matthews coefficient of 2.44 Å3 Da−1 and a solvent content of 49.64%. Initial phases for PhyH-DI were obtained from SeMet SAD data sets. These data will be useful for further studies of the structure–function relationship of PhyH-DI.
1. Introduction
Phytates (myo-inositol-1,2,3,4,5,6-hexakisphosphates) are the most abundant organic phosphorus compounds in most plants and soils; they are very stable and are potentially used by microorganisms, plants and animals (Mukhametzianova et al., 2012 ▸). Phytases represent a subgroup of phosphomonoesterases that initiate the stepwise hydrolysis of phytates into myo-inositol derivatives and phosphate (Ullah, 1988 ▸). Although the first research on phytases was reported in 1907, it has increased exponentially in the past two to three decades (Lei et al., 2012 ▸). Studies show that phytases have attracted considerable attention from both scientists and entrepreneurs in the areas of nutrition, environmental protection and biotechnology (Lei & Stahl, 2001 ▸). Phytases have multifarious applications and are used in the food and feed industries for improving nutritional quality, in breadmaking, in the promotion of plant growth, in the preparation of myo-inositol phosphates and in the reduction of phosphorus pollutant levels in the environment (Kumar et al., 2017 ▸; Jain et al., 2016 ▸). In 2010, world experts gathered for the first international phytase summit in Washington DC, which was indicative of the tremendous economic impact of phytases in the animal feed market. This inaugural summit facilitated and encouraged the exploration of a number of phytase-related issues ranging from the strengths and weaknesses of phytases to implications for their future use.
To date, four types of phytases have been characterized by their structural and catalytic properties: histidine acid phosphatases (HAPs), protein tyrosine phosphatases (PTPs) or cysteine phytases (CPs), purple acid phosphatases (PAPs) and β-propeller phytases (BPPs) (Lei et al., 2012 ▸; Lim et al., 2007 ▸). Among the phytases that have been identified, BPPs are considered to be the most abundant phytases in nature and are assumed to play a leading role in phytate phosphorus cycling in soil and water. The BPPs were found to be widely distributed in the genomes of a number of microbes according to the NCBI databases and were named for the molecular structure of the protein, which consists mostly of β-structural shields and resembles a six-bladed propeller (Mukhametzianova et al., 2012 ▸). To date, all known BPPs are from bacteria, in particular Bacillus spp. (Zhang et al., 2011 ▸). BPPs, which have an optimum pH in the range 6.0–8.0, belong to the alkaline phytases and have several different biological properties, such as phytate specificity, resistance to proteolysis, high catalytic efficiency and high thermostability (Cheng & Lim, 2006 ▸). Calcium plays an important role in the catalytic activity and thermostability of BPPs. These properties make them more versatile than other types of phytase.
PhyH-DI is a BPP from Bacillus sp. HJB17 that belongs to the 3-phytases (EC 3.1.3.8). PhyH-DI shares 69–85% sequence identity with 3-phytase from Rheinheimera and has 47–55% amino-acid similarity to 3-phytase from Idiomarina. These results indicate that this type of protein has always existed in extreme environments and has special catalytic functions. PhyH-DI has catalytic activity towards the phytate intermediate d-Ins(1,4,5,6)P4 (InsP4), but it has no specific activity towards InsP6 and cannot hydrolyze other IPPs such as d-Ins(2)P1, d-Ins(1,4)P2, d-Ins(1,4,5)P3, d-Ins(1,4,5,6)P4 and Ins(1,3,4,5,6)P5 (Li et al., 2011 ▸). Additionally, PhyH-DI was found to hydrolyze both InsP6 and InsP4 when acting synergistically (1.2–2.5-fold increase) with BPPs (such as PhyH-DII, PhyP and 168PhyA) and HAPs (Li et al., 2011 ▸; Lu et al., 2014 ▸). Thus, we presume that PhyH-DI can improve the catalytic efficiency of other single-domain phytases and is responsible for their higher activity when acting synergistically. We conjecture that dual-domain BPPs may have evolved from a single domain and persisted owing to improved phosphate hydrolysis. Specific structural features of PhyH-DI are important for regulation of the phytase activity of the dual-domain BPP and will provide valid information on its synergistic function with other phytases. Structure–sequence analysis of the PDB revealed that PhyH-DI shares 32% sequence similarity with a thermostable phytase (TS-Phy) from B. amyloliquefaciens (Ha et al., 2000 ▸) and 38% structure identity with B. subtilis phytase (Zeng et al., 2011 ▸). The structure of TS-Phy (Ha et al., 2000 ▸) determined at 2.1 Å resolution showed that the binding of different amounts of calcium ion could increase the thermostability and initiate the catalytic activity of the enzyme. The catalytic mechanism and properties of TS-Phy were then delineated by the crystal structure of this enzyme in complex with inorganic phosphate (Shin et al., 2001 ▸), which indicated that two phosphates and four calcium ions are tightly bound at the active site. These provided a convincing catalytic mechanism for BPPs. Another study (Zeng et al., 2011 ▸) revealed an interaction between a phytate analogue and divalent metal ions in B. subtilis phytase. It was conjectured that associated metal ions in the active site could be of crucial importance for substrate binding. This study provided a new perspective for understanding the catalytic function of phytases, especially BPPs.
To elucidate the mechanisms of the specific function in the synergy with other single-domain phytases, further exploration of the structure of PhyH-DI was performed. These studies will provide information on the unique biochemical features of PhyH-DI. Here, we present the expression, purification, crystallization and preliminary X-ray diffraction analysis of native and selenomethionine-substituted PhyH-DI from Bacillus sp. HJB17.
2. Materials and methods
2.1. Macromolecule production
The expression vector pET-22b(+)-PhyH-DI was reconstructed with minor modifications using the procedure described by Li et al. (2011 ▸). Briefly, the gene fragment for PhyH-DI, a domain (residues 41–318; 278 amino acids) of PhyH (HM003049.1), was amplified using the PCR method and cloned into the EcoRI–XhoI sites of a pET-22(+) plasmid to construct the recombinant plasmid [pET-22b(+)-PhyH-DI]. The recombinant plasmid was transformed into Escherichia coli strain BL21 (DE3) competent cells. Positive transformants were grown in 5 ml LB medium pH 7.0 containing 100 µg ml−1 ampicillin overnight at 310 K and subcultured into 1 l LB medium. When the OD600 reached 0.6–0.8, protein expression was induced by the addition of 0.2 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 16–18 h at 291 K. The cells were harvested and the bacterial pellet was resuspended in lysis buffer (20 mM Tris–HCl pH 8.0, 0.5 M NaCl, 10% glycerol) with 1 mM phenylmethanesulfonyl fluoride (PMSF) and the cells were lysed by sonication.
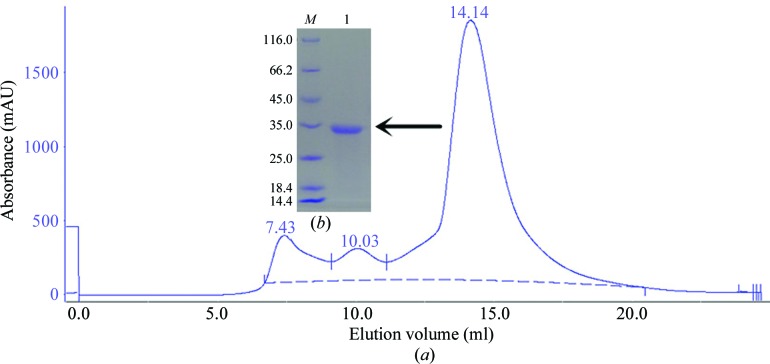
The clear supernatant that was obtained after centrifugation at 13 000 rev min−1 for 30 min at 277 K was subjected to Ni2+-chelating chromatography using the following procedure. The column was equilibrated with buffer A (20 mM Tris–HCl pH 8.0, 500 mM NaCl, 1 mM CaCl2), washed with buffer B (20 mM Tris–HCl pH 8.0, 500 mM NaCl, 20 mM imidazole, 1 mM CaCl2) and eluted with buffer C (20 mM Tris–HCl pH 8.0, 500 mM NaCl, 250 mM imidazole, 1 mM CaCl2). The protein was polished by size-exclusion chromatography (SEC) on a Superdex G200 column (GE Healthcare, USA) with buffer D (20 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM CaCl2). The target peak (Fig. 1 ▸ a) was assessed by SDS–PAGE (Fig. 1 ▸ b). Macromolecule-production information is summarized in Table 1 ▸.
Figure 1.
Purification of PhyH-DI using a gel-filtration column and SDS–PAGE analysis. (a) Purification profile of PhyH-DI, which eluted as a symmetrical peak from a Superdex G200 SEC column. (b) 15% SDS–PAGE stained with Coomassie Brilliant Blue. Lane M, protein marker (labelled in kDa); lane 1, PhyH-DI corresponding to the peak on the gel-filtration profile (0.5 mg ml−1).
Table 1. Macromolecule-production information.
| Source organism | Bacillus sp. HJB17 |
| DNA source | GenBank accession No. HM003049.1 |
| Forward primer | 5′-GCCCGGAATTCGGCAGTCCAACCTTTATCGGTC-3′ |
| Reverse primer | 5′-GCCGTGCTCGAGCTCCGGCTTCGCCTGCAGCAC-3′ |
| Cloning vector | pET-22b(+) |
| Expression vector | pET-22b(+) |
| Expression host | E. coli BL21 (DE3) |
| Complete amino-acid sequence of the construct produced | MDIGINSDPNSAVQPLSVNASHFHKVQLAGQDYQLFTTTTALQIRQGNRELAQQAGQFSRLVLQPLDTDALLLAAMDINSNTLFLWRFSTKQTPSLQLLQRRLISSRVVDDLCFYHSTENQQLSLFLLGGRGGADQLLLQQQQQWLAQPVVIRELNIPYDSTACVVDQTAGALYIAEADRAIWRYQAEPEADEGRSLVQVNKPFGQLQGEVKALQTLSDGSLLALEEAPARLLHINSDGQLRSAIAVPALAEASGLAVSMQGNTATAYISTEDAGAVQQLAVVLQAKPELEHHHHHH |
Selenomethionine-subsituted PhyH-DI (SeMet PhyH-DI) was expressed under conditions of methionine-pathway inhibition and was generated by the following procedure. Briefly, a single colony of E. coli BL21 (DE3) cells containing the pET-22b(+)-PhyH-DI vector was used to inoculate medium containing 100 mg ml−1 ampicillin, and the cells were resuspended in M9 medium (carbon source glucose) at 0.4%(m/v) with 100 mg ml−1 ampicillin and incubated at 310 K until the OD600 reached 0.6–0.8. Specific amino acids (lysine, phenylalanine, threonine, isoleucine, leucine and valine at 80 mg l−1, selenomethionine at 60 mg l−1 and glycine, alanine, proline, cysteine, tyrosine, tryptophan, histidine, aspartic acid, glutamic acid, serine, asparagine, glutamine and arginine at 30 mg l−1) were added to the cultures. Induction with 0.2 mM IPTG was performed 15 min after the addition of the amino acids; the cultures were incubated overnight (16–20 h) and the cells were harvested. The expression and purification of SeMet PhyH-DI was performed in a similar manner to that of PhyH-DI. The concentration of the purified native PhyH-DI and SeMet PhyH-DI proteins was analyzed using the Bradford method and they were used for crystallization studies.
2.2. Crystallization
Both native PhyH-DI and SeMet PhyH-DI were concentrated to 5 mg ml−1. Crystallization screening of PhyH-DI was initially performed at 289 K with commercially available kits from Hampton Research (Crystal Screen, Crystal Screen 2, Index, PEGRx 1, PEGRx 2, PEG/Ion and PEG/Ion 2) using the sitting-drop vapour-diffusion method in a 48-well Double Sample Crystallization Plate. The drop size was 2 µl, consisting of 1 µl protein solution mixed with an equal amount of reservoir solution, and was equilibrated against 80 µl reservoir solution. After the initial crystallization condition had been obtained, optimization of the crystallization condition was performed by fine-tuning the pH in the range 7.5–9.5 and the PEG 3350 concentration in the range 25–35% using the hanging-drop vapour-diffusion method in a 16-well Crystallization Plate at 289 K. Subsequently, the quality of the crystals was improved using the streak-seeding method. A suitable single crystal for data collection was finally obtained after 50 d in a condition consisting of 0.2 M sodium chloride, 0.1 M Tris pH 8.5, 25%(w/v) PEG 3350. SeMet PhyH-DI crystals were grown at a protein concentration of 1.5 mg ml−1 in a buffer consisting of 0.2 M sodium chloride, 0.1 M Tris pH 9.0, 30%(w/v) PEG 3350. Crystallization information is provided in Table 2 ▸.
Table 2. Crystallization.
| Method | Vapour diffusion |
| Plate type | 48-well Double Sample Crystallization Plate (screening), 16-well Crystallization Plate (final) |
| Temperature (K) | 289 |
| Protein concentration (mg ml−1) | 1–5 |
| Buffer composition of protein solution | 20 mM Tris pH 8.0, 100 mM NaCl |
| Composition of reservoir solution | 0.2 M sodium chloride, 0.1 M Tris pH 8.5, 25%(w/v) PEG 3350 (screening); 0.2 M sodium chloride, 0.1 M Tris pH 9.0, 30%(w/v) PEG 3350 (final) |
| Volume and ratio of drop (µl) | 2, 1:1 |
| Volume of reservoir (µl) | 80 (screening), 400 (final) |
2.3. Data collection and processing
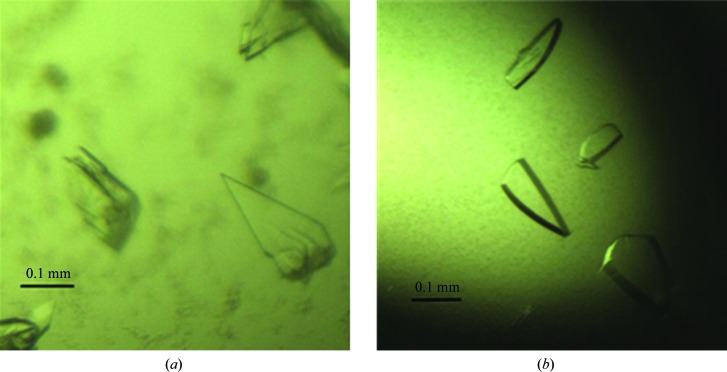
The harvested crystals (Fig. 2 ▸) were soaked for a few seconds in reservoir solution containing 15%(v/v) glycerol and were quickly mounted on a goniometer in a nitrogen stream at 100 K. Data collection was performed with a step size of 1.0° using a MAR165 CCD detector on beamline 3W1A at BSRF, Institute of High Energy Physics, Chinese Academy of Sciences. Data were indexed, integrated and scaled with the HKL-2000 suite (Otwinowski & Minor, 1997 ▸) and processed using the CCP4 suite (Winn et al., 2011 ▸). Initial phases and selenium sites were found with SHELXD (Sheldrick, 2015 ▸). The data-collection statistics are summarized in Table 3 ▸.
Figure 2.
Crystals used for data collection. (a) Native PhyH-DI. (b) SeMet PhyH-DI.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| PhyH-DI | SeMet PhyH-DI | |
|---|---|---|
| Diffraction source | 3W1A, BSRF | 3W1A, BSRF |
| Wavelength (Å) | 1.0000 | 0.9792 |
| Temperature (K) | 100 | 100 |
| Detector | MAR CCD | MAR CCD |
| Crystal-to-detector distance (mm) | 180 | 180 |
| Rotation range per image (°) | 1 | 1 |
| Total rotation range (°) | 243 | 1080 |
| Exposure time per image (s) | 90 | 45 |
| Space group | C121 | C2221 |
| a, b, c (Å) | 156.84, 45.54, 97.64 | 94.71, 97.03, 69.16 |
| α, β, γ (°) | 90.00, 125.86, 90.00 | 90.00, 90.00, 90.00 |
| Mosaicity (°) | 1.489 | 0.858 |
| Resolution range (Å) | 50.0–3.00 (3.05–3.00) | 50.00–2.70 (2.75–2.70) |
| Total No. of reflections | 58014 | 376627 |
| No. of unique reflections | 11541 | 8935 |
| Completeness (%) | 99.8 (100) | 99.9 (99.3) |
| Multiplicity | 5.0 (5.0) | 41.6 (31.2) |
| 〈I/σ(I)〉 | 9.61 (2.49) | 21.16 (3.38) |
| R r.i.m. (%) | 20.01 (70.88) | 20.95 (68.63) |
| Overall B factor from Wilson plot (Å2) | 41.0 | 44.2 |
3. Results and discussion
Native PhyH-DI and SeMet PhyH-DI were expressed, purified and crystallized. After Ni2+-chelating chromatography and size-exclusion chromatography (SEC) steps, PhyH-DI eluted as a symmetrical peak from a Superdex G200 SEC column and the target peak was observed at 14.14 ml (Fig. 1 ▸ a). PhyH-DI was obtained at high purity; it was was subjected to SDS–PAGE and had an apparent molecular weight of 32.5 kDa (Fig. 1 ▸ b).
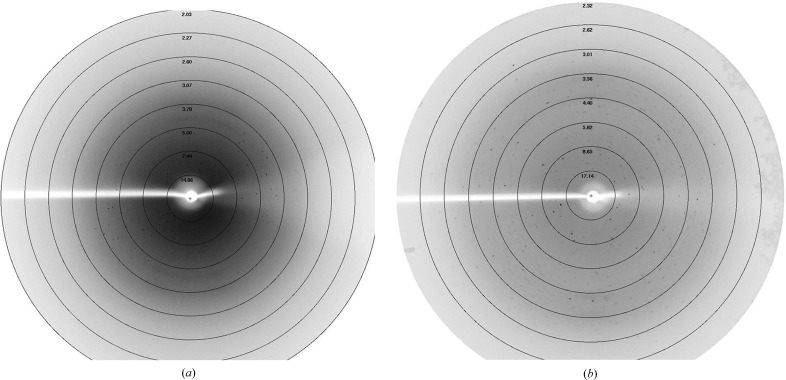
Diffraction data sets were collected from native and selenomethionine-substituted crystals to resolution limits of 3.0 and 2.70 Å, respectively (Fig. 3 ▸). The crystal of native PhyH-DI belonged to space group C121, with unit-cell parameters a = 156.84, b = 45.54, c = 97.64 Å, α = 90.00, β = 125.86, γ = 90.00°. Processing of the data using the CCP4 suite showed that two molecules of PhyH-DI are estimated to be present in the asymmetric unit, with a corresponding Matthews coefficient of 2.17 Å3 Da−1 and a solvent content of 43.26%. We then overexpressed SeMet PhyH-DI and obtained a crystal that belonged to space group C2221, with unit-cell parameters a = 94.71, b = 97.03, c = 69.16 Å, α = β = γ = 90.00°. The asymmetric unit contained one molecule, with a corresponding Matthews coefficient of 2.44 Å3 Da−1 and solvent content of 49.64%. Initial phases for PhyH-DI were obtained from SeMet SAD data sets, and three expected selenium sites were found with SHELXD.
Figure 3.
X-ray diffraction images of the crystals. (a) Native PhyH-DI. (b) SeMet PhyH-DI.
In this study, we successfully expressed, purified and crystallized PhyH-DI. Diffraction data sets were collected, indexed, integrated and scaled with the HKL-2000 suite. In an attempt to obtain the structure of PhyH-DI, molecular replacement was performed using the structures of a thermostable phytase from B. amyloliquefaciens (PDB entry 1cvm; Ha et al., 2000 ▸) and of 3-phytase from B. subtilis (PDB entry 3amr; Zeng et al., 2011 ▸) as templates. However, no solution was found, which was likely to be owing to significant differences between the three-dimensional structures of PhyH-DI and these proteins. We prepared selenomethionine-substituted crystals of PhyH-DI and collected several data sets at the Se peak wavelength (λ = 0.9792 Å) in order to obtain accurate phases for structure determination using anomalous dispersion techniques. PhyH-DI can act synergistically with other single-domain phytases to release phosphate and is responsible for increased hydrolytic activity (Li et al., 2011 ▸). Therefore, specific structural features of the intact PhyH-DI will provide important information about the functional relationship between single-domain phytases and PhyH-DI. Diffraction data statistics for native and SeMet PhyH-DI crystals are given in Table 3 ▸. Currently, efforts are under way to build the model of PhyH-DI. This initial information and the selenomethionine-substituted crystals will make a significant contribution to structural and functional determinations in the future.
Acknowledgments
We thank our coworkers at the BSRF for their help in data collection and Dr Zengqiang Gao for his help in data processing.
Funding Statement
This work was funded by The Fundamental Research Funds for the Central Universities grant 2015ZCQ-LY-02. National Natural Science Foundation of China grant 31070651.
References
- Cheng, C. & Lim, B. L. (2006). Arch. Microbiol. 185, 1–13. [DOI] [PubMed]
- Ha, N.-C., Oh, B.-C., Shin, S., Kim, H.-J., Oh, T.-K., Kim, Y.-O., Choi, K.-Y. & Oh, B.-H. (2000). Nature Struct. Biol. 7, 147–153. [DOI] [PubMed]
- Jain, J., Sapna & Singh, B. (2016). Process Biochem. 51, 159–169.
- Kumar, V., Yadav, A. N., Verma, P., Sangwan, P., Saxena, A., Kumar, K. & Singh, B. (2017). Int. J. Biol. Macromol. 98, 595–609. [DOI] [PubMed]
- Lei, X. & Stahl, C. (2001). Appl. Microbiol. Biotechnol. 57, 474–481. [DOI] [PubMed]
- Lei, X. G., Weaver, J. D., Mullaney, E., Ullah, A. H. & Azain, M. J. (2012). Annu. Rev. Anim. Biosci. 1, 283–309. [DOI] [PubMed]
- Li, Z., Huang, H., Yang, P., Yuan, T., Shi, P., Zhao, J., Meng, K. & Yao, B. (2011). FEBS J. 278, 3032–3040. [DOI] [PubMed]
- Lim, B. L., Yeung, P., Cheng, C. & Hill, J. E. (2007). ISME J. 1, 321–330. [DOI] [PubMed]
- Lu, F., Guo, G., Li, Q., Feng, D., Liu, Y., Huang, H., Yang, P., Gao, W. & Yao, B. (2014). Acta Cryst. F70, 1671–1674. [DOI] [PMC free article] [PubMed]
- Mukhametzianova, A. D., Akhmetova, A. I. & Sharipova, M. R. (2012). Mikrobiologiia, 81, 291–300. [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Shin, S., Ha, N.-C., Oh, B.-C., Oh, T.-K. & Oh, B.-H. (2001). Structure, 9, 851–858. [DOI] [PubMed]
- Ullah, A. H. (1988). Prep. Biochem. 18, 459–471. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Zeng, Y., Ko, T., Lai, H., Cheng, Y., Wu, T., Ma, Y., Chen, C., Yang, C., Cheng, K., Huang, C., Guo, R. & Liu, J. (2011). J. Mol. Biol. 409, 214–224. [DOI] [PubMed]
- Zhang, R., Yang, P., Huang, H., Shi, P., Yuan, T. & Yao, B. (2011). Curr. Microbiol. 63, 408–415. [DOI] [PubMed]